Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.458
Peer-review started: September 4, 2023
First decision: November 22, 2023
Revised: December 2, 2023
Accepted: December 20, 2023
Article in press: December 20, 2023
Published online: February 15, 2024
Gastric cancer (GC) is a prevalent malignant tumor of the gastrointestinal system. ZNF710 is a transcription factor (TF), and zinc finger protein 710 (ZNF710)-AS1-201 is an immune-related long noncoding RNA (lncRNA) that is upregulated in GC cells.
To assess the correlation between ZNF710-AS1-201 and immune microenvironment features and to investigate the roles of ZNF710-AS1-201 in the invasion and metastasis processes of GC cells.
We obtained data from The Cancer Genome Atlas and Wujin Hospital. We assessed cell growth, migration, invasion, and programmed cell death using cell counting kit-8, EdU, scratch, Transwell, and flow cytometry assays. Quantitative real-time polymerase chain reaction (qRT-PCR) was used to identify the potential downstream targets of ZNF710-AS1-201.
In GC tissues with low ZNF710-AS1-201 expression, immunoassays detected significant infiltration of various antitumor immune cells, such as memory CD8 T cells and activated CD4 T cells. In the low-expression group, the half-maximal inhibitory concentrations (IC50s) of 5-fluorouracil, cisplatin, gemcitabine, and trametinib were lower, whereas the IC50s of dasatinib and vorinostat were higher. The malignant degree of GC was higher and the stage was later in the high-expression group. Additionally, patients with high expression of ZNF710-AS1-201 had lower overall survival and disease-free survival rates. In vitro, the overexpression of ZNF710-AS1-201 greatly enhanced growth, metastasis, and infiltration while suppressing cell death in HGC-27 cells. In contrast, the reduced expression of ZNF710-AS1-201 greatly hindered cell growth, enhanced apoptosis, and suppressed the metastasis and invasion of MKN-45 cells. The expression changes in ZNF710 were significant, but the corresponding changes in isocitrate dehydrogenase-2, Semaphorin 4B, ARHGAP10, RGMB, hsa-miR-93-5p, and ZNF710-AS1-202 were not consistent or statistically significant after overexpression or knockdown of ZNF710-AS1-201, as determined by qRT-PCR.
Immune-related lncRNA ZNF710-AS1-201 facilitates the metastasis and invasion of GC cells. It appears that ZNF710-AS1-201 and ZNF710 have potential as effective targets for therapeutic intervention in GC. Nevertheless, it is still necessary to determine the specific targets of the ZNF710 TF.
Core Tip: In the field of oncology, there is significant interest in long noncoding RNAs (lncRNAs). They have a significant impact on the immune microenvironments of tumors and immunotherapy. In addition, lncRNAs regulate many crucial mechanisms of cancer immunity, including the presentation of antigens and the exhaustion of T cells. According to our prior investigation, the immune-associated lncRNA zinc finger protein 710 (ZNF710)-AS1-201 has the potential to function as an indicator of GC patient prognosis (recurrence, metastasis and survival). The primary focus of this study was to assess the correlation between ZNF710-AS1-201 and the immune microenvironment and to investigate the roles of ZNF710-AS1-201 in the cellular growth and metastasis processes within GC cells.
- Citation: Ding W, Chen WW, Wang YQ, Xu XZ, Wang YB, Yan YM, Tan YL. Immune-related long noncoding RNA zinc finger protein 710-AS1-201 promotes the metastasis and invasion of gastric cancer cells. World J Gastrointest Oncol 2024; 16(2): 458-474
- URL: https://www.wjgnet.com/1948-5204/full/v16/i2/458.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i2.458
Gastric cancer (GC) is a prevalent malignant tumor affecting the gastrointestinal tract. At present, surgical excision is the favored approach for treating stomach cancer[1]. The prognosis of GC is mainly influenced by the recurrence and spread of the disease after surgery[2]. To prevent the recurrence and spread of tumors after surgery, adjuvant chemotherapy, radiotherapy, targeted therapy, and immunotherapy are employed[3]. The progression of GC is significantly influenced by recent advancements in comprehending the immune microenvironment[4]. Compared to traditional chemotherapy and radiotherapy, immunotherapy is more effective and less toxic[5]. The current methods of immunotherapy consist immune checkpoint inhibitors, adoptive cell therapy, vascular endothelial growth factor inhibitors, cancer vaccines, and chimeric antigen receptor T cell therapy[6]. Immune checkpoint therapy has yielded surprising results[7]. The related drugs include pembrolizumab, nivolumab, avelumab and so on[8]. Nevertheless, the overall prognosis for advanced GC remains poor. Currently, there is a pressing requirement to identify additional, more efficient therapeutic targets.
In the field of oncology, there is a significant interest in long noncoding RNAs (lncRNAs). It has a significant impact on the immune microenvironments of tumors and immunotherapy[9,10]. In addition, lncRNAs regulate many crucial mechanisms of cancer immunity, including the presentation of antigens and the exhaustion of T cells[11]. According to our prior investigation, the immune-associated lncRNA zinc finger protein 710 (ZNF710)-AS1-201 has the potential to function as an indicator of patient prognosis (recurrence, metastasis and survival) in cases of GC[12]. The study illustrated that patients with high ZNF710-AS1-201 expression in tumor tissues had poorer disease-free survival (DFS) than patients with low ZNF710-AS1-201 expression in both The Cancer Genome Atlas (TCGA) data and our data. ZNF710-AS1-201 is one of the transcripts of the ZNF710-AS1 gene, which acts as the opposite strand to ZNF710. The primary focus of this study was to assess the correlation between ZNF710-AS1-201 and the immune microenvironment and to investigate the roles of ZNF710-AS1-201 in the cellular growth and metastasis processes within GC cells.
The RNAseq data of all patients in the TCGA-stomach adenocarcinoma database for stomach cancer were acquired from The UCSC Xena website (https://xenabrowser.net/datapages/). To standardize the transcriptome sequencing data, the utilization of fragments per kilobase of transcript per million mapped reads was implemented. According to the median ZNF710-AS1-201 expression level, the patients were categorized into two groups: High expression (HExp) and low expression (LExp) groups.
To compare the infiltration of immune cells among each sample, we employed single-sample gene set enrichment analysis using the R package “GSVA (Ver.1.44.5)”[13]. We acquired the feature genes of various immune cells from the TISIDB website (http://cis.hku.hk/TISIDB/). Additionally, we utilized the R package “estimate (Ver. 1.0.13)” to assess the stromal and immune cells within every sample, and a measurement was conducted. Wilcoxon analysis with the R package “stats (Ver. 4.2.0)” was utilized to compare the infiltration of various immune cells, including stromal and immune cells, as well as immune checkpoints, between the HExp and LExp groups.
The R package “oncoPredict (Ver. 1.0.13)” was utilized to compute the half-maximal inhibitory concentrations (IC50s) of chemotherapy and targeted medications for every individual[14]. We compared the IC50s of 19 antitumor drugs (5-fluorouracil, afatinib, lapatinib, axitinib, cisplatin, crizotinib, dabrafenib, dasatinib, epirubicin, gefitinib, gemcitabine, irinotecan, lapatinib, nilotinib, oxaliplatin, paclitaxel, sorafenib, trametinib and vorinostat) between the HExp and LExp groups using Wilcoxon analysis.
The R package “limma (Ver. 1.3.54.2)” was used to screen for differentially expressed genes (DEGs) between the HExp and LExp groups, with a false discovery rate threshold (FDR) of less than 0.05 and a log2|fold change (FC)| greater than 1. Utilizing the R package “clusterProfiler (Ver. 4.4.4)” to assess these DEGs, we performed an enrichment analysis using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases.
A total of 68 patients who underwent radical surgery for GC in Changzhou Wujin People’s Hospital were enrolled from January 2014 to November 2014. Below are the criteria for inclusion. First, the pathological diagnosis was gastric adenocarcinoma, and no distant metastasis was found on imaging. Second, none of the patients received preoperative chemotherapy or targeted therapy. Finally, all follow-up data were complete. Tumor and adjacent tissues were collected immediately after tumor isolation and stored in liquid nitrogen.
The HGC-27 (CL-0107) and MKN-45 (CL-0292) cell lines of GC, obtained from Procell Life Science and Technology Co. (Wuhan, China), were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Procella, Wuhan, China) at 37 ℃ in a humid atmosphere consisting of 5% CO2 and 95% air. RPMI-1640 contained 10% fetal bovine serum (FBS) (Procella, Wuhan, China) and 1% penicillin-streptomycin solution (Procell, Wuhan, China) as the medium.
Yuanzi Biotech Co. (Shanghai, China) synthesized the overexpression and shRNA plasmids of ZNF710-AS1-201. The most efficient shRNA of the three was selected based on the experimental findings. For all cell transfections, Lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States) was administered according to the instructions provided by Thermo Fisher for Lipofectamine 3000 Transfection Reagent.
