Published online Apr 15, 2024. doi: 10.4251/wjgo.v16.i4.1119
Peer-review started: December 28, 2023
First decision: January 4, 2024
Revised: January 16, 2024
Accepted: February 18, 2024
Article in press: February 18, 2024
Published online: April 15, 2024
Processing time: 104 Days and 21.6 Hours
Colorectal cancer (CRC) is one of the most prevalent malignancies worldwide, being the third most commonly diagnosed malignancy and the second leading cause of cancer-related deaths globally. Despite the progress in screening, early diagnosis, and treatment, approximately 20%-25% of CRC patients still present with metastatic disease at the time of their initial diagnosis. Furthermore, the burden of disease is still expected to increase, especially in individuals younger than 50 years old, among whom early-onset CRC incidence has been increasing. Screening and early detection are pivotal to improve CRC-related outcomes. It is well established that CRC screening not only reduces incidence, but also decreases deaths from CRC. Diverse screening strategies have proven effective in decreasing both CRC incidence and mortality, though variations in efficacy have been reported across the literature. However, uncertainties persist regarding the optimal screening method, age intervals and periodicity. Moreover, adherence to CRC screening remains globally low. In recent years, emerging technologies, notably artificial intelligence, and non-invasive biomarkers, have been developed to overcome these barriers. However, controversy exists over the actual impact of some of the new discoveries on CRC-related outcomes and how to effectively integrate them into daily practice. In this review, we aim to cover the current evidence surrounding CRC screening. We will further critically assess novel approaches under investigation, in an effort to differentiate promising inno
Core Tip: Despite progress in screening and early diagnosis, a significant proportion (approximately 20%-25%) of patients diagnosed with colorectal cancer still exhibit metastatic disease at the time of their initial diagnosis. Various screening tests are available, differing in invasiveness and preparation requirements. Nevertheless, adherence rates remain suboptimal. While new and promising methods are emerging to address these challenges, further research is needed before its integration in clinical practice.
- Citation: Lopes SR, Martins C, Santos IC, Teixeira M, Gamito É, Alves AL. Colorectal cancer screening: A review of current knowledge and progress in research. World J Gastrointest Oncol 2024; 16(4): 1119-1133
- URL: https://www.wjgnet.com/1948-5204/full/v16/i4/1119.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i4.1119
Colorectal cancer (CRC) is one of the most prevalent malignancies worldwide, being the third most commonly diagnosed malignancy, accounting for 10.0% of total cases, and the second leading cause of cancer-related deaths globally, contributing to 9.4% of the total cancer deaths[1,2]. The lifetime risk of developing CRC is approximately 1 in 23 (4.4%) for men and 1 in 25 (4.1%) for women[3]. CRC represents a global health concern, prompting the implementation of various public health policies, with screening programs emerging as one of the most impactful measures.
In this comprehensive review, the authors evaluate the extant knowledge regarding CRC screening, encompassing the currently endorsed tests, as well as promising technologies alongside their respective evidential bases. An extensive electronic was conducted across PubMed, Cochrane, and ISI Web of Science to identify relevant studies published between January 2000 and December 2023. Preference was given to peer-reviewed articles from highly ranked journals written in English, employing the search terms: “Colorectal cancer”, “screening”, “marker” or “biomarker” and “artificial intelligence”. Unpublished data from abstracts, contained in volumes from various congresses or conferences, were excluded from the analysis.
Amongst gastrointestinal cancers, CRC stands out with the highest incidence and mortality rates[4]. In the year 2020 alone, more than 1.9 million new cases of CRC and 935000 associated deaths were estimated to occur[1]. Incidence and mortality rates are increasing worldwide. There is however considerable geographical variation both between regions, according to the aggregated geographic regions defined by the UN Population Division, and within the same nation among different population groups[5]. Notably, when comparing age-standardized incidence rates, CRC incidence is 4-fold higher in developed countries than in developing nations. Regions such as Europe, Australia/New Zealand and North America rank among the highest incidence rates for colon cancer, while Africa and South Central Asia generally report lower incidence rates. Rectal cancer incidence rates display a similar regional distribution, although Eastern Asia also exhibits one the highest rates[1]. As countries undergo socio-economic development, there is a concurrent increase in CRC incidence, likely linked to lifestyle and dietary shifts more aligned with Western patterns, which led to CRC being considered by some as a marker of socioeconomic development. These changes involve an increase in sedentarism and excess body weight, both independently associated with CRC risk. Additional risk factors include excessive alcohol consumption, increased intake of red and processed meat, smoking and a low fiber intake[3-8]. As for mortality, this difference is mitigated by the higher fatality in transitioning countries[1]. These findings likely reflect disparities in healthcare access and the influence of existing national health policies, encompassing population-based interventions targeting risk factors and screening programs. In fact, in many low human development index (HDI) countries, screening remains largely opportunistic. Nevertheless, caution is warranted in interpreting these data due to deficiencies in registries related to incidence and mortality in certain regions, notably in low HDI countries in Africa and various parts of Asia[4].
Despite global trends and the expected increase on disease burden, some countries do have reported declines in CRC incidence. This decrease in incidence is due not only to population interventions related to lifestyle changes, but also to screening. However, it is noteworthy that even in high HDI countries with improved CRC incidence outcomes, rates of early-onset CRC are increasing by 1% to 4% annually[1,9,10]. The rising incidence in younger age cohorts is not only but mainly attributable to dietary patterns, excess body weight and lifestyle factors. Currently, these ages are not included in screening protocols.
Notably, screening seems to the main responsible for the accelerated progress since the early 2000s[11-14]. CRC screening not only reduces incidence, but also decreases deaths from CRC in adults older than 50 years of age who were at average risk for CRC. Various screening strategies have effectively decreased CRC incidence and mortality, with efficacy varying across the literature. Reductions in cancer incidence range from 39% to 60%, and reduction in CRC mortality from 55% to 80%, compared to no screening[15-17].
In alignment with the principles defined by Wilson and Junger in their landmark publication nearly 60 years ago, principles that persist as relevant and foundational to contemporary policy tools, CRC is deemed amenable to screening for several reasons[18,19]. Firstly, it constitutes a significant health concern, as evidenced by the aforementioned incidence and mortality rates. Furthermore, despite its heterogeneity and complex pathophysiology, CRC predominantly develops from a preclinical precursor, the adenoma. The progression from early adenoma to invasive cancer takes years to occur[20,21], providing a window for early detection and intervention. Additionally, there are screening tests available that seem to be suitable and acceptable to the population, which is of utmost importance as adherence and compliance are pivotal for screening effectiveness. Lastly, numerous studies have demonstrated that CRC screening is cost-effective[22-27].
However, uncertainties persist regarding the optimal screening methods, age intervals and periodicity. Moreover, adherence to CRC screening remains globally low, even in most developed countries, ranging from 19% in Croatia and the Czech Republic, 60% in the United States of America to 69% in the Basque region of Spain[28-30]. Concerns also arise regarding limited human and physical resources, particularly with new evidence suggesting the need to reconsider screening age, and how to improve adherence to screening programs. In response to these challenges, new methods are being designed to overcome barriers and improve CRC screening.
An in-depth understanding of CRC pathophysiology has proven essential for the implementation of screening and serves as the rationale for the development of tests. For example, the initial stool-based tests, which continue to be employed, are based in the identification of blood in stool, originating from vessels disrupted on the surface of tumors or adenomas. Subsequent advances in research and evolving insights into carcinogenesis have facilitated the refinement of existing tests and the development of novel and more effective screening modalities. Notably, the recognition that molecular alterations found in tumors and pre-cancerous lesions can also be detected in stool due to the natural exfoliation of colonocytes into the lumen has allowed for the development of tests targeting stool DNA. Additionally, other molecular markers, including messenger RNA, methylation of gene promoters, non-coding RNA molecules, as well as specific proteins, have been investigated and may constitute potential targets for testing. Furthermore, recent discoveries regarding the role of the intestinal microbiota lay the foundation for the development of microbial biomarkers[31].
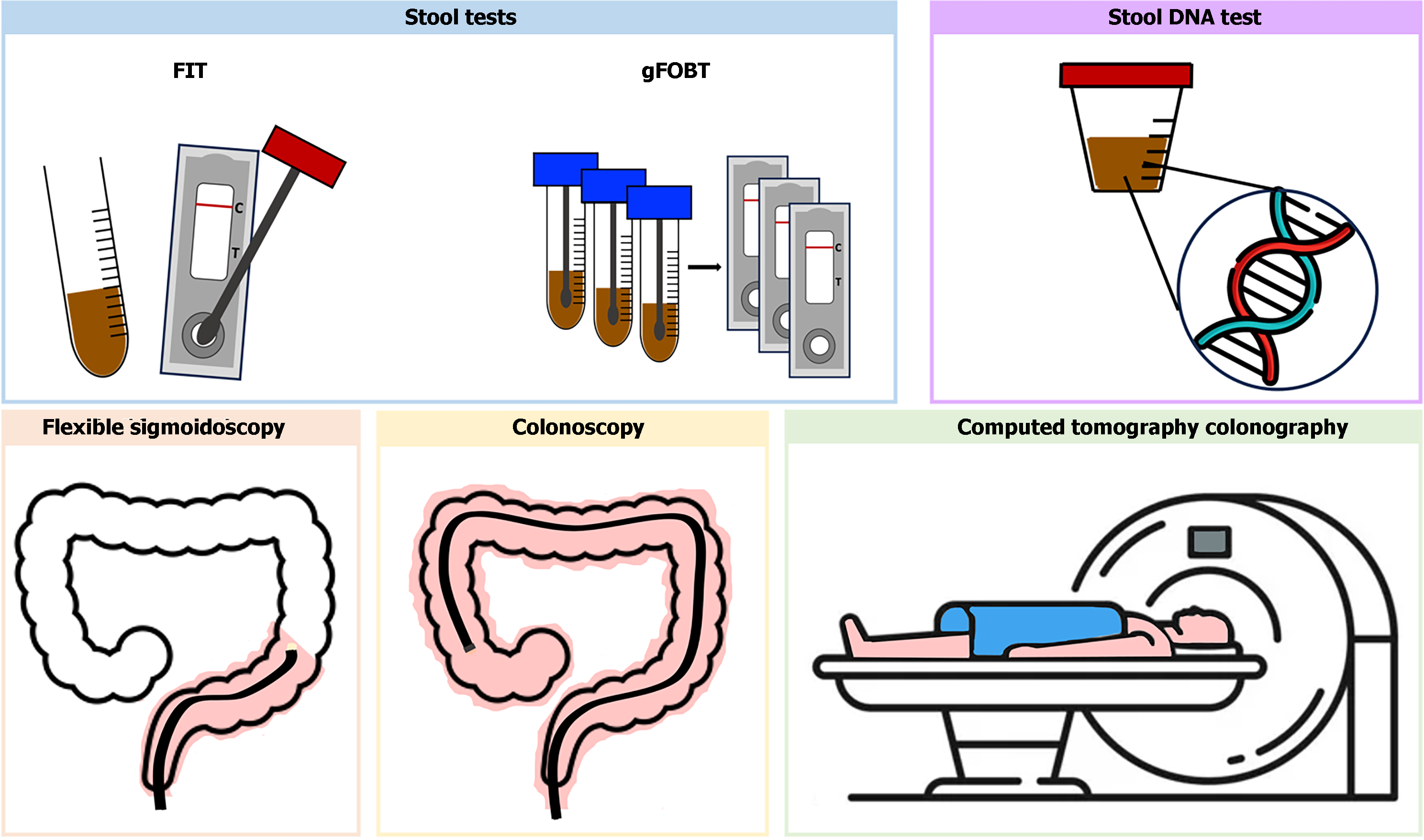
The screening tests currently recommended can be categorized into stool-based tests, blood-based tests, and imaging-tests, encompassing both indirect and direct visualization methods (Figure 1).
