Published online Feb 15, 2010. doi: 10.4251/wjgo.v2.i2.117
Revised: August 5, 2009
Accepted: August 12, 2009
Published online: February 15, 2010
AIM: To identify genetic polymorphisms in the promoter region of the human base excision repair gene NEIL1 in gastric cancer patients.
METHODS: The NEIL1 promoter region in DNA from 80 Japanese patients with gastric cancer was searched for genetic polymorphisms by polymerase chain reaction-single-strand conformation polymorphism and subsequent sequencing analyses.
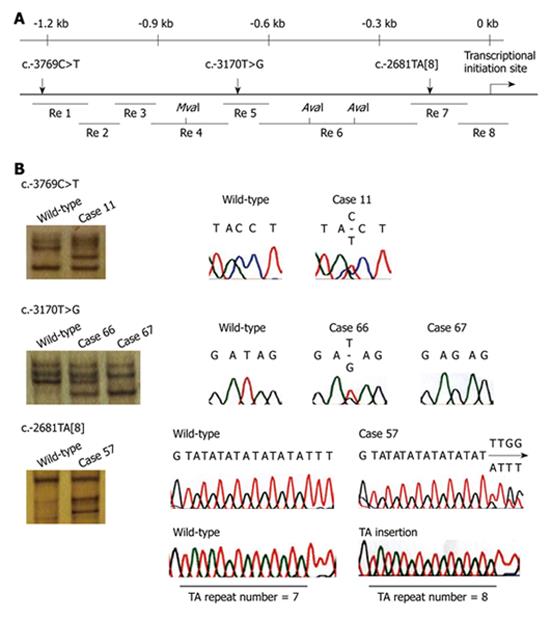
RESULTS: Three novel genetic polymorphisms, i.e. c.-3769C>T, c.-3170T>G, and c.-2681TA[8], were identified in the NEIL1 promoter region at an allele frequency of 0.6%, 9.4%, and 4.4%, respectively, in Japanese gastric cancer patients.
CONCLUSION: Three NEIL1 promoter polymorphisms detected in this study may be of importance in gastric carcinogenesis.
-
Citation: Goto M, Shinmura K, Tao H, Tsugane S, Sugimura H. Three novel
NEIL1 promoter polymorphisms in gastric cancer patients. World J Gastrointest Oncol 2010; 2(2): 117-120 - URL: https://www.wjgnet.com/1948-5204/full/v2/i2/117.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v2.i2.117
Stomach tissue is exposed to oxidative stress, including inflammation induced by Helicobacter pylori infection, sodium chloride, and smoking[1-5]. Since severe oxidative stress leads to accumulation of huge amounts of damaged bases[6-9], maintenance of a system to repair damaged bases in the stomach is thought to be important. The base excision repair protein NEIL1 has activity that is capable of removing oxidatively damaged bases, including thymine glycol, 5-hydroxyuracil, urea, formamidopyrimidine-A, and formamidopyrimidine-G, which have been shown to cause mutagenesis and cell death[10-14]. We have recently demonstrated somatic inactivating NEIL1 mutations and reduced NEIL1 expression in a subset of gastric cancers, suggesting that reduced NEIL1 activity is involved in gastric carcinogenesis[15]. In a recent investigation of the NEIL1 expression system an approximately 1.2 kb sequence upstream of the transcriptional initiation site of the NEIL1 gene was shown to have promoter activity by a luciferase reporter assay in human cells[16]. However, since no genetic polymorphisms have been reported in the promoter region thus far and genetic polymorphisms in the region may be of importance in gastric carcinogenesis, we tried searching DNA extracted from the blood of 80 gastric cancer patients for NEIL1 promoter polymorphisms.
Blood samples from 80 gastric cancer patients were obtained from hospitals in Nagano Prefecture, Japan, and genomic DNA was extracted from them with a DNA Extractor WB Kit (Wako, Osaka, Japan)[17]. The baseline characteristics of the patients have been described previously[17]. This study was approved by the Institutional Review Boards of Hamamatsu University School of Medicine and the National Cancer Center.