TRIzol reagent (TaKaRa, Dalian, China) was used to extract RNA from HGC-27 and MKN-45 cells following the supplier's instructions, resulting in a total amount of RNA. The RNA sample was converted into complementary DNA using a kit from Moloney-murine leukemia virus (Promega Biotech, Beijing, China). To assess the RNA expression level in GC cells, SYBR Green Master Mix from Takara (Dalian, China) was utilized. Glyceraldehyde-3-phosphate dehydrogenase was employed as an endogenous control. The calculation of RNA expression was determined using the 2−ΔΔCT approach. Table 1 displays the introductory details.
| Name | Primer sequence (5’ to 3’) |
| ZNF710-AS1-201-F | ACCGCAGGCACTTTGAAGA |
| ZNF710-AS1-201-R | AAGGGACATCAGAGGGGAG |
| Bcl-2-F | GGTGGGGTCATGTGTGTGG |
| Bcl-2-R | CGGTTCAGGTACTCAGTCATCC |
| Bax-F | CCCGAGAGGTCTTTTTCCGAG |
| Bax-R | CCAGCCCATGATGGTTCTGAT |
| ZNF710-F | AACAGGAGGTCTATGAGGTTTCT |
| ZNF710-R | CGCTGAGGTCGATCATCTTGA |
| IDH2-F | CGCCACTATGCCGACAAAAG |
| IDH2-R | ACTGCCAGATAATACGGGTCA |
| SEMA4B-F | GAGCGGCCATTCCTCAGATTC |
| SEMA4B-R | CACCCACGTACAGGGTCCT |
| ARHGAP10-F | CCCAGCGGAAGTTTGCTCAT |
| ARHGAP10-R | ACAGCTCCAAGTTGCTCTTTTC |
| RGMB-F | CCAGCCCAATGTCGAATCCA |
| RGMB-R | GGTCACTGATACCCAACACGG |
| miR-93-5p-F | CGCAAAGTGCTGTTCGTGC |
| miR-93-5p-R | AGTGCAGGGTCCGAGGTATT |
| ZNF710-AS1-202-F | GTCCGATCACAGTTCACTAC |
| ZNF710-AS1-202-R | ATACCAAACCAGGCAACATA |
The cell suspension (100 μL/well) was cultured in 96-well plates and incubated for 24, 48, and 72 h in the incubators. Next, we added 10 μL of cell counting kit-8 (CCK-8) (Beyotime, Shanghai, China) solution to every well. An enzyme label instrument (BioTek, Beijing, China) was used to measure the absorbance at 450 nm.
The Cell-Light EdU Apollo567 In Vitro kit (RiboBio, China) was utilized to conduct EdU incorporation assays. The exponential increase was calculated for vascular smooth muscle cells plated in 96-well dishes at a density of 4 × 103 cells per well. Following 24 h of the specified treatment, each well was exposed to 100 μL of 50 μM EdU medium for a duration of 2 h. Subsequently, the cells were immobilized in a PBS solution containing 4% paraformaldehyde for a period of 30 min. Next, a solution containing 2 milligrams per milliliter of glycine was introduced for a duration of 5 min. Following rinsing with phosphate buffered saline (PBS), the cells were subjected to an incubation period of 30 min with a 1 × Apollo staining solution. After discarding the staining solution, the cells were rinsed with PBS containing 0.5% Triton X-100 for 10 min. Following another round of washing with PBS, 1 × Hoechst 33342 was introduced and incubated at room temperature for 30 min. Following washing with PBS, fluorescence microscopy was used to observe the positive cells.
The evaluation of cell apoptosis was conducted through a flow cytometry test (BD, New Jersey, United States). After being rinsed twice in PBS, the cells were then suspended in 1 × binding buffer. For the control group, 400 microliters of 1 × binding buffer was utilized, while for the remaining treatment groups, 100 microliters of 1 × binding buffer was employed. Then, 100 microliters was extracted from the aforementioned solution and transferred to the flow tube. Five microliters of Fluorescein isothiocyanate-Annexin V and five microliters of propidium iodide (at a concentration of 50 micrograms per milliliter) was added. The sample was left at room temperature (25 ℃) and away from light for a duration of 15 min. Next, 400 μL of 1 × binding buffer was added to each tube and mixed well. Analysis using flow cytometry was conducted within one hour. FlowJo software (BD, New Jersey, United States) was utilized to analyze the data.
To evaluate the cell metastasis characteristics, a scratch assay was conducted in vitro by creating a linear scratch across the center of the layer of cultured cells. Pictures were captured at regular intervals (every 24 h), and the gathered data were utilized to analyze the experimental outcomes.
The invasion of cells was observed by employing the Transwell assay with Transwell kits from Corning, United States. The Matrigel adhesive was melted overnight at a temperature of 4 ℃ and then mixed with the serum-free culture after dilution. The diluted Matrigel adhesive was applied onto the upper chamber and left in the incubator for 1-1.5 h until the adhesive hardened. After terminating digestion, the cells were centrifuged while waiting for the Matrigel gum to solidify. To adjust the cell density to 5 × 105 cells/mL, the cells were suspended in medium that included 2% FBS. The upper chamber was inoculated with 200 μL of cell suspension, with 3 replicates per group. The lower chamber was supplemented with 800 μL of medium containing 20% FBS and cultured at 37 ℃ and 5% CO2 for 48 h. After removing the chamber, the medium was discarded, and the chamber was washed twice with PBS. Then, the cells were fixed with 4% paraformaldehyde at room temperature for 30 min. The paraformaldehyde was disposed of and rinsed two times with PBS. After being thoroughly dried, the chamber was treated with crystal violet stain for 15 min. The excess crystal violet dye was washed away with PBS. A microscope (Olympus, Japan) was used to count the cells.
WB was used to analyze the protein expression levels associated with apoptosis. Protein concentrations were measured with a bicinchoninic acid protein assay kit from Thermo, United States. Bax (Abcam, ab32503, 1:1000), Bcl2 (Abcam, ab182858, 1:2000), and β-actin (Affinity, AF7018, 1:3000) were left to react with the membrane overnight at 4 ℃. The blots were visualized using the Millipore Enterochromaffin-like detection system.
All analyses were conducted using GraphPad Prism (Version 9.4.1) and R (Version 4.2.0). mean ± SD deviation values were used to present all continuous variable results. The t test was used if the variance was homogeneous, and the Mann-Whitney test was used if the variance was not homogeneous. Categorical variables were displayed using ratios. Differences between components were tested using chi-square tests. The grayscale values of bands on WB images were calculated using ImageJ from NIH (Bethesda, MD). The screening criteria for a significant difference were P < 0.05, P < 0.01, or P < 0.001.
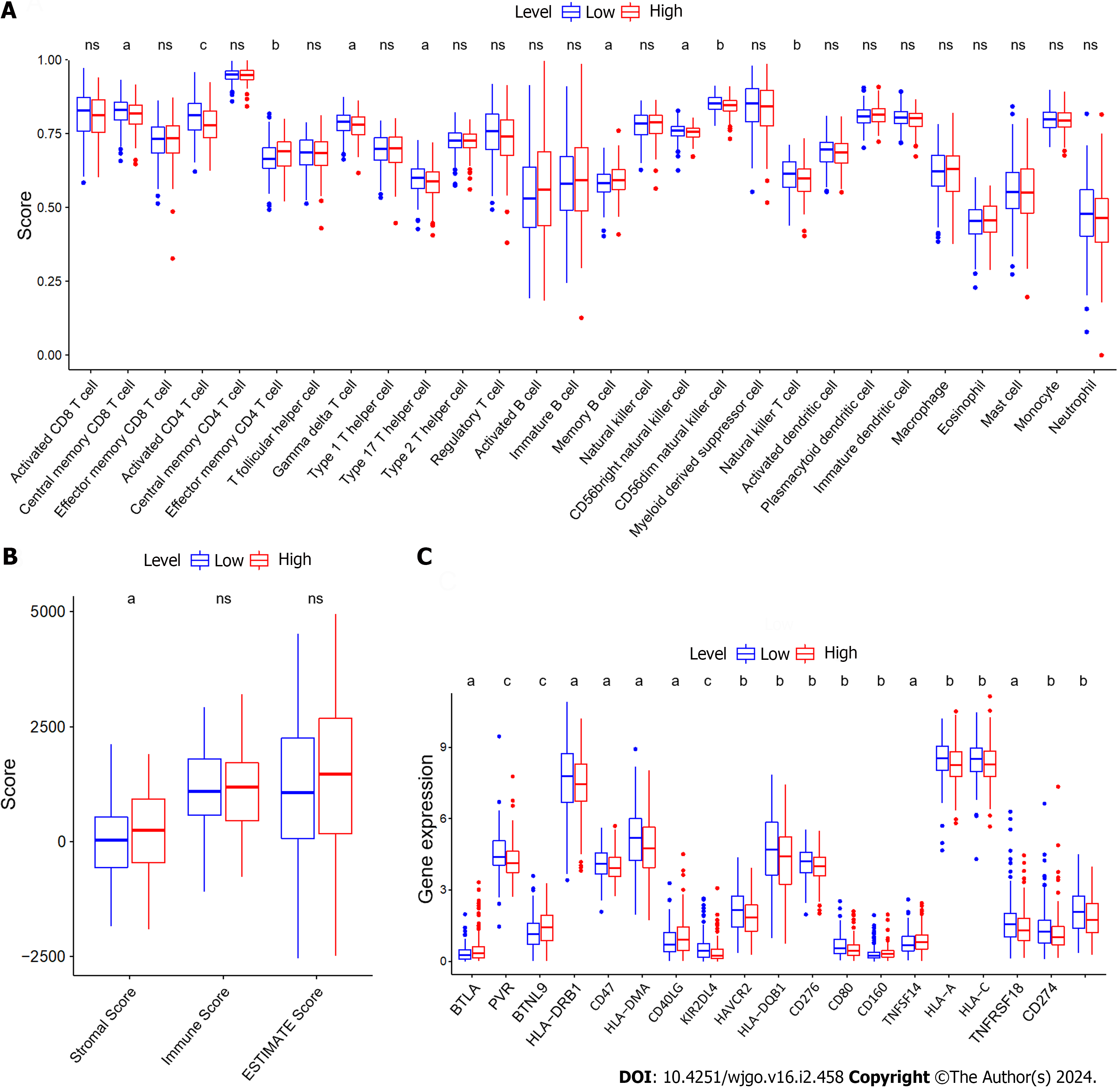
To assess the relationship between ZNF710-AS1-200 and the immune microenvironment of GC patients, we analyzed the infiltration of immune cells in each sample. The findings demonstrated that the level of infiltration by central memory CD8 T cells, activated CD4 T cells, gamma delta T cells, type 17 T helper cells, CD56bright natural killer cells, CD56dim natural killer cells, and natural killer T cells was elevated in GC samples exhibiting reduced expression of ZNF710-AS1-201 (Figure 1A). Conversely, GC samples exhibiting high levels of ZNF710-AS1-201 displayed an increased infiltration level of effector memory CD4 T cells and memory B cells. Therefore, there was no significant variation in immune scores observed between the two groups exhibiting high or LExp (Figure 1B). Remarkably, the stromal scores was increased the high-expression cohort. Figure 1C demonstrates that B and T lymphocyte attenuator, butyrophilin-like 9, CD40LG, CD160, and tumor necrosis factor superfamily member 14 exhibited elevated levels of expression in GC samples with HExp of ZNF710-AS1-201. Conversely, PVR, HLA-DRB1, CD47, HLA-DMA, KIR2DL4, HAVCR2, HLA-DQB1, CD276, CD80, HLA-A, HLA-C, TNFRSF18, CD274, and CD86 displayed increased expression in GC samples with LExp of ZNF710-AS1-201.
