Published online Dec 15, 2023. doi: 10.4239/wjd.v14.i12.1766
Peer-review started: October 7, 2023
First decision: October 17, 2023
Revised: October 24, 2023
Accepted: November 17, 2023
Article in press: November 17, 2023
Published online: December 15, 2023
The global prevalence of type 2 diabetes mellitus (T2DM) is increasing. T2DM is associated with alterations of the gut microbiota, which can be affected by age, illness, and genetics. Previous studies revealed that there are discriminating microbiota compositions between the Dai and the Han populations. However, the specific gut microbiota differences between the two populations have not been elucidated.
To compare the gut microbiota differences in subjects with and without T2DM in the Dai and Han populations.
A total of 35 subjects of the Han population (including 15 healthy children, 8 adult healthy controls, and 12 adult T2DM patients) and 32 subjects of the Dai popu
No significant difference in alpha diversity was observed between healthy children and adults. The diversity of gut microbiota was decreased in T2DM patients compared to the healthy adults in both the Dai and Han populations. There was a significant difference in gut microbiota between healthy children and healthy adults in the Han population with an increased abundance of Bacteroidetes and decreased Firmicutes in children. However, this difference was less in the Dai population. Significant increases in Bacteroidetes in the Han population and Proteobacteria in the Dai population and decreases in Firmicutes in both the Han and Dai population were observed in T2DM patients compared to healthy adults. Linear discriminant analysis Effect Size analysis also showed that the gut microbiota was different between the Han and Dai populations in heathy children, adults, and T2DM patients. Four bacteria were consistently increased and two consistently decreased in the Han population compared to the Dai population.
Differences in gut microbiota were found between the Han and Dai populations. A significant increase in Bacteroidetes was related to the occurrence of T2DM in the Han population, while a significant increase in Proteobacteria was related to the occurrence of T2DM in the Dai population.
Core Tip: This study revealed that gut microbiota in the Han population is significantly different from the Dai population in healthy children, healthy adults, and patients with type 2 diabetes mellitus (T2DM). There was a significant difference in gut microbiota between healthy children and healthy adults in the Han population, but the difference was less in the Dai population. A significant increase in Bacteroidetes was observed in T2DM patients in the Han population, while a significant increase in Proteobacteria was observed in T2DM patients in the Dai population when compared to healthy controls.
- Citation: Tang LT, Feng L, Cao HY, Shi R, Luo BB, Zhang YB, Liu YM, Zhang J, Li SY. Comparative study of type 2 diabetes mellitus-associated gut microbiota between the Dai and Han populations. World J Diabetes 2023; 14(12): 1766-1783
- URL: https://www.wjgnet.com/1948-9358/full/v14/i12/1766.htm
- DOI: https://dx.doi.org/10.4239/wjd.v14.i12.1766
As the world population and the proportion of elderly people increases, type 2 diabetes mellitus (T2DM), a prevalent metabolic disorder, has become a major global public health problem[1]. It is clinically characterized by hyperinsulinemia, insulin resistance, and islet cell damage, which can reach 50% at the time of diagnosis[2]. Individuals with T2DM are highly susceptible to vascular and neurological consequences in addition to life, psychological, and financial stress[3]. In 2021, 537 million people were diagnosed with diabetes, which is expected to increase to 643 million by 2030 and to 783 million by 2045[4]. Although the etiology and pathogenesis of T2DM are still unclear, recent studies have shown that gut microbiota may play key roles[5-8]. Identifying the features of gut microbiota associated with T2DM could help to better understand the pathogenesis of T2DM and prevent or delay the onset of the disease.
The gut microbiota is a complex microecological community composed of more than 100 trillion microorganisms[9-11]. However, it can be affected by various factors, such as age, diet, illness, environment, and genetics[12-14]. Different ethnic groups have a wide variety of dietary patterns, lifestyles, and geographical environments, which can lead to different presentations of gut microbiota and T2DM[15]. Previous studies revealed that there are discriminating microbiota compositions between the Dai and Han populations[16-18]. However, due to the heterogeneity among different ethnic groups, the results of these studies are difficult to replicate. Therefore, it is necessary to carry out analyses of gut microbiota in various ethnic groups with different genetic backgrounds in China.
The Dai ethnic group is a unique minority in Yunnan, China. They have lived in the valley area for generations and generally have endogamous marriages[19]. Due to the different genetic backgrounds, unique lifestyles, and geographical environments[19-22] of the Dai and Han populations, we hypothesized that there may be some underlying differences in the gut microbiota of the two populations. This study was designed to compare the gut microbiota in subjects with and without T2DM in the Dai and Han populations in Yunnan, China. The results of this investigation will help elucidate the underlying differences of gut microbiota between the Dai and Han populations and determine the association between gut microbiota and T2DM prevalence.
Healthy children, adult T2DM patients, and ethnically matched healthy adults from the Han and Dai populations living in the same area were recruited and enrolled in this study. Patients with T2DM met the following diagnostic criteria[23]: (1) Fasting blood glucose ≥ 7.0 mmol/L; or (2) Hemoglobin A1c ≥ 6.5%. The enrolled T2DM patients were newly diagnosed and drug-naïve. Subjects who had been treated with antibiotics in the previous 3 mo, were pregnant or lactating, or had inflammatory bowel disease were excluded from the study. The T2DM patients and healthy adults in each population were age-matched (P > 0.05). The enrolled subjects have completed the Study Questionnaires to provide personal details and information about health, diet, smoking activity and lifestyle at the time of sample collection. The study was approved by the ethics committee of the Sixth Affiliated Hospital of Kunming Medical University (approval no. 2023-kmykdx6f-66). All participants provided written informed consent.
Fasting venous blood samples were collected from all participants in the morning. After centrifugation (3000 rpm) at 4 °C for 10 min, the serum was immediately extracted and aliquoted. All blood and serum samples were stored at -80 °C until further biochemical analysis was performed. Stool samples were collected into 5-mL disposable sterile tubes within 30 min of discharge and kept at -80 °C until further processing was conducted.
Total genomic DNA samples were extracted using the OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s instructions. The quantity and quality of extracted DNA were measured using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and agarose gel electrophoresis, respectively. Prepared DNA samples were stored at -20 °C.
The V3-V4 region of 16S rRNA was amplified by polymerase chain reaction (PCR) with the primers 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). The following thermal cycling conditions were utilized: initial denaturation at 98 °C for 5 min; 25 cycles consisting of denaturation at 98 °C for 30 s, annealing at 53 °C for 30 s, and extension at 72 °C for 45 s; and a final extension of 5 min at 72 °C. PCR amplicons were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, United States). After the individual quantification step, amplicons were pooled in equal amounts, and pair-end 2 × 250 bp sequencing was performed using the Illlumina NovaSeq platform with NovaSeq 6000 SP Reagent Kit (500 cycles) at Shanghai Personal Biotechnology Co., Ltd (Shanghai, China).
Microbiome bioinformatics were performed with QIIME2 2019.4 with slight modifications according to the official tutorials (https://docs.qiime2.org/2019.4/tutorials/). Briefly, raw sequence data were demultiplexed using the demux plugin followed by primer cutting with cutadapt plugin. Sequences were then quality filtered, denoised, merged, and chimera removed using the DADA2 plugin. Non-singleton amplicon sequence variants were aligned with mafft and used to construct a phylogeny with fasttree2. Taxonomy was assigned to amplicon sequence variants using the classify-sklearn naïve Bayes taxonomy classifier in feature-classifier plugin. Microbiota comparisons were performed using principal coordinate analysis (PCoA), principal component analysis (PCA), and permutational multivariate analysis of variance (PERMANOVA), diversity index estimate and composition of microbiome analysis.