Stool-based tests represent the predominant approach globally for CRC screening, including high-sensitivity guaiac fecal occult blood testing (gFOBT), the fecal immunochemical test (FIT), stool DNA-FIT (sDNA-FIT).
gFOBT: gFOBT is a qualitative test that assesses the oxidative conversion of a colorless compound (guaiac) to a colored one in the presence of the pseudoperoxidase activity of hemoglobin[32]. Large adenomas and tumors exhibit a higher frequency of bleeding than smaller lesions[33], and in an intermittent fashion. Moreover, symptomatic tumors bleed more frequently than asymptomatic tumors, which are the intended target of screening[34]. Only high-sensitivity gFOBT is recommended for CRC screening. Reported sensitivity for advanced colorectal neoplasia and CRC ranges from 7% to 21%, and 50% to 75%, respectively, while specificity for advanced neoplasia ranges from 96% to 99%[35]. A systematic review including five randomized controlled trials (RCT) demonstrated reductions in CRC incidence and mortality with gFOBT, whether performed annual or biannual[35-40]. The Minnesota Study has provided robust evidence for the efficacy of screening with FOBT. Results of 18 years of follow-up reported a cumulative 18-year CRC mortality 33% lower in the annually screened group than in the control group. The group subjected to biennial screening demonstrated a 21% lower CRC mortality than did the control group. CRC incidence was reduced by 20% with annual screening vs 17% with biennial screening over 18 years of follow-up[41,42]. gFOBT is convenient for home use, enabling the collection of samples by mail and is cost-effective compared to no screening[26]. However, several factors may impact the result, as the degree of fecal hydration and the storage or fecal flora may impact hemoglobin degradation. It also requires more than one bowel movement, diet alterations and avoidance of some drugs to minimize false positives. Due to these considerations, gFOBT has been replaced by FIT. gFOBT were not sensitive to detect advanced adenoma[35].
FIT: FIT uses an antibody against the human globin moiety of heme to evaluate for the presence of occult blood. It has largely replaced gFOBT, since it requires only one stool sample, with no need for diet alterations or medication, thereby enhancing adherence. Observational studies have reported superior sensitivity in detecting CRC and advanced adenomas to both standard and high sensitivity gFOBT with comparable specificity. A meta-analysis of nineteen cohort studies revealed a sensitivity of 79% and specificity of 94% with a one-time FIT, utilizing a cutoff of 20 mg of hemoglobin per gram of stool[43], the FDA-approved threshold for a positive FIT. Another systematic review and meta-analysis including nineteen studies and utilizing the quantitative cutoff of 10 mg/g, demonstrated higher sensitivity (91%) and lower specificity (90%) for FIT in detecting CRC, as expected at lower cutoffs. No RCT has assessed the impact of FIT screening on CRC incidence and mortality. Observational cohort studies have reported a reduction in CRC mortality ranging from 22 to 62%, and a decrease in CRC incidence from 10% to 21% in the context of biannual FIT screening[44-47]. These findings are based on one-time application compared with colonoscopy. However, it is important to note that an annual or biennial FIT test exhibits a higher cumulative rate of detecting CRC and precursor neoplasia than a single FIT. In fact, aside from being cost-effective compared to no screening[23], a cost-effectiveness modeling study revealed that the number of gained life-years with a screening strategy involving annual FIT is comparable to that achieved with a colonoscopy every 10 years[47]. Annual screening is optimally cost-effective when using FIT[48]. Regarding detection of advanced adenomas, a systematic review and meta-analysis including thirty-one cross-sectional studies that utilized screening colonoscopy as the reference standard reported a sensitivity of 25%[49].
Multitarget stool DNA (mts DNA): The mtsDNA test combines a FIT with assays for several molecular marker, namely mutant KRAS and β-actin, and also abnormally methylated regions of DNA from advanced adenomas or CRC associated with colorectal carcinogenesis, including methylated bone morphogenetic protein 3 (BMP3) and methylated N-Myc downstream-regulated gene 4 protein (NDRG4). It is the first approved stool DNA test for CRC screening. As for FIT, there are no RCT on the impact of mtsDNA test on CRC incidence and mortality. In a prospective study comparing mtsDNA to FIT in individuals at average-risk undergoing colonoscopy, mtsDNA exhibited higher sensitivity for detection of CRC (92% vs 74%) and advanced adenoma (42% vs 24%), but lower specificity for detection of CRC or advanced lesions (87% vs 95%)[50]. However, cost effectiveness studies concluded that both FIT and colonoscopy were more cost-effective[51,52]. Doubts persist regarding the management of patients with positive test results and a negative colonoscopy, given its lower specificity. Currently recommendations suggest not submitting patients to further procedures and maintain the recommended screening intervals[53]. Another disadvantage of mtsDNA testing lies in the complexity of stool collection[54]. The recommended interval for repeating is 3 years, based on simulations models, due to the absence of studies on the performance of mtsDNA testing with repeat testing.
Colonoscopy: Colonoscopy detects not only early-stage cancers, but also identifies and allows for the excision of precancerous lesions within the same procedure. It is most commonly indicated following positive results from other less-invasive tests. However, it can also be used as a first-line test in some countries, like the United States of America, where colonoscopy is recommended every 10 years as a screening modality in average-risk individuals. There are no RCT assessing the effectiveness of colonoscopy in reducing CRC incidence and mortality in average-risk patients, but several cohort studies have demonstrated an impact on CRC incidence and mortality for both proximal and distal cancers[38,55-62]. In a prospective cohort study including 89000 health care professionals with over 24 years of follow-up found, a 68% reduction in CRC mortality and 43% reduction in CRC were reported for those who underwent colonoscopy compared to those who did not[60]. Similarly, a study including 24820 United State veterans reported a reduction in CRC mortality of 61% with screening colonoscopy[55]. The National Polyp Study, which included 1.418 average-risk patients, reported a 53% reduction in CRC mortality with colonoscopy screening[63]. Systematic reviews indicate that a colonoscopy performed every 10 years is cost-effective compared to no screening[23,25]. In fact, in the United States, a colonoscopy performed every ten years was deemed optimal in terms of cost-effectiveness. However, making further generalizations regarding the cost-effectiveness of colonoscopy compared to other methods is challenging due to differences in cost assumptions found across the literature. Factors such as the inclusion of anesthesiologist assistance and non-medical costs contribute to variations in the results of different models[25]. It is also noteworthy that colonoscopies can miss lesions. A systematic review of tandem colonoscopy studies reported miss rates of 26% for adenomas, 9% for advanced adenomas and 27% for serrated polyps[64]. Furthermore, being an invasive procedure, it carries risks of complications and necessitates bowel preparations, potentially compromising adherence. RCTs have demonstrated that adherence to colonoscopy is lower than for FIT[65]. Additionally, uncertainties persist regarding which polyps have the potential to develop into cancer, leading to surveillance colonoscopies at shorter intervals, with the associated burden and costs and an uncertain benefit[66].
Flexible sigmoidoscopy: Flexible sigmoidoscopy (FS) enables direct visualization of left colon, allowing for the detection of CRC and its precursor lesions and their removal. However, if adenomas are identified, a subsequent colonoscopy is required. Several studies have demonstrated the efficacy of FS in reducing CRC mortality and incidence. Two large RCTs conducted in United Kingdom and Italy including 170,432 and 34,292 individuals aged 55-64 years, respectively, compared a one-time FS with no screening. The results revealing a reduction in CRC incidence by 23% and 18%, and in CRC mortality by 31% and 22%, respectively[67,68]. A systematic review, based on four RCTs involving 458002 participants, found that a one or two-time FS were consistently associated with a decrease in CRC incidence [incidence rate ratio (IRR): 0.78; 95% confidence interval (95%CI): 0.74-0.83], equivalent to 28 to 47 fewer CRC cases per 100 000 person-years, and CRC-specific mortality (IRR: 0.74; 95%CI, 0.68-0.80), with 10 to 17 fewer CRC deaths per 100000 person-years, compared with no screening over an 11 to 17-year follow-up period[32]. However, a long-term follow up of the NORCCAP trial in Norway, including 98678 individuals, initially reporting a 20% reduction in CRC incidence and 27% reduction in CRC mortality, found no sustained reduction in CRC incidence or mortality with FS screening in women after 15 years of follow-up[69]. Additionally, screening with FS also has practical barriers, such as resource requirements similar to colonoscopy, limited examination of the entire colon, the need to perform a colonoscopy in case polyps are found and the lack of sedation. These factors have led to decreased utilization of screening FS in some countries and its discontinuation from guidelines[70], being currently reserved for individuals unwilling to undergo colonoscopy or FIT[53].