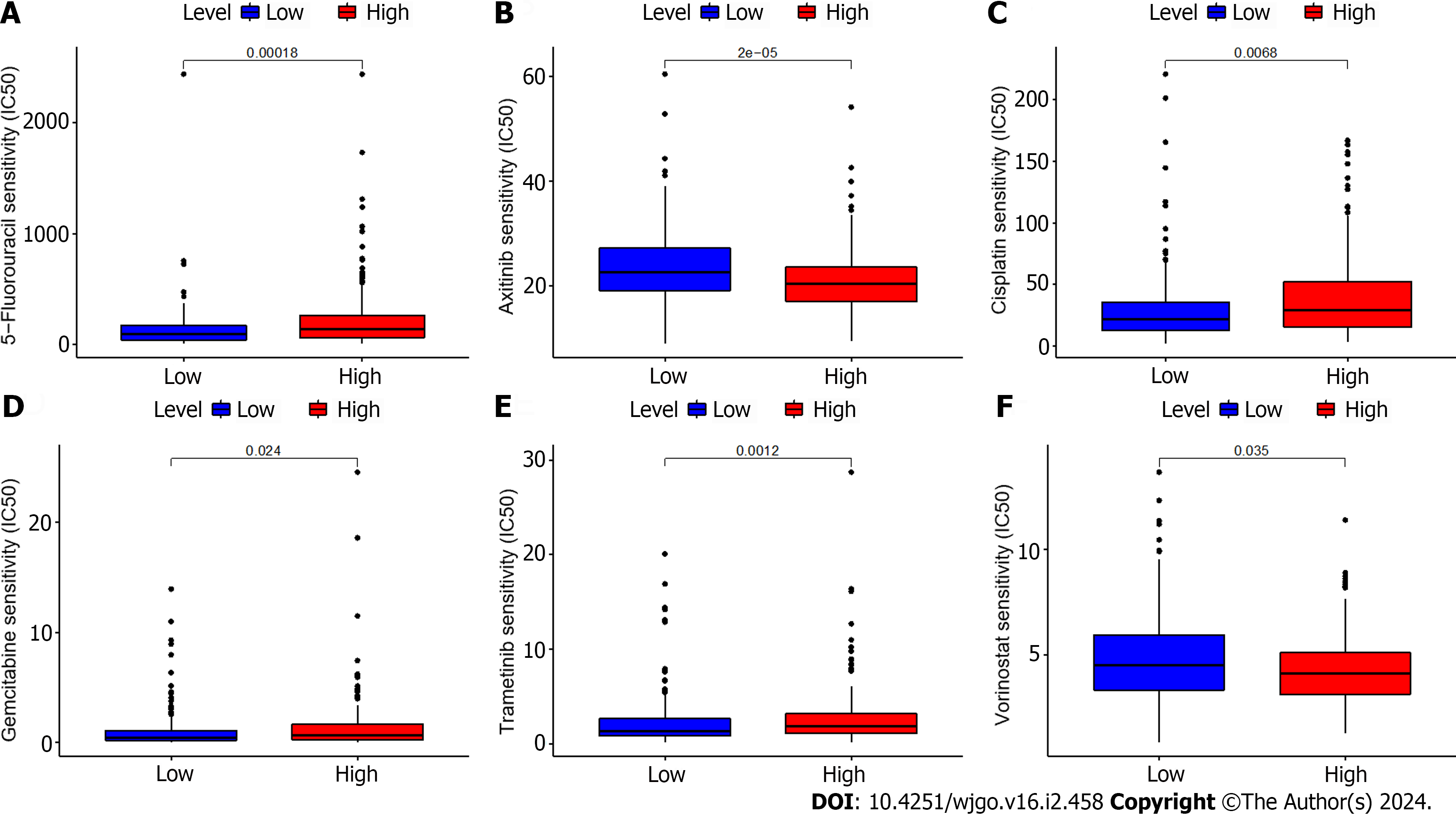
We additionally evaluated the correlation between the expression of ZNF710-AS1-201 and the responsiveness to antitumor drugs in GC. Figure 2 demonstrates that the IC50s of 5-fluorouracil, cisplatin, gemcitabine, and trametinib were reduced in the low-expression group, whereas the IC50s of dasatinib and vorinostat were elevated. The HExp and LExp groups showed no notable disparity in the IC50s of other medications.
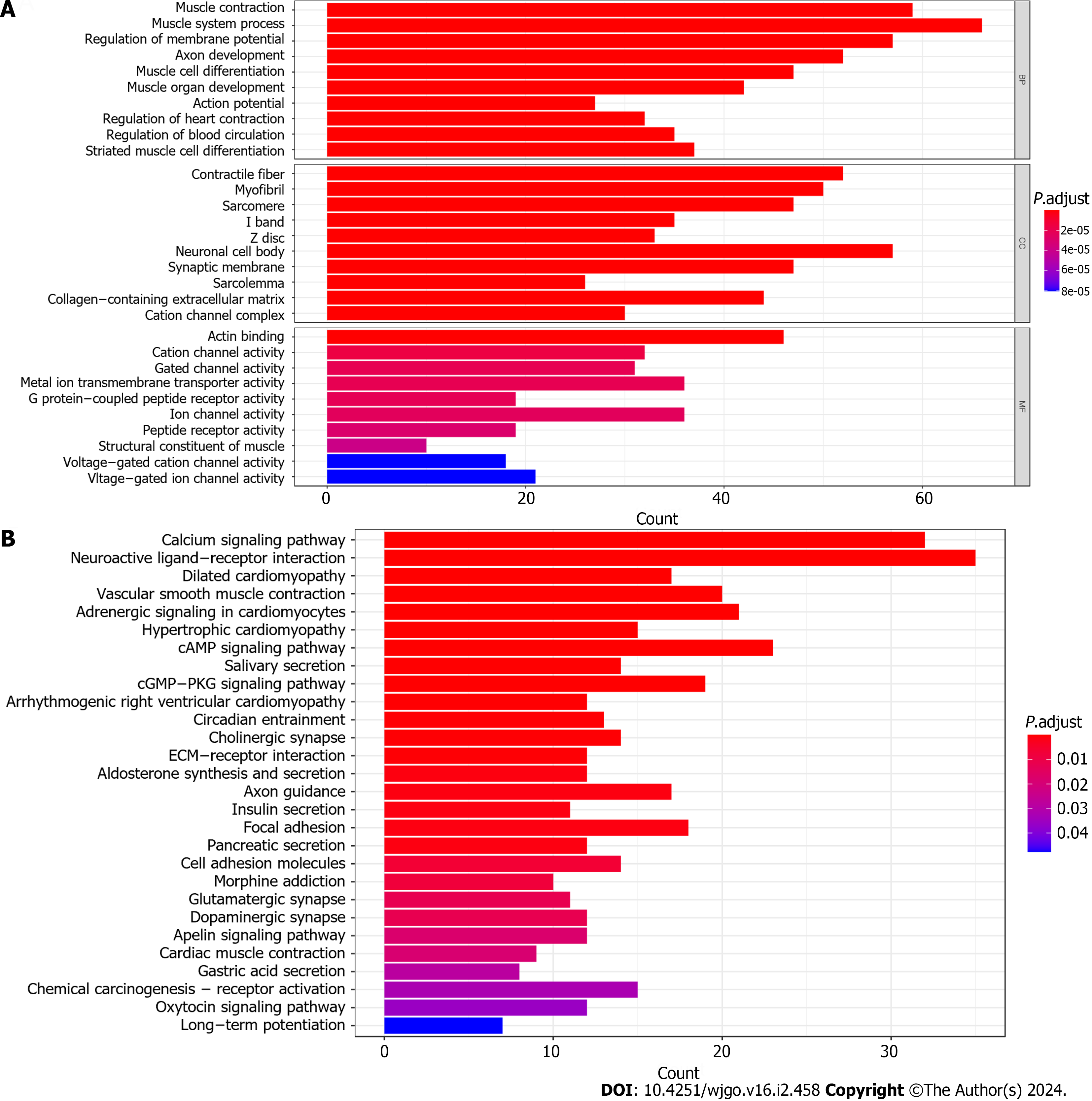
Initially, we examined the disparity between the groups with high and LExp levels and identified 647 genes that exhibited differential expression (Supplementary Table 1). Then, we conducted FEA of these DEGs. In conclusion, KEGG analysis revealed significant enrichment in 28 mechanisms, whereas GO analysis showed significant enrichment in 565 functions (Figure 3). In the GO analysis, there was a significant enrichment of collagen-containing extracellular matrix, actin binding, G protein−coupled peptide receptor activity, ion channel activity, and others (Figure 3A). Significant enrichment of gastric acid secretion, chemical carcinogenesis-receptor activation, and other pathways was observed in the KEGG analysis (Figure 3B).