PCR-SSCP analysis was used to examine the DNA samples for genetic polymorphisms in the NEIL1 promoter region. An approximately 1.2 kb 5' upstream sequence that was shown to have promoter activity in a previous study[16] was divided into 8 regions (Figure 1A), and each region was amplified by PCR with HotStarTaq DNA polymerase (QIAGEN, Valencia, CA, USA). The primer sets used were: 5'-CAAATATTGCAGTCTGAAAGGGG-3' and 5'-GAAACTGATCAAGACAGGGGC-3' for region 1, 5'-GTTTTCTAATGCAGAGGTCTGG-3' and 5'-TACAGGGATAAGCCACTA CGC-3' for region 2, 5'-CCTCCTGATATGATGCAATTC-3' and 5'-CACTCCCAGCTGATTTTTG-3' for region 3, 5'-ATGGTGAAACCCCGTCTCTAC-3' and 5'-TGCTGGGAATTAGATCTAAAGGC-3' for region 4, 5'-AGCACCTAGGAAGTATCCCTG-3' and 5'-GTCTCAGCCAGTTGTTGTTTG-3' for region 5, 5'-CAAATTGAGAATGTGATGCAGC-3' and 5'-CAGATTTCCCCAATTGTCCC-3' for region 6, 5'-TGACCCATGATTGTAGCCTG-3' and 5'-GAGGTTTCGCCT TGTTGG-3' for region 7, and 5'-GAGGCGGGCAGATTACTT G-3' and 5'-CTCACTGCAGCC TCCACTTC-3' for region 8. The PCR products of regions 4 and 6 were digested with restriction enzymes MvaI (New England Biolabs, Beverly, MA, USA) and AvaI (New England Biolabs), respectively, in order to adjust their size to < 230 bp before SSCP. The PCR products of all regions were diluted with two volumes of loading solution, and after applying them to 8% polyacrylamide gels in the presence or absence of 5% glycerol, the products were electrophoresed at room temperature and 4°C and detected by silver staining. PCR products exhibiting an abnormally shifted band in the SSCP analysis were directly sequenced with a BigDye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Tokyo, Japan) and an ABI 3100 Genetic Analyzer (Applied Biosystems). A PCR product of region 7 was also sequenced after subcloning into a pGEM-T Easy vector (Promega, Madison, WI, USA). The reference nucleotide sequence is accession number NM_024608. Deviation of the genotype distribution from Hardy-Weinberg equilibrium (HWE) was tested by using SNPAlyze software (Dynacom, Yokohama, Japan).
We searched for genetic polymorphisms in the region containing NEIL1 promoter activity by PCR-SSCP analysis using blood samples derived from 80 gastric cancer patients. Three genetic polymorphisms, c.-3769C>T, c.-3170T>G, and c.-2681TA[8], were identified in the NEIL1 promoter region at an allele frequency of 0.6%, 9.4%, and 4.4%, respectively (Figure 1B). The distribution of the genotypes of these polymorphisms was in HWE. Examination of the frequency of the polymorphisms revealed a homozygote for the variant allele of only one of the three polymorphisms, c.-3170T>G, and in only one patient, indicating that the three polymorphisms in the NEIL1 promoter are rare genetic polymorphisms.
In this study, we found three novel promoter polymorphisms, c.-3769C>T, c.-3170T>G, and c.-2681TA[8]. None of these polymorphisms has previously been reported or registered in the database of the single nucleotide polymorphism (dbSNP) homepage of the National Center for Biotechnology Information web site (web site : http://www.ncbi.nlm.nih.gov/SNP/) or the database of the Japanese single nucleotide polymorphism homepage (http://snp.ims.u-tokyo.ac.jp/), indicating that they are novel genetic polymorphisms.
Interestingly, when we used Genomatix software (http://www.genomatix.de/matinspector.html) to search for transcription factors that putatively bind to the sequence containing these polymorphism sites, a sequence containing c.-3170T was predicted to bind to GATA binding factors and a sequence containing c.-2681TA[7] was predicted to bind to a GZF1, a TATA-binding protein and LIM homeodomain factors. The change from c.-3170T to c.-3170G eliminates the binding site for GATA binding factors. On the other hand, although the change from c.-2681TA[7] to c.-2681TA[8] would appear to retain the sequence of binding sites for the GZF1, TATA-binding protein, and LIM homeodomain factors, there are examples of a change in the number of repetitive sequences in a promoter being associated with a difference in the expression level[18]. Thus, these nucleotide changes may be associated with a difference in the NEIL1 expression level. Moreover, since some factors involved in the regulation of the transcription level in human cells remain unknown, the three NEIL1 promoter polymorphisms may be associated with differences in the NEIL1 expression level by binding to factors that have yet to be identified.