Data were analyzed with SPSS Software (Version 29.0; IBM Corp., Armonk, NY, United States). Continuous data were expressed as mean ± SD. Comparisons between groups were performed using one-way analysis of variance and Student’s t test. Cases with missing data for analysis were omitted, and the remaining data were analyzed. P < 0.05 was considered statistically significant.
A total of 35 subjects from the Han population (including 15 healthy children, 8 adult healthy controls, and 12 T2DM patients) and 32 subjects from the Dai population (including 10 healthy children, 10 adult healthy controls, and 12 T2DM patients) were enrolled in this study. The main demographics and blood biochemical indexes of the subjects are presented in Table 1. The levels of total bilirubin, direct bilirubin, indirect bilirubin, total cholesterol, hemoglobin A1c, gamma glutamyl transferase, and hypersensitive C-reactive protein were significantly higher in the T2DM patients in the Han population compared to the T2DM patients in the Dai population. Low density lipoprotein cholesterol and apolipoprotein A1 were significantly lower in T2DM patients in the Han population compared to T2DM patients in the Dai population. The levels of triglycerides, total bilirubin, indirect bilirubin, and gamma glutamyl transferase were sig
| Parameter | Han population | Dai population | ||||
| Healthy adults, n = 8 | T2DM patients, n = 12 | Healthy children, n = 15 | Healthy adults, n = 10 | T2DM patients, n = 12 | Healthy children, n = 10 | |
| Sex as male/female | 3/5 | 8/4 | 7/8 | 5/5 | 9/3 | 4/6 |
| Age in yr | 50.63 ± 5.85 | 55.27 ± 11.76 | 5.00 ± 2.101 | 45.70 ± 14.662 | 55.92 ± 9.07 | 5.50 ± 1.183 |
| FBG in mmol/L | 4.85 ± 0.38 | 9.98 ± 5.891 | 3.23 ± 0.99 | 5.62 ± 1.36 | 7.74 ± 2.393 | 4.52 ± 0.29 |
| HbA1c as % | 5.55 ± 0.24 | 8.55 ± 2.591 | 5.23 ± 0.15 | 5.58 ± 0.19 | 7.24 ± 0.773,4 | 5.19 ± 0.35 |
| TG in mmol/L | 1.90 ± 0.63 | 2.22 ± 1.54 | 0.97 ± 0.241 | 3.90 ± 4.102 | 2.48 ± 1.91 | 1.12 ± 0.453 |
| TC in mmol/L | 5.15 ± 0.61 | 3.76 ± 1.451 | 4.01 ± 0.88 | 5.51 ± 1.04 | 4.85 ± 1.264 | 3.91 ± 0.573 |
| HDL-C in mmol/L | 1.28 ± 0.28 | 0.97 ± 0.221 | 1.48 ± 0.34 | 1.23 ± 0.28 | 1.26 ± 0.40 | 1.39 ± 0.42 |
| LDL-C in mmol/L | 3.12 ± 0.64 | 1.82 ± 0.711 | 2.16 ± 0.631 | 2.98 ± 1.23 | 2.75 ± 1.054 | 2.16 ± 0.38 |
| APO-A1 in g/L | 1.75 ± 0.27 | 1.30 ± 0.301 | 1.75 ± 0.36 | 1.83 ± 0.23 | 1.74 ± 0.374 | 1.61 ± 0.36 |
| APO-B in g/L | 1.04 ± 0.15 | 0.68 ± 0.181 | 0.72 ± 0.191 | 0.98 ± 0.24 | 0.94 ± 0.29 | 0.66 ± 0.093 |
| WBC as 109/L | 6.06 ± 1.44 | 6.41 ± 1.87 | 6.95 ± 1.11 | 6.79 ± 1.43 | 7.07 ± 2.09 | 8.32 ± 2.38 |
| RBC as 1012/L | 5.19 ± 0.66 | 4.82 ± 0.57 | 4.89 ± 0.27 | 5.09 ± 0.65 | 5.05 ± 0.57 | 4.90 ± 0.42 |
| TBil in μmol/L | 6.84 ± 1.59 | 14.19 ± 9.351 | 6.07 ± 4.48 | 8.71 ± 5.082 | 6.19 ± 1.984 | 5.74 ± 2.38 |
| DBil in μmol/L | 3.84 ± 0.79 | 6.31 ± 2.921 | 2.92 ± 2.34 | 3.96 ± 2.05 | 3.35 ± 1.024 | 3.13 ± 1.40 |
| IBil in μmol/L | 3.00 ± 0.85 | 7.88 ± 6.511 | 3.15 ± 2.28 | 4.75 ± 3.052 | 2.84 ± 1.344 | 2.61 ± 1.413 |
| ALT in U/L | 22.50 ± 11.33 | 31.36 ± 26.34 | 15.80 ± 14.85 | 25.50 ± 15.68 | 26.10 ± 17.22 | 13.70 ± 14.96 |
| AST in U/L | 21.13 ± 4.58 | 27.36 ± 25.61 | 31.50 ± 7.181 | 23.26 ± 8.71 | 23.14 ± 8.64 | 27.40 ± 6.45 |
| GGT in U/L | 34.75 ± 24.40 | 51.82 ± 90.65 | 9.67 ± 2.941 | 70.40 ± 66.342 | 38.67 ± 22.804 | 13.50 ± 8.153 |
| BUN in mmol/L | 5.63 ± 1.40 | 4.59 ± 1.70 | 3.68 ± 0.821 | 5.12 ± 1.51 | 5.23 ± 2.18 | 3.75 ± 0.91 |
| Cr in μmol/L | 70.38 ± 8.75 | 69.55 ± 20.92 | 32.17 ± 7.601 | 68.20 ± 17.69 | 85.50 ± 30.21 | 32.00 ± 4.003 |
| Hcy in μmol/L | 15.63 ± 3.45 | 12.35 ± 1.551 | 14.20 ± 3.12 | 13.59 ± 3.57 | 16.69 ± 3.474 | 11.55 ± 1.535 |
| hs-CRP in mg/L | 1.85 ± 3.35 | 3.46 ± 5.23 | 2.84 ± 6.665 | 1.84 ± 1.57 | 1.83 ± 1.474 | 0.47 ± 0.503 |
The total amount of data comprised 4505369 reads, with an average of 68736 reads per sample. For the 35 Han individuals, the total amount of data included 2132603 reads, with an average of 60931 reads per sample. For the 32 Dai individuals, the total amount of data included 2472766 reads, with an average of 77274 reads per sample. By clustering analysis at a 97% similarity, 22890 operational taxonomic units were identified.