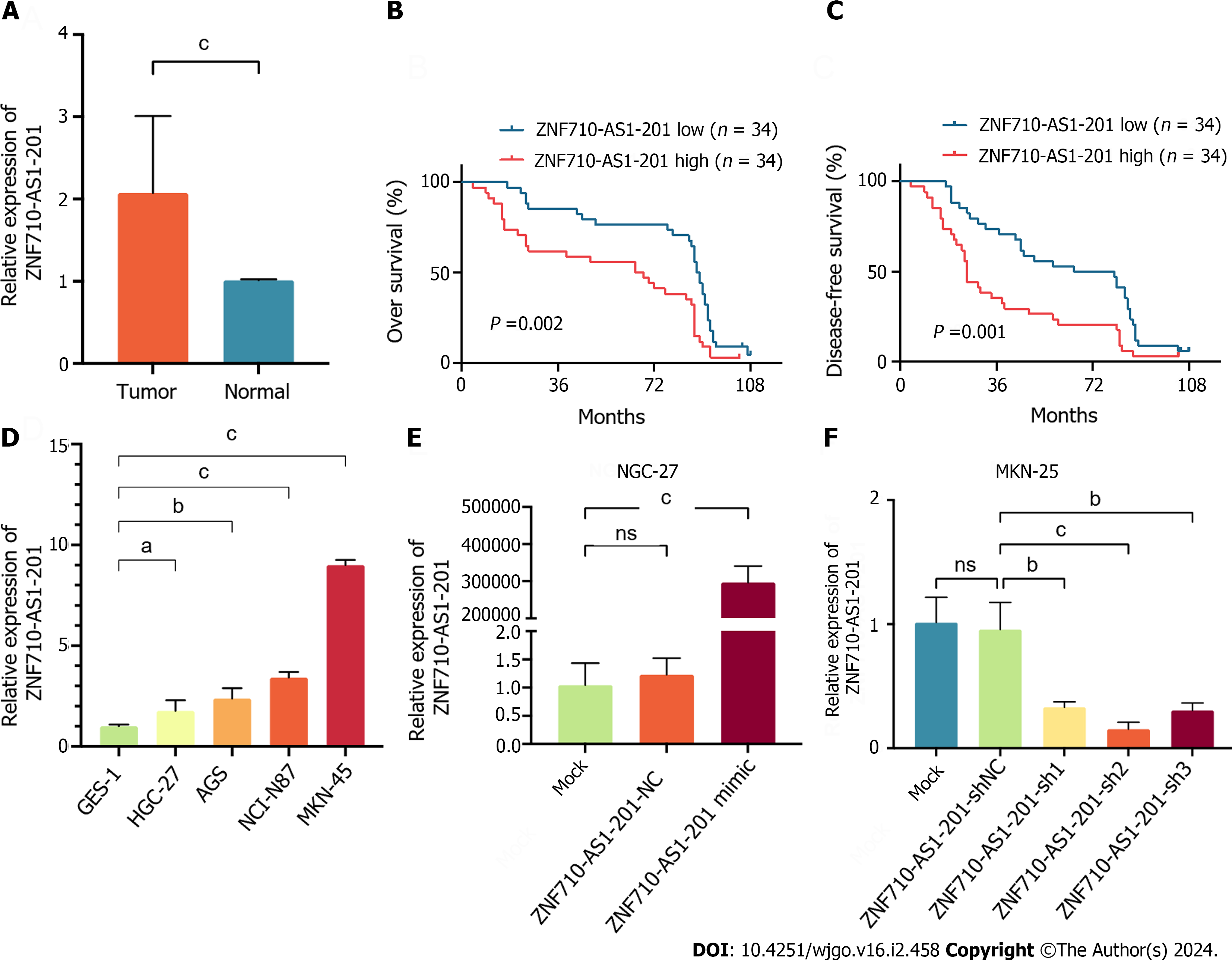
Clinical specimens were gathered to measure the expression level of ZNF710-AS1-201 in the samples. The findings indicated a notable increase in the expression level of ZNF710-AS1-201 in GC tissues compared to adjacent tissues (Figure 4A). Subsequently, we divided patients into high-expression and low-expression groups based on the expression level of ZNF710-AS1-201 in cancer tissues. The comparison of the clinicopathological features of patients between the low and HExp groups is shown in Table 2. The malignant degree of GC was higher and the stage was later in the high-expression group. Additionally, patients with HExp of ZNF710-AS1-201 had lower OS and DFS (Figure 4B and C). Consistent with this, the expression level of ZNF710-AS1-201 in GC cell lines (HGC-27, AGS, NCI-N87, and MKN-45) was elevated compared to that in the normal gastric mucosa cell line GES-1 (Figure 4D). To examine the biological role of ZNF710-AS1-201 in GC, HGC-27 cells were transfected with a plasmid overexpressing ZNF710-AS1-201 and an empty plasmid, while MKN-45 cells were transfected with a plasmid containing ZNF710-AS1-201 shRNA and an empty plasmid. The results demonstrated a significant increase in ZNF710-AS1-201 expression following the transfection of ZNF710-AS1-201 mimic in HGC-27 cells. Conversely, in MKN-45 cells, ZNF710-AS1-201 expression was markedly decreased upon transfection with ZNF710-AS1-201 shRNA (Figure 4E and F).