Computed tomography colonography: Computed tomography colonography (CTC) enables visualization of the entire colon and rectum. It requires bowel preparation, ingestion of a radiopaque agent and the use of CO2 insufflation via a rectal balloon catheter. Currently, it is performed predominantly in individuals unable to undergo colonoscopy, although it is recognized as the first-line screening test in select centers. There are no studies evaluating the impact of CTC on CRC incidence or mortality. Two large trials have compared the diagnostic yield of CTC with colonoscopy performed on the same day[71,72]. In a study involving 1233 average-risk individuals, CTC demonstrated a sensitivity of 92% with a specificity of 96% for adenomas > 10 mm, and sensitivity of 86% with specificity of 80% for adenomas > 6 mm[71]. The National CT Colonography Trial, sponsored by the American College of Radiology Imaging Network, included 2600 average-risk individuals, reported a sensitivity of 84% and specificity of 85% for detecting adenomas or CRC, and a sensitivity of 70% with a specificity of 86% for adenomas > 6 mm[72]. However, it is noteworthy that CTC exhibits significantly lower sensitivity in detecting sessile serrated lesions. In an RCT comparing CTC with colonoscopy for population screening, 982 individuals underwent CTC, detection of high-risk sessile serrated lesions (dysplastic and/or ≥ 10 mm) was significantly lower with CTC (0.8%) compared to individuals undergoing colonoscopy (4.3%)[73]. Concerns persist regarding lesions smaller than 6 mm, which were not reported in the aforementioned studies, with doubts about the clinical significance of such lesions. Cost-effectiveness analysis found that CTC screening is more cost-effective than no screening[74-76], although studies comparing with other screening tests are heterogeneous[77,78]. Furthermore, CTC also reports on extracolonic findings, which can be identified in up to 66% of individuals[61], though the benefit of such information remains uncertain and gives rise to concerns regarding the potential for overdiagnosis and overtreatment. In the United States, the United States Preventive Services Task Force recommends testing every 5 years.
The colon capsule (CC) is a wireless, disposable pill-sized camera capsule designed to be ingested, capturing images during its transit through the intestine. It is a minimally invasive and painless imaging system that allows exploration of the colon without the need for sedation and gas insufflation but requiring bowel preparation. It was first introduced in 2006 and since then several advances have been made that improved the diagnostic yield, namely an increased and adaptive capsule frame rate, widened angle of view, new software to estimate polyp size and improved data recording[79]. Studies of CC impact on CRC screening are limited. The majority are related to test characteristics compared to colonoscopy and none has evaluated the efficacy in reducing CRC incidence and mortality. In a prospective study of 695 average-risk individuals who underwent capsule colonoscopy followed by colonoscopy, a 100% sensitivity for CRC was found. The sensitivity and specificity for detecting adenomas larger than 6mm was 88% and 82%, respectively, and for adenomas larger than 10mm was 92% and 95%, respectively. However, like CTC, CC performed poorly for sessile serrated lesions, accounting for 26% of false-negative results[80]. Other prospective study comparing CC and CTC involving 320 individuals found that the sensitivity of CTC and CC for polyps larger than 6 mm was 26% and 80%, respectively, and that for polyps larger than 10 mm was 50% and 96%, respectively[81]. Caution is advised while critically revising these results because not only individuals with inadequate bowel preparation and transit time were excluded, but also only experienced gastroenterologist read all capsules, not reflecting the usual care setting in which screening occurs[82]. Currently, CC is approved for individuals with incomplete colonoscopy or evidence of lower gastrointestinal bleeding, but not for CRC screening[55].
Various novel circulating biomarkers are currently under investigation and the most promising are being proposed as potential screening tests for CRC. Both blood and stool tests are minimally invasive and require little patient preparation. Stool-based biomarker tests, like all other fecal screening modalities, have compliance issues because adherence over time decreases. These include methylation markers, circulating microRNA (miRNA), plasma proteins and cytokines (Table 1).
| Tests | CRC vs control | Advanced adenoma vs control | Ref. | ||||
| Sample | AUC | Sensitivity | Specificity | References | Sensitivity | ||
| mSEPT9 | Blood | 0.930 | 0.68 | 0.80 | 87 | 0.11 | [86] |
| SDC2 | Stool | 0.933 | 0.90 | 0.91 | 94 | 0.33 | [94] |
| SFRP | Stool | 0.957 | 0.79 | 0.93 | 95 | 0.43 | [96] |
| MiRNA | |||||||
| Single miRNA (n = 31) | Blood | 0.590-0.943 | 46%-93% | 41%-93% | 101 | 0.69 | [104] |
| Panels with X4 miRNA (miR-29a, miR-92a, miR-601, and miR-760) | 0.943 | 0.83 | 0.93 | ||||
| CA11-19 | Blood | - | 0.98 | 0.84 | 107 | 0.4 | [107] |
| DC-SIGN/DC-SIGNR | Blood | 0.988 | 0.99 | 0.95 | 108 | - | - |
| Non-coding RNA | - | - | |||||
| NEAT1_v1 | Blood | 0.732 | 0.57 | 0.87 | 110 | ||
| NEAT1_v2 | 0.845 | 0.83 | 0.83 | ||||
| CLIP4 | Stool | 0.961 | 0.90 | 0.88 | 111 | 0.78 | [111] |
| SALL4 | Blood | 0.916 | 0.86 | 0.86 | 114 | - | - |
| Cytokines IL-8 | Blood | 0.920 | 0.70 | 0.91 | 116 | - | - |
Methylated septin 9 gene: DNA-methylation biomarkers are a promising method in CRC screening, not only because methylation is one the most prevalent epigenetic alteration in CRC, but it also occurs in the early stages, allowing for detection of early-stage CRC[83]. Methylation markers can be found in blood, stool and in some of them in both samples. The Methylated septin 9 gene test is the only approved blood-based test for CRC detection. The Septin 9 gene (SEPT9) is a tumor suppressor gene that encodes septin 9, a protein early mutated in the CRC pathway in almost all CRC. DNA methylation is the most prevalent epigenetic alteration that occurs in the early stages of carcinogenesis. It is a polymerase chain reaction (PCR) based qualitative test that uses a blood sample to detect methylation of the promoter region of septin 9 DNA[83,84]. In a prospective study including 7941 average-risk individuals scheduled for screening with colonoscopy, the SEPT9 test demonstrated a sensitivity of 48% and specificity of 92% for CRC detection[85]. Subsequent retesting of samples using a next-generation assay revealed an improved sensitivity of 59% for early-stage CRC and 87% for later-stage CRC, with an overall specificity of 79%[86,87]. A systematic review including 39 studies reported a sensitivity of 62% and specificity of 90% for CRC detection[88]. Regarding the detection of advanced adenomas, SEPT9 test reported a notably low sensitivity of 11%[86]. Thus, due to lack of sensitivity and lack of evidence showing morbidity or mortality benefit, its approval is currently limited for individuals who refuse other CRC screening methods.
Syndecan-2 gene: Another most promising methylation marker is the SDC2 encodes for the syndecan-2 protein, which functions as an integral membrane protein and has tumor suppressor effects on cell signaling, migration, and proliferation, as well as angiogenesis. Hypermethylation of SDC2 has been associated mostly to CRC, but also to head and neck squamous cell carcinoma[89,90]. In a prospective study involving 139 patients with CRC, SDC2 methylation of DNA (mSDC2) in blood samples showed a sensitivity for detecting CRC of 87% and specificity of 92%. The sensitivity at stage I was 92.3%, indicating the potential of SDC2 methylation as a blood-based DNA test for early detection of CRC[91,92]. Subsequent studies assessing stool samples were performed, with three studies reporting sensitivities for CRC of 77%-90% and specificities of 88%-98%[93,94]. A recent systematic review has highlighted that SDC2 displays a reduced sensitivity dependent on CRC staging, with sensitivities of 83-91% for CRC stages I-II and 90%-100% for stage III-IV CRC[94], nonetheless still superior to the only currently approved SEPT9 test, although head-to-head studies are lacking.
Secreted frizzled-related protein: Secreted frizzled-related protein (SFRP) is another biomarker holding great promise. Researchers have found that methylation and consequent loss of SFRP gene expression leads to the activation of Wnt pathway, one of most important mechanisms for tumorigenesis and cancer development, with both SFRP1 gene and SFRP2 methylation being found in patients with CRC[95]. A meta-analysis including 37 studies reported a sensitivity of 79% and specificity of 93% for stool samples. Although the specificity of the SFRP2 methylation is also high for colorectal adenomas (94%), it is found to have a sensitivity of only 43%[96]. Though the results from these independent test cohorts confirm the SFRP2 potential as a screening marker, none of these studies tested their assays in validation cohorts.
miRNA: MiRNA are a class of small, non-coding RNAs molecules which play a pivotal role in gene expression regulation. They act as tumor suppressor genes or oncogenes, interfering with various cellular processes crucial to cancer development and progression[97-100]. Increasing evidence supports the diagnostic value of miRNAs in CRC detection, confirming their potential to be used as primary CRC screening tests. MiRNA can be used singled or in combination, increasing specificity. In a systematic review encompassing 34 studies comprising 3454 CRC cases and 2556 controls, dysregulation of 617 plasma miRNAs was observed. Notably, a panel of four miRNAs (miR-29a, miR-92a, miR-601, and miR-760) demonstrated the highest area under the curve at 0.943, achieving 83% sensitivity and 93% specificity[101]. A more recent meta-analysis comprising 35 studies with 3258 CRC patients and 2683 controls reported a sensitivity and specificity of 80%[102]. MiRNAs also exhibit promise in detecting early-stage CRC, with miR-506 and miR-4316 effectively discriminating between patients with early-stage CRC and healthy individuals[103]. Furthermore, miRNA also appear to display high sensitivities for detecting precancer lesions, which is a limitation frequently encountered with other biomarkers currently under investigation. A systematic review and meta-analysis revealed that miR-60 and miR-760 had sensitivities of 83% and 72% for detecting advanced adenomas, with a specificity of 69% and 62%, respectively[104]. Despite the promising results, certain constrain the application of miRNAs in CRC screening. Firstly, not all miRNAs are specific of CRC. Additionally, not all identified miRNAs markers have undergone subsequently validation by independent groups. In fact, some contradictory results have been published. For instance, miR-21 it noted in one study as a highly accurate indicator of early CRC with a sensitivity of 96% and a specificity of 92%[105], was considered inappropriate for clinical practice in another study due to a sensitivity of 79% and specificity of 48%[106]. Lastly, the optimal detection method for miRNAs, whether through PCR, microarray, or next generation sequencing, has not been determined.
CA11-19 glycoprotein and DC-SIGN/ DC-SIGNR: Serologic levels of specific proteins have been associated with CRC. In a comprehensive systematic review, the most promising circulating proteins identified were CA11-19 glycoprotein and DC-SIGN/ DC-SIGNR[100].