Most common genetic polymorphisms have been registered in various genetic polymorphism databases, such as dbSNP. However, as shown in this study, there appear to be many genetic polymorphisms that still have not been registered in databases, the reason being that many unregistered genetic polymorphisms are rare. Since finding rare and novel polymorphisms requires many human samples and repeating this kind of study, the NEIL1 data presented in this study are very valuable for future studies, such as searches for alleles that increase the risk of diseases and allele-specific expression analyses. Furthermore, NEIL1 protein plays a very important role in excision repair of oxidatively damaged bases, which have been implicated in a wide variety of human cancer. Promoter polymorphisms in some DNA repair genes, including XRCC1, MLH1, and MSH2, have recently been reported to be associated with an increased risk of cancer[19-21]. Like these examples, the novel NEIL1 promoter polymorphisms identified by screening gastric cancer patients in this study may be associated with increased risk of gastric cancer. If so, this information should be of value in management to prevent the development of gastric cancer in individuals with the risk allele. We are therefore planning to examine the NEIL1 promoter polymorphisms in the framework of a gastric cancer case-control study in the future, and we are also planning to investigate the effect of the polymorphisms on NEIL1 promoter activity.
The human base excision repair protein NEIL1 has activity that is capable of removing oxidatively damaged bases, such as thymine glycol and 5-hydroxyuracil. We have recently demonstrated somatic inactivatingNEIL1mutations and reduced NEIL1 expression in a subset of gastric cancers, suggesting that reduced NEIL1 activity is involved in gastric carcinogenesis. In the present study, we searched for genetic polymorphisms in the promoter region of the humanNEIL1gene in gastric cancer patients and succeeded in identifying three novel genetic polymorphisms.
Oxidized-DNA-base lesions have been implicated in carcinogenesis, and base excision repair proteins are involved in the repair of such lesions. The research frontier in the area of studying the relationship between the base excision repair genes and carcinogenesis lies in the discovery of genetic variants in the genes that are associated with increased cancer risk.
Stomach tissue is exposed to oxidative stress, including inflammation induced byHelicobacter pyloriinfection, sodium chloride, and smoking, and promoter polymorphisms in some DNA repair genes have been reported to be associated with increased cancer risk. However, there have been no reports of studies that have examined associations betweenNEIL1promoter polymorphisms and gastric cancer risk. The identification of three novelNEIL1promoter polymorphisms in this study should be of value for future research in this field.
TheNEIL1data presented in this study will be useful for various future studies, such as studies that evaluate the effects ofNEIL1polymorphisms on the risk of disease, haplotype analyses, and allele-specific expression analyses.
This study investigated genetic polymorphisms in the humanNEIL1gene and identified three polymorphisms in the promoter region of the gene. Although the study did not determine whether the polymorphisms are specific for gastric cancer patients, and no functional analysis of the polymorphisms was performed, the polymorphisms identified are indeed novel, and the results may facilitate future research in this field. In conclusion, the results of this study are somewhat valuable, and the paper appears to be worth publishing as a brief communication.
