Published online Aug 15, 2021. doi: 10.4239/wjd.v12.i8.1200
Peer-review started: March 1, 2021
First decision: April 20, 2021
Revised: May 4, 2021
Accepted: July 9, 2021
Article in press: July 9, 2021
Published online: August 15, 2021
Type 2 diabetes mellitus (T2DM) is a metabolic disorder that currently affects more than 400 million worldwide and is projected to cause 552 million cases by the year 2030. Long-term vascular complications, such as coronary artery disease, myocardial infarction, stroke, are the leading causes of morbidity and mortality among diabetic patients. The recent advances in genome-wide technologies have given a powerful impetus to the study of risk markers for multifactorial diseases. To date, the role of genetic and epigenetic factors in modulating susceptibility to T2DM and its vascular complications is being successfully studied that provides the accumulation of genomic knowledge. In the future, this will provide an opportunity to reveal the pathogenetic pathways in the development of the disease and allow to predict the macrovascular complications in T2DM patients. This review is focused on the evidence of the role of genetic variants and epigenetic changes in the development of macrovascular pathology in diabetic patients.
Core Tip: Type 2 diabetes mellitus (T2DM) is often associated with life-threatening macrovascular complications which may lead to an eye injury, kidney failure, and reduction in life expectancy in patients with diabetes. This review is focused on genetic and epigenetic risk factors for macrovascular complications development in patients with T2DM.
- Citation: Tonyan ZN, Nasykhova YA, Danilova MM, Glotov AS. Genetics of macrovascular complications in type 2 diabetes. World J Diabetes 2021; 12(8): 1200-1219
- URL: https://www.wjgnet.com/1948-9358/full/v12/i8/1200.htm
- DOI: https://dx.doi.org/10.4239/wjd.v12.i8.1200
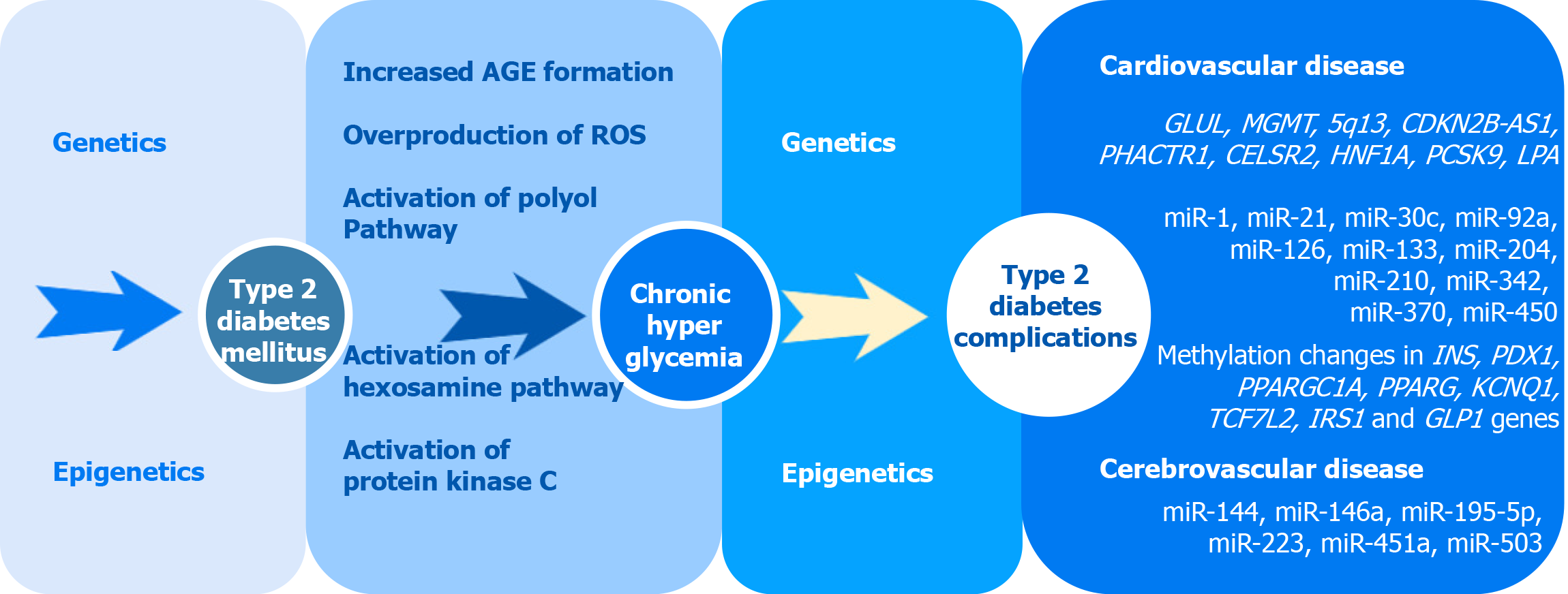
Type 2 diabetes mellitus (T2DM) is a metabolic disorder that currently affects more than 400 million worldwide and is projected to cause 552 million cases by the year 2030[1]. Long-term vascular complications, such as coronary artery disease (CAD), myocardial infarction (MI), stroke, are the leading causes of morbidity and mortality among diabetic patients[2]. T2DM is strongly associated with both microvascular and macrovascular complications which may lead to an eye injury, kidney failure, and reduction in life expectancy in patients with diabetes. Diabetic microvascular (involving small vessels such as capillaries) and macrovascular (involving large vessels such as arteries and veins) complications have similar etiologic characteristics[3]. Chronic hyperglycemia plays a major role in the development of microvascular and macrovascular pathology in diabetic patients through several molecular mechanisms, including overproduction of reactive oxygen species (ROS), advanced glycation end-products formation (AGE), activation of protein kinase C as well as polyol and hexosamine pathways[2] (Figure 1). The key pathological mechanism of macrovascular complications is assumed to be an injury to the vascular endothelium. The altered glucose metabolism inhibits the enzyme responsible for NO production and increases production of ROS[3]. In combination with endothelial cell insulin resistance, it causes endothelial dysfunction manifesting itself in increased expression of adhesion molecules and further changes[4]. Another factor involved in the development and progression of diabetic macrovascular complications is impaired platelet function which may lead to increased risks for thrombus formation and atherosclerosis progression[5]. A number of studies, including family- and twin-based studies, demonstrated the role of a genetic component in both T2DM and macrovascular pathology[6]. The development of high-throughput and affordable genotyping technologies, statistical tools, and computational software allowed remarkable progress over the past decade in the search for genetic associations of complex disorders such as T2DM, CAD, MI, stroke[7,8]. However, the pathogenetic mechanisms leading to macrovascular complications in individuals with diabetes are not yet fully understood. Moreover, the question of how genetic susceptibility interacts with environmental factors and epigenetic factors remains unsolved. In this review, we summarize the evidence for genetic variants and epigenetic factors involved in the development of macrovascular pathology in T2DM and discuss the pathogenetic mechanisms underlying their development in T2DM.
CAD represents the manifestation of atherosclerosis in the coronary arteries which supply the myocardium with oxygen and other nutrients and is the leading cause of morbidity and mortality worldwide due to serious complications like MI[9]. Diabetes mellitus is associated with increased risk factor of CAD, independent of other risk factors such as hypertension, hyperlipidemia, and tobacco smoking. Patients with T2DM have a 2-3 times higher rate of cardiovascular disease as compared to people without T2DM[10]. To date, genomic research led to the identification of more than 150 common genetic risk loci of CAD and MI[8]. And some of these variants were demonstrated to be significantly associated with cardiovascular diseases (CVD) in individuals with diabetes[11,12]. Several studies that analyzed T2DM or CAD using the mendelian randomization (MR) approach, commonly used for testing causal associations between a risk factor and outcome of interest, were published. The results of these studies provided genetic evidence that higher BMI and hyperglycemia had a positive causal association with CAD[13]. Early studies in the genetics of T2DM and CAD identified several shared loci associated with both diseases[14]. Some studies were performed to evaluate whether CAD-susceptibility loci identified by genome-wide association (GWA) studies of the general population also contributed to CAD in type 2 diabetes. The association between 9p21 Locus (the variants rs2383206 and rs10757278) and CAD in individuals with type 2 diabetes was shown in a case-control study performed by Doria et al[11] in 2008. Previous GWA studies reported the independent association of this locus with CAD[15,16]. The association of rs10757274 with MI risk was later replicated in a Chinese study[17]. In 2011, Qi et al[12] genotyped 15 genetic markers in three cohorts of diabetic patients: the prospective Nurses’ Health Study (309 CAD cases and 544 controls) and Health Professional Follow-up Study (345 CAD cases and 451 controls), and the cross-sectional Joslin Heart Study (422 CAD cases and 435 controls). Five single-nucleotide polymorphisms, rs4977574 (CDKN2A/2B), rs12526453 (PHACTR1), rs646776 (CELSR2-PSRC1-SORT1), rs2259816 (HNF1A), and rs11206510 (PCSK9) showed directionally consistent association with CAD in the 3 studies[12].
A total of 1517 CAD cases and 2671 CAD-negative controls, all with type 2 diabetes, were included in the 3-stage genome-wide analysis performed by Qi et al[12]. A previously unknown genetic variant rs10911021 in the region of the GLUL gene on chromosome 1q25 was found to be associated with CAD. The GLUL encodes glutamate-ammonia ligase (also known as glutamine synthase) which catalyzes the conversion of glutamic acid and ammonia into glutamine. Evidence from experimental and human studies points to glutamine/glutamic acid metabolism contribute to the regulation of insulin secretion and glucose metabolism. According to the results of this GWAS, the minor allele had a protective effect. The authors also observed that the risk homozygous genotype of rs10911021 was associated with a 32% lower expression level of the nearest downstream gene GLUL compared to the protective allele homozygous genotype in endothelial cells. The identified variant was not associated with the risk of type 2 diabetes in the DIAGRAM database. No association between the risk variant and serum fasting insulin, HOMA-IR, or 2 h-glucose was found in the MAGIC database. This suggests that the pathways underlying the association of the variant with CAD are distinct from those involved in the etiology of type 2 diabetes and insulin-resistance mechanisms[18]. The association of the variant rs10911021 with CVD in T2DM was confirmed in several follow-up studies[19,20]. In 2016 Shah et al conducted a GWA study of cardiovascular mortality in the ACCORD intensive arm and found two loci at 10q26 and 5q13 specifically associated with cardiovascular mortality. The lead variant (rs9299870) was shown to be associated with a 3.6-fold increased risk of cardiovascular death. The variant rs9299870 is located in intron 1 of the MGMT gene. The MGMT encodes for O-6-methylguanine-DNA methyltransferase that is involved in cellular defense against mutagenesis and toxicity from alkylating agents, and in gene methylation[21,22]. The other locus is located on chromosome 5, upstream and proximal to three long intergenic noncoding (LINC) RNAs (LINC1335, LINC1333, and LINC1331) and associated with NSA2 expression. The lead variant (rs57922) was associated with a 2.7-fold increased risk of cardiovascular death[23]. A GWAS of CAD was conducted in the UK Biobank in the cohort that included 15666 unrelated individuals (3968 CAD cases and 11698 controls) of white British ancestry with diabetes. Significant evidence for association of the previously well-established LPA locus (rs74617384) and locus at 9p21 (rs10811652) with CAD was reported. Moreover, some other variants previously associated with CAD showed similar effects in patients with and without diabetes[24]. In a recent GWAS, a systematic assessment of genetic overlap between CAD and T2DM was performed on a large cohort (66643 subjects). The results of the study demonstrated that none of the previously characterized CAD loci had a specific effect on CAD in T2DM individuals[25]. The results of this study, indicated that the increased risk of CAD in diabetic patients could be explained not only by known genetic variants with a large effect but by the other risk factors, including epigenetic changes, that may contribute to the pathogenesis of T2DM, should be considered. Genes associated with macrovascular complications of T2DM are summarized in Table 1.