CA11-19 is a 701 amino acid glycoprotein. In a single-center study involving 522 average-risk individuals who underwent colonoscopy, with 131 diagnosed with CRC, CA11-19 achieved a notable diagnostic performance, with a sensitivity of 98% and specificity of 84%. However, its sensitivity for adenomas was comparatively lower at 40%[107]. While head-to-head studies are lacking and direct comparisons cannot be made from studies with distinct methodologies, CA11-19 appears to exhibit higher efficacy in detecting adenomas compared to other emerging biomarkers. Further prospective studies with larger samples sizes are necessary to validate these results clarify the role of CA11-19 in CRC screening.
DC-SIGN/ DC-SIGNR are membrane-bound C type leptins. In a single-center study including a 290-patient cohort, sDC-SIGN and sDC-SIGNR reported sensitivities of 88% and 62%, respectively, and specificities of 56% and 98%, respectively. Combining the two markers increased the diagnostic yield, achieving a sensitivity of 99% and a specificity of 95%. The authors concluded that DC-SIGN and DC-SIGNR may serve as independent markers for CRC screening[108]. Further studies are needed to validate these results, but sDC-SIGN and sDC-SIGNR appear to be promising biological markers for the CRC screening.
Non-coding RNA nuclear-enriched abundant transcript 1 and 2: The non-coding RNA nuclear-enriched abundant transcript 1 (NEAT1) is also elevated in peripheral blood from patients with CRC[109]. This genomic region encodes two transcripts, NEAT1_v1 and NEAT1_v2. Subsequent investigations by the same authors focused on evaluating the diagnostic utility of whole blood NEAT1 in CRC found NEAT1_v2 to be more sensitive and specific biomarker in comparison to NEAT1_v1, with reported values of 70% vs 69% for sensitivity and 96% vs 79% for specificity, respectively[110].
CAP-Gly domain containing linker protein: CAP-Gly domain containing linker protein (CLIP4), a member of CLIP family, is also emerging as a promising biomarker. It is involved in plus-end binding of microtubule and in immune response-related biological processes, cell migration and viability in certain cancer metastases. Recent studies have demonstrated a sensitivity of 77%-90% and specificity of 88%-99%[111,112]. Further validation is necessary and studies comparing with other existing molecular diagnostic tests are lacking.
Sal-like protein 4: Another promising biomarker under investigation is sal-like protein 4 (SALL4), an oncogene belonging to the family of zinc-finger transcription factors. In a prospective study involving 51 CRC patients, determination of SALL4 in blood samples exhibited sensitivities of 96% and specificity of 95%[113]. In a separated prospective study including 151 CRC patients, the reported sensitivity for CRC diagnosis was 86%, with a specificity of 86%[114]. Although these results are experimental and need further validation, they hold considerable promise.
Cytokines: There is substantial evidence supporting the involvement of cytokines in the pathophysiology of malignancies, exerting influence at various stages of carcinogenesis, such as regulating angiogenesis and activating signaling pathways that lead to cancer cell proliferation. Among the cytokines investigated in CRC, interleukine-8 (IL-8) is the most studied. It is a member of CXC. It has the capability to induce angiogenesis and activate the MAPK signaling, facilitating the proliferation of tumor cells[115]. In a meta-analysis comprising eighteen studies, including five diagnostic studies, IL-8 exhibited a sensitivity of 70% and specificity of 91%[116]. Despite the limited number of studies and inherent limitations, namely variations in cutoffs and methods employed, these findings suggest that IL-8 is a potential effective tool in CRC screening, and further studies are highly anticipated.
In recent years, increased data suggest intestinal microbiota plays a pivotal role in carcinogenesis. In fact, CRC patients exhibit changes in microbiota and fecal metabolome, indicating potential applications in CRC screening and diagnosis[117]. A recent systematic review including 28 studies indicated optimal diagnostic performance with a sensitivity of 88% and specificity of 94% for CRC diagnosis[118]. Fusobacterium nucleatum (Fn), Lachnoclostridium gene marker (named as
Microbial biomarkers also appear to be effective in detecting colorectal adenomas. In the same study, the diagnostic yield of all biomarkers for detecting colorectal adenomas was assessed, with m3 demonstrating the best diagnostic yield[120]. The authors later suggest that microbial biomarker may predict the risk of adenoma recurrence. In a prospective study involving 161 average-risk individuals during a 10-year follow-up, elevated levels of Fn, m3, and Ch in follow-up stools were significantly associated with adenoma recurrence. Combining these three markers resulted in an 81% sensitivity for adenoma recurrence, while FIT alone or in combination did not predict adenoma recurrence or further improved the diagnostic performance[121].
Certain studies also suggest that stool-based microbial markers exhibit superior sensitivity compared to stool-based tumor markers, but head-to-head studies are awaited to confirm this[122]. Concerns have been raised regarding variations in microbiota among different populations due to diet-induced changes and lifestyle, potentially compromising universal use. In a multi-cohort analysis of 526 metagenomic samples from China, Austria, America, German and France, seven CRC-enriched bacteria were consistently present (Bacteroides fragilis, Fn, Porphyromonas asaccharolytica, Parvimonas micra, Prevotella intermedia, Alistipes finegoldii, and Thermanaerovibrio acidaminovorans)[123]. These results suggest that microbial biomarkers are robust across populations with distinct lifestyle and dietary patterns, indicating potential for universal use. Despite the promise of microbial biomarkers, substantial limitations persist, mostly related to the limited number of studies and small sample sizes, warning further validation and the lack of standardization in the collection and processing of the samples. All studies used different analytical methods to identify bacteria, either sequencing rRNA, qPCR, or whole genome sequencing. Furthermore, studies addressing cost-effectiveness are also lacking. Nonetheless, available data suggest the potential of microbial biomarkers for CRC screening and adenoma detection and recurrence.
Despite numerous studies evaluating their diagnostic performance, the majority of these promising screening methods have not undergone cost-effectiveness analysis. Several factors have impeded the assessment of the economic viability of these screening methods. Consensus on the optimal technology for certain tests is lacking, and in other cases, despite consensus, the corresponding technology is still in an early stage of development. These factors, coupled with limited sample sizes, hinder universal validation, thereby impeding cost-effectiveness analysis.
Nevertheless, certain strategies can already be identified that have the potential to decrease costs while preserving efficacy. One such approach involves emphasizing the detection of precancerous lesions rather than exclusively focusing on CRC detection. It is noteworthy that specific biomarker studies have exclusively concentrated on CRC screening, while others have assessed the diagnostic yield for both CRC and precursor lesions. Some of these tests have reported lower sensitivities for the detection of precursors. An illustrative example is the SEPT9 test, whose diminished sensitivity has led to its classification as cost-ineffective and its exclusion from current screening guidelines. Another strategy involves the integration of multiple molecular biomarkers in a single test, potentially enhancing efficacy with reduced additional costs[124]. However, the optimal combination of molecular biomarkers that maximizes both screening sensitivity and specificity remains undetermined. Moreover, the costs associated with PCR sequencing techniques and large-scale DNA sequencing are anticipated to continue decreasing, thereby reducing the overall cost of the analysis.
Artificial intelligence (AI) is the field of computer sciences dedicated to the development of software machines capable of executing cognitive tasks that typically require human-level intelligence. The core principles of AI include machine learning (ML), a subfield that empowers a machine to enhance its effectiveness through experiential training, and deep learning, a subset of ML that employs artificial neural networks resembling the human brain, enabling in autonomous learning and decision-making capabilities[125]. Regarding CRC screening, the integration of AI into Medicine has resulted in the publication of numerous studies with growing evidence substantiating its efficacy[126-128]. Nevertheless, the broad adoption of AI medical devices in clinical settings, particularly those associated with endoscopy, remains limited[129].
In the context of CRC screening, endoscopy has attracted significant attention, being the subject of more than 10 high-quality RCTs. Despite improvements in endoscopic technology, adenoma missed rates remain as high as 26%, with sessile serrated lesions, proximal advanced adenomas and flat adenomas (34%) posing particular challenges for endoscopists[65]. Moreover, certain adenomas are not recognized by endoscopists even though they were visualized, with reported rates of 14%[130]. In response to these challenges, computer-aided detection (CADe) and computer-aided diagnosis (CADx) were designed to assist endoscopists in the detection and characterization of polyps during colonoscopy, to mitigate missed adenomas and increase the adenoma detection rate (ADR). A meta-analysis of six RCTs including 4354 patients demonstrated a 44% increase in ADR with CADe[131]. This enhancement was most pronounced in the detection of diminutive (< 5 mm) adenomas, whose clinical significance remains uncertain. As for larger polyps, the results are not so consistent, but four of the seven meta-analyses specifically analyzing > 10 mm adenomas found a statistically significant improvement in detection with CADe[132]. Concerns have been raised regarding false positives and their potential impact on withdrawal time. Despite overall high false positive rates, with a post hoc analysis of an RCT revealing an average of 27 per colonoscopy, the resultant increase in total withdrawal time was a negligible 1%[133]. Additional concerns were raised related to the use of CADe potentially leading to an elevated detection of polyps with uncertain clinical significance and subsequently escalating the rates of polypectomies and histopathological examinations. In fact, Effectiveness alone is insufficient for evaluating the suitability of AI implementation in clinical practice. Cost-effectiveness is an essential consideration, with limited studies available, including only one considering the United States healthcare system. However, microsimulations studies suggest that while CADe may initially increase healthcare costs by detecting more adenomas, the long-term balance could be achieved through savings in cancer treatment costs[134,135]. In fact, the same microsimulation study suggested the use of CADe could contribute to a 5% reduction in CRC incidence and 3% in death, compared to standard colonoscopy-based screening.
As for CADx, it is based on optical diagnosis, a method that predicts histopathology of a polyp based on its appearance, allowing for appropriate treatment measures aligned with the predicted histology, with the potential to reduce costs[136,137]. However, optical diagnosis requires specific training and competence, with less than half of endoscopists demonstrating willingness to perform optical diagnosis[138]. CADx has the potential to surpass these barriers. Large prospective studies have demonstrated that CADx can accurately differentiate diminutive polyps, achieving over 90% negative predictive value and over 80% sensitivity and specificity for adenoma identification, reducing polypectomies and pathology-related costs[139,140]. Thus, use of CADx may minimize the number of polypectomies which CADe increases. Even though CADx is currently used for distinguishing between neoplasia and non-neoplasia, ongoing studies are exploring its efficacy in assessing dysplasia and the degree of submucosal invasion. Two large-scale prospective studies have raised doubts about the utility of CADx, indicating no improvement in adenoma identification sensitivity compared to standard optical diagnosis[141,142], suggesting the benefits may be limited to nontrained endoscopists on optical diagnosis. Furthermore, CADx may face additional resistance among endoscopists because of legal reasons. As for CADe, studies assessing cost-effectiveness of CADx are lacking.
