Copyright
©The Author(s) 2024.
World J Gastrointest Oncol. Feb 15, 2024; 16(2): 436-457
Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.436
Published online Feb 15, 2024. doi: 10.4251/wjgo.v16.i2.436
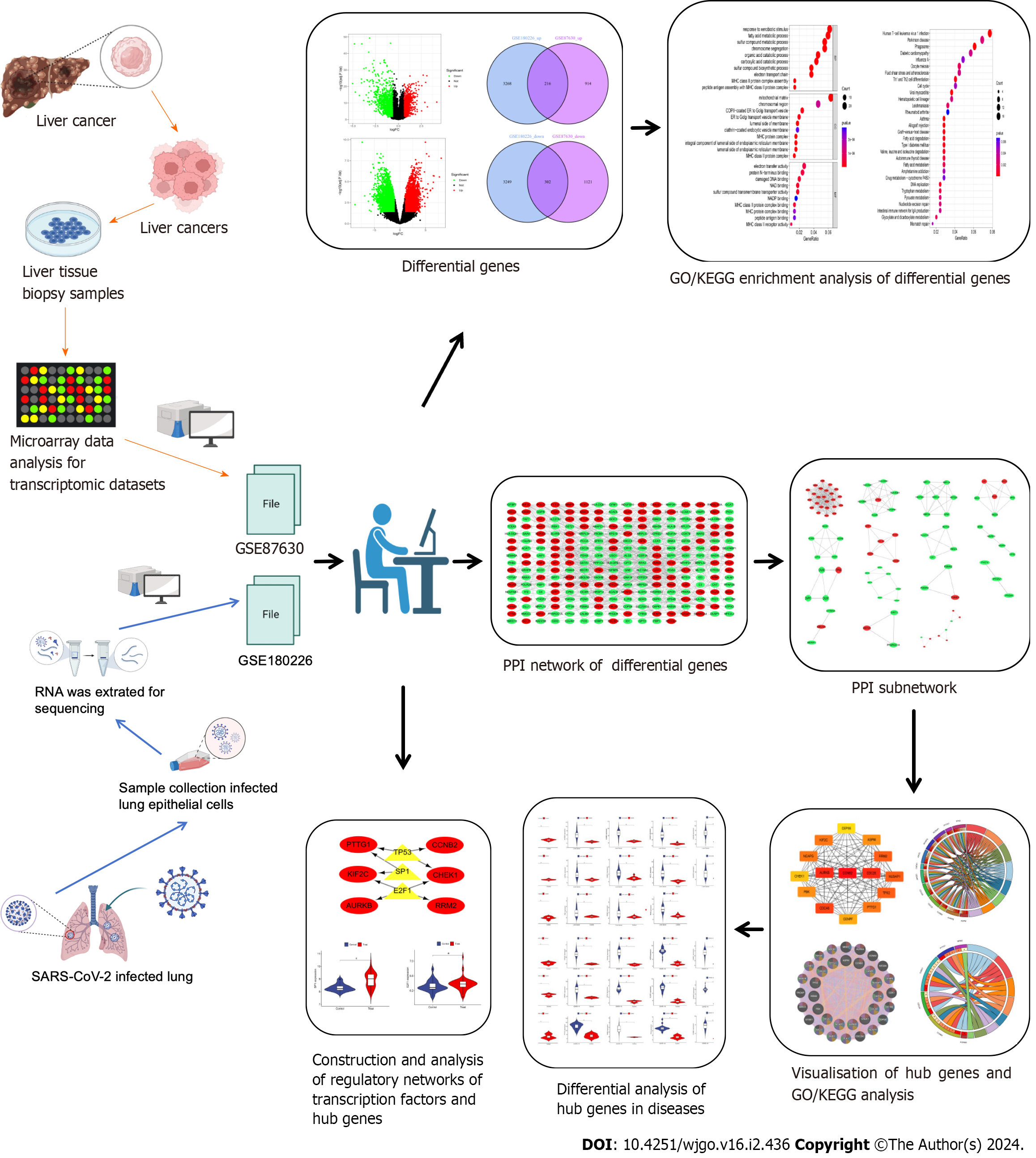
Figure 1 The predominant methodical workflow in the current study.
GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; SARS-CoV-2: Severe acute respiratory syndrome coronavirus-2; PPI: Protein–protein interaction.
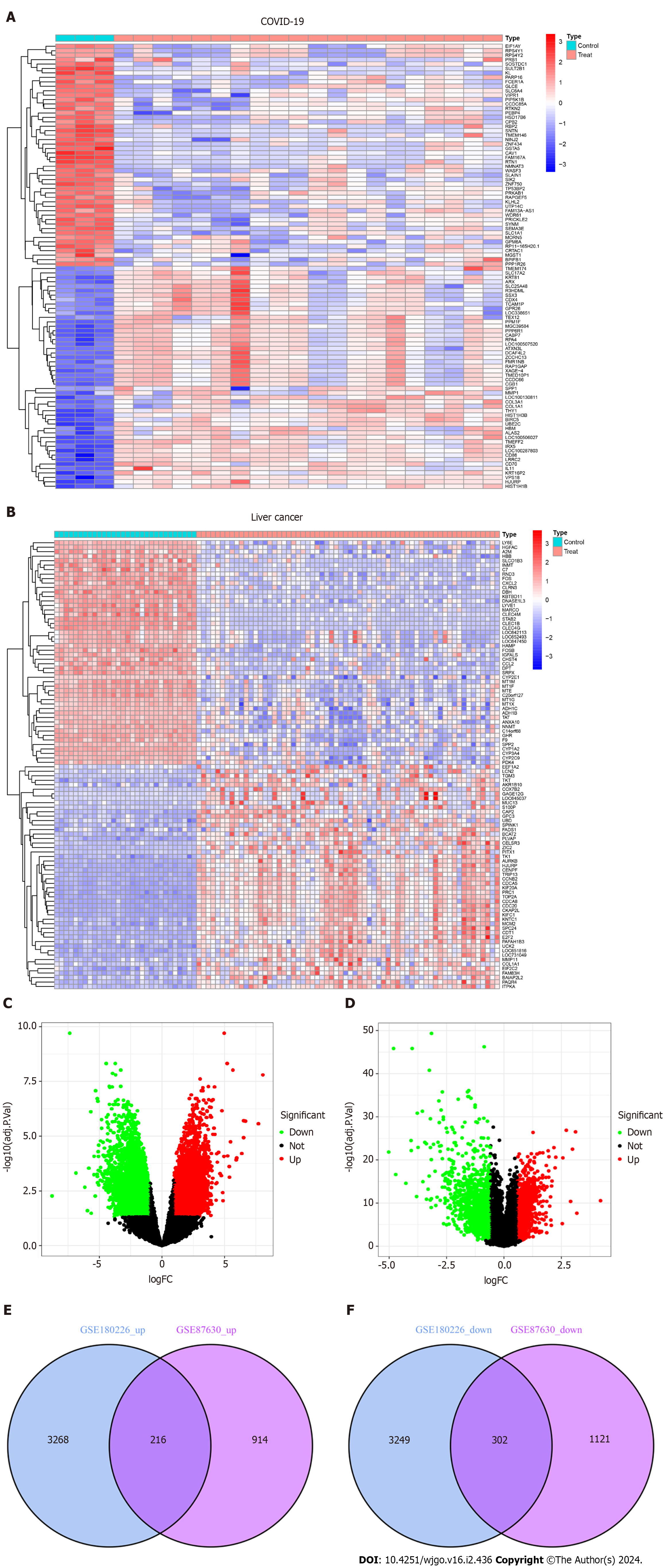
Figure 2 Volcanic diagram, heat map and Venn diagram.