| Features | ZNF710-AS1-201 | P value | |
| Low expression (n = 34) | High expression (n = 34) | ||
| Age, yr | 65.62 ± 6.85 | 67.85 ± 8.25 | 0.228 |
| Sex, male (%) | 19 (55.9) | 22 (64.7) | 0.457 |
| CEA, U/mL | 18.32 ± 69.96 | 11.09 ± 24.58 | 0.4151 |
| Neutrophil count, × 109/L | 3.56 ± 0.97 | 4.67 ± 2.08 | 0.0061,a |
| Lymphocyte count, × 109/L | 1.89 ± 0.62 | 1.42 ± 0.47 | < 0.001a |
| Monocyte count, × 109/L | 0.38 ± 0.18 | 0.44 ± 0.24 | 0.240 |
| Differentiation degree, III-IV (%) | 18 (52.9) | 26 (76.5) | 0.042a |
| T stage, III-IV (%) | 15 (44.1) | 28 (82.4) | 0.001a |
| Lymphatic invasion, positive (%) | 15 (44.1) | 24 (70.6) | 0.044a |
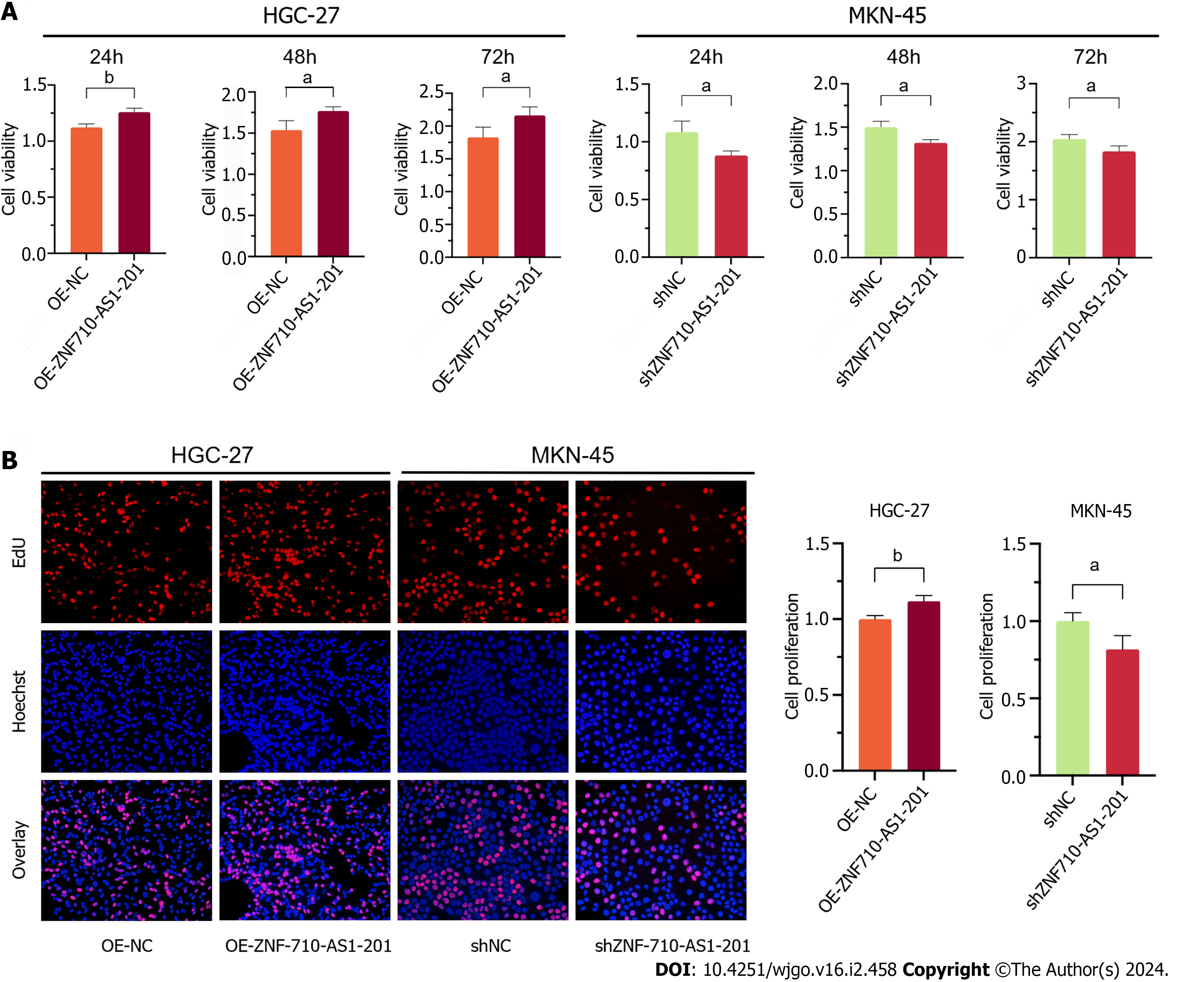
Cell viability was assessed using the CCK-8 assay. After 24 h, 48 h, and 72 h of transfection with the ZNF710-AS1-201 overexpression plasmid, the viability of HGC-27 cells was significantly increased (Figure 5A). The viability of MKN-45 cells decreased when transfected with ZNF710-AS1-201 shRNA plasmid for 24, 48, and 72 h (Figure 5A). Furthermore, the EdU assay was utilized to assess cellular proliferation. The proliferation of HGC-27 cells was enhanced following transfection of the plasmid overexpressing ZNF710-AS1-201 for 72 h (Figure 5B). The growth rate of MKN-45 cells decreased after treatment with the ZNF710-AS1-201 shRNA plasmid for 72 h (Figure 5B).
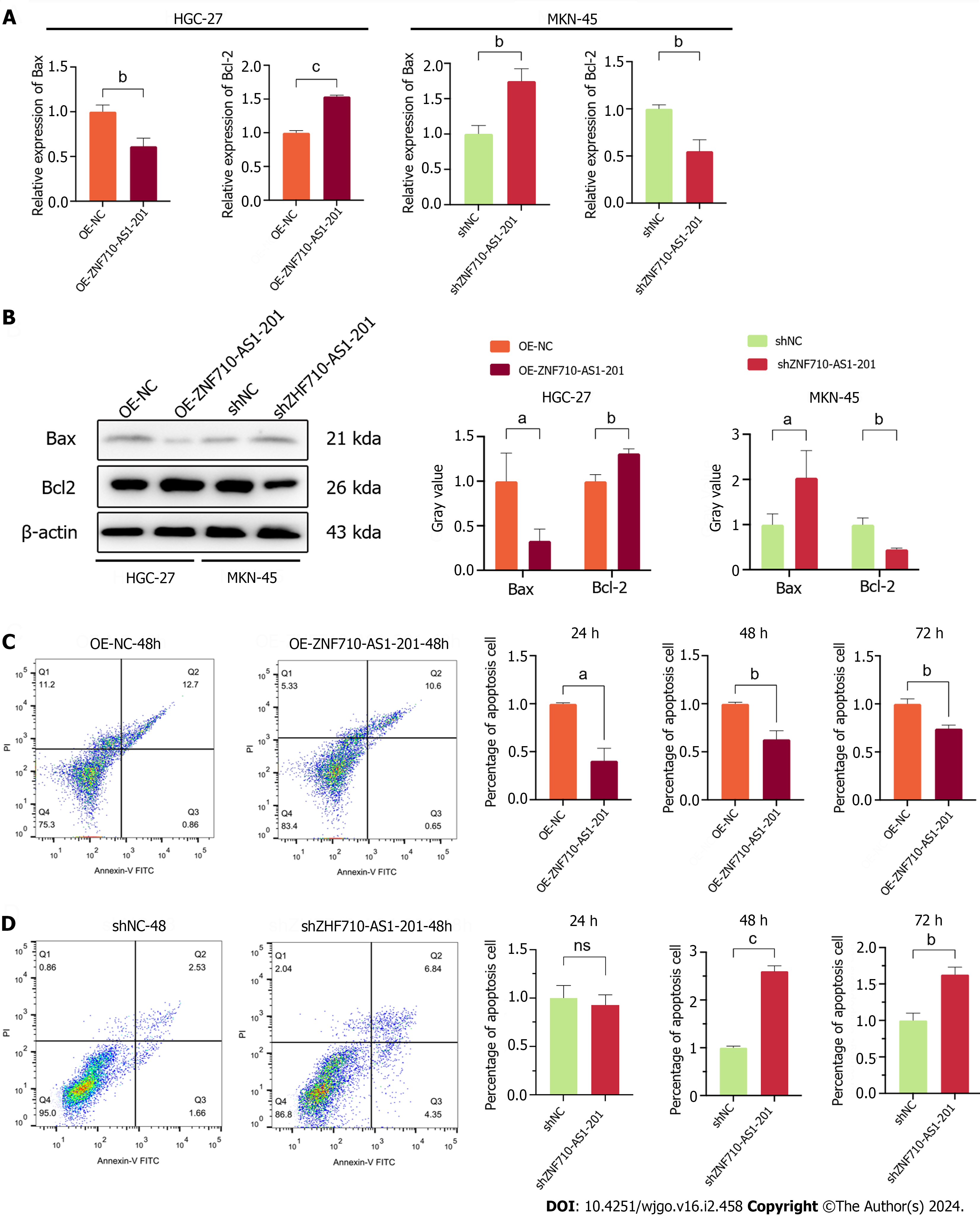
The quantification of proteins related to apoptosis and the assessment of cell apoptosis were conducted using qRT-PCR, western blot, and flow cytometry. qRT-PCR and Western blot analysis revealed that ZNF710-AS1-201 overexpression in HGC-27 cells significantly decreased the expression of Bax and increased the expression of Bcl-2 (Figure 6A and B). In contrast, ZNF710-AS1-201 knockdown in MKN-45 cells led to an increase in Bax levels and a decrease in Bcl-2 levels (Figure 6A and B). The flow cytometry assay results showed that the overexpression of ZNF710-AS1-200 significantly reduced HGC-27 cell apoptosis after 24 h, 48 h, and 72 h (Figure 6C). In contrast, knockdown of ZNF710-AS1-201 significantly enhanced MKN-45 cell apoptosis at 48 h and 72 h (Figure 6D).