AI applications in CRC screening are not limited to endoscopy. Regarding other methods, such as blood-based markers currently under investigation, Wan et al[143] proposed a ML method using tumor-derived cell-free DNA that achieved a 85% sensitivity and specificity for CRC detection. As for miRNAs, several studies have applied AI to identify potential methylated miRNAs using predictive models[144,145]. In one study, a predictive model demonstrated an 85% sensitivity and 90% specificity for identifying patients with CRC and advanced adenomas.
The primary objective of CRC screening is to reduce both the incidence and mortality of CRC and facilitate early detection and intervention to enhance patient prognosis and outcomes. Despite the array of currently available screening tests, each varying in invasiveness and preparation requirements, adherence rates remain suboptimal.
Non-invasive biomarkers present a potential screening avenue due to their minimal invasiveness and limited patient preparation demands, potentially improving compliance. However, existing evidence is based on small sample sizes and lacks validation by independent groups. The optimal screening periodicity is also yet to be determined. Regarding AI, despite some controversy surrounding the clinical benefits of the increased ADR, compelling high-quality studies endorse its utilization. Nevertheless, cost-effectiveness studies are lacking. While the enthusiasm for these emerging technologies is justified, consideration must be given to countries with lower development indices, where resources are constrained and screening is still opportunistic in most settings.
In summary, despite promising evidence supporting the potential enhancement of CRC screening through non-invasive biomarkers and AI, the current body of evidence is not robust enough for widespread implementation in clinical practice. Future studies should go beyond a singular focus on diagnostic yield and statistical performance. Standardizing methods and enrolling large cohorts are needed to comprehensively assess the potential of these markers. Furthermore, in a world of limited resources, critical cost-effectiveness studies are necessary for the practical implementation of any screening method. A scarcity of these essential studies may impede the progress of these promising innovations from mere novelties to tangible progress in CRC screening.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology & hepatology
Country/Territory of origin: Portugal
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gragnaniello V, Italy; Morya AK, India S-Editor: Chen YL L-Editor: A P-Editor: Li X
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 63411] [Article Influence: 15852.8] [Reference Citation Analysis (174)] |
| 2. | Siegel RL, Wagle NS, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 2023;73:233-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1465] [Reference Citation Analysis (3)] |
| 3. | Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2268] [Cited by in RCA: 3236] [Article Influence: 647.2] [Reference Citation Analysis (2)] |
| 4. | Wang S, Zheng R, Li J, Zeng H, Li L, Chen R, Sun K, Han B, Bray F, Wei W, He J. Global, regional, and national lifetime risks of developing and dying from gastrointestinal cancers in 185 countries: a population-based systematic analysis of GLOBOCAN. Lancet Gastroenterol Hepatol. 2024;9:229-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 24] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 5. | Baidoun F, Elshiwy K, Elkeraie Y, Merjaneh Z, Khoudari G, Sarmini MT, Gad M, Al-Husseini M, Saad A. Colorectal Cancer Epidemiology: Recent Trends and Impact on Outcomes. Curr Drug Targets. 2021;22:998-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 147] [Article Influence: 36.8] [Reference Citation Analysis (2)] |
| 6. | Cancer: Disease Control Priorities, Third Edition (Volume 3). Washington (DC): The International Bank for Reconstruction and Development / The World Bank; 2015-Nov-1 . [PubMed] |
| 7. | Fidler MM, Soerjomataram I, Bray F. A global view on cancer incidence and national levels of the human development index. Int J Cancer. 2016;139:2436-2446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 211] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 8. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 857] [Cited by in RCA: 1198] [Article Influence: 239.6] [Reference Citation Analysis (0)] |
| 9. | Bailey CE, Hu CY, You YN, Bednarski BK, Rodriguez-Bigas MA, Skibber JM, Cantor SB, Chang GJ. Increasing disparities in the age-related incidences of colon and rectal cancers in the United States, 1975-2010. JAMA Surg. 2015;150:17-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 764] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 10. | Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol. 2022;7:262-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 367] [Article Influence: 122.3] [Reference Citation Analysis (6)] |
| 11. | Schreuders EH, Ruco A, Rabeneck L, Schoen RE, Sung JJ, Young GP, Kuipers EJ. Colorectal cancer screening: a global overview of existing programmes. Gut. 2015;64:1637-1649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 674] [Cited by in RCA: 907] [Article Influence: 90.7] [Reference Citation Analysis (0)] |
| 12. | Siegel RL, Ward EM, Jemal A. Trends in colorectal cancer incidence rates in the United States by tumor location and stage, 1992-2008. Cancer Epidemiol Biomarkers Prev. 2012;21:411-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 13. | Keum N, Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol. 2019;16:713-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 1544] [Article Influence: 257.3] [Reference Citation Analysis (2)] |
| 14. | Jodal HC, Helsingen LM, Anderson JC, Lytvyn L, Vandvik PO, Emilsson L. Colorectal cancer screening with faecal testing, sigmoidoscopy or colonoscopy: a systematic review and network meta-analysis. BMJ Open. 2019;9:e032773. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 15. | Khandker RK, Dulski JD, Kilpatrick JB, Ellis RP, Mitchell JB, Baine WB. A decision model and cost-effectiveness analysis of colorectal cancer screening and surveillance guidelines for average-risk adults. Int J Technol Assess Health Care. 2000;16:799-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Frazier AL, Colditz GA, Fuchs CS, Kuntz KM. Cost-effectiveness of screening for colorectal cancer in the general population. JAMA. 2000;284:1954-1961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 420] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 17. | Sonnenberg A, Delcò F, Inadomi JM. Cost-effectiveness of colonoscopy in screening for colorectal cancer. Ann Intern Med. 2000;133:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 323] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 18. | Principles and practice of screening for disease. J R Coll Gen Pract. 1968;16:318. |
| 19. | Andermann A, Blancquaert I, Beauchamp S, Déry V. Revisiting Wilson and Jungner in the genomic age: a review of screening criteria over the past 40 years. Bull World Health Organ. 2008;86:317-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 567] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 20. | Kuntz KM, Lansdorp-Vogelaar I, Rutter CM, Knudsen AB, van Ballegooijen M, Savarino JE, Feuer EJ, Zauber AG. A systematic comparison of microsimulation models of colorectal cancer: the role of assumptions about adenoma progression. Med Decis Making. 2011;31:530-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Brenner H, Hoffmeister M, Stegmaier C, Brenner G, Altenhofen L, Haug U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: estimates based on 840,149 screening colonoscopies. Gut. 2007;56:1585-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 303] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 22. | Pignone M, Saha S, Hoerger T, Mandelblatt J. Cost-effectiveness analyses of colorectal cancer screening: a systematic review for the U.S. Preventive Services Task Force. Ann Intern Med. 2002;137:96-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 376] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 23. | Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19:521-531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 231] [Article Influence: 77.0] [Reference Citation Analysis (1)] |
| 24. | Khalili F, Najafi B, Mansour-Ghanaei F, Yousefi M, Abdollahzad H, Motlagh A. Cost-Effectiveness Analysis of Colorectal Cancer Screening: A Systematic Review. Risk Manag Healthc Policy. 2020;13:1499-1512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 25. | Lansdorp-Vogelaar I, Knudsen AB, Brenner H. Cost-effectiveness of colorectal cancer screening. Epidemiol Rev. 2011;33:88-100. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 192] [Cited by in RCA: 217] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 26. | Ran T, Cheng CY, Misselwitz B, Brenner H, Ubels J, Schlander M. Cost-Effectiveness of Colorectal Cancer Screening Strategies-A Systematic Review. Clin Gastroenterol Hepatol. 2019;17:1969-1981.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 27. | Dan YY, Chuah BY, Koh DC, Yeoh KG. Screening based on risk for colorectal cancer is the most cost-effective approach. Clin Gastroenterol Hepatol. 2012;10:266-71.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 28. | Portillo I, Arana-Arri E, Idigoras I, Bilbao I, Martínez-Indart L, Bujanda L, Gutierrez-Ibarluzea I. Colorectal and interval cancers of the Colorectal Cancer Screening Program in the Basque Country (Spain). World J Gastroenterol. 2017;23:2731-2742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 30] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 29. | Peterse EFP, Meester RGS, de Jonge L, Omidvari AH, Alarid-Escudero F, Knudsen AB, Zauber AG, Lansdorp-Vogelaar I. Comparing the Cost-Effectiveness of Innovative Colorectal Cancer Screening Tests. J Natl Cancer Inst. 2021;113:154-161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 30. | Feldman M, Friedman L, Sleisenger M. Sleisenger & Fordtran’s Gastrointestinal and Liver Disease: Pathophysiology, Diagnosis, Management. Philadelphia: Saunders, 2002: 1524-1526. |