| Gene symbol | Region | dbSNP ID | Risk allele | Condition | Ref. | |
| 1 | GLUL | 1q25 | rs10911021 | C | CAD | Qi et al[18], Beaney et al[19], and Look AHEAD Research Group[20] |
| 2 | MGMT | 10q26.3 | rs9299870 | A | Cardiovascular mortality | Shah et al[23] |
| 3 | Intergenic | 5q13 | rs57922 | T | Cardiovascular mortality | Shah et al[23] |
| 4 | CDKN2B-AS1 | 9p21 | rs10757274 | G | MI | Doria et al[11], and Zhang et al[17] |
| 5 | CDKN2B-AS1 | 9p21 | rs2383206 | G | MI | Doria et al[11], and Zhang et al[17] |
| 6 | CDKN2B-AS1 | 9p21 | rs4977574 | G | CAD | Qi et al[12] |
| 7 | PHACTR1 | 6p24 | rs12526453 | C | CAD | Qi et al[12] |
| 8 | CELSR2 | 1p21 | rs646776 | T | CAD | Qi et al[12] |
| 9 | HNF1A | 12q24 | rs2259816 | T | CAD | Qi et al[12] |
| 10 | PCSK9 | 1p32 | rs11206510 | T | CAD | Qi et al[12] |
| 11 | LPA | 6q25 | rs74617384 | T | CAD | Fall et al[24] |
| 12 | CDKN2B-AS1 | 9p21 | rs10811652 | C | CAD | Fall et al[24] |
Cerebrovascular diseases (CeVD) include a variety of medical conditions that affect the blood vessels of the brain and cerebral circulation. About 20%-40% of patients with type 2 diabetes suffer from cerebral blood vessel diseases. The mechanisms of the CeVD development in type 2 diabetes are complex and not fully understood, but the underlying process is associated with atherosclerotic changes in the cerebral arteries[26,27]. Individuals with diabetes develop dyslipidemia characterized by small dense low-density lipoproteins (LDLs), reduced high-density lipoproteins, and increased triacylglycerol levels[28]. Epidemiologic studies showed that type 2 diabetes was associated with a 2- to 5-fold increased risk of ischemic CeVD[29,30]. To date, information on the role of genetic variants associated with CeVD in individuals with T2DM is extremely limited. To explore the effects of genetic predisposition to T2DM, hyperglycemia, insulin resistance, and β-cell dysfunction on the risk of stroke subtypes and related cerebrovascular phenotypes, the MR analysis was recently performed by Georgakis et al[31]. Results of the study provided genetic evidence for a causal effect of T2DM and elevated HbA1c levels in the pre-diabetic range on the risk of an ischemic stroke, large artery stroke, carotid plaque, and small vessel stroke. Genetic predisposition to insulin resistance was found to be associated with large artery and small vessel stroke, whereas predisposition to β-cell dysfunction was associated with small vessel stroke[31]. Further studies are needed to clarify etiological mechanisms of the CeVD-T2DM association and genetic variants involved in it.
Investigation of the epigenetic pathways will allow us to get closer to a complete understanding of the causes and mechanisms of T2DM complications development. Based on the results of previous studies, the main epigenetic changes, contributing to the occurrence and progress of T2DM and its complications, include alteration of microRNA (miRNA) expression, DNA methylation, and histone acetylation.
Increasing evidence demonstrated an impact of epigenetics on the development of T2DM macrovascular complications. Several theories were proposed, focusing on miRNAs, histone modifications, and DNA methylation.miRNAs are small noncoding RNAs involved in the post-transcriptional regulation of gene expression. Previous researches showed changes in the miRNA expression profile in T2DM patients[32]. A series of recent studies indicated that miRNA expression profiling might contribute to the identification of miRNAs with prognostic value for the early detection of macrovascular complications of diabetes (Table 2).
| miRNA | miRNA detection method | Condition | Source | Country | Ref. |
| miR-92a | qRT-PCR | Acute coronary syndrome | Serum | China | Wang et al[69] |
| miR-30c | qRT-PCR | CAD | Plasma | China | Luo et al[70] |
| miR-126 | qRT-PCR | CAD | Whole blood | Bahrain | Al-Kafaji et al[33] |
| miR-126, miR-210 | qRT-PCR | CAD | Plasma | Egypt | Amr et al[34] |
| miR-370 | qRT-PCR | CAD | Serum | Egypt | Motawae et al[80] |
| miR-21 | qRT-PCR | In-stent restenosis CAD | Plasma | China | Guan et al[59] |
| miR-342, miR-450 | qRT-PCR | CAD | Serum | Egypt | Seleem et al[81] |
| miR-1, miR-133 | qRT-PCR | CAD | Whole blood | Bahrain | Al-Muhtaresh et al[87] |
| miR-204 | qRT-PCR | Coronary artery calcification | Plasma | China | Ding et al[94] |
| miR-21 | qRT-PCR | Acute heart failure | Serum | Turkey | Al-Hayali et al[54] |
| miR-144, miR-223 | qRT-PCR | Ischemic stroke | Platelet, plasma | China | Yang et al[88] |
| miR-223, miR-146a | qRT-PCR | Ischemic stroke | Platelet | China | Duan et al[97] |
| miR-195-5p, miR-451a | qRT-PCR | Transient ischemic attack | Serum | Italy | Giordano et al[106] |
| miR-503 | qRT-PCR | Ischemic stroke | Plasma | Iran | Sheikhbahaei et al[118] |
| miR-223 | qRT-PCR | Ischemic stroke | PBMC | China | Long et al[99] |
For example, based on the results of the recent research by Al-Kafaji et al[33], miR-126 was differentially expressed in T2DM patients with CAD, T2DM patients without macrovascular complications, and healthy control subjects. An inverse correlation with LDL was also demonstrated in the first group of patients[33]. These findings were successfully replicated in more recent work[34]. MiR-126 also was found to have lower expression levels in diabetic patients with CAD compared to T2DM patients without MVC in this study. The authors additionally showed that a significantly higher expression level of miR-210 was a risk factor for developing MVC and might potentially serve as a biomarker for CAD. MiR-126 is endothelial cell-specific and is known to directly inhibit negative regulators of the VEGF (vascular endothelial growth factor) and to affect vascular integrity and angiogenesis[35,36], which indicates the possible involvement of miR-126 in the pathogenesis of CAD. As was previously reported in the literature, the decreased expression level of miR-126 is significantly associated with CAD risk[37] and elevated levels of LDL cholesterol in CAD patients[38]. Furthermore, many studies provided evidence for the implication of miR-126 in T2DM pathogenesis. A number of authors identified low circulating miR-126 in T2DM individuals with CAD[33,34,39] and without MVC[40].
MiR-210 expression is induced during hypoxia in normal and transformed cells. Aberrant expression of miR-210 was detected in many pathological conditions such as tumor progression, MI, cutaneous ischemic wounds[41]. This can be explained by a wide range of miR-210 targets involved in the processes of angiogenesis, DNA repair, mitochondrial metabolism, and cell survival[42]. There is some evidence that the expression of miR-210 is significantly elevated in stable atherosclerotic plaques[43]. This can be explained by the protective effect of miR-210 that provides fibrous cap stability and prevents plaque rupture[44]. MiR-210 may also play a role in atherosclerosis progression through inhibiting endothelial apoptosis and regulating cell proliferation and differentiation during hypoxia, thus protecting a heart from damage[45,46]. Increased miR-210 expression is required for endothelial cell survival and migration during oxygen deficiency[47]. According to Hu et al[48], angiogenesis stimulation and apoptosis inhibition by miR-210 allowed to improve cardiac function and to reduce negative consequences of MI in mice. Another study demonstrated that miR-210 overexpression protected limbs from muscular and vascular ischemic damage in transgenic mouse strain[49,50]. Several studies suggest that miR-210 may be considered as a potential therapeutic target for ischemic conditions, in particular for ischemic heart disease[44,48], and also serve as biomarkers for peripheral artery disease[51]. Results concerning aberrant miR-210 expression in T2DM patients also showed tissue-dependent expression changes. The miR-210 level was reduced in adipose tissue of diabetic individuals according to Pek et al[52]. However, its circulating level was higher compared to normal control in peripheral blood of newly diagnosed T2DM patients based on the results of another research[53]. Since miR-210 is involved in multiple hypoxia-regulated metabolic pathways, its differential expression was detected in many diseases. This leaves us with an open question of the potential use of miR-210 as a single biomarker. A possible way out is to determine co-expressed miRNAs for a more accurate prediction of a particular disease or comp
An association between miR-21 expression profile and T2DM MVC was confirmed in 2 independent studies. It was found out that miR-21 was overexpressed in diabetic patients with CAD or with heart failure or without any MVC compared to the control group[54]. However, a still unsolved question is whether miR-21 may be used as a potential biomarker for MVC. Numerous studies were conducted to investigate miR-21 expression levels in T2DM patients. Most of them demonstrated significantly elevated amounts of circulating miR-21 in plasma and serum in subjects with T2DM[55-58]. At this stage, authors suggest that combined analysis of miR-21 expression, galectin-3, and N-terminal pro-brain natriuretic peptide can help to overcome the limitations and to improve the predictive value of miR-21[54]. Nevertheless, despite the low specificity of miR-21 as an independent prognostic marker of MVC, there is some evidence to suggest miR-21 expression profiling may be used for predicting the occurrence of in-stent restenosis for CAD patients after percutaneous coronary intervention[59]. MiR-21 also was found to be upregulated in patients with acute coronary syndrome and could be a possible candidate of a biomarker for CAD prediction[60].
Previous studies in mice showed that downregulation of miR-92a expression had a positive effect on the state of the vascular wall, attenuated inflammation of endo
The association of circulating miR-30c with the risk of MVC of T2DM was demonstrated in the study performed by Luo et al[70]. The expression levels of circulating miR-30c were significantly downregulated in patients with T2DM complicated by CAD. Authors also found out that decreased circulating miR-30c was associated with severe coronary artery lesions[70]. These data confirmed the previous findings, according to which downregulation of miR-30c-5p in macrophage-derived microparticles might result in the development of early atherosclerosis[71]. A decrease in expression of miR-30c also contributes to the pathogenesis of diabetic cardiomyopathy[72]. Moreover, miR-30c was shown to be associated with an increased risk of recurrent ischemic events in intracranial atherosclerotic disease[73]. Stimulation of miR-30c expression was proposed as a therapy for the prevention of atherosclerosis[71] and diabetic cardiomyopathy[72].
One of the first reports demonstrating the participation of miR-370 in the pathogenesis of atherosclerosis showed that patients suffering from a stable form of CAD had a high miR-370 level in peripheral blood mononuclear cells (PBMC). Based on the results, the authors suggested miR-370 could be used to determine individuals at risk for acute coronary syndromes[74]. The results were replicated in another study, according to which the expression levels of miR-370 in PBMC were upregulated compared to controls. This study also revealed that miR-370 was involved in atherosclerosis development through targeting FOXO1 gene[75]. Genetic variations and epigenetic modifications of FOXO1 are known to be associated with atherosclerotic plaque formation[76,77]. Upregulation of circulating miR-370 expression in plasma of CAD patients was found by several authors[78,79]. At the same time, decreased expression was shown in the peripheral blood in pre-atherosclerotic subjects[63]. Concerning diabetic patients, expression of miRNA 370 was higher in the T2DM+CAD group while compared to T2DM patients without MVC[80].