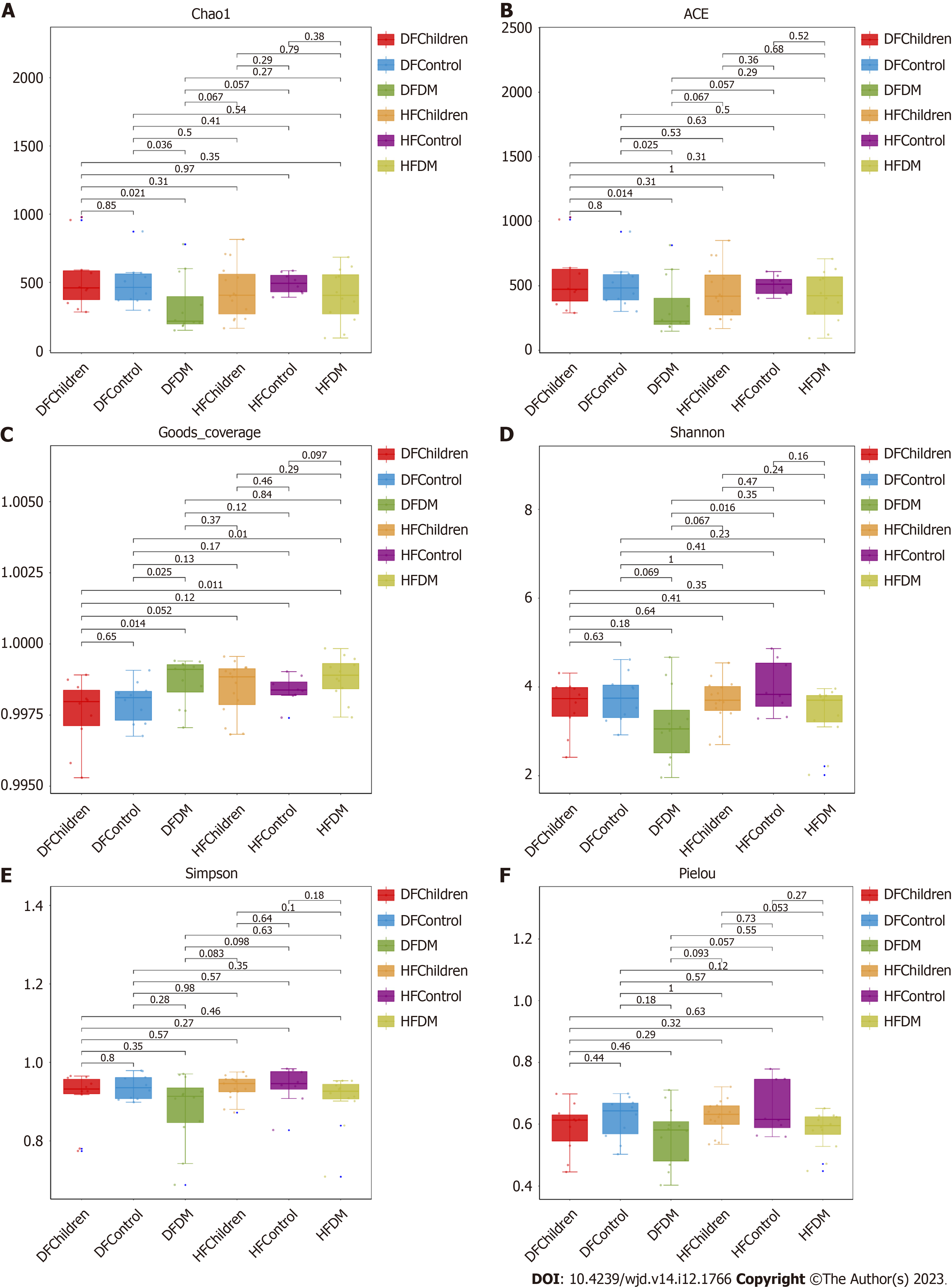
Six indices (Chao1, abundance-based coverage estimator, Good’s coverage, Shannon, Simpson, and Pielou) were compared between all the population groups to evaluate the alpha diversity in gut microbiota. No significant difference in alpha diversity was observed between healthy children and adults. However, the diversity of the gut microbiota was decreased in T2DM patients compared to healthy adults in both the Dai and Han populations (Figure 1).
PCA scores plot showed that there’s differences among groups (Supplementary Figure 1). Further PCoA and PER
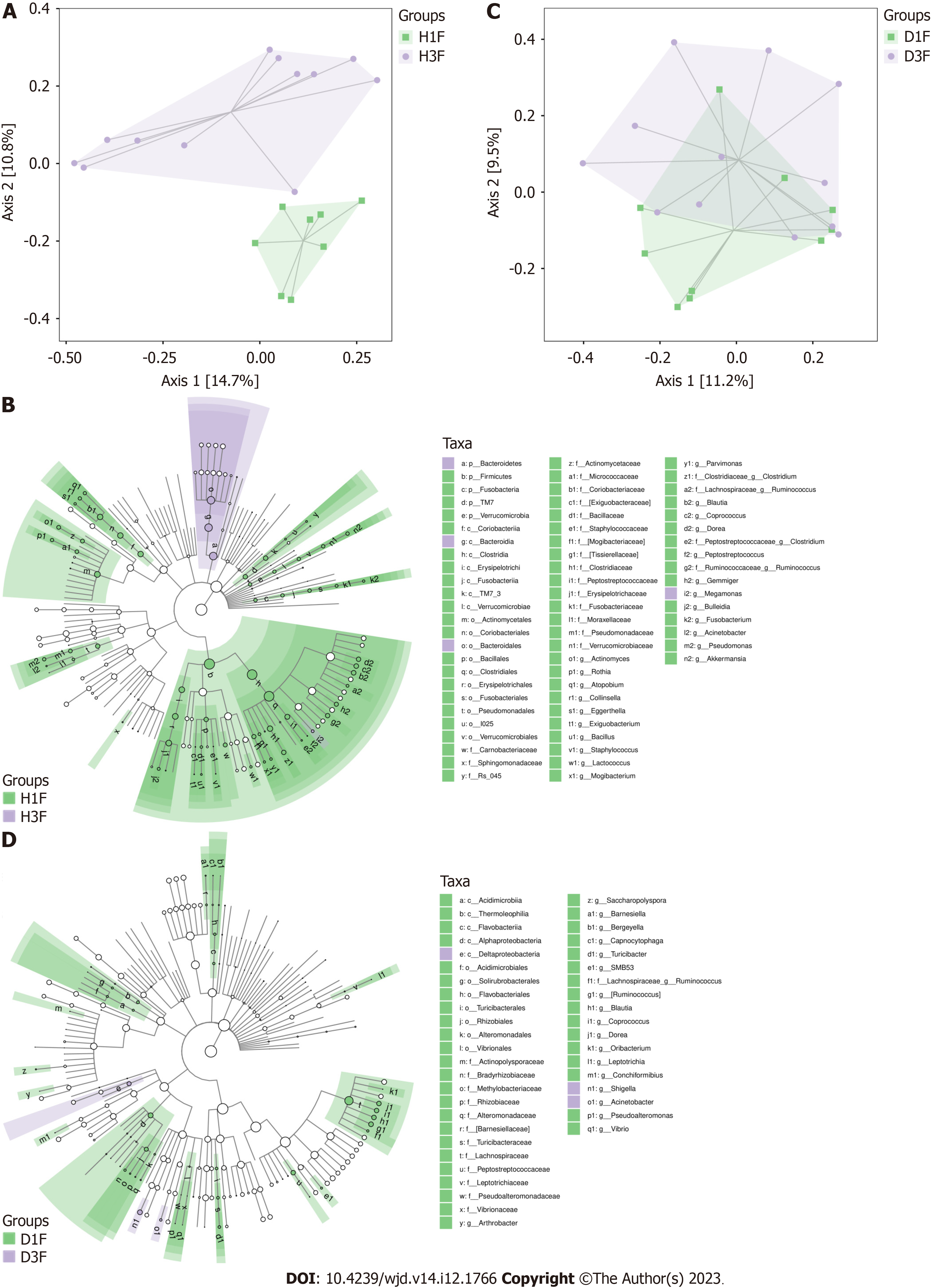
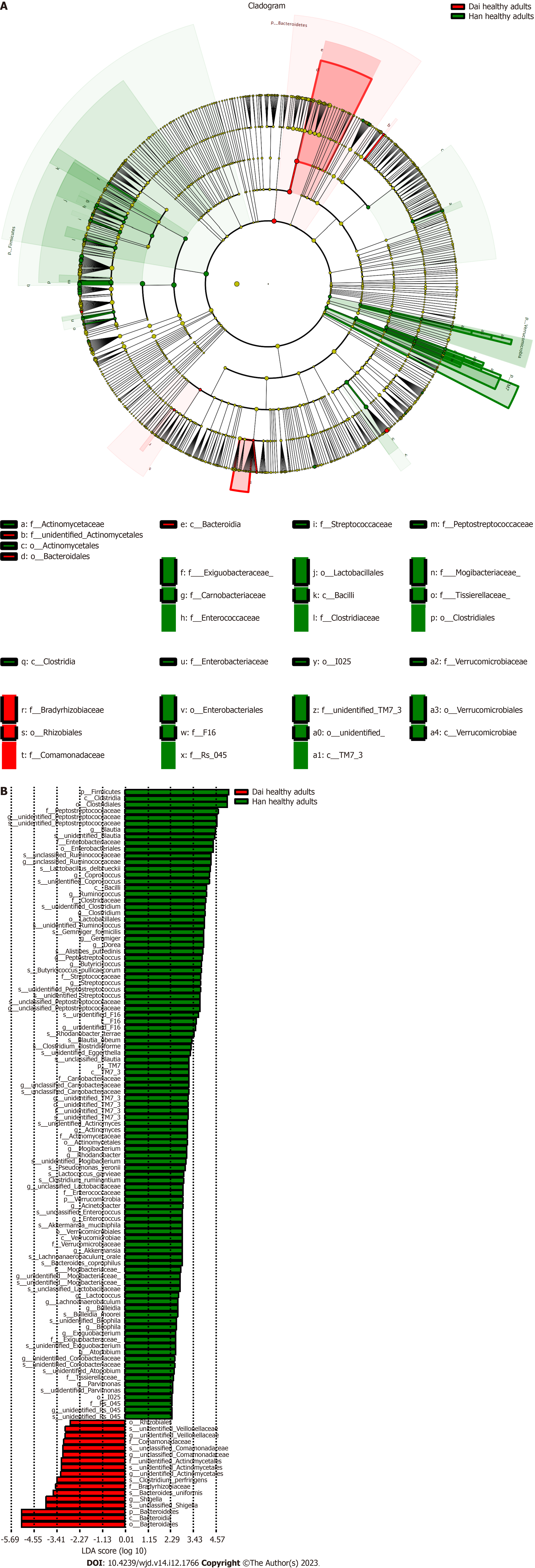