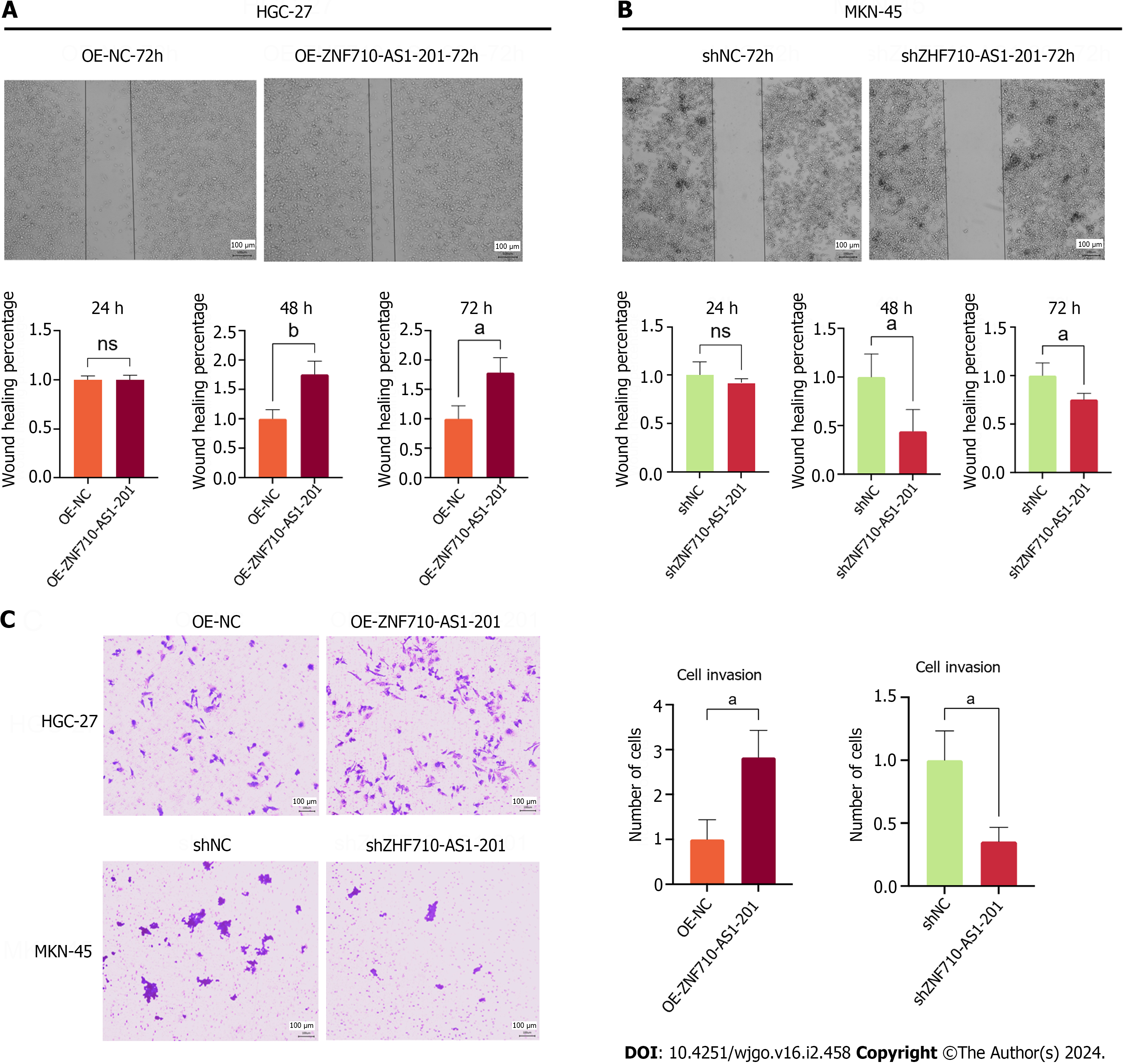
The study utilized the scratch assay to measure cell metastasis. The findings indicated that the overexpression of ZNF710-AS1-201 significantly enhanced the metastasis of HGC-27 cells after 48 and 72 h (Figure 7A). In contrast, the knockdown of ZNF710-AS1-201 significantly inhibited the metastasis of MKN-45 cells at 48 h and 72 h (Figure 7B). The study utilized the transwell assay to quantify cell invasion. The findings indicated that the overexpression of ZNF710-AS1-201 significantly enhanced HGC-27 cell invasion after 72 h (Figure 7C). In contrast, the invasion of MKN-45 cells was significantly inhibited by knockdown of ZNF710-AS1-201 for 72 h (Figure 7C).
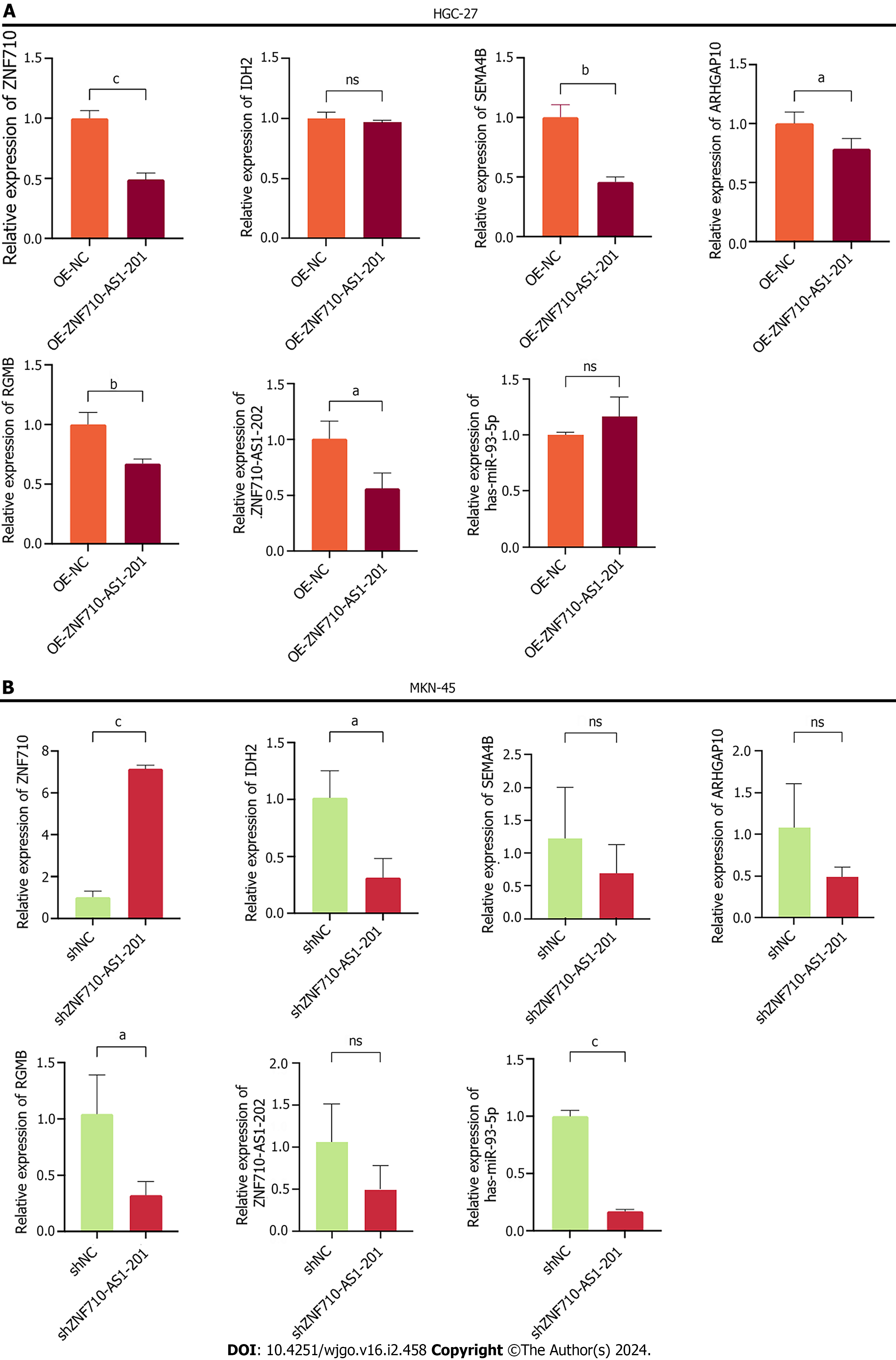
After conducting a thorough search in the StarBase database (https//starbase.sysu.edu.cn/), we identified the potential targets [ZNF710, isocitrate dehydrogenase-2 (IDH2), Semaphorin 4B (SEMA4B), ARHGAP10, RGMB, hsa-miR-93-5p, and ZNF710-AS1-202] for ZNF710-AS1-201. Additionally, we employed qRT-PCR to analyze the expression levels of these potential targets following cell transfection. The findings indicated that after overexpressing ZNF710-AS1-201 in HGC-27 cells, there were no significant changes observed in the expression levels of IDH2 and miR-93-5p. Moreover, in MKN-45 cells with decreased expression of ZNF710-AS1-201, the expression levels of SEMA4B, ARHGAP10, and ZNF710-AS1-202 also showed no significant changes. Additionally, whether ZNF710-AS1-202 was overexpressed or knocked down in GC cells, there was a consistent decrease in the expression levels of RGMB. However, the only expression change that was statistically significant and logically coherent was that for ZNF710 (Figure 8A and B).
This study focused on examining the contribution of ZNF710-AS1-201, an immune-related lncRNA, in facilitating the advancement and spread of GC. In line with prior research, GC tissues with low levels of ZNF710-AS1-201 exhibited significant infiltration of various antitumor immune cells[15-18], such as memory CD8 T cells, activated CD4 T cells, tumor-infiltrating gamma delta T cells, natural killer cells, and natural killer T cells. Furthermore, there was a significant presence of memory B cells in GC tissues exhibiting elevated levels of ZNF710-AS1-201 expression. These findings align with the outcomes of prior research indicating that memory B cells have the potential to facilitate the development of cancer[19]. Nevertheless, the recruitment and activation of these immune cells by ZNF710-AS1-201 are still uncertain. While the ESTIMATE scores of GC samples with elevated expression of ZNF710-AS1-201 were greater than those of samples with LExp, the observed disparity did not reach statistical significance. In contrast, the HExp group exhibited a considerably elevated stromal score. The reason for this could be an escalation in the remodeling of the extracellular matrix, which has the potential to stimulate the growth of tumors, their spread to other parts of the body, and the formation of new blood vessels[20].