The recent research performed by Seleem et al[81] confirmed the importance of miR-450 and miR-342 in developing T2DM MVC by demonstrating an aberrant expression in patients suffering from T2DM complicated by CAD with clots. Downregulation of miR-450 in a mouse model with diabetic cardiomyopathy was previously reported in the literature[82]. The stimulating effect of miR-342-3p on the fibroblast growth factor 11 gene (FGF11) is known to promote the proliferation and migration of endothelial cells. Hyperinsulinemia results in the instability of interaction of miR-342 and FGF11, leading to vascular dysfunction in diabetic subjects[83]. Moreover, the upregulation of miR-342-5p in early atherosclerotic lesions in mice was discovered by Wei et al[84]. The authors proposed a possible use of miR-342-5p as a target for the therapy of atherosclerotic lesions.
MiR-1 and miR-133 also deserve attention as possible biomarkers for MVC development in T2DM patients. MiR-1 is known to be selectively expressed in heart muscle and to regulate cardiomyocyte growth responses[85]. MiR-133 also was shown to be expressed in cardiac muscle and to regulate cardiomyocyte proliferation through targeting CCND2 gene[86]. MiR-1 and miR-133 both were significantly associated with CAD risk in T2DM patients according to Al-Muhtaresh et al[87]. Although the association was stronger for miR-1 after adjustment for some anthropometric and clinical characteristics, authors proposed the use of a combined assessment of the expression levels of miR-1 and miR-133 to improve the diagnostic power for MVC prediction[87]. An elevated miR-1 expression level in individuals with CAD compared to healthy subjects was also shown in the previous research[88]. At the same time, the results were not replicated in another study, where miR-1 expression level did not show a significant difference between CAD patients and controls[89]. MiR-133b, in turn, was downregulated in CAD individuals according to Kumar et al[60].
MiR-204 is known to regulate cardiomyocyte proliferation through targeting the Jarid2 pathway[90,91]. Recently, there were several studies of miR-204 association with CAD that demonstrated significant downregulation of miR-204 expression in atherosclerotic plaques[92,93]. Interestingly, the decreased level of miR-204 expression in T2DM patients was associated with coronary artery calcification[94]. The contribution of miR-204 downregulation to vascular smooth muscle cell calcification also was previously shown by another group of authors in mice[95].
MiR-223 as well may be a substantial factor in the development of T2DM. As known, it is important for the regulation of GLUT4 expression and glucose uptake in the heart[96]. The correlation between miR-223 expression level and susceptibility to MVC of T2DM was described several times by now but remains controversial. For instance, in the study performed by Duan et al[97], platelet and plasma expression of miR-223 was significantly altered in diabetic patients with and without ischemic stroke compared to healthy controls, but there was no difference between T2DM and T2DM + ischemic stroke groups. Also, no differences were found between patients with ischemic stroke and controls[97]. According to another study, expression of miR-223 was decreased in diabetic subjects, but these changes were more noticeable in T2DM patients with ischemic stroke[98]. The lack of association between expression of miR-223-3p in PBMC and risk of ischemic stroke was confirmed afterward by Long et al[99], although expression level in PBMC of T2DM patients was also significantly decreased. An interesting finding regarding miR-223-3p was described recently. MiR-223-3p was found to be upregulated in atherosclerotic plaques in individuals with unstable CAD[100]. This fact allows suggesting the potential use of miR-223-3p as a biomarker for acute coronary events in diabetic subjects.
MiR-144 plays an important role in the T2DM pathogenesis, since one of its target genes, IRS1, encodes insulin receptor substrate 1[101]. IRS1/PI3K/AKT signaling pathway is involved in the translocation of glucose transporter type 4 into the plasma membrane[102]. MiR-144 may thus regulate glucose uptake by skeletal muscle, cardiac muscle, and adipose tissue. At the same time, there is a proven association between miR-144 and predisposition to CAD. A study by Chen et al[103] concluded that increased plasma level of miR-144 might serve as a promising biomarker for CAD and its severity. This association can be partially explained by the recent research that demonstrated the involvement of miR-144 in cholesterol and oxysterol metabolism in male CAD mice[104]. Moreover, altered expression of miR-144 was shown to be a risk factor for ischemic stroke in T2DM patients[98]. Upregulated miR-144 expression was detected in samples from individuals with large-vessel stroke in the study by Tan et al[105].
According to the results of recent research, miR-451a and miR-195-5p were found to be upregulated in diabetic patients after a transient ischemic attack in comparison to control patients and non-diabetic patients after a transient ischemic attack[106]. The earlier study also demonstrated an increased expression level of miR-451a in the serum of subjects with ischemic stroke[107]. It is worth noting that aberrant expression of miR-451a was also found in T2DM[108] and CAD patients[109]. MiR-195, in turn, was downregulated in diabetic subjects[110].
One of the most promising miRNAs for consideration as a biomarker for stroke in diabetic patients is miR-146a. Previous research showed a significant reduction in miR-146a expression level in whole blood and serum of patients with acute ischemic stroke[111,112]. The data contradict the results of the previous study that showed no difference in the expression of miR-146a when comparing stroke patients with controls, although expression in plasma and platelet was downregulated in diabetic individuals with stroke[97]. This inconsistency can be explained by the different material in which expression was analyzed. There are several theories about the mechanism of miR-146a involvement in stroke pathogenesis. Li et al[111] suggest upregulation of the miR-146a target, Fbxl10, protects neurons from ischemic damage. Also, researchers demonstrated a different mechanism for miR-146a involvement in stroke recovery. MiR-146a was shown to repress its other targets TRAF6 and IRAK1 which are components of TRAF6/NF-κB signaling pathway responsible for cellular responses to stress. This makes the expression of miR-146a important for the proliferation, migration, and angiogenesis ability of endothelial progenitor cells[113]. Upregulation of miR-146a also had an anti-inflammatory effect and prevented oxidative stress in mice after hemorrhagic stroke[114]. At the same time, alongside the protective potential in stroke, miR-146a was shown to participate in the process of atherogenesis[115]. For instance, miR-146a was upregulated in human atherosclerotic plaques in the Tampere Vascular Study[43]. Cheng et al[116] demonstrated knockout of miR-146a in the vasculature might contribute to atherogenesis by endothelial activation. Nguyen and co-authors described atherosclerosis progression resulting from a miR-146a-mediated decrease in cell migration and macrophage entrapment in vascular intima[117].
The data obtained by Sheikhbahaei et al[118] allows raising the question of the potential use of miR-503 as a biomarker for ischemic stroke in T2DM patients. The study also provides evidence that miR-503 may serve as an indicator of short-term outcomes of ischemic stroke in diabetic individuals. Another group of authors earlier demonstrated upregulated miR-503 in ischemic limbs of diabetic mice. Circulating miR-503 was also elevated in T2DM patients[119]. The mechanism of influence on the vascular wall was explained by the inhibiting of vascular smooth muscle cells migration and proliferation through targeting the INSR gene[120]. The same effect was demonstrated afterward in HUVECs in high glucose conditions[121].
With the expansion of knowledge of epigenetic mechanisms, the question of the importance of DNA methylation and histone acetylation in the pathogenesis of MVC of T2DM arises. The main mechanism in the development of MVC of T2DM is atherosclerotic lesions of major arteries. There is wide evidence of a key role of epigenetics in atherosclerotic plaque formation. The association of DNA methylation and histone acetylation with atherosclerosis was discussed in detail in a recent review by Lee. In particular, the authors described a cascade that begins with endothelial dysfunction through Dnmt1 overexpression mediated by DNA methylation changes in mice with disturbed blood flow and after leads to infiltration into the intima and subintima by macrophages[122,123]. A significant DNA hypomethylation was detected in human atherosclerotic lesions and ApoE knock-out mice[124]. The same processes are affected by histone acetylation, as illustrated by numerous studies[123]. Histone acetylation is connected with the expression of matrix metalloproteinases which play an important role in extracellular matrix destruction that leads to plaque rupture[125,126]. Histone modifications were also reported to contribute to oxLDL mediated expression of inflammatory IL-8 and monocyte-chemoattractant protein-1 in endothelial cells[127], increasing the endothelium permeability and subsequent infiltration by macrophages. Another process in atherogenesis is related to increased H3K9 and H3K27 acetylation, which is associated with smooth muscle cell proliferation, migration, and stabilization of plaque[128].
The influence of DNA methylation changes on T2DM pathogenesis was extensively studied. A recent literature review[129] provides information on hypermethylated genes responsible for beta cell function in diabetic patients. These genes include promoters of INS, encoding insulin[130], PDX1, responsible for beta cell development and regeneration[131,132], PPARGC1A, significantly associated with type 2 diabetes during increased physical activity[133,134], and GLP1R, encoding receptor of gluca
The inflammatory process also plays an important role in the development of diabetes[136]. Macrophage activation is mediated by epigenetic changes and contributes to chronic inflammation and the pathogenesis of diabetes complications. The main mechanisms of epigenetic modification of macrophage activity were described in detail in a review article by Ahmed et al[137]. The authors described a hyperlipidemia-induces pathway of macrophage activation through DNA methylation and suppression of anti-inflammatory genes in macrophages of diabetic rats with hindlimb ischemia[138] and obese mice[139]. Another reported activation pathway was through increased histone methylation of the promoter of RelA (or p65) subunit of NF-κB (nuclear factor-κappa beta), a regulator of innate immune cell responses[140-142]. The research also concluded hypoxia stimulated macrophage activation through histone acetylation.
Overweight and obesity are associated with insulin resistance and metabolic syndrome. It is a well-known risk factor for T2DM and its complications. Several recent studies indicated the altered level of DNA methylation in the adipose tissue of diabetic patients. Genes with aberrant methylation status were mainly related to carbohydrate and lipid metabolism, insulin resistance, inflammation, and cell cycle regulation[143]. For instance, methylation of PPARG, KCNQ1, TCF7L2, and IRS1 was different in adipose tissue of individuals with T2DM and subjects from the control group[144]. DNA methylation level was also increased in B cells from obese and T2DM patients and natural killer cells from diabetic patients, and associated with insulin resistance[145,146].
Epigenetic changes may also mediate the impact of environmental factors on the development of T2DM, in addition to controlling the regulation of gene expression. For example, transient spikes of hyperglycemia result in epigenetic changes in the promoter of RelA subunit of the nuclear factor κB in aortic endothelial cells in mice, leading to upregulated RelA expression, associated with endothelial cell inflammation[147,148].
Prophylactic measures play an important role in preventing the development of MVC in T2DM patients and include correction of hyperglycemia, dyslipidemia, anti-hypertensive therapy, and lifestyle modification with diet, physical activity, and smoking cessation[149]. Main standards for hypoglycemic therapy were described by the American Diabetes Association and European Association for the Study of Diabetes[150,151]. The effectiveness of antiglycemic therapy largely depends on genetic factors, which was repeatedly shown by many pharmacogenetic candidate gene studies and GWAS. The data on the main polymorphisms affecting the effectiveness of both metformin and new oral hypoglycemic drugs were summarized in a recent review by Mannino[152]. Interindividual variability in response to different classes of antihypertensive drugs and adverse reactions may also be partially explained by genetic polymorphisms[153]. Pharmacogenetics is of particular importance in the correction of dyslipidemia in T2DM due to muscle toxicity of statins, observed in about 10% of patients[154]. However, the results of many pharmacogenetic studies conducted in small cohorts were not replicated in larger populations. Also, the inconsistency may be associated with the ethnicity of the population analyzed, which must be taken into account when prescribing personalized therapy[155].