These data indicated that the difference in the gut microbiota between healthy children and healthy adults was greater in the Han population than in the Dai population and suggest that the influence of age, diet, or lifestyle on the gut microbiota is greater in the Han population than the Dai population.
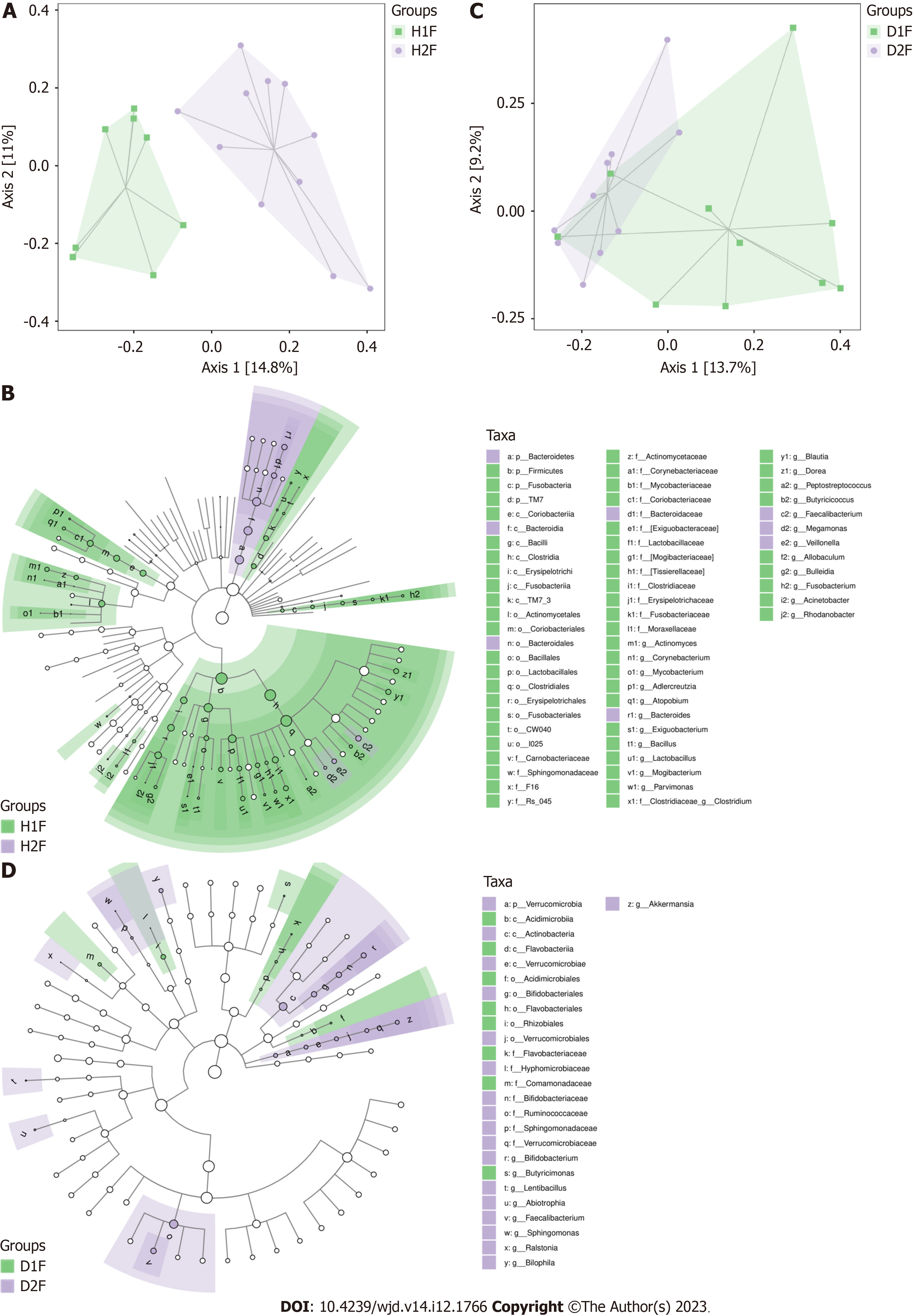
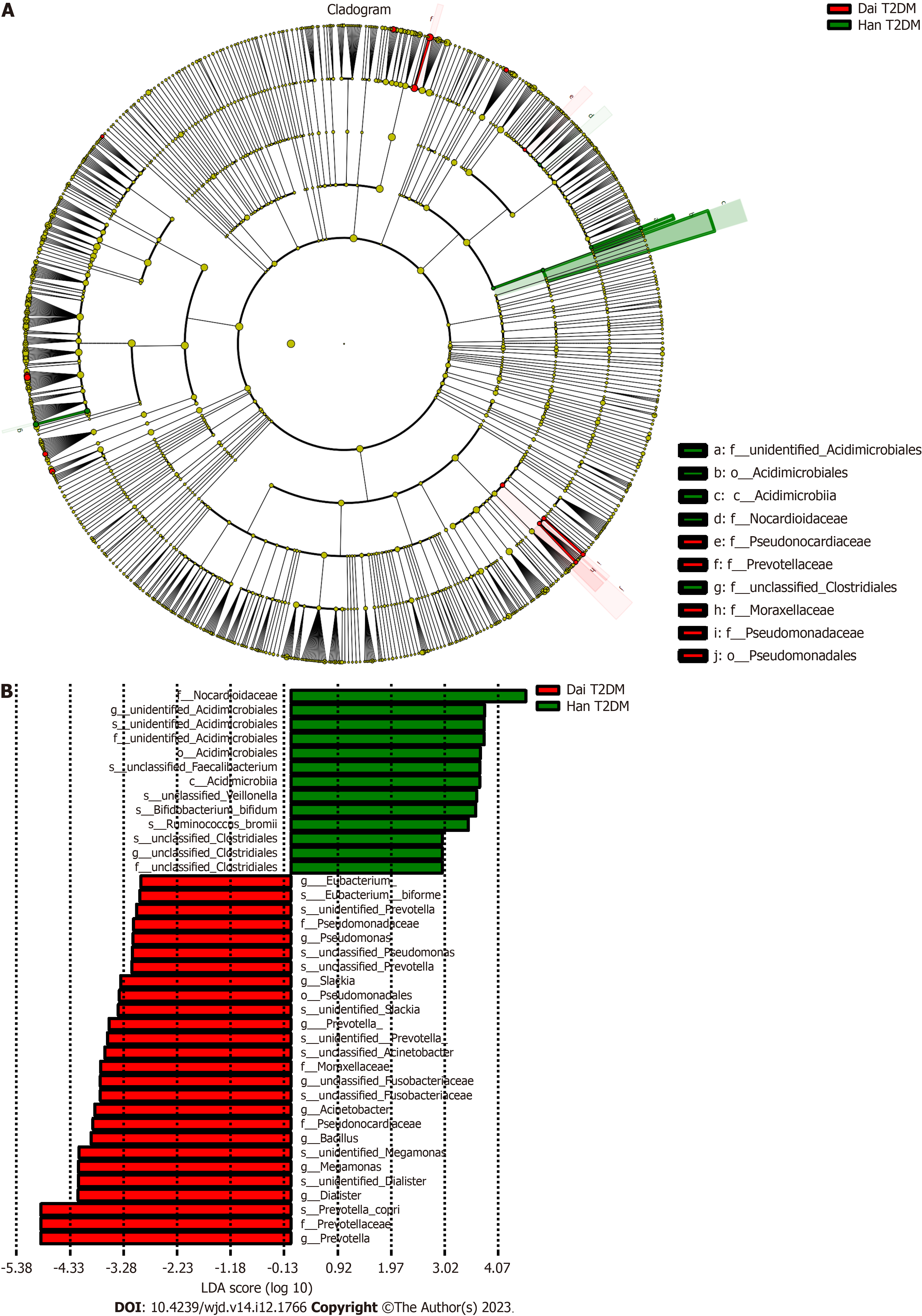
An obvious distinction between healthy adults and T2DM patients in the Han population was observed on the PCoA plot (Figure 3A). The gut microbiota composition of healthy adults and T2DM patients in the Han population was presented in a cladogram (Figure 3B). Compared with healthy adults, the T2DM patients from the Han population had an increased abundance of Bacteroidetes, Bacteroidales, Megamonas, and Bacteroidia within the Bacteroidetes phylum (Table 2).
| Taxa | Abundance, T2DM patients vs healthy adults | LDA score | P value |
| Bacteria. Bacteroidetes | 5.353 | 4.939 | 0.037 |
| Bacteria. Bacteroidetes. Bacteroidia. Bacteroidales | 5.353 | 4.939 | 0.037 |
| Bacteria. Firmicutes. Clostridia. Clostridiales. Veillonellaceae. Megamonas | 4.347 | 3.760 | 0.045 |
| Bacteria. Bacteroidetes. Bacteroidia | 5.353 | 4.939 | 0.037 |
There were no observable differences in the gut microbiota between healthy adults and T2DM patients in the Dai population after the PCoA (Figure 3C) and LEfSe analysis (Figure 3D). Compared with healthy adults, the T2DM patients from the Dai population had an increased abundance of Deltaproteobacteria, Shigella, and Acinetobacter within the Proteobacteria phylum (Table 3).