Chemotherapy, targeted therapy, and immunotherapy are the present methods used for conservatively treating advanced GC[21]. By analyzing immune checkpoints and drug sensitivity, we have the ability to choose drugs that exhibit high sensitivity and immunotherapies targeting highly expressed immune checkpoints. Currently, the first-line chemotherapy treatment for GC is the combination of cisplatin and 5-fluorouracil[22]. The survival of patients with human epidermal growth factor receptor 2-positive GC is improved by adding trastuzumab to the initial chemotherapy[23]. Furthermore, pembrolizumab, an immune checkpoint inhibitor, has been authorized in various contexts based on the presence of programmed death ligand 1[24]. According to prior studies, axitinib exhibited strong antineoplastic effects on human GC, either by itself or when combined with 5-fluorouracil or cisplatin[25]. Considering these results and those of our study, the combination of axitinib and 5-fluorouracil may demonstrate enhanced antitumor impacts in individuals exhibiting elevated ZNF710-AS1-201 expression.
Based on the enrichment analysis, we identified several biological processes and mechanisms that are associated with tumors, such as collagen-containing extracellular matrix, actin binding, G protein-coupled peptide receptor activity and ion channel activity. According to the study conducted by Chen et al[26], the presence of collagen in the tumor microenvironment is an independent predictor of lymphatic metastasis in early GC. Jia et al[27] discovered that Anillin, a protein that binds to actin, facilitated the advancement of GC both in laboratory settings and in mice. Yan et al[28] demonstrated that the abnormal activation of G-protein-coupled receptors stimulated the growth and spread of GC cells, altered the tumor microenvironment, and enhanced immune evasion[28]. In GC, Chen et al[29] observed modified expression and/or malfunction of ion channels/transporters, which is believed to have a significant impact on tumor cell proliferation, metastasis, invasion, and apoptosis. In summary, ZNF710-AS1-201 might exert its influence on GC via these pathways, thereby facilitating the onset and progression of GC.
In a recent study, Li and colleagues found that ZNF710-AS1-202 played a role in enhancing cell growth and suppressing cell death in clear cell renal cell carcinoma through the reverse regulation of ZNF710[30]. Nevertheless, there is no existing report on the regulatory impact of ZNF710-AS1-201 on tumors. The current research examined the influence of ZNF710-AS1-201 on the promotion of tumors in GC. In HGC-27 cells, we observed that the upregulation of ZNF710-AS1-201 stimulated cell growth, hindered cell death, and enhanced cell metastasis and infiltration. Conversely, in MKN-45 cells, the downregulation of ZNF710-AS1-201 suppressed cell proliferation, promoted cell death, and impeded cell metastasis and invasion.
qRT-PCR was used to validate possible targets for ZNF710-AS1-201. IDH2, also known as isocitrate dehydrogenase 2, is an isocitrate dehydrogenase that relies on NADP and is located in the mitochondria. Through inhibition of matrix metalloproteinase 7, it has been discovered to impede the invasion of GC cells[31]. However, a significant reduction in IDH2 mRNA expression after ZNF710-AS1-201 overexpression appeared in MKN-45 cells. ARHGAP10 can promote gastric tumor metastasis[32]. Nonetheless, the upregulation of ZNF710-AS1-201 suppressed the expression of ARHGAP10 mRNA in HGC-27 cells. In their study, Jian and colleagues demonstrated that the invasion of non-small cell lung cancer was hindered by SEMA4B through the suppression of matrix metallopeptidase 9 activation, which was dependent on phosphoinositide 3-kinase[33]. According to the study conducted by Li et al[34], the overexpression of miR-93-5p was found to enhance the growth, movement, infiltration, and resistance to chemotherapy in SGC-7901 cells. In line with these perspectives, the mRNA expression of SEMA4B was inhibited when ZNF710-AS1-201 was overexpressed in HGC-27 cells, whereas the expression of miR-93-5p was enhanced when ZNF710-AS1-201 was knocked down in MKN-45 cells. Further investigation is needed to explore the role of these genes in GC, even though they might be controlled by ZNF710-AS1-201.
ZNF710-AS1-201 had a negative regulatory effect on the expression level of ZNF710 in both HGC-27 and MKN-45 cells. However, this result only showed that ZNF710 is negatively regulated by ZNF710-AS1-201. The function of ZNF710 in GC is currently not understood. ZNF710, as stated in the UniProt repository, is a transcription factor (TF)/cofactor of the Kruppel Cys2-His2-type zinc-finger protein[35]. ZNF710 may function as a TF to regulate the occurrence and development of GC. TFs are key factors in gene regulation[36]. TFs can regulate the migration of tumor cells through specific signaling pathways[37]. In their study, Guo and colleagues discovered that the TF RUNX2 stimulated the yes-associated protein 1 signaling pathway, contributing to the development of GC[38]. In their study, Zhao and colleagues found that the growth and metastasis of GC cells were suppressed by the TF kruppel-like factor 15 through the regulation of the TFAP2A-AS1/Neonatal Ichthyosis Sclerosing Cholangitis axis[39]. This demonstrated that transcription regulators had a controlling function in the metastasis and invasion of stomach cancer.
The findings from this study indicate that ZNF710-AS1-201 facilitated growth, metastasis and invasion while suppressing cell death in GC cells. These findings suggest that ZNF710-AS1-201 and ZNF710 have potential as effective targets for therapeutic intervention in GC. Nevertheless, these findings have not been observed in living organisms. Furthermore, rescue experiments are needed to demonstrate the interaction mechanism between ZNF710-AS1-201 and ZNF710. Moreover, it is still necessary to determine the specific targets of the ZNF710 TF.
Gastric cancer (GC) is a prevalent malignant tumor of the gastrointestinal system. ZNF710 is a transcription factor, and ZNF710-AS1-201 is an immune-related long noncoding RNA (lncRNA) that is upregulated in GC cells.
The research is motivated by the pressing challenges posed by GC and the need for a deeper understanding of its underlying molecular mechanisms. Specifically, investigating the functional role of lncRNA ZNF710-AS1-201 and exploring the dynamics of the immune microenvironment are crucial for unraveling the complexities of GC progression. The study aims to contribute valuable insights that may have implications for precision therapy approaches in addressing this formidable disease.
The primary objectives of this research are to elucidate the functional significance of lncRNA ZNF710-AS1-201 in GC, analyze its impact on cellular processes and pathways, investigate its potential as a biomarker, and comprehensively characterize the immune microenvironment in GC tissues. By achieving these objectives, the study aims to provide a foundation for developing targeted therapeutic strategies, advancing our understanding of GC pathogenesis, and contributing to the broader field of cancer research.
This research employed a multidisciplinary approach, integrating molecular biology techniques, bioinformatics analyses, and in vitro experiments. The study involved the profiling of lncRNA ZNF710-AS1-201 expression in GC tissues, functional assays to assess its impact on cell behavior, and bioinformatics tools to unravel potential interacting pathways. Additionally, immune microenvironment analysis was conducted to explore the relationship between lncRNA expression and immune responses in GC. These methods collectively provided a comprehensive platform for investigating the role of ZNF710-AS1-201 in GC progression.
The research revealed a significant upregulation of lncRNA ZNF710-AS1-201 in GC tissues compared to adjacent normal tissues. Functional assays demonstrated that ZNF710-AS1-201 overexpression promoted cell proliferation, migration, and invasion, suggesting its potential oncogenic role. Bioinformatics analyses unveiled the involvement of ZNF710-AS1-201 in key cancer-related pathways. Moreover, immune microenvironment analysis indicated a correlation between ZNF710-AS1-201 expression and immune cell infiltration, providing insights into its potential influence on the tumor immune response in GC.
In conclusion, this study elucidates the crucial role of lncRNA ZNF710-AS1-201 in GC progression, highlighting its potential as a diagnostic and therapeutic target. The findings underscore the significance of ZNF710-AS1-201 in modulating cellular processes and its association with cancer-related pathways. Furthermore, insights into the immune microenvironment suggest its impact on the tumor immune response. Overall, these results contribute to our understanding of the molecular mechanisms underlying GC and open avenues for future research and targeted therapeutic interventions.
The study opens new perspectives for further investigation into the intricate mechanisms of lncRNA ZNF710-AS1-201 in GC. Future research should explore its potential as a biomarker for early diagnosis and prognosis. Additionally, understanding its interactions within the tumor microenvironment and elucidating its role in therapeutic resistance could pave the way for innovative treatment strategies. Collaborative efforts integrating multi-omics approaches may provide a comprehensive view of ZNF710-AS1-201’s involvement in GC, offering valuable insights for precision medicine and targeted therapies.