Coronavirus disease 2019 (COVID-19) pandemic made it clear that the presence of metabolic syndrome and its components (T2DM, hypertension, and obesity) significantly aggravated the severity of infectious diseases[156,157]. The most common comorbidity in COVID-19 patients is hypertension, followed by T2DM, which are observed in 30% and 19% of patients respectively[158]. Diabetic patients with COVID-19 are more likely to suffer from CVD and associated vascular complications which lead to a poor prognosis and an increased risk of in-hospital death[159]. The high incidence of MVC in COVID-19 patients with T2DM and hypertension can be explained by the virus targeting the endothelium[160]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is known to bind to angiotensin-converting enzyme 2, encoded by ACE2 gene, for entering cells[161]. Previous researches showed that variants in the ACE2 and its expression level might influence the severity of the disease[162,163]. Also, the question of the possible influence of ACE2 expression on the clinical outcome during therapy with ACE inhibitors and angiotensin II receptor blocker remains open. A recent study by Sardu et al[164] showed that anti-hypertensive drugs did not affect the clinical outcomes in COVID-19 patients. Moreover, a combination of those drugs with anti-inflammatory and immune therapies might even improve the prognosis. ACE2 is expressed in lungs, kidneys, heart, and, importantly, in pancreatic islets, namely, in β-cells producing insulin[165,166]. Besides, there is evidence of the participation of ACE2 in glucose metabolism. Ace2-knockout mice had β-cells defects in research performed by Bernardi et al[167], although the defect was compensated by energy shift to glucose utilization. ACE2 deletion also led to cardiovascular dysfunction, which was demonstrated in studies in mice[162,168,169]. At the same time, ACE2 overexpression resulted in improvement of β-cell function[170]. The association between ACE2 polymorphisms and SARS-CoV-2 susceptibility or severity of outcomes was investigated in numerous previous studies. For instance, ACE2 variants were shown to be protective against COVID-19 in African and Eastern Mediterranean populations, but no association was found in American and European cohorts[171]. Three rare variants in ACE2 were found to have a possible impact on the disease severity in patients of Russian ancestry[172]. A recent analysis of a large genomic dataset allowed to identify 17 potentially protective polymorphisms in ACE2 gene and 9 variants increasing susceptibility[173]. Interestingly, the ACE2 gene promoter region contains a binding site for hepatocyte nuclear factor 1 alpha that induces ACE2 expression in pancreatic islet cells[174]. Polymorphisms within the promoter region of ACE2 may thus also contribute to the severity of disease, especially in diabetic patients, and should be considered as possible variants predisposing to susceptibility to SARS-CoV-2 and severe course of COVID-19 disease[175].
Type 2 diabetes is associated with vascular complications of both small and large vessels, which seriously impair the overall quality of life and can result in lower life expectancy. The discovery of genetic determinants of T2DM complications would advance the development of personalized treatment of diabetic patients and significantly reduce adverse outcomes. Despite the recent progress in the discovery of new genetic and epigenetic determinants of T2DM and its complications, the pathogenetic mechanisms of their participation remain largely unknown. An additional challenge for genetic studies of complex diseases is to establish the causal relationship of the genes involved in pathogenesis and their interactions in the development of the underlying disease and comorbid pathologies. Further research in the large independent cohorts, deep phenotyping of participants, and functional studies are needed to reveal pathogenetic pathways underlying the disorders. It is especially important to pay attention to genetic and epigenetic factors during pregnancy or in cases when a diabetic individual has a comorbid disease. As for pregnancy, the frequency of MI was shown to be 3-4 times higher during the peripartum period[176]. As was discussed above, the presence of an acute infectious pathology can significantly increase the risks of diabetes complications. The study of the development of macrovascular complications in diabetic patients with SARS-CoV-2 infection may be of particular interest.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Verma AK S-Editor: Gao CC L-Editor: Filipodia P-Editor: Wang LL
| 1. | Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diabetes Res Clin Pract. 2011;94:311-321. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Cole JB, Florez JC. Genetics of diabetes mellitus and diabetes complications. Nat Rev Nephrol. 2020;16:377-390. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Cade WT. Diabetes-related microvascular and macrovascular diseases in the physical therapy setting. Phys Ther. 2008;88:1322-1335. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Rask-Madsen C, King GL. Vascular complications of diabetes: mechanisms of injury and protective factors. Cell Metab. 2013;17:20-33. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Grant PJ. Diabetes mellitus as a prothrombotic condition. J Intern Med. 2007;262:157-172. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Rich SS. Mapping genes in diabetes. Genetic epidemiological perspective. Diabetes. 1990;39:1315-1319. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Malik R, Chauhan G, Traylor M, Sargurupremraj M, Okada Y, Mishra A, Rutten-Jacobs L, Giese AK, van der Laan SW, Gretarsdottir S, Anderson CD, Chong M, Adams HHH, Ago T, Almgren P, Amouyel P, Ay H, Bartz TM, Benavente OR, Bevan S, Boncoraglio GB, Brown RD Jr, Butterworth AS, Carrera C, Carty CL, Chasman DI, Chen WM, Cole JW, Correa A, Cotlarciuc I, Cruchaga C, Danesh J, de Bakker PIW, DeStefano AL, den Hoed M, Duan Q, Engelter ST, Falcone GJ, Gottesman RF, Grewal RP, Gudnason V, Gustafsson S, Haessler J, Harris TB, Hassan A, Havulinna AS, Heckbert SR, Holliday EG, Howard G, Hsu FC, Hyacinth HI, Ikram MA, Ingelsson E, Irvin MR, Jian X, Jiménez-Conde J, Johnson JA, Jukema JW, Kanai M, Keene KL, Kissela BM, Kleindorfer DO, Kooperberg C, Kubo M, Lange LA, Langefeld CD, Langenberg C, Launer LJ, Lee JM, Lemmens R, Leys D, Lewis CM, Lin WY, Lindgren AG, Lorentzen E, Magnusson PK, Maguire J, Manichaikul A, McArdle PF, Meschia JF, Mitchell BD, Mosley TH, Nalls MA, Ninomiya T, O'Donnell MJ, Psaty BM, Pulit SL, Rannikmäe K, Reiner AP, Rexrode KM, Rice K, Rich SS, Ridker PM, Rost NS, Rothwell PM, Rotter JI, Rundek T, Sacco RL, Sakaue S, Sale MM, Salomaa V, Sapkota BR, Schmidt R, Schmidt CO, Schminke U, Sharma P, Slowik A, Sudlow CLM, Tanislav C, Tatlisumak T, Taylor KD, Thijs VNS, Thorleifsson G, Thorsteinsdottir U, Tiedt S, Trompet S, Tzourio C, van Duijn CM, Walters M, Wareham NJ, Wassertheil-Smoller S, Wilson JG, Wiggins KL, Yang Q, Yusuf S; AFGen Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium; International Genomics of Blood Pressure (iGEN-BP) Consortium; INVENT Consortium; STARNET; Bis JC, Pastinen T, Ruusalepp A, Schadt EE, Koplev S, Björkegren JLM, Codoni V, Civelek M, Smith NL, Trégouët DA, Christophersen IE, Roselli C, Lubitz SA, Ellinor PT, Tai ES, Kooner JS, Kato N, He J, van der Harst P, Elliott P, Chambers JC, Takeuchi F, Johnson AD; BioBank Japan Cooperative Hospital Group; COMPASS Consortium; EPIC-CVD Consortium; EPIC-InterAct Consortium; International Stroke Genetics Consortium (ISGC); METASTROKE Consortium; Neurology Working Group of the CHARGE Consortium; NINDS Stroke Genetics Network (SiGN); UK Young Lacunar DNA Study; MEGASTROKE Consortium; Sanghera DK, Melander O, Jern C, Strbian D, Fernandez-Cadenas I, Longstreth WT Jr, Rolfs A, Hata J, Woo D, Rosand J, Pare G, Hopewell JC, Saleheen D, Stefansson K, Worrall BB, Kittner SJ, Seshadri S, Fornage M, Markus HS, Howson JMM, Kamatani Y, Debette S, Dichgans M. Multiancestry genome-wide association study of 520,000 subjects identifies 32 Loci associated with stroke and stroke subtypes. Nat Genet. 2018;50:524-537. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Erdmann J, Kessler T, Munoz Venegas L, Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. 2018;114:1241-1257. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117-171. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | World Health Organization. Global report on diabetes. [cited 1 March 2021]. In: World Health Organization [Internet]. 2016: 1-83. Available from: https://www.who.int/publications/i/item/9789241565257. [Cited in This Article: ] |