| Taxa | Abundance, T2DM patients vs healthy adults | LDA score | P value |
| Bacteria. Proteobacteria. Gammaproteobacteria. Pseudomonadales. Moraxellaceae. Acinetobacter | 2.047 | 2.754 | 0.004 |
| Bacteria. Proteobacteria. Gammaproteobacteria. Enterobacteriales. Enterobacteriaceae. Shigella | 4.711 | 4.288 | 0.021 |
| Bacteria. Proteobacteria. Deltaproteobacteria | 3.199 | 2.764 | 0.044 |
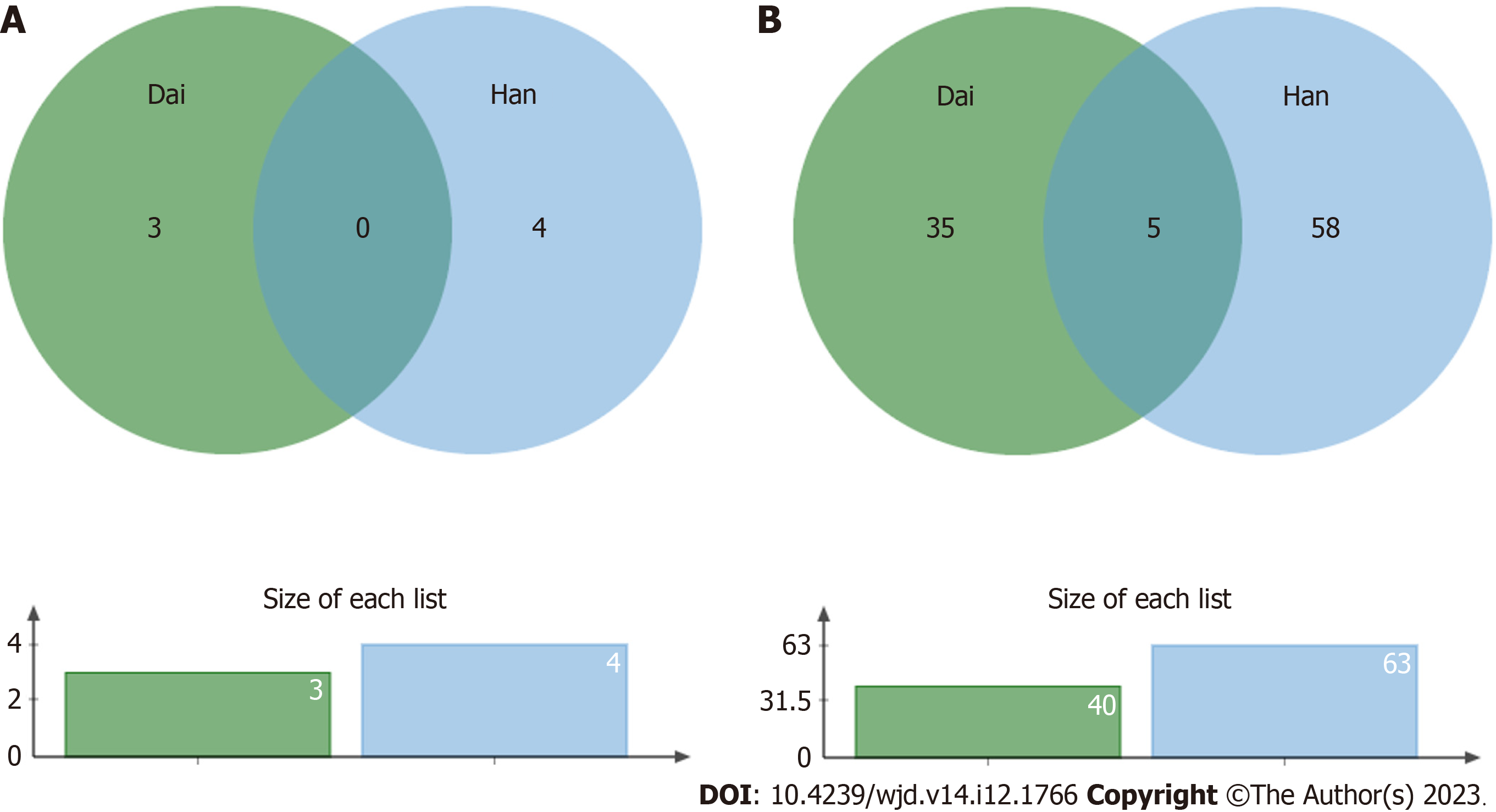
There were several bacterial types with a decreased abundance in T2DM patients in the Han population (63 total types) and in the Dai population (40 total types). Five of these bacteria, including Dorea, Peptostreptococcaceae, Blautia, Ruminococcus, and Coprococcus, were decreased in both populations (Figure 4).
To investigate the differences of gut microbiota between the Han and Dai populations during the transition from healthy children to healthy adults and T2DM, the LEfSe analysis was performed (Figures 5-7). Compared to the Dai population, the abundances of Coprococcus, Streptococcus, and Lactococcus within the phylum Firmicutes were consistently higher in the Han population. We also observed that the abundances of Rhizobiales within the phylum Proteobacteria and Veillonellaceae within the phylum Firmicutes were significantly lower in the Han population (Table 4).
| Differential gut microbiota | Healthy children | Healthy adults | T2DM patients |
| Han population | |||
| d__Bacteria.p__Firmicutes.c__Clostridia.o__Clostridiales.f__Lachnospiraceae.g__Coprococcus.s__unidentified_Coprococcus | 0.542a | 0.259a | 0.461 |
| d__Bacteria.p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Streptococcaceae.g__Streptococcus.s__unclassified_Streptococcus | 0.000a | 0.800a | 1.000 |
| d__Bacteria.p__Firmicutes.c__Clostridia.o__Clostridiales.f__Lachnospiraceae.g__Coprococcus | 0.521a | 0.286a | 0.475 |
| d__Bacteria.p__Firmicutes.c__Bacilli.o__Lactobacillales.f__Streptococcaceae.g__Lactococcus | 0.009a | 0.019a | 0.083 |
| Dai population | |||
| d__Bacteria.p__Proteobacteria.c__Alphaproteobacteria.o__Rhizobiales | 4.840a | 1.159a | 1.636 |
| d__Bacteria.p__Firmicutes.c__Clostridia.o__Clostridiales.f__Veillonellaceae.g__unclassified_Veillonellaceae | 8.800a | 12.000a | 1.688 |
The relationship between gut microbiota and T2DM is becoming increasingly important. In the past decade, studies have supported the role of gut microbiota in the pathogenesis of T2DM[24-28]. Some researchers have reported that there are discriminating microbiota compositions between the Han and the Tibetans populations[16-18] and also different among the different ethnicities: Han, Zang, Bai, Hani, Dai, and Miao (including both healthy urban and rural residents of each ethnicity)[29]. However, the underlying differences of the gut microbiota between the Han and Dai populations have not been elucidated. Here, we performed a comparative analysis of the gut microbiota in subjects with and without T2DM from the Dai and Han populations in Yunnan Province, China. To the best of our knowledge, this is the first time to compare the T2DM-associated gut microbiota between Han and Dai populations.
The alpha diversity of the gut microbiota, which reflects the abundance, evenness, and richness[30], might vary between ethnic groups in part due to the varied prevalence of T2DM among ethnic groups[31]. Interestingly, our study showed that there was no significant difference in alpha diversity between the Han and Dai populations, suggesting that the abundance, evenness, and richness of the gut microbiota were not significantly different between the Han and Dai populations. However, the diversity of gut microbiota was decreased in T2DM patients compared to healthy adults in both the Han and Dai populations (Figure 1), which is consistent with the previous results in different populations of the world, including other populations in China[32-34].
The gut microbiota is associated with the age of host[35]. Despite being matched for age (significance less than 0.05), there was a large deviation from the mean value of the age of healthy adults with diabetes in Dai who were actually enrolled in the analysis. We also selected some of these samples with matched age means for subset analysis, and the results were consistent with the existing result. To determine the influence of age on gut microbiota, we also conducted a comparison of gut microbiota between healthy children and healthy adults in the Han and Dai populations. These findings suggested that the difference in the gut microbiota between healthy children and healthy adults was greater in the Han population than the Dai population. The observed higher relative abundances of genus Bacteroides in children and higher relative abundances of genus Blautia in adults were consistent with the previous studies[36].
Many researchers have reported that the gut microbiota diversity is affected by T2DM[26,37]. After comparing the gut microbiota between healthy adults and T2DM patients in the Han population, we observed a significant difference in the gut microbiota. However, there was no clear distinction between healthy adults and T2DM patients in the Dai population. The underlying reason behind this warrant further investigation. Moreover, our data showed that the T2DM patients of the Dai population possessed a distinctive microbiota composition characterized by a high abundance of Proteobacteria, which is consistent with the previous results[38]. Recent evidence has also shown that Proteobacteria in gut microbial dysbiosis is essential for metabolic disorders[39]. Interestingly, we found that T2DM patients from the Han population had an increase in Bacteroidetes, Bacteroidales, Megamonas and Bacteroidia within the phylum Bacteroidetes. This discovery conflicted with the results of some earlier studies[32,40]. The possible reason might be that the proportion of Bacteroidales abundance can be altered by high-calorie diets[41], which is also a possible cause of T2DM.