The authors would like to acknowledge Professor Li Yang for guidance and support.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Plaza MA, Spain; Watanabe T, Japan S-Editor: Qu XL L-Editor: A P-Editor: Cai YX
| 1. | Sun H, Wang H, Pan H, Zuo Y, Zhao R, Huang R, Xue Y, Song H. CD19 (+) B Cell Combined with Prognostic Nutritional Index Predicts the Clinical Outcomes of Patients with Gastric Cancer Who Underwent Surgery. Cancers (Basel). 2023;15. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Zhang T, Wang B, Su F, Gu B, Xiang L, Gao L, Zheng P, Li XM, Chen H. TCF7L2 promotes anoikis resistance and metastasis of gastric cancer by transcriptionally activating PLAUR. Int J Biol Sci. 2022;18:4560-4577. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264-279. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Chen Y, Jia K, Sun Y, Zhang C, Li Y, Zhang L, Chen Z, Zhang J, Hu Y, Yuan J, Zhao X, Gong J, Dong B, Zhang X, Li J, Shen L. Predicting response to immunotherapy in gastric cancer via multi-dimensional analyses of the tumour immune microenvironment. Nat Commun. 2022;13:4851. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Xiang Y, Gong M, Deng Y, Wang H, Ye D. T cell effects and mechanisms in immunotherapy of head and neck tumors. Cell Commun Signal. 2023;21:49. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Jin X, Liu Z, Yang D, Yin K, Chang X. Recent Progress and Future Perspectives of Immunotherapy in Advanced Gastric Cancer. Front Immunol. 2022;13:948647. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Li K, Zhang A, Li X, Zhang H, Zhao L. Advances in clinical immunotherapy for gastric cancer. Biochim Biophys Acta Rev Cancer. 2021;1876:188615. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Bang YJ, Ruiz EY, Van Cutsem E, Lee KW, Wyrwicz L, Schenker M, Alsina M, Ryu MH, Chung HC, Evesque L, Al-Batran SE, Park SH, Lichinitser M, Boku N, Moehler MH, Hong J, Xiong H, Hallwachs R, Conti I, Taieb J. Phase III, randomised trial of avelumab vs physician's choice of chemotherapy as third-line treatment of patients with advanced gastric or gastro-oesophageal junction cancer: primary analysis of JAVELIN Gastric 300. Ann Oncol. 2018;29:2052-2060. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Chen L, Deng J. Role of non-coding RNA in immune microenvironment and anticancer therapy of gastric cancer. J Mol Med (Berl). 2022;100:1703-1719. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Park EG, Pyo SJ, Cui Y, Yoon SH, Nam JW. Tumor immune microenvironment lncRNAs. Brief Bioinform. 2022;23. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Wu M, Fu P, Qu L, Liu J, Lin A. Long Noncoding RNAs, New Critical Regulators in Cancer Immunity. Front Oncol. 2020;10:550987. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Ding W, Sun P, Tan Y, Jiang H, Xi C, Zhuang L, Xu Y, Xu X. Construction of a Prognostic Immune-Related LncRNA Risk Model for Gastric Cancer. J Oncol. 2022;2022:5137627. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Han J, Khatwani N, Searles TG, Turk MJ, Angeles CV. Memory CD8(+) T cell responses to cancer. Semin Immunol. 2020;49:101435. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Borst J, Ahrends T, Bąbała N, Melief CJM, Kastenmüller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18:635-647. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Saura-Esteller J, de Jong M, King LA, Ensing E, Winograd B, de Gruijl TD, Parren PWHI, van der Vliet HJ. Gamma Delta T-Cell Based Cancer Immunotherapy: Past-Present-Future. Front Immunol. 2022;13:915837. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Kucuksezer UC, Aktas Cetin E, Esen F, Tahrali I, Akdeniz N, Gelmez MY, Deniz G. The Role of Natural Killer Cells in Autoimmune Diseases. Front Immunol. 2021;12:622306. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Downs-Canner SM, Meier J, Vincent BG, Serody JS. B Cell Function in the Tumor Microenvironment. Annu Rev Immunol. 2022;40:169-193. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Oya Y, Hayakawa Y, Koike K. Tumor microenvironment in gastric cancers. Cancer Sci. 2020;111:2696-2707. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol. 2021;18:473-487. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Wang FH, Zhang XT, Li YF, Tang L, Qu XJ, Ying JE, Zhang J, Sun LY, Lin RB, Qiu H, Wang C, Qiu MZ, Cai MY, Wu Q, Liu H, Guan WL, Zhou AP, Zhang YJ, Liu TS, Bi F, Yuan XL, Rao SX, Xin Y, Sheng WQ, Xu HM, Li GX, Ji JF, Zhou ZW, Liang H, Zhang YQ, Jin J, Shen L, Li J, Xu RH. The Chinese Society of Clinical Oncology (CSCO): Clinical guidelines for the diagnosis and treatment of gastric cancer, 2021. Cancer Commun (Lond). 2021;41:747-795. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Shitara K, Bang YJ, Iwasa S, Sugimoto N, Ryu MH, Sakai D, Chung HC, Kawakami H, Yabusaki H, Lee J, Saito K, Kawaguchi Y, Kamio T, Kojima A, Sugihara M, Yamaguchi K; DESTINY-Gastric01 Investigators. Trastuzumab Deruxtecan in Previously Treated HER2-Positive Gastric Cancer. N Engl J Med. 2020;382:2419-2430. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Patel TH, Cecchini M. Targeted Therapies in Advanced Gastric Cancer. Curr Treat Options Oncol. 2020;21:70. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | He Q, Gao J, Ge S, Wang T, Li Y, Peng Z, Shen L. Axitinib alone or in combination with chemotherapeutic drugs exerts potent antitumor activity against human gastric cancer cells in vitro and in vivo. J Cancer Res Clin Oncol. 2014;140:1575-1583. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Chen D, Chen G, Jiang W, Fu M, Liu W, Sui J, Xu S, Liu Z, Zheng X, Chi L, Lin D, Li K, Chen W, Zuo N, Lu J, Chen J, Li G, Zhuo S, Yan J. Association of the Collagen Signature in the Tumor Microenvironment With Lymph Node Metastasis in Early Gastric Cancer. JAMA Surg. 2019;154:e185249. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Jia H, Yu F, Li B, Gao Z. Actin-binding protein Anillin promotes the progression of gastric cancer in vitro and in mice. J Clin Lab Anal. 2021;35:e23635. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Yan H, Zhang JL, Leung KT, Lo KW, Yu J, To KF, Kang W. An Update of G-Protein-Coupled Receptor Signaling and Its Deregulation in Gastric Carcinogenesis. Cancers (Basel). 2023;15. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Chen J, Zhang M, Ma Z, Yuan D, Zhu J, Tuo B, Li T, Liu X. Alteration and dysfunction of ion channels/transporters in a hypoxic microenvironment results in the development and progression of gastric cancer. Cell Oncol (Dordr). 2021;44:739-749. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Li G, Xie M, Huang Z, Li H, Li P, Zhang Z, Ding Y, Jia Z, Yang J. Overexpression of antisense long noncoding RNA ZNF710AS1202 promotes cell proliferation and inhibits apoptosis of clear cell renal cell carcinoma via regulation of ZNF710 expression. Mol Med Rep. 2020;21:2502-2512. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Wu D. Isocitrate dehydrogenase 2 inhibits gastric cancer cell invasion via matrix metalloproteinase 7. Tumour Biol. 2016;37:5225-5230. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Wang Z, Yao L, Li Y, Hao B, Wang M, Wang J, Gu W, Zhan H, Liu G, Wu Q. miR3373p inhibits gastric tumor metastasis by targeting ARHGAP10. Mol Med Rep. 2020;21:705-719. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Jian H, Zhao Y, Liu B, Lu S. SEMA4B inhibits growth of non-small cell lung cancer in vitro and in vivo. Cell Signal. 2015;27:1208-1213. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Li L, Zhao J, Huang S, Wang Y, Zhu L, Cao Y, Xiong J, Deng J. MiR-93-5p promotes gastric cancer-cell progression via inactivation of the Hippo signaling pathway. Gene. 2018;641:240-247. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506-D515. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Long P, Zhang L, Huang B, Chen Q, Liu H. Integrating genome sequence and structural data for statistical learning to predict transcription factor binding sites. Nucleic Acids Res. 2020;48:12604-12617. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Hamidi AA, Taghehchian N, Basirat Z, Zangouei AS, Moghbeli M. MicroRNAs as the critical regulators of cell migration and invasion in thyroid cancer. Biomark Res. 2022;10:40. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Guo Z, Zhou K, Wang Q, Huang Y, Ji J, Peng Y, Zhang X, Zheng T, Zhang Z, Chong D, Yang Z. The transcription factor RUNX2 fuels YAP1 signaling and gastric cancer tumorigenesis. Cancer Sci. 2021;112:3533-3544. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Zhao X, Chen L, Wu J, You J, Hong Q, Ye F. Transcription factor KLF15 inhibits the proliferation and migration of gastric cancer cells via regulating the TFAP2A-AS1/NISCH axis. Biol Direct. 2021;16:21. [PubMed] [DOI] [Cited in This Article: ] |