| 11. | Doria A, Wojcik J, Xu R, Gervino EV, Hauser TH, Johnstone MT, Nolan D, Hu FB, Warram JH. Interaction between poor glycemic control and 9p21 Locus on risk of coronary artery disease in type 2 diabetes. JAMA. 2008;300:2389-2397. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Qi L, Parast L, Cai T, Powers C, Gervino EV, Hauser TH, Hu FB, Doria A. Genetic susceptibility to coronary heart disease in type 2 diabetes: 3 independent studies. J Am Coll Cardiol. 2011;58:2675-2682. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Frayling TM, Stoneman CE. Mendelian randomisation in type 2 diabetes and coronary artery disease. Curr Opin Genet Dev. 2018;50:111-120. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Goodarzi MO, Rotter JI. Genetics Insights in the Relationship Between Type 2 Diabetes and Coronary Heart Disease. Circ Res. 2020;126:1526-1548. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491-1493. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488-1491. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Zhang LW, Li JP, Duan FF, Liu ZK, Zhan SY, Hu YH, Jiang J, Zhang Y, Huo Y, Chen DF. Interaction of type 2 diabetes mellitus with chromosome 9p21 rs10757274 polymorphism on the risk of myocardial infarction: a case-control study in Chinese population. BMC Cardiovasc Disord. 2014;14:170. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Qi L, Qi Q, Prudente S, Mendonca C, Andreozzi F, di Pietro N, Sturma M, Novelli V, Mannino GC, Formoso G, Gervino EV, Hauser TH, Muehlschlegel JD, Niewczas MA, Krolewski AS, Biolo G, Pandolfi A, Rimm E, Sesti G, Trischitta V, Hu F, Doria A. Association between a genetic variant related to glutamic acid metabolism and coronary heart disease in individuals with type 2 diabetes. JAMA. 2013;310:821-828. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Beaney KE, Cooper JA, McLachlan S, Wannamethee SG, Jefferis BJ, Whincup P, Ben-Shlomo Y, Price JF, Kumari M, Wong A, Ong K, Hardy R, Kuh D, Kivimaki M, Kangas AJ, Soininen P, Ala-Korpela M, Drenos F, Humphries SE; UCLEB consortium. Variant rs10911021 that associates with coronary heart disease in type 2 diabetes, is associated with lower concentrations of circulating HDL cholesterol and large HDL particles but not with amino acids. Cardiovasc Diabetol. 2016;15:115. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Look AHEAD Research Group. Prospective Association of GLUL rs10911021 With Cardiovascular Morbidity and Mortality Among Individuals With Type 2 Diabetes: The Look AHEAD Study. Diabetes. 2016;65:297-302. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Fagerberg L, Hallström BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjöstedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlén M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397-406. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Lin D, Xiao Y, Huang B, Wu X, Chen C, Liang Y, Zeng. O-6-methylguanine DNA methyltransferase is a favorable biomarker with proliferation suppressive potential in Breast Cancer. J Cancer. 2020;11:6326-6336. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Shah HS, Gao H, Morieri ML, Skupien J, Marvel S, Paré G, Mannino GC, Buranasupkajorn P, Mendonca C, Hastings T, Marcovina SM, Sigal RJ, Gerstein HC, Wagner MJ, Motsinger-Reif AA, Buse JB, Kraft P, Mychaleckyj JC, Doria A. Genetic Predictors of Cardiovascular Mortality During Intensive Glycemic Control in Type 2 Diabetes: Findings From the ACCORD Clinical Trial. Diabetes Care. 2016;39:1915-1924. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Fall T, Gustafsson S, Orho-Melander M, Ingelsson E. Genome-wide association study of coronary artery disease among individuals with diabetes: the UK Biobank. Diabetologia. 2018;61:2174-2179. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | van Zuydam NR, Ladenvall C, Voight BF, Strawbridge RJ, Fernandez-Tajes J, Rayner NW, Robertson NR, Mahajan A, Vlachopoulou E, Goel A, Kleber ME, Nelson CP, Kwee LC, Esko T, Mihailov E, Mägi R, Milani L, Fischer K, Kanoni S, Kumar J, Song C, Hartiala JA, Pedersen NL, Perola M, Gieger C, Peters A, Qu L, Willems SM, Doney ASF, Morris AD, Zheng Y, Sesti G, Hu FB, Qi L, Laakso M, Thorsteinsdottir U, Grallert H, van Duijn C, Reilly MP, Ingelsson E, Deloukas P, Kathiresan S, Metspalu A, Shah SH, Sinisalo J, Salomaa V, Hamsten A, Samani NJ, März W, Hazen SL, Watkins H, Saleheen D, Morris AP, Colhoun HM, Groop L, McCarthy MI, Palmer CNA; SUMMIT Steering Committee; CARDIOGRAMplusC4D Steering Committee*. Genetic Predisposition to Coronary Artery Disease in Type 2 Diabetes Mellitus. Circ Genom Precis Med. 2020;13:e002769. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Yang R, Pedersen NL, Bao C, Xu W, Xu H, Song R, Qi X. Type 2 diabetes in midlife and risk of cerebrovascular disease in late life: a prospective nested case-control study in a nationwide Swedish twin cohort. Diabetologia. 2019;62:1403-1411. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Air EL, Kissela BM. Diabetes, the metabolic syndrome, and ischemic stroke: epidemiology and possible mechanisms. Diabetes Care. 2007;30:3131-3140. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Lee M, Saver JL, Hong KS, Song S, Chang KH, Ovbiagele B. Effect of pre-diabetes on future risk of stroke: meta-analysis. BMJ. 2012;344:e3564. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Kissela B, Air E. Diabetes: impact on stroke risk and poststroke recovery. Semin Neurol. 2006;26:100-107. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Larsson SC, Wallin A, Håkansson N, Stackelberg O, Bäck M, Wolk A. Type 1 and type 2 diabetes mellitus and incidence of seven cardiovascular diseases. Int J Cardiol. 2018;262:66-70. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Georgakis MK, Harshfield EL, Malik R, Franceschini N, Langenberg C, Wareham NJ, Markus HS, Dichgans M. Diabetes Mellitus, Glycemic Traits, and Cerebrovascular Disease: A Mendelian Randomization Study. Neurology. 2021;96:e1732-e1742. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Kim M, Zhang X. The Profiling and Role of miRNAs in Diabetes Mellitus. J Diabetes Clin Res. 2019;1:5-23. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Al-Kafaji G, Al-Mahroos G, Abdulla Al-Muhtaresh H, Sabry MA, Abdul Razzak R, Salem AH. Circulating endothelium-enriched microRNA-126 as a potential biomarker for coronary artery disease in type 2 diabetes mellitus patients. Biomarkers. 2017;22:268-278. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Amr KS, Abdelmawgoud H, Ali ZY, Shehata S, Raslan HM. Potential value of circulating microRNA-126 and microRNA-210 as biomarkers for type 2 diabetes with coronary artery disease. Br J Biomed Sci. 2018;75:82-87. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261-271. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272-284. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Wang X, Lian Y, Wen X, Guo J, Wang Z, Jiang S, Hu Y. Expression of miR-126 and its potential function in coronary artery disease. Afr Health Sci. 2017;17:474-480. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Sun X, Zhang M, Sanagawa A, Mori C, Ito S, Iwaki S, Satoh H, Fujii S. Circulating microRNA-126 in patients with coronary artery disease: correlation with LDL cholesterol. Thromb J. 2012;10:16. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Rezk NA, Sabbah NA, Saad MS. Role of MicroRNA 126 in screening, diagnosis, and prognosis of diabetic patients in Egypt. IUBMB Life. 2016;68:452-458. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Liu Y, Gao G, Yang C, Zhou K, Shen B, Liang H, Jiang X. The role of circulating microRNA-126 (miR-126): a novel biomarker for screening prediabetes and newly diagnosed type 2 diabetes mellitus. Int J Mol Sci. 2014;15:10567-10577. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: the master hypoxamir. Microcirculation. 2012;19:215-223. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94-100. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Raitoharju E, Lyytikäinen LP, Levula M, Oksala N, Mennander A, Tarkka M, Klopp N, Illig T, Kähönen M, Karhunen PJ, Laaksonen R, Lehtimäki T. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211-217. [PubMed] [DOI] [Cited in This Article: ] |
| 44. | Eken SM, Jin H, Chernogubova E, Li Y, Simon N, Sun C, Korzunowicz G, Busch A, Bäcklund A, Österholm C, Razuvaev A, Renné T, Eckstein HH, Pelisek J, Eriksson P, González Díez M, Perisic Matic L, Schellinger IN, Raaz U, Leeper NJ, Hansson GK, Paulsson-Berne G, Hedin U, Maegdefessel L. MicroRNA-210 Enhances Fibrous Cap Stability in Advanced Atherosclerotic Lesions. Circ Res. 2017;120:633-644. [PubMed] [DOI] [Cited in This Article: ] |
| 45. | Li Y, Yang C, Zhang L, Yang P. MicroRNA-210 induces endothelial cell apoptosis by directly targeting PDK1 in the setting of atherosclerosis. Cell Mol Biol Lett. 2017;22:3. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Guan Y, Song X, Sun W, Wang Y, Liu B. Effect of Hypoxia-Induced MicroRNA-210 Expression on Cardiovascular Disease and the Underlying Mechanism. Oxid Med Cell Longev. 2019;2019:4727283. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878-15883. [PubMed] [DOI] [Cited in This Article: ] |
| 48. | Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, Robbins RC, Wu JC. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124-S131. [PubMed] [DOI] [Cited in This Article: ] |
| 49. | Zaccagnini G, Maimone B, Di Stefano V, Fasanaro P, Greco S, Perfetti A, Capogrossi MC, Gaetano C, Martelli F. Hypoxia-induced miR-210 modulates tissue response to acute peripheral ischemia. Antioxid Redox Signal. 2014;21:1177-1188. [PubMed] [DOI] [Cited in This Article: ] |
| 50. | Zaccagnini G, Maimone B, Fuschi P, Maselli D, Spinetti G, Gaetano C, Martelli F. Overexpression of miR-210 and its significance in ischemic tissue damage. Sci Rep. 2017;7:9563. [PubMed] [DOI] [Cited in This Article: ] |
| 51. | Li T, Cao H, Zhuang J, Wan J, Guan M, Yu B, Li X, Zhang W. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2011;412:66-70. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Pek SL, Sum CF, Lin MX, Cheng AK, Wong MT, Lim SC, Tavintharan S. Circulating and visceral adipose miR-100 is down-regulated in patients with obesity and Type 2 diabetes. Mol Cell Endocrinol. 2016;427:112-123. [PubMed] [DOI] [Cited in This Article: ] |
| 53. | Li X, Jia Z, Zhao X, Xu M, Chen M. Expression of miR-210 in the peripheral blood of patients with newly diagnosed type 2 diabetes mellitus and its effect on the number and function of endothelial progenitor cells. Microvasc Res. 2020;131:104032. [PubMed] [DOI] [Cited in This Article: ] |
| 54. | Al-Hayali MA, Sozer V, Durmus S, Erdenen F, Altunoglu E, Gelisgen R, Atukeren P, Atak PG, Uzun H. Clinical Value of Circulating Microribonucleic Acids miR-1 and miR-21 in Evaluating the Diagnosis of Acute Heart Failure in Asymptomatic Type 2 Diabetic Patients. Biomolecules. 2019;9. [PubMed] [DOI] [Cited in This Article: ] |
| 55. | Seyhan AA, Nunez Lopez YO, Xie H, Yi F, Mathews C, Pasarica M, Pratley RE. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: a pilot cross-sectional study. Sci Rep. 2016;6:31479. [PubMed] [DOI] [Cited in This Article: ] |