Although we have identified that both age and T2DM influence the gut microbiota, it is unknown which has a greater effect. We explored the differences in bacteria between healthy children to healthy adults and healthy adults to T2DM in both ethnic groups. The results showed that the differences of healthy children between the Han and Dai population were still significant in healthy adults. However, these changes in T2DM patients were not statistically significant. These results demonstrated that the differences were influenced more by age than T2DM during the transition from healthy children to healthy adults and T2DM patients in both the Han and Dai populations.
Several limitations of this study should be taken into account. First, the sample size was relatively small, which limits the generalizability of the findings. It should be confirmed in a larger scale of samples in the future. Second, the effect of gender, food, and smoking activity were not investigated in the study. Third, the metabolic profile requires further investigation to confirm the relationship between the imbalance of metabolism and gut microbiota alterations.
Through the comparative analysis, this study found significant differences in the gut microbiota in the Han and Dai populations, and these differences were influenced to a greater degree by age than by T2DM. Our findings may provide additional insight for further study of gut microbiota dysbiosis-related diseases in the Han and Dai populations.
Previous studies revealed that there are discriminating microbiota compositions between the Han and Dai populations. However, the underlying differences in the gut microbiota between the Han and Dai populations have not yet been elucidated.
We compared the differences in the gut microbiota in subjects with and without type 2 diabetes mellitus (T2DM) in the Han and Dai populations to explore the pathogenic mechanism of T2DM.
To identify the differences in the gut microbiota related to the occurrence of T2DM in the Han and Dai populations.
A total of 35 subjects of the Han population (15 healthy children, 8 adult healthy controls, and 12 adult T2DM patients) and 32 subjects of the Dai population (10 healthy children, 10 adult healthy controls, and 12 adult T2DM patients) were enrolled in this study. Fasting venous blood samples were collected from all the subjects for biochemical analysis. Fecal samples were collected from all the subjects for DNA extraction and 16S rRNA sequencing, which was followed by analyses of the gut microbiota composition.
Fasting plasma glucose levels and hemoglobin A1c were significantly increased in the T2DM patients. The gut microbiota of the Han population was significantly different from the Dai population in healthy children, healthy adults, and T2DM patients. Significant increases in Bacteroidetes were observed in T2DM patients from the Han population, while significant increases in Proteobacteria were observed in T2DM patients in the Dai population.
We observed significant differences in the gut microbiota in the Han and Dai populations, and these differences were influenced to a greater degree by age than by T2DM.
Our findings may provide additional insight for further study of the gut microbiota dysbiosis-related diseases in the Han and Dai populations. Future research should include a larger scale of samples and an investigation of the metabolic profile in order to confirm the relationship between an imbalance of the metabolism and gut microbiota alterations.
The authors would like to thank all the collaborators for their efforts and also thank parents for allowing participation of their children in this study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Endocrinology and metabolism
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Joda BA, Iraq; Popovic DS, Serbia; Horowitz M, Australia S-Editor: Wang JJ L-Editor: A P-Editor: Xu ZH
| 1. | Soares Andrade CA, Shahin B, Dede O, Akpeji AO, Ajene CL, Albano Israel FE, Varga O. The burden of type 2 diabetes mellitus in states of the European Union and United Kingdom at the national and subnational levels: A systematic review. Obes Rev. 2023;24:e13593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 2. | Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med. 2008;359:1577-1589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5314] [Cited by in F6Publishing: 5035] [Article Influence: 314.7] [Reference Citation Analysis (0)] |
| 3. | Papatheodorou K, Banach M, Bekiari E, Rizzo M, Edmonds M. Complications of Diabetes 2017. J Diabetes Res. 2018;2018:3086167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 245] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 4. | Su J, Luo Y, Hu S, Tang L, Ouyang S. Advances in Research on Type 2 Diabetes Mellitus Targets and Therapeutic Agents. Int J Mol Sci. 2023;24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 5. | Hamjane N, Mechita MB, Nourouti NG, Barakat A. Gut microbiota dysbiosis -associated obesity and its involvement in cardiovascular diseases and type 2 diabetes. A systematic review. Microvasc Res. 2023;151:104601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 8] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 6. | Ma Q, Li Y, Li P, Wang M, Wang J, Tang Z, Wang T, Luo L, Wang C, Zhao B. Research progress in the relationship between type 2 diabetes mellitus and intestinal flora. Biomed Pharmacother. 2019;117:109138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 180] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 7. | Wu J, Yang K, Fan H, Wei M, Xiong Q. Targeting the gut microbiota and its metabolites for type 2 diabetes mellitus. Front Endocrinol (Lausanne). 2023;14:1114424. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 8. | Zhou Z, Sun B, Yu D, Zhu C. Gut Microbiota: An Important Player in Type 2 Diabetes Mellitus. Front Cell Infect Microbiol. 2022;12:834485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 68] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 9. | Zhou YD, Liang FX, Tian HR, Luo D, Wang YY, Yang SR. Mechanisms of gut microbiota-immune-host interaction on glucose regulation in type 2 diabetes. Front Microbiol. 2023;14:1121695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 10. | Bajinka O, Tan Y, Darboe A, Ighaede-Edwards IG, Abdelhalim KA. The gut microbiota pathway mechanisms of diabetes. AMB Express. 2023;13:16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 11. | Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355-1359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3481] [Cited by in F6Publishing: 2999] [Article Influence: 166.6] [Reference Citation Analysis (0)] |
| 12. | Brooks AW, Priya S, Blekhman R, Bordenstein SR. Gut microbiota diversity across ethnicities in the United States. PLoS Biol. 2018;16:e2006842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 186] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 13. | Deschasaux M, Bouter KE, Prodan A, Levin E, Groen AK, Herrema H, Tremaroli V, Bakker GJ, Attaye I, Pinto-Sietsma SJ, van Raalte DH, Snijder MB, Nicolaou M, Peters R, Zwinderman AH, Bäckhed F, Nieuwdorp M. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med. 2018;24:1526-1531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 304] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 14. | Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4098] [Cited by in F6Publishing: 4152] [Article Influence: 319.4] [Reference Citation Analysis (0)] |