| 31. | Wu CW, Sung JJ. Colorectal cancer screening: are stool and blood based tests good enough? Chin Clin Oncol. 2013;2:8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 32. | Lin JS, Perdue LA, Henrikson NB, Bean SI, Blasi PR. Screening for Colorectal Cancer: Updated Evidence Report and Systematic Review for the US Preventive Services Task Force. JAMA. 2021;325:1978-1998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 67] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 33. | Ahlquist DA, McGill DB, Schwartz S, Taylor WF, Owen RA. Fecal blood levels in health and disease. A study using HemoQuant. N Engl J Med. 1985;312:1422-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 122] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 34. | Ahlquist DA, McGill DB, Fleming JL, Schwartz S, Wieand HS, Rubin J, Moertel CG. Patterns of occult bleeding in asymptomatic colorectal cancer. Cancer. 1989;63:1826-1830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 35. | Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, Church TR. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369:1106-1114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 640] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 36. | Lindholm E, Brevinge H, Haglind E. Survival benefit in a randomized clinical trial of faecal occult blood screening for colorectal cancer. Br J Surg. 2008;95:1029-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 213] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 37. | Faivre J, Dancourt V, Lejeune C, Tazi MA, Lamour J, Gerard D, Dassonville F, Bonithon-Kopp C. Reduction in colorectal cancer mortality by fecal occult blood screening in a French controlled study. Gastroenterology. 2004;126:1674-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 290] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 38. | Nishihara R, Wu K, Lochhead P, Morikawa T, Liao X, Qian ZR, Inamura K, Kim SA, Kuchiba A, Yamauchi M, Imamura Y, Willett WC, Rosner BA, Fuchs CS, Giovannucci E, Ogino S, Chan AT. Long-term colorectal-cancer incidence and mortality after lower endoscopy. N Engl J Med. 2013;369:1095-1105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 968] [Cited by in RCA: 1147] [Article Influence: 95.6] [Reference Citation Analysis (0)] |
| 39. | Hampton FJ, MacFadyen UM, Mayberry JF. Variations in results of simultaneous ambulatory esophageal pH monitoring. Dig Dis Sci. 1992;37:506-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 176] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 40. | Mandel JS, Church TR, Ederer F, Bond JH. Colorectal cancer mortality: effectiveness of biennial screening for fecal occult blood. J Natl Cancer Inst. 1999;91:434-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 529] [Cited by in RCA: 506] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 41. | Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, Snover DC, Schuman LM. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 940] [Cited by in RCA: 962] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 42. | Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Intern Med. 2014;160:171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 389] [Cited by in RCA: 483] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 43. | Levin TR, Corley DA, Jensen CD, Schottinger JE, Quinn VP, Zauber AG, Lee JK, Zhao WK, Udaltsova N, Ghai NR, Lee AT, Quesenberry CP, Fireman BH, Doubeni CA. Effects of Organized Colorectal Cancer Screening on Cancer Incidence and Mortality in a Large Community-Based Population. Gastroenterology. 2018;155:1383-1391.e5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 367] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 44. | Zorzi M, Fedeli U, Schievano E, Bovo E, Guzzinati S, Baracco S, Fedato C, Saugo M, Dei Tos AP. Impact on colorectal cancer mortality of screening programmes based on the faecal immunochemical test. Gut. 2015;64:784-790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 221] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 45. | Giorgi Rossi P, Vicentini M, Sacchettini C, Di Felice E, Caroli S, Ferrari F, Mangone L, Pezzarossi A, Roncaglia F, Campari C, Sassatelli R, Sacchero R, Sereni G, Paterlini L, Zappa M. Impact of Screening Program on Incidence of Colorectal Cancer: A Cohort Study in Italy. Am J Gastroenterol. 2015;110:1359-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 46. | Chiu HM, Chen SL, Yen AM, Chiu SY, Fann JC, Lee YC, Pan SL, Wu MS, Liao CS, Chen HH, Koong SL, Chiou ST. Effectiveness of fecal immunochemical testing in reducing colorectal cancer mortality from the One Million Taiwanese Screening Program. Cancer. 2015;121:3221-3229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 217] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 47. | Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, Pabiniak C, Johanson C, Fischer SE, Lansdorp-Vogelaar I, Kuntz KM. Estimation of Benefits, Burden, and Harms of Colorectal Cancer Screening Strategies: Modeling Study for the US Preventive Services Task Force. JAMA. 2016;315:2595-2609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 377] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 48. | Pokharel R, Lin YS, McFerran E, O'Mahony JF. A Systematic Review of Cost-Effectiveness Analyses of Colorectal Cancer Screening in Europe: Have Studies Included Optimal Screening Intensities? Appl Health Econ Health Policy. 2023;21:701-717. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 49. | Imperiale TF, Gruber RN, Stump TE, Emmett TW, Monahan PO. Performance Characteristics of Fecal Immunochemical Tests for Colorectal Cancer and Advanced Adenomatous Polyps: A Systematic Review and Meta-analysis. Ann Intern Med. 2019;170:319-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 50. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, Berger BM. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370:1287-1297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 1224] [Article Influence: 111.3] [Reference Citation Analysis (1)] |
| 51. | Naber SK, Knudsen AB, Zauber AG, Rutter CM, Fischer SE, Pabiniak CJ, Soto B, Kuntz KM, Lansdorp-Vogelaar I. Cost-effectiveness of a multitarget stool DNA test for colorectal cancer screening of Medicare beneficiaries. PLoS One. 2019;14:e0220234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 52. | Ladabaum U, Mannalithara A. Comparative Effectiveness and Cost Effectiveness of a Multitarget Stool DNA Test to Screen for Colorectal Neoplasia. Gastroenterology. 2016;151:427-439.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 53. | Shaukat A, Marsh TL, Crockett SD, Syngal S, Bresalier RS, Brenner DE. Low Prevalence of Screen-Detected Colorectal Cancer in an Average-Risk Population: The New Normal. Clin Gastroenterol Hepatol. 2022;20:2650-2652.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | Loktionov A. Biomarkers for detecting colorectal cancer non-invasively: DNA, RNA or proteins? World J Gastrointest Oncol. 2020;12:124-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 93] [Cited by in RCA: 78] [Article Influence: 15.6] [Reference Citation Analysis (7)] |
| 55. | Kahi CJ, Imperiale TF, Juliar BE, Rex DK. Effect of screening colonoscopy on colorectal cancer incidence and mortality. Clin Gastroenterol Hepatol. 2009;7:770-5; quiz 711. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 299] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 56. | Manser CN, Bachmann LM, Brunner J, Hunold F, Bauerfeind P, Marbet UA. Colonoscopy screening markedly reduces the occurrence of colon carcinomas and carcinoma-related death: a closed cohort study. Gastrointest Endosc. 2012;76:110-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 57. | Doubeni CA, Corley DA, Quinn VP, Jensen CD, Zauber AG, Goodman M, Johnson JR, Mehta SJ, Becerra TA, Zhao WK, Schottinger J, Doria-Rose VP, Levin TR, Weiss NS, Fletcher RH. Effectiveness of screening colonoscopy in reducing the risk of death from right and left colon cancer: a large community-based study. Gut. 2018;67:291-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 179] [Cited by in RCA: 281] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 58. | Kahi CJ, Pohl H, Myers LJ, Mobarek D, Robertson DJ, Imperiale TF. Colonoscopy and Colorectal Cancer Mortality in the Veterans Affairs Health Care System: A Case-Control Study. Ann Intern Med. 2018;168:481-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 59. | Brenner H, Chang-Claude J, Jansen L, Knebel P, Stock C, Hoffmeister M. Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology. 2014;146:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 60. | Lin OS, Kozarek RA, Cha JM. Impact of sigmoidoscopy and colonoscopy on colorectal cancer incidence and mortality: an evidence-based review of published prospective and retrospective studies. Intest Res. 2014;12:268-274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 61. | Baxter NN, Warren JL, Barrett MJ, Stukel TA, Doria-Rose VP. Association between colonoscopy and colorectal cancer mortality in a US cohort according to site of cancer and colonoscopist specialty. J Clin Oncol. 2012;30:2664-2669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 270] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 62. | Doubeni CA, Weinmann S, Adams K, Kamineni A, Buist DS, Ash AS, Rutter CM, Doria-Rose VP, Corley DA, Greenlee RT, Chubak J, Williams A, Kroll-Desrosiers AR, Johnson E, Webster J, Richert-Boe K, Levin TR, Fletcher RH, Weiss NS. Screening colonoscopy and risk for incident late-stage colorectal cancer diagnosis in average-risk adults: a nested case-control study. Ann Intern Med. 2013;158:312-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 63. | Zauber AG, Winawer SJ, O'Brien MJ, Lansdorp-Vogelaar I, van Ballegooijen M, Hankey BF, Shi W, Bond JH, Schapiro M, Panish JF, Stewart ET, Waye JD. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1952] [Cited by in RCA: 2272] [Article Influence: 174.8] [Reference Citation Analysis (1)] |
| 64. | Zhao S, Wang S, Pan P, Xia T, Chang X, Yang X, Guo L, Meng Q, Yang F, Qian W, Xu Z, Wang Y, Wang Z, Gu L, Wang R, Jia F, Yao J, Li Z, Bai Y. Magnitude, Risk Factors, and Factors Associated With Adenoma Miss Rate of Tandem Colonoscopy: A Systematic Review and Meta-analysis. Gastroenterology. 2019;156:1661-1674.e11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 361] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 65. | Inadomi JM, Vijan S, Janz NK, Fagerlin A, Thomas JP, Lin YV, Muñoz R, Lau C, Somsouk M, El-Nachef N, Hayward RA. Adherence to colorectal cancer screening: a randomized clinical trial of competing strategies. Arch Intern Med. 2012;172:575-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 452] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 66. | Knudsen AB, Rutter CM, Peterse EFP, Lietz AP, Seguin CL, Meester RGS, Perdue LA, Lin JS, Siegel RL, Doria-Rose VP, Feuer EJ, Zauber AG, Kuntz KM, Lansdorp-Vogelaar I. Colorectal Cancer Screening: An Updated Modeling Study for the US Preventive Services Task Force. JAMA. 2021;325:1998-2011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 206] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 67. | Atkin WS, Edwards R, Kralj-Hans I, Wooldrage K, Hart AR, Northover JM, Parkin DM, Wardle J, Duffy SW, Cuzick J; UK Flexible Sigmoidoscopy Trial Investigators. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010;375:1624-1633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1134] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 68. | Segnan N, Armaroli P, Bonelli L, Risio M, Sciallero S, Zappa M, Andreoni B, Arrigoni A, Bisanti L, Casella C, Crosta C, Falcini F, Ferrero F, Giacomin A, Giuliani O, Santarelli A, Visioli CB, Zanetti R, Atkin WS, Senore C; SCORE Working Group. Once-only sigmoidoscopy in colorectal cancer screening: follow-up findings of the Italian Randomized Controlled Trial--SCORE. J Natl Cancer Inst. 2011;103:1310-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 451] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 69. | Holme Ø, Løberg M, Kalager M, Bretthauer M, Hernán MA, Aas E, Eide TJ, Skovlund E, Lekven J, Schneede J, Tveit KM, Vatn M, Ursin G, Hoff G; NORCCAP Study Group†. Long-Term Effectiveness of Sigmoidoscopy Screening on Colorectal Cancer Incidence and Mortality in Women and Men: A Randomized Trial. Ann Intern Med. 2018;168:775-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 70. | Cookies on GOV. UK. Public Health England. Bowel Cancer Screening: Programme Overview. 2021. [cited 1 January 2024]. Available from: https://www.gov.uk/guidance/bowel-cancer-screening-programme-overview. |