| 56. | Nunez Lopez YO, Garufi G, Seyhan AA. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol Biosyst. 2016;13:106-121. [PubMed] [DOI] [Cited in This Article: ] |
| 57. | Andoorfar S, Hosseini Tafreshi SA, Rezvani Z. Assessment of the expression level of miRNA molecules using a semi-quantitative RT-PCR approach. Mol Biol Rep. 2019;46:5057-5062. [PubMed] [DOI] [Cited in This Article: ] |
| 58. | La Sala L, Mrakic-Sposta S, Tagliabue E, Prattichizzo F, Micheloni S, Sangalli E, Specchia C, Uccellatore AC, Lupini S, Spinetti G, de Candia P, Ceriello A. Circulating microRNA-21 is an early predictor of ROS-mediated damage in subjects with high risk of developing diabetes and in drug-naïve T2D. Cardiovasc Diabetol. 2019;18:18. [PubMed] [DOI] [Cited in This Article: ] |
| 59. | Guan JJ, Zhang Y, Liu YJ. [Effects of miRNA-1,miRNA-21 in plasma on in-stent restenosis in patients with coronary heart disease and diabetes mellitus after percutaneous coronary intervention]. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2018;34:304-308 384. [PubMed] [DOI] [Cited in This Article: ] |
| 60. | Kumar D, Narang R, Sreenivas V, Rastogi V, Bhatia J, Saluja D, Srivastava K. Circulatory miR-133b and miR-21 as Novel Biomarkers in Early Prediction and Diagnosis of Coronary Artery Disease. Genes (Basel). 2020;11. [PubMed] [DOI] [Cited in This Article: ] |
| 61. | Loyer X, Potteaux S, Vion AC, Guérin CL, Boulkroun S, Rautou PE, Ramkhelawon B, Esposito B, Dalloz M, Paul JL, Julia P, Maccario J, Boulanger CM, Mallat Z, Tedgui A. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ Res. 2014;114:434-443. [PubMed] [DOI] [Cited in This Article: ] |
| 62. | Dutzmann J, Daniel JM, Bauersachs J, Sedding D. MiR-92a – a key player in cardiovascular remodeling. RNA Dis. 2014;1:e371. [DOI] [Cited in This Article: ] |
| 63. | Jiang Y, Wang HY, Li Y, Guo SH, Zhang L, Cai JH. Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Sci Rep. 2014;4:5026. [PubMed] [DOI] [Cited in This Article: ] |
| 64. | Niculescu LS, Simionescu N, Sanda GM, Carnuta MG, Stancu CS, Popescu AC, Popescu MR, Vlad A, Dimulescu DR, Simionescu M, Sima AV. MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS One. 2015;10:e0140958. [PubMed] [DOI] [Cited in This Article: ] |
| 65. | Wang Z, Zhang J, Zhang S, Yan S, Wang Z, Wang C, Zhang X. MiR30e and miR92a are related to atherosclerosis by targeting ABCA1. Mol Med Rep. 2019;19:3298-3304. [PubMed] [DOI] [Cited in This Article: ] |
| 66. | Parahuleva MS, Lipps C, Parviz B, Hölschermann H, Schieffer B, Schulz R, Euler G. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci Rep. 2018;8:7823. [PubMed] [DOI] [Cited in This Article: ] |
| 67. | Zhang Y, Cheng J, Chen F, Wu C, Zhang J, Ren X, Pan Y, Nie B, Li Q, Li Y. Circulating endothelial microparticles and miR-92a in acute myocardial infarction. Biosci Rep. 2017;37. [PubMed] [DOI] [Cited in This Article: ] |
| 68. | Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF, Jeyaseelan K. Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab. 2012;97:E2271-E2276. [PubMed] [DOI] [Cited in This Article: ] |
| 69. | Wang W, Li Z, Zheng Y, Yan M, Cui Y, Jiang J. Circulating microRNA-92a level predicts acute coronary syndrome in diabetic patients with coronary heart disease. Lipids Health Dis. 2019;18:22. [PubMed] [DOI] [Cited in This Article: ] |
| 70. | Luo M, Wang G, Xu C, Zeng M, Lin F, Wu J, Wan Q. Circulating miR-30c as a predictive biomarker of type 2 diabetes mellitus with coronary heart disease by regulating PAI-1/VN interactions. Life Sci. 2019;239:117092. [PubMed] [DOI] [Cited in This Article: ] |
| 71. | Ceolotto G, Giannella A, Albiero M, Kuppusamy M, Radu C, Simioni P, Garlaschelli K, Baragetti A, Catapano AL, Iori E, Fadini GP, Avogaro A, Vigili de Kreutzenberg S. miR-30c-5p regulates macrophage-mediated inflammation and pro-atherosclerosis pathways. Cardiovasc Res. 2017;113:1627-1638. [PubMed] [DOI] [Cited in This Article: ] |
| 72. | Chen C, Yang S, Li H, Yin Z, Fan J, Zhao Y, Gong W, Yan M, Wang DW. Mir30c Is Involved in Diabetic Cardiomyopathy through Regulation of Cardiac Autophagy via BECN1. Mol Ther Nucleic Acids. 2017;7:127-139. [PubMed] [DOI] [Cited in This Article: ] |
| 73. | Jiang H, Toscano JF, Song SS, Schlick KH, Dumitrascu OM, Pan J, Lyden PD, Saver JL, Gonzalez NR. Differential expression of circulating exosomal microRNAs in refractory intracranial atherosclerosis associated with antiangiogenesis. Sci Rep. 2019;9:19429. [PubMed] [DOI] [Cited in This Article: ] |
| 74. | Hoekstra M, van der Lans CA, Halvorsen B, Gullestad L, Kuiper J, Aukrust P, van Berkel TJ, Biessen EA. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem Biophys Res Commun. 2010;394:792-797. [PubMed] [DOI] [Cited in This Article: ] |
| 75. | Shi X, Chen X. Effect of microRNA-370 on coronary atherosclerosis and its underlying mechanism. Exp Ther Med. 2019;17:115-122. [PubMed] [DOI] [Cited in This Article: ] |
| 76. | Qiang L, Tsuchiya K, Kim-Muller JY, Lin HV, Welch C, Accili D. Increased atherosclerosis and endothelial dysfunction in mice bearing constitutively deacetylated alleles of Foxo1 gene. J Biol Chem. 2012;287:13944-13951. [PubMed] [DOI] [Cited in This Article: ] |
| 77. | Kedenko L, Lamina C, Kedenko I, Kollerits B, Kiesslich T, Iglseder B, Kronenberg F, Paulweber B. Genetic polymorphisms at SIRT1 and FOXO1 are associated with carotid atherosclerosis in the SAPHIR cohort. BMC Med Genet. 2014;15:112. [PubMed] [DOI] [Cited in This Article: ] |
| 78. | Gao W, He HW, Wang ZM, Zhao H, Lian XQ, Wang YS, Zhu J, Yan JJ, Zhang DG, Yang ZJ, Wang LS. Plasma levels of lipometabolism-related miR-122 and miR-370 are increased in patients with hyperlipidemia and associated with coronary artery disease. Lipids Health Dis. 2012;11:55. [PubMed] [DOI] [Cited in This Article: ] |
| 79. | Liu H, Yang N, Fei Z, Qiu J, Ma D, Liu X, Cai G, Li S. Analysis of plasma miR-208a and miR-370 expression levels for early diagnosis of coronary artery disease. Biomed Rep. 2016;5:332-336. [PubMed] [DOI] [Cited in This Article: ] |
| 80. | Motawae TM, Ismail MF, Shabayek MI, Seleem MM. MicroRNAs 9 and 370 Association with Biochemical Markers in T2D and CAD Complication of T2D. PLoS One. 2015;10:e0126957. [PubMed] [DOI] [Cited in This Article: ] |
| 81. | Seleem M, Shabayek M, Ewida HA. MicroRNAs 342 and 450 together with NOX-4 activity and their association with coronary artery disease in diabetes. Diabetes Metab Res Rev. 2019;35:e3130. [PubMed] [DOI] [Cited in This Article: ] |
| 82. | Chavali V, Tyagi SC, Mishra PK. Differential expression of dicer, miRNAs, and inflammatory markers in diabetic Ins2+/- Akita hearts. Cell Biochem Biophys. 2014;68:25-35. [PubMed] [DOI] [Cited in This Article: ] |
| 83. | Cheng S, Cui Y, Fan L, Mu X, Hua Y. T2DM inhibition of endothelial miR-342-3p facilitates angiogenic dysfunction via repression of FGF11 signaling. Biochem Biophys Res Commun. 2018;503:71-78. [PubMed] [DOI] [Cited in This Article: ] |
| 84. | Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, Heyll K, Corbalán-Campos J, Hartmann P, Thiemann A, Weber C, Schober A. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609-1619. [PubMed] [DOI] [Cited in This Article: ] |
| 85. | Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, Pu WT. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193-2204. [PubMed] [DOI] [Cited in This Article: ] |
| 86. | Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242-3254. [PubMed] [DOI] [Cited in This Article: ] |
| 87. | Al-Muhtaresh HA, Salem AH, Al-Kafaji G. Upregulation of Circulating Cardiomyocyte-Enriched miR-1 and miR-133 Associate with the Risk of Coronary Artery Disease in Type 2 Diabetes Patients and Serve as Potential Biomarkers. J Cardiovasc Transl Res. 2019;12:347-357. [PubMed] [DOI] [Cited in This Article: ] |
| 88. | Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486-491. [PubMed] [DOI] [Cited in This Article: ] |
| 89. | Nabiałek E, Wańha W, Kula D, Jadczyk T, Krajewska M, Kowalówka A, Dworowy S, Hrycek E, Włudarczyk W, Parma Z, Michalewska-Włudarczyk A, Pawłowski T, Ochała B, Jarząb B, Tendera M, Wojakowski W. Circulating microRNAs (miR-423-5p, miR-208a and miR-1) in acute myocardial infarction and stable coronary heart disease. Minerva Cardioangiol. 2013;61:627-637. [PubMed] [Cited in This Article: ] |
| 90. | Toyoda M, Shirato H, Nakajima K, Kojima M, Takahashi M, Kubota M, Suzuki-Migishima R, Motegi Y, Yokoyama M, Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev Cell. 2003;5:85-97. [PubMed] [DOI] [Cited in This Article: ] |
| 91. | Liang D, Li J, Wu Y, Zhen L, Li C, Qi M, Wang L, Deng F, Huang J, Lv F, Liu Y, Ma X, Yu Z, Zhang Y, Chen YH. miRNA-204 drives cardiomyocyte proliferation via targeting Jarid2. Int J Cardiol. 2015;201:38-48. [PubMed] [DOI] [Cited in This Article: ] |
| 92. | Torella D, Iaconetti C, Tarallo R, Marino F, Giurato G, Veneziano C, Aquila I, Scalise M, Mancuso T, Cianflone E, Valeriano C, Marotta P, Tammè L, Vicinanza C, Sasso FC, Cozzolino D, Torella M, Weisz A, Indolfi C. miRNA Regulation of the Hyperproliferative Phenotype of Vascular Smooth Muscle Cells in Diabetes. Diabetes. 2018;67:2554-2568. [PubMed] [DOI] [Cited in This Article: ] |
| 93. | Wang N, Yuan Y, Sun S, Liu G. microRNA-204-5p Participates in Atherosclerosis Via Targeting MMP-9. Open Med (Wars). 2020;15:231-239. [PubMed] [DOI] [Cited in This Article: ] |
| 94. | Ding YD, Pei YQ, Rui-Wang, Yang JX, Zhao YX, Liu XL, Shen H, Ma Q, Zhang S, Ge HL. Association of Plasma MiRNA-204 and the Presence and Severity of Coronary Artery Calcification in Patients With Type 2 Diabetes. Angiology. 2021;72:451-458. [PubMed] [DOI] [Cited in This Article: ] |
| 95. | Cui RR, Li SJ, Liu LJ, Yi L, Liang QH, Zhu X, Liu GY, Liu Y, Wu SS, Liao XB, Yuan LQ, Mao DA, Liao EY. MicroRNA-204 regulates vascular smooth muscle cell calcification in vitro and in vivo. Cardiovasc Res. 2012;96:320-329. [PubMed] [DOI] [Cited in This Article: ] |
| 96. | Lu H, Buchan RJ, Cook SA. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc Res. 2010;86:410-420. [PubMed] [DOI] [Cited in This Article: ] |