Peer reviewer: Tatsuo Kanda, MD, PhD, Division of Digestive and General Surgery, Graduate School of Medical and Dental Sciences, Niigata University, Niigata City 951-8510, Japan
S- Editor Li LF L- Editor Lalor PF E- Editor Lin YP
| 1. | Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, Ko GH, Park CK, Kasai H, Rhee KH. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996;56:1279-1282. [Cited in This Article: ] |
| 2. | Kanada R, Uchida T, Tsukamoto Y, Nguyen LT, Hijiya N, Matsuura K, Kodama M, Okimoto T, Murakami K, Fujioka T. Genotyping of the cagA gene of Helicobacter pylori on immunohistochemistry with East Asian CagA-specific antibody. Pathol Int. 2008;58:218-225. [Cited in This Article: ] |
| 3. | Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk: epidemiological and biological evidence. World J Gastroenterol. 2009;15:2204-2213. [Cited in This Article: ] |
| 4. | Farinati F, Cardin R, Degan P, Rugge M, Mario FD, Bonvicini P, Naccarato R. Oxidative DNA damage accumulation in gastric carcinogenesis. Gut. 1998;42:351-356. [Cited in This Article: ] |
| 5. | Trédaniel J, Boffetta P, Buiatti E, Saracci R, Hirsch A. Tobacco smoking and gastric cancer: review and meta-analysis. Int J Cancer. 1997;72:565-573. [Cited in This Article: ] |
| 6. | Ernst P. Review article: the role of inflammation in the pathogenesis of gastric cancer. Aliment Pharmacol Ther. 1999;13 Suppl 1:13-18. [Cited in This Article: ] |
| 7. | Goto M, Shinmura K, Igarashi H, Kobayashi M, Konno H, Yamada H, Iwaizumi M, Kageyama S, Tsuneyoshi T, Tsugane S. Altered expression of the human base excision repair gene NTH1 in gastric cancer. Carcinogenesis. 2009;30:1345-1352. [Cited in This Article: ] |
| 8. | Federico A, Morgillo F, Tuccillo C, Ciardiello F, Loguercio C. Chronic inflammation and oxidative stress in human carcinogenesis. Int J Cancer. 2007;121:2381-2386. [Cited in This Article: ] |
| 9. | Mena S, Ortega A, Estrela JM. Oxidative stress in environmental-induced carcinogenesis. Mutat Res. 2009;674:36-44. [Cited in This Article: ] |
| 10. | Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002;99:3523-3528. [Cited in This Article: ] |
| 11. | Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679-49684. [Cited in This Article: ] |
| 12. | Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst). 2003;2:581-591. [Cited in This Article: ] |
| 13. | Miller H, Fernandes AS, Zaika E, McTigue MM, Torres MC, Wente M, Iden CR, Grollman AP. Stereoselective excision of thymine glycol from oxidatively damaged DNA. Nucleic Acids Res. 2004;32:338-345. [Cited in This Article: ] |
| 14. | Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J Biol Chem. 2004;279:14464-14471. [Cited in This Article: ] |
| 15. | Shinmura K, Tao H, Goto M, Igarashi H, Taniguchi T, Maekawa M, Takezaki T, Sugimura H. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25:2311-2317. [Cited in This Article: ] |
| 16. | Das A, Hazra TK, Boldogh I, Mitra S, Bhakat KK. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J Biol Chem. 2005;280:35272-35280. [Cited in This Article: ] |
| 17. | Tsukino H, Hanaoka T, Otani T, Iwasaki M, Kobayashi M, Hara M, Natsukawa S, Shaura K, Koizumi Y, Kasuga Y. hOGG1 Ser326Cys polymorphism, interaction with environmental exposures, and gastric cancer risk in Japanese populations. Cancer Sci. 2004;95:977-983. [Cited in This Article: ] |
| 18. | Guillemette C, Millikan RC, Newman B, Housman DE. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000;60:950-956. [Cited in This Article: ] |
| 19. | Hu Z, Ma H, Lu D, Zhou J, Chen Y, Xu L, Zhu J, Huo X, Qian J, Wei Q. A promoter polymorphism (-77T>C) of DNA repair gene XRCC1 is associated with risk of lung cancer in relation to tobacco smoking. Pharmacogenet Genomics. 2005;15:457-463. [Cited in This Article: ] |
| 20. | Raptis S, Mrkonjic M, Green RC, Pethe VV, Monga N, Chan YM, Daftary D, Dicks E, Younghusband BH, Parfrey PS. MLH1 -93G>A promoter polymorphism and the risk of microsatellite-unstable colorectal cancer. J Natl Cancer Inst. 2007;99:463-474. [Cited in This Article: ] |
| 21. | Mrkonjic M, Raptis S, Green RC, Monga N, Daftary D, Dicks E, Younghusband HB, Parfrey PS, Gallinger SS, McLaughlin JR. MSH2 118T>C and MSH6 159C>T promoter polymorphisms and the risk of colorectal cancer. Carcinogenesis. 2007;28:2575-2580. [Cited in This Article: ] |