| 71. | Pickhardt PJ, Hassan C, Halligan S, Marmo R. Colorectal cancer: CT colonography and colonoscopy for detection--systematic review and meta-analysis. Radiology. 2011;259:393-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 356] [Cited by in RCA: 301] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 72. | Halligan S, Altman DG, Taylor SA, Mallett S, Deeks JJ, Bartram CI, Atkin W. CT colonography in the detection of colorectal polyps and cancer: systematic review, meta-analysis, and proposed minimum data set for study level reporting. Radiology. 2005;237:893-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 241] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 73. | IJspeert JE, Tutein Nolthenius CJ, Kuipers EJ, van Leerdam ME, Nio CY, Thomeer MG, Biermann K, van de Vijver MJ, Dekker E, Stoker J. CT-Colonography vs. Colonoscopy for Detection of High-Risk Sessile Serrated Polyps. Am J Gastroenterol. 2016;111:516-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 74. | Obaro AE, Burling DN, Plumb AA. Colon cancer screening with CT colonography: logistics, cost-effectiveness, efficiency and progress. Br J Radiol. 2018;91:20180307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 75. | Hassan C, Pickhardt PJ. Cost-effectiveness of CT colonography. Radiol Clin North Am. 2013;51:89-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 76. | Kriza C, Emmert M, Wahlster P, Niederländer C, Kolominsky-Rabas P. An international review of the main cost-effectiveness drivers of virtual colonography versus conventional colonoscopy for colorectal cancer screening: is the tide changing due to adherence? Eur J Radiol. 2013;82:e629-e636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 77. | Hanly P, Skally M, Fenlon H, Sharp L. Cost-effectiveness of computed tomography colonography in colorectal cancer screening: a systematic review. Int J Technol Assess Health Care. 2012;28:415-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 78. | Greuter MJ, Berkhof J, Fijneman RJ, Demirel E, Lew JB, Meijer GA, Stoker J, Coupé VM. The potential of imaging techniques as a screening tool for colorectal cancer: a cost-effectiveness analysis. Br J Radiol. 2016;89:20150910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 79. | Spada C, De Vincentis F, Cesaro P, Hassan C, Riccioni ME, Minelli Grazioli L, Bolivar S, Zurita A, Costamagna G. Accuracy and safety of second-generation PillCam COLON capsule for colorectal polyp detection. Therap Adv Gastroenterol. 2012;5:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 80. | Rex DK, Adler SN, Aisenberg J, Burch WC Jr, Carretero C, Chowers Y, Fein SA, Fern SE, Fernandez-Urien Sainz I, Fich A, Gal E, Horlander JC Sr, Isaacs KL, Kariv R, Lahat A, Leung WK, Malik PR, Morgan D, Papageorgiou N, Romeo DP, Shah SS, Waterman M. Accuracy of capsule colonoscopy in detecting colorectal polyps in a screening population. Gastroenterology. 2015;148:948-957.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 125] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 81. | Cash BD, Fleisher MR, Fern S, Rajan E, Haithcock R, Kastenberg DM, Pound D, Papageorgiou NP, Fernández-Urién I, Schmelkin IJ, Rex DK. Multicentre, prospective, randomised study comparing the diagnostic yield of colon capsule endoscopy versus CT colonography in a screening population (the TOPAZ study). Gut. 2021;70:2115-2122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 82. | Voska M, Zavoral M, Grega T, Majek O, Martinek J, Tacheci I, Benes M, Vojtechova G, Drastich P, Bures J, Spicak J, Buckova B, Ngo O, Suchanek S. Accuracy of Colon Capsule Endoscopy for Colorectal Neoplasia Detection in Individuals Referred for a Screening Colonoscopy. Gastroenterol Res Pract. 2019;2019:5975438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 83. | Anghel SA, Ioniță-Mîndrican CB, Luca I, Pop AL. Promising Epigenetic Biomarkers for the Early Detection of Colorectal Cancer: A Systematic Review. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 84. | Molnár B, Tóth K, Barták BK, Tulassay Z. Plasma methylated septin 9: a colorectal cancer screening marker. Expert Rev Mol Diagn. 2015;15:171-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 85. | Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N, He YQ, Han X, Hang J, Zhang J, Song L, Han Y, Sheng JQ. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol. 2015;30:830-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 86. | Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, Snover D, Day RW, Ransohoff DF; PRESEPT Clinical Study Steering Committee, Investigators and Study Team. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317-325. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 486] [Cited by in RCA: 585] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 87. | Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB, Weiss G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183-1191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 214] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 88. | Li B, Gan A, Chen X, Wang X, He W, Zhang X, Huang R, Zhou S, Song X, Xu A. Diagnostic Performance of DNA Hypermethylation Markers in Peripheral Blood for the Detection of Colorectal Cancer: A Meta-Analysis and Systematic Review. PLoS One. 2016;11:e0155095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 89. | Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, Kim TS, Kim NK, Chung HC, An S. Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn. 2013;15:498-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 90. | Zhao G, Li H, Yang Z, Wang Z, Xu M, Xiong S, Li S, Wu X, Liu X, Zhu Y, Ma Y, Fei S, Zheng M. Multiplex methylated DNA testing in plasma with high sensitivity and specificity for colorectal cancer screening. Cancer Med. 2019;8:5619-5628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 91. | Worm Ørntoft MB. Review of Blood-Based Colorectal Cancer Screening: How Far Are Circulating Cell-Free DNA Methylation Markers From Clinical Implementation? Clin Colorectal Cancer. 2018;17:e415-e433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 92. | Su WC, Kao WY, Chang TK, Tsai HL, Huang CW, Chen YC, Li CC, Hsieh YC, Yeh HJ, Chang CC, Wang JY. Stool DNA test targeting methylated syndecan-2 (SDC2) as a noninvasive screening method for colorectal cancer. Biosci Rep. 2021;41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 93. | Kim CW, Kim H, Kim HR, Kye BH, Kim HJ, Min BS, Oh TJ, An S, Lee SH. Colorectal cancer screening using a stool DNA-based SDC2 methylation test: a multicenter, prospective trial. BMC Gastroenterol. 2021;21:173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 94. | Oh TJ, Oh HI, Seo YY, Jeong D, Kim C, Kang HW, Han YD, Chung HC, Kim NK, An S. Feasibility of quantifying SDC2 methylation in stool DNA for early detection of colorectal cancer. Clin Epigenetics. 2017;9:126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 95. | Yu J, Xie Y, Li M, Zhou F, Zhong Z, Liu Y, Wang F, Qi J. Association between SFRP promoter hypermethylation and different types of cancer: A systematic review and meta-analysis. Oncol Lett. 2019;18:3481-3492. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 96. | Zhang H, Qi J, Wu YQ, Zhang P, Jiang J, Wang QX, Zhu YQ. Accuracy of early detection of colorectal tumours by stool methylation markers: a meta-analysis. World J Gastroenterol. 2014;20:14040-14050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 20] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 97. | Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 254] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 98. | Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 326] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 99. | Salarinia R, Sahebkar A, Peyvandi M, Mirzaei HR, Jaafari MR, Riahi MM, Ebrahimnejad H, Nahand JS, Hadjati J, Asrami MO, Fadaei S, Salehi R, Mirzaei H. Epi-Drugs and Epi-miRs: Moving Beyond Current Cancer Therapies. Curr Cancer Drug Targets. 2016;16:773-788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 108] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 100. | Nikolaou S, Qiu S, Fiorentino F, Rasheed S, Tekkis P, Kontovounisios C. Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol. 2018;22:481-498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (4)] |
| 101. | Carter JV, Galbraith NJ, Yang D, Burton JF, Walker SP, Galandiuk S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116:762-774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 102. | Zuo Z, Jiang Y, Zeng S, Li Y, Fan J, Guo Y, Tao H. The value of microRNAs as the novel biomarkers for colorectal cancer diagnosis: A meta-analysis. Pathol Res Pract. 2020;216:153130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 103. | Krawczyk P, Powrózek T, Olesiński T, Dmitruk A, Dziwota J, Kowalski D, Milanowski J. Evaluation of miR-506 and miR-4316 expression in early and non-invasive diagnosis of colorectal cancer. Int J Colorectal Dis. 2017;32:1057-1060. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 104. | Wang Q, Huang Z, Ni S, Xiao X, Xu Q, Wang L, Huang D, Tan C, Sheng W, Du X. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One. 2012;7:e44398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 183] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 105. | Ghareib AF, Mohamed RH, Abd El-Fatah AR, Saadawy SF. Assessment of Serum MicroRNA-21 Gene Expression for Diagnosis and Prognosis of Colorectal Cancer. J Gastrointest Cancer. 2020;51:818-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 106. | Choi HH, Cho YS, Choi JH, Kim HK, Kim SS, Chae HS. Stool-Based miR-92a and miR-144* as Noninvasive Biomarkers for Colorectal Cancer Screening. Oncology. 2019;97:173-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 107. | Overholt BF, Wheeler DJ, Jordan T, Fritsche HA. CA11-19: a tumor marker for the detection of colorectal cancer. Gastrointest Endosc. 2016;83:545-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 108. | Jiang Y, Zhang C, Chen K, Chen Z, Sun Z, Zhang Z, Ding D, Ren S, Zuo Y. The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PLoS One. 2014;9:e114748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 109. | Xu Y, Xu Q, Yang L, Ye X, Liu F, Wu F, Ni S, Tan C, Cai G, Meng X, Cai S, Du X. Identification and validation of a blood-based 18-gene expression signature in colorectal cancer. Clin Cancer Res. 2013;19:3039-3049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 110. | Wu Y, Yang L, Zhao J, Li C, Nie J, Liu F, Zhuo C, Zheng Y, Li B, Wang Z, Xu Y. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. 2015;14:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 122] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 111. | Cao Y, Zhao G, Cao Y, Chen Z, Liu X, Yuan M, Yang J, Wang X, Ma Y, Liu Z, Xiong S, Zheng M, Fei S. Feasibility of Methylated CLIP4 in Stool for Early Detection of Colorectal Cancer: A Training Study in Chinese Population. Front Oncol. 