| 97. | Duan X, Zhan Q, Song B, Zeng S, Zhou J, Long Y, Lu J, Li Z, Yuan M, Chen X, Yang Q, Xia J. Detection of platelet microRNA expression in patients with diabetes mellitus with or without ischemic stroke. J Diabetes Complications. 2014;28:705-710. [PubMed] [DOI] [Cited in This Article: ] |
| 98. | Yang S, Zhao J, Chen Y, Lei M. Biomarkers Associated with Ischemic Stroke in Diabetes Mellitus Patients. Cardiovasc Toxicol. 2016;16:213-222. [PubMed] [DOI] [Cited in This Article: ] |
| 99. | Long Y, Zhan Q, Yuan M, Duan X, Zhou J, Lu J, Li Z, Yu F, Zhou X, Yang Q, Xia J. The Expression of microRNA-223 and FAM5C in Cerebral Infarction Patients with Diabetes Mellitus. Cardiovasc Toxicol. 2017;17:42-48. [PubMed] [DOI] [Cited in This Article: ] |
| 100. | Singh S, de Ronde MWJ, Kok MGM, Beijk MA, De Winter RJ, van der Wal AC, Sondermeijer BM, Meijers JCM, Creemers EE, Pinto-Sietsma SJ. MiR-223-3p and miR-122-5p as circulating biomarkers for plaque instability. Open Heart. 2020;7. [PubMed] [DOI] [Cited in This Article: ] |
| 101. | Karolina DS, Armugam A, Tavintharan S, Wong MT, Lim SC, Sum CF, Jeyaseelan K. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6:e22839. [PubMed] [DOI] [Cited in This Article: ] |
| 102. | Thong FS, Dugani CB, Klip A. Turning signals on and off: GLUT4 traffic in the insulin-signaling highway. Physiology (Bethesda). 2005;20:271-284. [PubMed] [DOI] [Cited in This Article: ] |
| 103. | Chen B, Luo L, Wei X, Gong D, Jin L. Altered Plasma miR-144 as a Novel Biomarker for Coronary Artery Disease. Ann Clin Lab Sci. 2018;48:440-445. [PubMed] [Cited in This Article: ] |
| 104. | Cheng J, Cheng A, Clifford BL, Wu X, Hedin U, Maegdefessel L, Pamir N, Sallam T, Tarling EJ, de Aguiar Vallim TQ. MicroRNA-144 Silencing Protects Against Atherosclerosis in Male, but Not Female Mice. Arterioscler Thromb Vasc Biol. 2020;40:412-425. [PubMed] [DOI] [Cited in This Article: ] |
| 105. | Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4:e7689. [PubMed] [DOI] [Cited in This Article: ] |
| 106. | Giordano M, Trotta MC, Ciarambino T, D'Amico M, Galdiero M, Schettini F, Paternosto D, Salzillo M, Alfano R, Andreone V, Malatino LS, Biolo G, Paolisso G, Adinolfi LE. Circulating MiRNA-195-5p and -451a in Diabetic Patients with Transient and Acute Ischemic Stroke in the Emergency Department. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] |
| 107. | Li P, Teng F, Gao F, Zhang M, Wu J, Zhang C. Identification of circulating microRNAs as potential biomarkers for detecting acute ischemic stroke. Cell Mol Neurobiol. 2015;35:433-447. [PubMed] [DOI] [Cited in This Article: ] |
| 108. | Ding L, Ai D, Wu R, Zhang T, Jing L, Lu J, Zhong L. Identification of the differential expression of serum microRNA in type 2 diabetes. Biosci Biotechnol Biochem. 2016;80:461-465. [PubMed] [DOI] [Cited in This Article: ] |
| 109. | Li S, Lee C, Song J, Lu C, Liu J, Cui Y, Liang H, Cao C, Zhang F, Chen H. Circulating microRNAs as potential biomarkers for coronary plaque rupture. Oncotarget. 2017;8:48145-48156. [PubMed] [DOI] [Cited in This Article: ] |
| 110. | Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E, Xifra G, Martínez C, Ricart W, Rieusset J, Rome S, Karczewska-Kupczewska M, Straczkowski M, Fernández-Real JM. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37:1375-1383. [PubMed] [DOI] [Cited in This Article: ] |
| 111. | Li SH, Chen L, Pang XM, Su SY, Zhou X, Chen CY, Huang LG, Li JP, Liu JL. Decreased miR-146a expression in acute ischemic stroke directly targets the Fbxl10 mRNA and is involved in modulating apoptosis. Neurochem Int. 2017;107:156-167. [PubMed] [DOI] [Cited in This Article: ] |
| 112. | Kotb HG, Ibrahim AH, Mohamed EF, Ali OM, Hassanein N, Badawy D, Abdelatty Aly E. The expression of microRNA 146a in patients with ischemic stroke: an observational study. Int J Gen Med. 2019;12:273-278. [PubMed] [DOI] [Cited in This Article: ] |
| 113. | Su ZF, Sun ZW, Zhang Y, Wang S, Yu QG, Wu ZB. Regulatory effects of miR-146a/b on the function of endothelial progenitor cells in acute ischemic stroke in mice. Kaohsiung J Med Sci. 2017;33:369-378. [PubMed] [DOI] [Cited in This Article: ] |
| 114. | Qu X, Wang N, Cheng W, Xue Y, Chen W, Qi M. MicroRNA-146a protects against intracerebral hemorrhage by inhibiting inflammation and oxidative stress. Exp Ther Med. 2019;18:3920-3928. [PubMed] [DOI] [Cited in This Article: ] |
| 115. | Petrkova J, Borucka J, Kalab M, Klevcova P, Michalek J, Taborsky M, Petrek M. Increased Expression of miR-146a in Valvular Tissue From Patients With Aortic Valve Stenosis. Front Cardiovasc Med. 2019;6:86. [PubMed] [DOI] [Cited in This Article: ] |
| 116. | Cheng HS, Besla R, Li A, Chen Z, Shikatani EA, Nazari-Jahantigh M, Hammoutène A, Nguyen MA, Geoffrion M, Cai L, Khyzha N, Li T, MacParland SA, Husain M, Cybulsky MI, Boulanger CM, Temel RE, Schober A, Rayner KJ, Robbins CS, Fish JE. Paradoxical Suppression of Atherosclerosis in the Absence of microRNA-146a. Circ Res. 2017;121:354-367. [PubMed] [DOI] [Cited in This Article: ] |
| 117. | Nguyen MA, Karunakaran D, Geoffrion M, Cheng HS, Tandoc K, Perisic Matic L, Hedin U, Maegdefessel L, Fish JE, Rayner KJ. Extracellular Vesicles Secreted by Atherogenic Macrophages Transfer MicroRNA to Inhibit Cell Migration. Arterioscler Thromb Vasc Biol. 2018;38:49-63. [PubMed] [DOI] [Cited in This Article: ] |
| 118. | Sheikhbahaei S, Manizheh D, Mohammad S, Hasan TM, Saman N, Laleh R, Mahsa M, Sanaz AK, Shaghayegh HJ. Can MiR-503 be used as a marker in diabetic patients with ischemic stroke? BMC Endocr Disord. 2019;19:42. [PubMed] [DOI] [Cited in This Article: ] |
| 119. | Caporali A, Meloni M, Völlenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C. Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282-291. [PubMed] [DOI] [Cited in This Article: ] |
| 120. | Bi R, Ding F, He Y, Jiang L, Jiang Z, Mei J, Liu H. miR-503 inhibits platelet-derived growth factor-induced human aortic vascular smooth muscle cell proliferation and migration through targeting the insulin receptor. Biomed Pharmacother. 2016;84:1711-1716. [PubMed] [DOI] [Cited in This Article: ] |
| 121. | Hou LJ, Han JJ, Liu Y. Up-regulation of microRNA-503 by high glucose reduces the migration and proliferation but promotes the apoptosis of human umbilical vein endothelial cells by inhibiting the expression of insulin-like growth factor-1 receptor. Eur Rev Med Pharmacol Sci. 2018;22:3515-3523. [PubMed] [DOI] [Cited in This Article: ] |
| 122. | Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, Fan Y, Jordan IK, Jo H. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest. 2014;124:3187-3199. [PubMed] [DOI] [Cited in This Article: ] |
| 123. | Lee HT, Oh S, Ro DH, Yoo H, Kwon YW. The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis. J Lipid Atheroscler. 2020;9:419-434. [PubMed] [DOI] [Cited in This Article: ] |
| 124. | Hiltunen MO, Turunen MP, Häkkinen TP, Rutanen J, Hedman M, Mäkinen K, Turunen AM, Aalto-Setälä K, Ylä-Herttuala S. DNA hypomethylation and methyltransferase expression in atherosclerotic lesions. Vasc Med. 2002;7:5-11. [PubMed] [DOI] [Cited in This Article: ] |
| 125. | Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS One. 2009;4:e4864. [PubMed] [DOI] [Cited in This Article: ] |
| 126. | Newby AC. Metalloproteinases promote plaque rupture and myocardial infarction: A persuasive concept waiting for clinical translation. Matrix Biol. 2015;44-46:157-166. [PubMed] [DOI] [Cited in This Article: ] |
| 127. | Dje N'Guessan P, Riediger F, Vardarova K, Scharf S, Eitel J, Opitz B, Slevogt H, Weichert W, Hocke AC, Schmeck B, Suttorp N, Hippenstiel S. Statins control oxidized LDL-mediated histone modifications and gene expression in cultured human endothelial cells. Arterioscler Thromb Vasc Biol. 2009;29:380-386. [PubMed] [DOI] [Cited in This Article: ] |
| 128. | Greißel A, Culmes M, Burgkart R, Zimmermann A, Eckstein HH, Zernecke A, Pelisek J. Histone acetylation and methylation significantly change with severity of atherosclerosis in human carotid plaques. Cardiovasc Pathol. 2016;25:79-86. [PubMed] [DOI] [Cited in This Article: ] |
| 129. | Davegårdh C, García-Calzón S, Bacos K, Ling C. DNA methylation in the pathogenesis of type 2 diabetes in humans. Mol Metab. 2018;14:12-25. [PubMed] [DOI] [Cited in This Article: ] |
| 130. | Yang BT, Dayeh TA, Kirkpatrick CL, Taneera J, Kumar R, Groop L, Wollheim CB, Nitert MD, Ling C. Insulin promoter DNA methylation correlates negatively with insulin gene expression and positively with HbA(1c) levels in human pancreatic islets. Diabetologia. 2011;54:360-367. [PubMed] [DOI] [Cited in This Article: ] |
| 131. | Yang BT, Dayeh TA, Volkov PA, Kirkpatrick CL, Malmgren S, Jing X, Renström E, Wollheim CB, Nitert MD, Ling C. Increased DNA methylation and decreased expression of PDX-1 in pancreatic islets from patients with type 2 diabetes. Mol Endocrinol. 2012;26:1203-1212. [PubMed] [DOI] [Cited in This Article: ] |
| 132. | Zhu Y, Liu Q, Zhou Z, Ikeda Y. PDX1, Neurogenin-3, and MAFA: critical transcription regulators for beta cell development and regeneration. Stem Cell Res Ther. 2017;8:240. [PubMed] [DOI] [Cited in This Article: ] |
| 133. | Ling C, Del Guerra S, Lupi R, Rönn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Del Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615-622. [PubMed] [DOI] [Cited in This Article: ] |
| 134. | Lai CQ, Tucker KL, Parnell LD, Adiconis X, García-Bailo B, Griffith J, Meydani M, Ordovás JM. PPARGC1A variation associated with DNA damage, diabetes, and cardiovascular diseases: the Boston Puerto Rican Health Study. Diabetes. 2008;57:809-816. [PubMed] [DOI] [Cited in This Article: ] |
| 135. | Hall E, Dayeh T, Kirkpatrick CL, Wollheim CB, Dekker Nitert M, Ling C. DNA methylation of the glucagon-like peptide 1 receptor (GLP1R) in human pancreatic islets. BMC Med Genet. 2013;14:76. [PubMed] [DOI] [Cited in This Article: ] |
| 136. | Tsalamandris S, Antonopoulos AS, Oikonomou E, Papamikroulis GA, Vogiatzi G, Papaioannou S, Deftereos S, Tousoulis D. The Role of Inflammation in Diabetes: Current Concepts and Future Perspectives. Eur Cardiol. 2019;14:50-59. [PubMed] [DOI] [Cited in This Article: ] |
| 137. | Ahmed M, de Winther MPJ, Van den Bossche J. Epigenetic mechanisms of macrophage activation in type 2 diabetes. Immunobiology. 2017;222:937-943. [PubMed] [DOI] [Cited in This Article: ] |
| 138. | Babu M, Durga Devi T, Mäkinen P, Kaikkonen M, Lesch HP, Junttila S, Laiho A, Ghimire B, Gyenesei A, Ylä-Herttuala S. Differential Promoter Methylation of Macrophage Genes Is Associated With Impaired Vascular Growth in Ischemic Muscles of Hyperlipidemic and Type 2 Diabetic Mice: Genome-Wide Promoter Methylation Study. Circ Res. 2015;117:289-299. [PubMed] [DOI] [Cited in This Article: ] |
| 139. | Yang X, Wang X, Liu D, Yu L, Xue B, Shi H. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Mol Endocrinol. 2014;28:565-574. [PubMed] [DOI] [Cited in This Article: ] |
| 140. | Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, Calkin AC, Brownlee M, Cooper ME, El-Osta A. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58:1229-1236. [PubMed] [DOI] [Cited in This Article: ] |
| 141. | Ea CK, Baltimore D. Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106:18972-18977. [PubMed] [DOI] [Cited in This Article: ] |
| 142. | Dorrington MG, Fraser IDC. NF-κB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front Immunol. 2019;10:705. [PubMed] [DOI] [Cited in This Article: ] |
| 143. | Barajas-Olmos F, Centeno-Cruz F, Zerrweck C, Imaz-Rosshandler I, Martínez-Hernández A, Cordova EJ, Rangel-Escareño C, Gálvez F, Castillo A, Maydón H, Campos F, Maldonado-Pintado DG, Orozco L. Altered DNA methylation in liver and adipose tissues derived from individuals with obesity and type 2 diabetes. BMC Med Genet. 2018;19:28. [PubMed] [DOI] [Cited in This Article: ] |
| 144. | Nilsson E, Jansson PA, Perfilyev A, Volkov P, Pedersen M, Svensson MK, Poulsen P, Ribel-Madsen R, Pedersen NL, Almgren P, Fadista J, Rönn T, Klarlund Pedersen B, Scheele C, Vaag A, Ling C. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63:2962-2976. [PubMed] [DOI] [Cited in This Article: ] |
| 145. | Simar D, Versteyhe S, Donkin I, Liu J, Hesson L, Nylander V, Fossum A, Barrès R. DNA methylation is altered in B and NK lymphocytes in obese and type 2 diabetic human. Metabolism. 2014;63:1188-1197. [PubMed] [DOI] [Cited in This Article: ] |
| 146. | Zhao J, Goldberg J, Bremner JD, Vaccarino V. Global DNA methylation is associated with insulin resistance: a monozygotic twin study. Diabetes. 2012;61:542-546. [PubMed] [DOI] [Cited in This Article: ] |
| 147. | El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205:2409-2417. [PubMed] [DOI] [Cited in This Article: ] |
| 148. | Bijli KM, Fazal F, Rahman A. Regulation of Rela/p65 and endothelial cell inflammation by proline-rich tyrosine kinase 2. Am J Respir Cell Mol Biol. 2012;47:660-668. [PubMed] [DOI] [Cited in This Article: ] |
| 149. | Johansen OE, Birkeland KI. Preventing macrovascular disease in patients with type 2 diabetes mellitus. Am J Cardiovasc Drugs. 2003;3:283-297. [PubMed] [DOI] [Cited in This Article: ] |
| 150. | American Diabetes Association. Standards of Medical Care in Diabetes-2020 Abridged for Primary Care Providers. Clin Diabetes. 2020;38:10-38. [PubMed] [DOI] [Cited in This Article: ] |
| 151. | Cosentino F, Grant PJ, Aboyans V, Bailey CJ, Ceriello A, Delgado V, Federici M, Filippatos G, Grobbee DE, Hansen TB, Huikuri HV, Johansson I, Jüni P, Lettino M, Marx N, Mellbin LG, Östgren CJ, Rocca B, Roffi M, Sattar N, Seferović PM, Sousa-Uva M, Valensi P, Wheeler DC; ESC Scientific Document Group. 2019 ESC Guidelines on diabetes, pre-diabetes, and cardiovascular diseases developed in collaboration with the EASD. Eur Heart J. 2020;41:255-323. [PubMed] [DOI] [Cited in This Article: ] |
| 152. | Mannino GC, Andreozzi F, Sesti G. Pharmacogenetics of type 2 diabetes mellitus, the route toward tailored medicine. Diabetes Metab Res Rev. 2019;35:e3109. [PubMed] [DOI] [Cited in This Article: ] |
| 153. | Fontana V, Luizon MR, Sandrim VC. An update on the pharmacogenetics of treating hypertension. J Hum Hypertens. 2015;29:283-291. [PubMed] [DOI] [Cited in This Article: ] |
| 154. | Kitzmiller JP, Mikulik EB, Dauki AM, Murkherjee C, Luzum JA. Pharmacogenomics of statins: understanding susceptibility to adverse effects. Pharmgenomics Pers Med. 2016;9:97-106. [PubMed] [DOI] [Cited in This Article: ] |
| 155. | Nasykhova YA, Tonyan ZN, Mikhailova AA, Danilova MM, Glotov AS. Pharmacogenetics of Type 2 Diabetes-Progress and Prospects. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] |
| 156. | Yanai H. Metabolic Syndrome and COVID-19. Cardiol Res. 2020;11:360-365. [PubMed] [DOI] [Cited in This Article: ] |
| 157. | Gregory JM, Slaughter JC, Duffus SH, Smith TJ, LeStourgeon LM, Jaser SS, McCoy AB, Luther JM, Giovannetti ER, Boeder S, Pettus JH, Moore DJ. COVID-19 Severity Is Tripled in the Diabetes Community: A Prospective Analysis of the Pandemic's Impact in Type 1 and Type 2 Diabetes. Diabetes Care. 2021;44:526-532. [PubMed] [DOI] [Cited in This Article: ] |
| 158. | Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054-1062. [PubMed] [DOI] [Cited in This Article: ] |
| 159. | Apicella M, Campopiano MC, Mantuano M, Mazoni L, Coppelli A, Del Prato S. COVID-19 in people with diabetes: understanding the reasons for worse outcomes. Lancet Diabetes Endocrinol. 2020;8:782-792. [PubMed] [DOI] [Cited in This Article: ] |
| 160. | Sardu C, Gambardella J, Morelli MB, Wang X, Marfella R, Santulli G. Hypertension, Thrombosis, Kidney Failure, and Diabetes: Is COVID-19 an Endothelial Disease? J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] |
| 161. | Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281-292.e6. [PubMed] [DOI] [Cited in This Article: ] |
| 162. | Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. 2020;53:425-435. [PubMed] [DOI] [Cited in This Article: ] |
| 163. | Bakhshandeh B, Sorboni SG, Javanmard AR, Mottaghi SS, Mehrabi MR, Sorouri F, Abbasi A, Jahanafrooz Z. Variants in ACE2; potential influences on virus infection and COVID-19 severity. Infect Genet Evol. 2021;90:104773. [PubMed] [DOI] [Cited in This Article: ] |
| 164. | Sardu C, Maggi P, Messina V, Iuliano P, Sardu A, Iovinella V, Paolisso G, Marfella R. Could Anti-Hypertensive Drug Therapy Affect the Clinical Prognosis of Hypertensive Patients With COVID-19 Infection? J Am Heart Assoc. 2020;9:e016948. [PubMed] [DOI] [Cited in This Article: ] |
| 165. | Yang JK, Lin SS, Ji XJ, Guo LM. Binding of SARS coronavirus to its receptor damages islets and causes acute diabetes. Acta Diabetol. 2010;47:193-199. [PubMed] [DOI] [Cited in This Article: ] |
| 166. | Fignani D, Licata G, Brusco N, Nigi L, Grieco GE, Marselli L, Overbergh L, Gysemans C, Colli ML, Marchetti P, Mathieu C, Eizirik DL, Sebastiani G, Dotta F. SARS-CoV-2 Receptor Angiotensin I-Converting Enzyme Type 2 (ACE2) Is Expressed in Human Pancreatic β-Cells and in the Human Pancreas Microvasculature. Front Endocrinol (Lausanne). 2020;11:596898. [PubMed] [DOI] [Cited in This Article: ] |
| 167. | Bernardi S, Tikellis C, Candido R, Tsorotes D, Pickering RJ, Bossi F, Carretta R, Fabris B, Cooper ME, Thomas MC. ACE2 deficiency shifts energy metabolism towards glucose utilization. Metabolism. 2015;64:406-415. [PubMed] [DOI] [Cited in This Article: ] |
| 168. | Yamamoto K, Ohishi M, Katsuya T, Ito N, Ikushima M, Kaibe M, Tatara Y, Shiota A, Sugano S, Takeda S, Rakugi H, Ogihara T. Deletion of angiotensin-converting enzyme 2 accelerates pressure overload-induced cardiac dysfunction by increasing local angiotensin II. Hypertension. 2006;47:718-726. [PubMed] [DOI] [Cited in This Article: ] |
| 169. | Rabelo LA, Todiras M, Nunes-Souza V, Qadri F, Szijártó IA, Gollasch M, Penninger JM, Bader M, Santos RA, Alenina N. Genetic Deletion of ACE2 Induces Vascular Dysfunction in C57BL/6 Mice: Role of Nitric Oxide Imbalance and Oxidative Stress. PLoS One. 2016;11:e0150255. [PubMed] [DOI] [Cited in This Article: ] |
| 170. | Bindom SM, Hans CP, Xia H, Boulares AH, Lazartigues E. Angiotensin I-converting enzyme type 2 (ACE2) gene therapy improves glycemic control in diabetic mice. Diabetes. 2010;59:2540-2548. [PubMed] [DOI] [Cited in This Article: ] |
| 171. | Karahalil B, Elkama A. The impact of ACE2 gene polymorphism in the development of COVID-19 disease. Gazi Med J. 2020;31. [DOI] [Cited in This Article: ] |
| 172. | Shikov AE, Barbitoff YA, Glotov AS, Danilova MM, Tonyan ZN, Nasykhova YA, Mikhailova AA, Bespalova ON, Kalinin RS, Mirzorustamova AM, Kogan IY, Baranov VS, Chernov AN, Pavlovich DM, Azarenko SV, Fedyakov MA, Tsay VV, Eismont YA, Romanova OV, Hobotnikov DN, Vologzhanin DA, Mosenko SV, Ponomareva TA, Talts YA, Anisenkova AU, Lisovets DG, Sarana AM, Urazov SP, Scherbak SG, Glotov OS. Analysis of the Spectrum of ACE2 Variation Suggests a Possible Influence of Rare and Common Variants on Susceptibility to COVID-19 and Severity of Outcome. Front Genet. 2020;11:551220. [PubMed] [DOI] [Cited in This Article: ] |
| 173. | Stawiski EW, Diwanji D, Suryamohan K, Gupta R, Fellouse FA, Sathirapongsasuti JF, Liu J, Jiang YP, Ratan A, Mis M, Santhosh D, Somasekar S, Mohan S, Phalke S, Kuriakose B, Antony A, Junutula JR, Schuster SC, Jura N, Seshagiri S. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. 2020 Preprint. Available from: bioRxiv:2020.04.07.024752. [DOI] [Cited in This Article: ] |
| 174. | Pedersen KB, Chhabra KH, Nguyen VK, Xia H, Lazartigues E. The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. Biochim Biophys Acta. 2013;1829:1225-1235. [PubMed] [DOI] [Cited in This Article: ] |
| 175. | Chaudhary M. COVID-19 susceptibility: potential of ACE2 polymorphisms. Egypt J Med Human Genet. 2020;21:54. [DOI] [Cited in This Article: ] |
| 176. | Ali S, Dornhorst A. Diabetes in pregnancy: health risks and management. Postgrad Med J. 2011;87:417-427. [PubMed] [DOI] [Cited in This Article: ] |