| 15. | Alvarez-Silva C, Kashani A, Hansen TH, Pinna NK, Anjana RM, Dutta A, Saxena S, Støy J, Kampmann U, Nielsen T, Jørgensen T, Gnanaprakash V, Gnanavadivel R, Sukumaran A, Rani CSS, Færch K, Radha V, Balasubramanyam M, Nair GB, Das B, Vestergaard H, Hansen T, Mande SS, Mohan V, Arumugam M, Pedersen O. Trans-ethnic gut microbiota signatures of type 2 diabetes in Denmark and India. Genome Med. 2021;13:37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 16. | Huan H, Ren T, Xu L, Hu H, Liu C. Compositional distinction of gut microbiota between Han Chinese and Tibetan populations with liver cirrhosis. PeerJ. 2021;9:e12142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 17. | Li K, Dan Z, Gesang L, Wang H, Zhou Y, Du Y, Ren Y, Shi Y, Nie Y. Comparative Analysis of Gut Microbiota of Native Tibetan and Han Populations Living at Different Altitudes. PLoS One. 2016;11:e0155863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 18. | Zhu LL, Ma ZJ, Ren M, Wei YM, Liao YH, Shen YL, Fan SM, Li L, Wu QX, Gao ZS, Song JF, Ma YL. Distinct Features of Gut Microbiota in High-Altitude Tibetan and Middle-Altitude Han Hypertensive Patients. Cardiol Res Pract. 2020;2020:1957843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Zhang L, Sun Y, Zhang X, Shan X, Li J, Yao Y, Shu Y, Lin K, Huang X, Yang Z, Chu J, Huang L, Sun H. Three Novel Genetic Variants in the FAM110D, CACNA1A, and NLRP12 Genes Are Associated With Susceptibility to Hypertension Among Dai People. Am J Hypertens. 2021;34:874-879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 20. | Zhang F, Amat G, Tang Y, Chen R, Tian X, Hu W, Chen C, Shen S, Xie Y. NUDT15 Genetic Variants in Chinese Han, Uighur, Kirghiz, and Dai Nationalities. Front Pediatr. 2022;10:832363. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 21. | He M, Lin K, Huang Y, Zhou L, Yang Q, Li S, Jiang W. Prevalence and Molecular Study of G6PD Deficiency in the Dai and Jingpo Ethnic Groups in the Dehong Prefecture of the Yunnan Province. Hum Hered. 2018;83:55-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 22. | Yang J, Li W, Zhang Z, Shen J, Zhang N, Yang M, Yu Y. PSCArs2294008 T polymorphism increases the risk of bladder cancer in Bai, Dai, and Han ethnicity in China and a potential mechanism. Genes Genomics. 2018;40:531-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 23. | Chinese Elderly Type 2 Diabetes Prevention and Treatment of Clinical Guidelines Writing Group; Geriatric Endocrinology and Metabolism Branch of Chinese Geriatric Society; Geriatric Endocrinology and Metabolism Branch of Chinese Geriatric Health Care Society; Geriatric Professional Committee of Beijing Medical Award Foundation; National Clinical Medical Research Center for Geriatric Diseases (PLA General Hospital). [Clinical guidelines for prevention and treatment of type 2 diabetes mellitus in the elderly in China (2022 edition)]. Zhonghua Nei Ke Za Zhi. 2022;61:12-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 47] [Reference Citation Analysis (0)] |
| 24. | Brunkwall L, Orho-Melander M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: from current human evidence to future possibilities. Diabetologia. 2017;60:943-951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 221] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 25. | Chen L, Guo L, Feng S, Wang C, Cui Z, Wang S, Lu Q, Chang H, Hang B, Snijders AM, Mao JH, Lu Y, Ding D. Fecal microbiota transplantation ameliorates type 2 diabetes via metabolic remodeling of the gut microbiota in db/db mice. BMJ Open Diabetes Res Care. 2023;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 26. | Carrizales-Sánchez AK, Tamez-Rivera O, Rodríguez-Gutiérrez NA, Elizondo-Montemayor L, Gradilla-Hernández MS, García-Rivas G, Pacheco A, Senés-Guerrero C. Characterization of gut microbiota associated with metabolic syndrome and type-2 diabetes mellitus in Mexican pediatric subjects. BMC Pediatr. 2023;23:210. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 27. | Huang X, Qiu Y, Gao Y, Zhou R, Hu Q, He Z, Lv Y, Wang X, Chen W, Deng Y, An Z, Zhang H, Mo Z, Lin R. Gut microbiota mediate melatonin signalling in association with type 2 diabetes. Diabetologia. 2022;65:1627-1641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 28. | Xia F, Wen LP, Ge BC, Li YX, Li FP, Zhou BJ. Gut microbiota as a target for prevention and treatment of type 2 diabetes: Mechanisms and dietary natural products. World J Diabetes. 2021;12:1146-1163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 20] [Article Influence: 6.7] [Reference Citation Analysis (4)] |
| 29. | Sun Y, Zuo T, Cheung CP, Gu W, Wan Y, Zhang F, Chen N, Zhan H, Yeoh YK, Niu J, Du Y, Wen Y, Yu J, Sung JJY, Chan PKS, Chan FKL, Wang K, Ng SC, Miao Y. Population-Level Configurations of Gut Mycobiome Across 6 Ethnicities in Urban and Rural China. Gastroenterology. 2021;160:272-286.e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 30. | Wei T, Jia Y, Xue W, Ma M, Wu W. Nutritional Effects of the Enteral Nutritional Formula on Regulation of Gut Microbiota and Metabolic Level in Type 2 Diabetes Mellitus Mice. Diabetes Metab Syndr Obes. 2021;14:1855-1869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 31. | Pham TM, Carpenter JR, Morris TP, Sharma M, Petersen I. Ethnic Differences in the Prevalence of Type 2 Diabetes Diagnoses in the UK: Cross-Sectional Analysis of the Health Improvement Network Primary Care Database. Clin Epidemiol. 2019;11:1081-1088. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 32. | Larsen N, Vogensen FK, van den Berg FW, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1783] [Cited by in F6Publishing: 1907] [Article Influence: 136.2] [Reference Citation Analysis (0)] |
| 33. | Wang Y, Luo X, Mao X, Tao Y, Ran X, Zhao H, Xiong J, Li L. Gut microbiome analysis of type 2 diabetic patients from the Chinese minority ethnic groups the Uygurs and Kazaks. PLoS One. 2017;12:e0172774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 34. | Li Q, Chang Y, Zhang K, Chen H, Tao S, Zhang Z. Implication of the gut microbiome composition of type 2 diabetic patients from northern China. Sci Rep. 2020;10:5450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 35. | Li Y, Chen T, Li Y, Tang Y, Huang Z. Gut microbiota are associated with sex and age of host: Evidence from semi-provisioned rhesus macaques in southwest Guangxi, China. Ecol Evol. 2021;11:8096-8122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 36. | Radjabzadeh D, Boer CG, Beth SA, van der Wal P, Kiefte-De Jong JC, Jansen MAE, Konstantinov SR, Peppelenbosch MP, Hays JP, Jaddoe VWV, Ikram MA, Rivadeneira F, van Meurs JBJ, Uitterlinden AG, Medina-Gomez C, Moll HA, Kraaij R. Diversity, compositional and functional differences between gut microbiota of children and adults. Sci Rep. 2020;10:1040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 37. | Takagi T, Naito Y, Kashiwagi S, Uchiyama K, Mizushima K, Kamada K, Ishikawa T, Inoue R, Okuda K, Tsujimoto Y, Ohnogi H, Itoh Y. Changes in the Gut Microbiota are Associated with Hypertension, Hyperlipidemia, and Type 2 Diabetes Mellitus in Japanese Subjects. Nutrients. 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 38. | Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1476] [Cited by in F6Publishing: 1989] [Article Influence: 221.0] [Reference Citation Analysis (0)] |
| 39. | Kwan SY, Sabotta CM, Joon A, Wei P, Petty LE, Below JE, Wu X, Zhang J, Jenq RR, Hawk ET, McCormick JB, Fisher-Hoch SP, Beretta L. Gut Microbiome Alterations Associated with Diabetes in Mexican Americans in South Texas. mSystems. 2022;7:e0003322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 40. | Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, Chen Y, Ji L. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8:e71108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 481] [Cited by in F6Publishing: 544] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 41. | Daniel H, Gholami AM, Berry D, Desmarchelier C, Hahne H, Loh G, Mondot S, Lepage P, Rothballer M, Walker A, Böhm C, Wenning M, Wagner M, Blaut M, Schmitt-Kopplin P, Kuster B, Haller D, Clavel T. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014;8:295-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 476] [Article Influence: 43.3] [Reference Citation Analysis (0)] |