2021;11:647066. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 112. | Constâncio V, Nunes SP, Henrique R, Jerónimo C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 138] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 113. | Ardalan Khales S, Abbaszadegan MR, Abdollahi A, Raeisossadati R, Tousi MF, Forghanifard MM. SALL4 as a new biomarker for early colorectal cancers. J Cancer Res Clin Oncol. 2015;141:229-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 114. | Wu HK, Liu C, Fan XX, Wang H, Zhou L. Spalt-like transcription factor 4 as a potential diagnostic and prognostic marker of colorectal cancer. Cancer Biomark. 2017;20:191-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 115. | Sgourakis G, Papapanagiotou A, Kontovounisios C, Karamouzis MV, Dedemadi G, Goumas C, Karaliotas C, Papavassiliou AG. The combined use of serum neurotensin and IL-8 as screening markers for colorectal cancer. Tumour Biol. 2014;35:5993-6002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 116. | Xia W, Chen W, Zhang Z, Wu D, Wu P, Chen Z, Li C, Huang J. Prognostic value, clinicopathologic features and diagnostic accuracy of interleukin-8 in colorectal cancer: a meta-analysis. PLoS One. 2015;10:e0123484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 117. | Nakatsu G, Li X, Zhou H, Sheng J, Wong SH, Wu WK, Ng SC, Tsoi H, Dong Y, Zhang N, He Y, Kang Q, Cao L, Wang K, Zhang J, Liang Q, Yu J, Sung JJ. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:8727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 558] [Cited by in RCA: 505] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 118. | Zwezerijnen-Jiwa FH, Sivov H, Paizs P, Zafeiropoulou K, Kinross J. A systematic review of microbiome-derived biomarkers for early colorectal cancer detection. Neoplasia. 2023;36:100868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 119. | Peng BJ, Cao CY, Li W, Zhou YJ, Zhang Y, Nie YQ, Cao YW, Li YY. Diagnostic Performance of Intestinal Fusobacterium nucleatum in Colorectal Cancer: A Meta-Analysis. Chin Med J (Engl). 2018;131:1349-1356. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 120. | Liang JQ, Wong SH, Szeto CH, Chu ES, Lau HC, Chen Y, Fang J, Yu J, Sung JJ. Fecal microbial DNA markers serve for screening colorectal neoplasm in asymptomatic subjects. J Gastroenterol Hepatol. 2021;36:1035-1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 121. | Liang JQ, Zeng Y, Kwok G, Cheung CP, Suen BY, Ching JYL, To KF, Yu J, Chan FKL, Ng SC. Novel microbiome signatures for non-invasive diagnosis of adenoma recurrence after colonoscopic polypectomy. Aliment Pharmacol Ther. 2022;55:847-855. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 122. | Chan FKL, Wong MCS, Chan AT, East JE, Chiu HM, Makharia GK, Weller D, Ooi CJ, Limsrivilai J, Saito Y, Hang DV, Emery JD, Makmun D, Wu K, Ali RAR, Ng SC. Joint Asian Pacific Association of Gastroenterology (APAGE)-Asian Pacific Society of Digestive Endoscopy (APSDE) clinical practice guidelines on the use of non-invasive biomarkers for diagnosis of colorectal neoplasia. Gut. 2023;72:1240-1254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 123. | Dai Z, Coker OO, Nakatsu G, Wu WKK, Zhao L, Chen Z, Chan FKL, Kristiansen K, Sung JJY, Wong SH, Yu J. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6:70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 250] [Cited by in RCA: 348] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 124. | Kamel F, Eltarhoni K, Nisar P, Soloviev M. Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 125. | Mitsala A, Tsalikidis C, Pitiakoudis M, Simopoulos C, Tsaroucha AK. Artificial Intelligence in Colorectal Cancer Screening, Diagnosis and Treatment. A New Era. Curr Oncol. 2021;28:1581-1607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 126. | Zhu W, Xie L, Han J, Guo X. The Application of Deep Learning in Cancer Prognosis Prediction. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 241] [Cited by in RCA: 153] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 127. | Chahal D, Byrne MF. A primer on artificial intelligence and its application to endoscopy. Gastrointest Endosc. 2020;92:813-820.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 128. | Goyal H, Mann R, Gandhi Z, Perisetti A, Ali A, Aman Ali K, Sharma N, Saligram S, Tharian B, Inamdar S. Scope of Artificial Intelligence in Screening and Diagnosis of Colorectal Cancer. J Clin Med. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 129. | Mori Y, Neumann H, Misawa M, Kudo SE, Bretthauer M. Artificial intelligence in colonoscopy - Now on the market. What's next? J Gastroenterol Hepatol. 2021;36:7-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 130. | Yamada M, Saito Y, Imaoka H, Saiko M, Yamada S, Kondo H, Takamaru H, Sakamoto T, Sese J, Kuchiba A, Shibata T, Hamamoto R. Development of a real-time endoscopic image diagnosis support system using deep learning technology in colonoscopy. Sci Rep. 2019;9:14465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 131. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 303] [Article Influence: 75.8] [Reference Citation Analysis (1)] |
| 132. | Young E, Edwards L, Singh R. The Role of Artificial Intelligence in Colorectal Cancer Screening: Lesion Detection and Lesion Characterization. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 133. | Nehme F, Coronel E, Barringer DA, Romero LG, Shafi MA, Ross WA, Ge PS. Performance and attitudes toward real-time computer-aided polyp detection during colonoscopy in a large tertiary referral center in the United States. Gastrointest Endosc. 2023;98:100-109.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 134. | Areia M, Mori Y, Correale L, Repici A, Bretthauer M, Sharma P, Taveira F, Spadaccini M, Antonelli G, Ebigbo A, Kudo SE, Arribas J, Barua I, Kaminski MF, Messmann H, Rex DK, Dinis-Ribeiro M, Hassan C. Cost-effectiveness of artificial intelligence for screening colonoscopy: a modelling study. Lancet Digit Health. 2022;4:e436-e444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 109] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 135. | Mori Y, Wang P, Løberg M, Misawa M, Repici A, Spadaccini M, Correale L, Antonelli G, Yu H, Gong D, Ishiyama M, Kudo SE, Kamba S, Sumiyama K, Saito Y, Nishino H, Liu P, Glissen Brown JR, Mansour NM, Gross SA, Kalager M, Bretthauer M, Rex DK, Sharma P, Berzin TM, Hassan C. Impact of Artificial Intelligence on Colonoscopy Surveillance After Polyp Removal: A Pooled Analysis of Randomized Trials. Clin Gastroenterol Hepatol. 2023;21:949-959.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 30] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 136. | Rex DK, Kahi C, O'Brien M, Levin TR, Pohl H, Rastogi A, Burgart L, Imperiale T, Ladabaum U, Cohen J, Lieberman DA. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc. 2011;73:419-422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 460] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 137. | Houwen BBSL, Hassan C, Coupé VMH, Greuter MJE, Hazewinkel Y, Vleugels JLA, Antonelli G, Bustamante-Balén M, Coron E, Cortas GA, Dinis-Ribeiro M, Dobru DE, East JE, Iacucci M, Jover R, Kuvaev R, Neumann H, Pellisé M, Puig I, Rutter MD, Saunders B, Tate DJ, Mori Y, Longcroft-Wheaton G, Bisschops R, Dekker E. Definition of competence standards for optical diagnosis of diminutive colorectal polyps: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy. 2022;54:88-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 138. | Wadhwa V, Alagappan M, Gonzalez A, Gupta K, Brown JRG, Cohen J, Sawhney M, Pleskow D, Berzin TM. Physician sentiment toward artificial intelligence (AI) in colonoscopic practice: a survey of US gastroenterologists. Endosc Int Open. 2020;8:E1379-E1384. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 139. | Mori Y, Kudo SE, Misawa M, Saito Y, Ikematsu H, Hotta K, Ohtsuka K, Urushibara F, Kataoka S, Ogawa Y, Maeda Y, Takeda K, Nakamura H, Ichimasa K, Kudo T, Hayashi T, Wakamura K, Ishida F, Inoue H, Itoh H, Oda M, Mori K. Real-Time Use of Artificial Intelligence in Identification of Diminutive Polyps During Colonoscopy: A Prospective Study. Ann Intern Med. 2018;169:357-366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 412] [Cited by in RCA: 348] [Article Influence: 49.7] [Reference Citation Analysis (1)] |
| 140. | Horiuchi H, Tamai N, Kamba S, Inomata H, Ohya TR, Sumiyama K. Real-time computer-aided diagnosis of diminutive rectosigmoid polyps using an auto-fluorescence imaging system and novel color intensity analysis software. Scand J Gastroenterol. 2019;54:800-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 141. | Rondonotti E, Hassan C, Tamanini G, Antonelli G, Andrisani G, Leonetti G, Paggi S, Amato A, Scardino G, Di Paolo D, Mandelli G, Lenoci N, Terreni N, Andrealli A, Maselli R, Spadaccini M, Galtieri PA, Correale L, Repici A, Di Matteo FM, Ambrosiani L, Filippi E, Sharma P, Radaelli F. Artificial intelligence-assisted optical diagnosis for the resect-and-discard strategy in clinical practice: the Artificial intelligence BLI Characterization (ABC) study. Endoscopy. 2023;55:14-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 67] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 142. | Barua I, Wieszczy P, Kudo SE, Misawa M, Holme Ø, Gulati S, Williams S, Mori K, Itoh H, Takishima K, Mochizuki K, Miyata Y, Mochida K, Akimoto Y, Kuroki T, Morita Y, Shiina O, Kato S, Nemoto T, Hayee B, Patel M, Gunasingam N, Kent A, Emmanuel A, Munck C, Nilsen JA, Hvattum SA, Frigstad SO, Tandberg P, Løberg M, Kalager M, Haji A, Bretthauer M, Mori Y. Real-Time Artificial Intelligence-Based Optical Diagnosis of Neoplastic Polyps during Colonoscopy. NEJM Evid. 2022;1:EVIDoa2200003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 143. | Wan N, Weinberg D, Liu TY, Niehaus K, Ariazi EA, Delubac D, Kannan A, White B, Bailey M, Bertin M, Boley N, Bowen D, Cregg J, Drake AM, Ennis R, Fransen S, Gafni E, Hansen L, Liu Y, Otte GL, Pecson J, Rice B, Sanderson GE, Sharma A, St John J, Tang C, Tzou A, Young L, Putcha G, Haque IS. Machine learning enables detection of early-stage colorectal cancer by whole-genome sequencing of plasma cell-free DNA. BMC Cancer. 2019;19:832. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 144. | Kel A, Boyarskikh U, Stegmaier P, Leskov LS, Sokolov AV, Yevshin I, Mandrik N, Stelmashenko D, Koschmann J, Kel-Margoulis O, Krull M, Martínez-Cardús A, Moran S, Esteller M, Kolpakov F, Filipenko M, Wingender E. Walking pathways with positive feedback loops reveal DNA methylation biomarkers of colorectal cancer. BMC Bioinformatics. 2019;20:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 145. | Herreros-Villanueva M, Duran-Sanchon S, Martín AC, Pérez-Palacios R, Vila-Navarro E, Marcuello M, Diaz-Centeno M, Cubiella J, Diez MS, Bujanda L, Lanas A, Jover R, Hernández V, Quintero E, José Lozano J, García-Cougil M, Martínez-Arranz I, Castells A, Gironella M, Arroyo R. Plasma MicroRNA Signature Validation for Early Detection of Colorectal Cancer. Clin Transl Gastroenterol. 2019;10:e00003. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |