Copyright
©The Author(s) 2025.
World J Gastrointest Oncol. Apr 15, 2025; 17(4): 103113
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103113
Published online Apr 15, 2025. doi: 10.4251/wjgo.v17.i4.103113
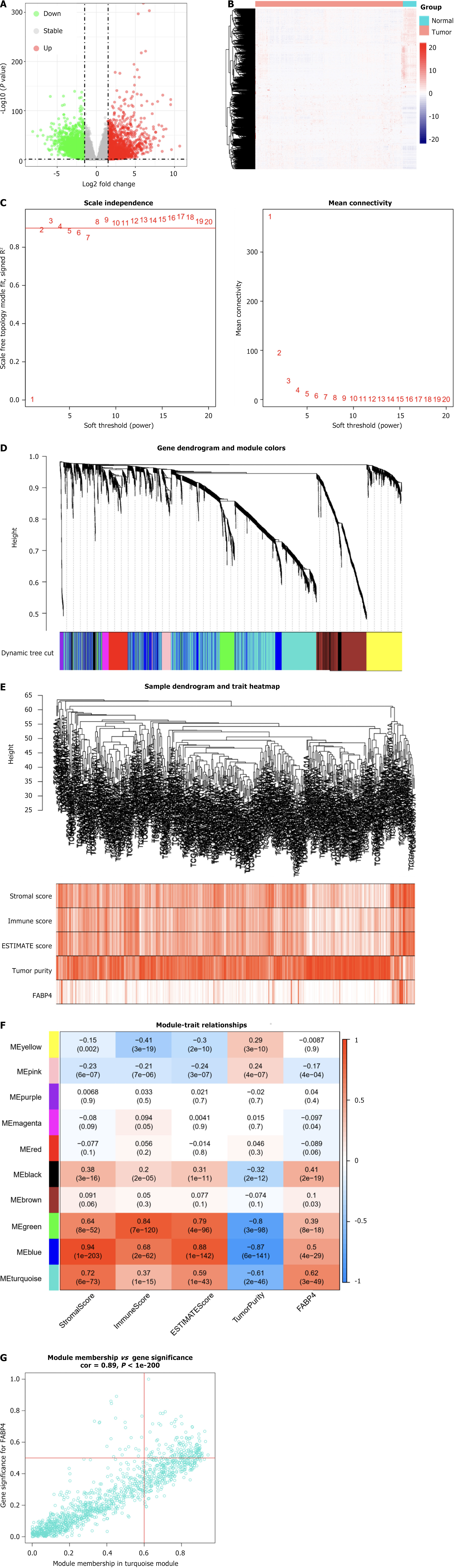
Figure 3 Screen for immune modules and genes related to fatty acid-binding protein 4 in colon adenocarcinoma tissues.
A: Volcano plot of differentially expressed genes (DEGs) between colon adenocarcinoma (COAD) tissues and normal tissues in The Cancer Genome Atlas (TCGA) database; B: Heatmap of DEGs between COAD tissues and normal tissues in TCGA database; C: Calculation of the scale-free fit indices of various soft-thresholding powers (β) and analysis of the mean connectivity of various soft-thresholding powers (β); D: DEGs were clustered based on the dissimilarity measure (1-TOM) and divided into 10 modules; E: Clustering dendrogram of COAD patients; F: A correlation heatmap between module eigengenes and immune parameters (fatty acid-binding protein 4 was used as the main research object) in patients with COAD; G: Scatter plot of the turquoise module eigengenes. COAD: Colon adenocarcinoma; DEGs: Differentially expressed genes; TCGA: The Cancer Genome Atlas.
- Citation: Zhang Y, Zhu WL, Wu M, Gao TY, Hu HX, Xu ZY. Using bioinformatics methods to elucidate fatty acid-binding protein 4 as a potential biomarker for colon adenocarcinoma. World J Gastrointest Oncol 2025; 17(4): 103113
- URL: https://www.wjgnet.com/1948-5204/full/v17/i4/103113.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v17.i4.103113