A: Volcano map of GSE180226; B: Volcano map of GSE87630. Up-regulated genes are colored in red; down-regulated genes are colored in green; C: Cluster analysis of GSE180226; D: Cluster analysis of GSE87630; E: Two datasets showed 216 up-regulated shared differentially expressed genes (DEGs); F: Two datasets showed 302 down-regulated shared DEGs. COVID-19: Coronavirus disease 2019.
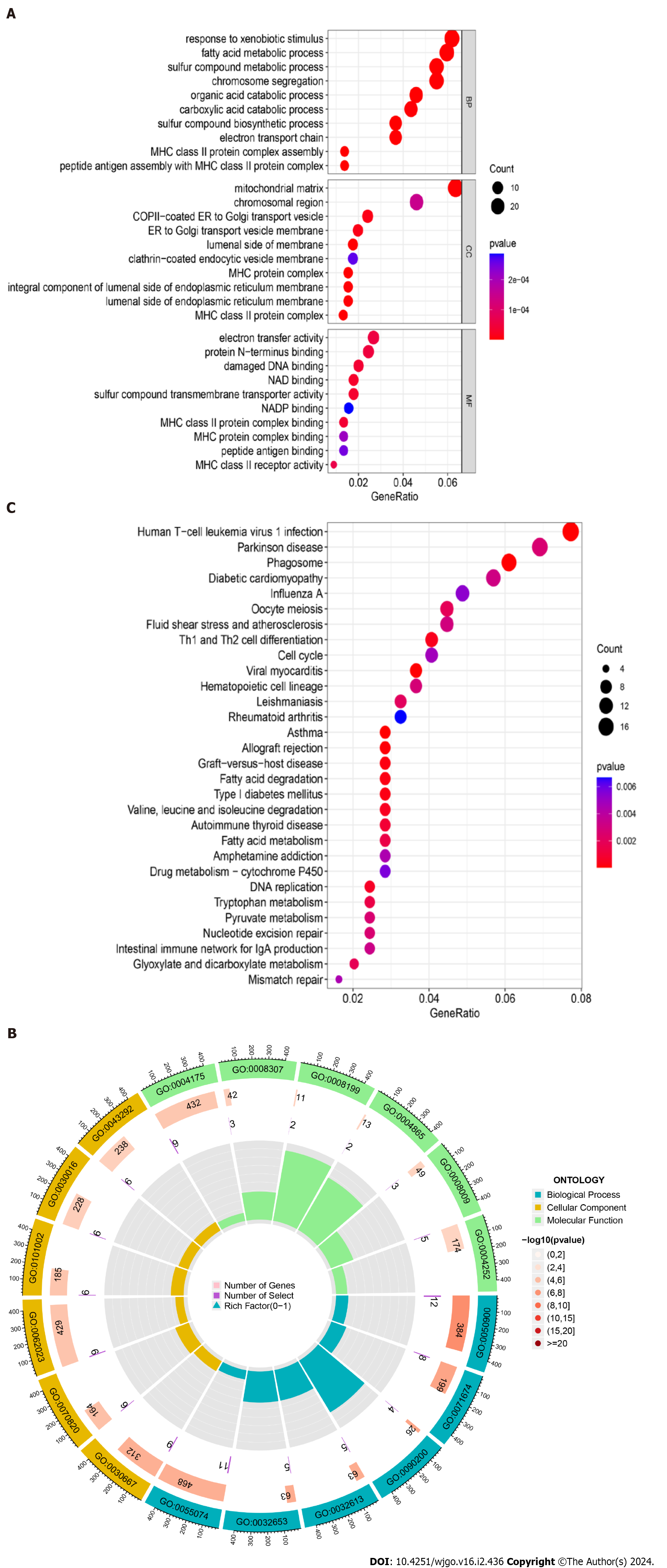
Figure 3 Gene Ontology functional annotation and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of shared differentially expressed genes from two datasets.
A: Annotated bubble chart of Gene Ontology (GO) functions for shared differentially expressed genes (DEGs); B: Annotated circle diagram of the GO function for shared DEGs; C: Bubble plot of the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis for shared DEGs.
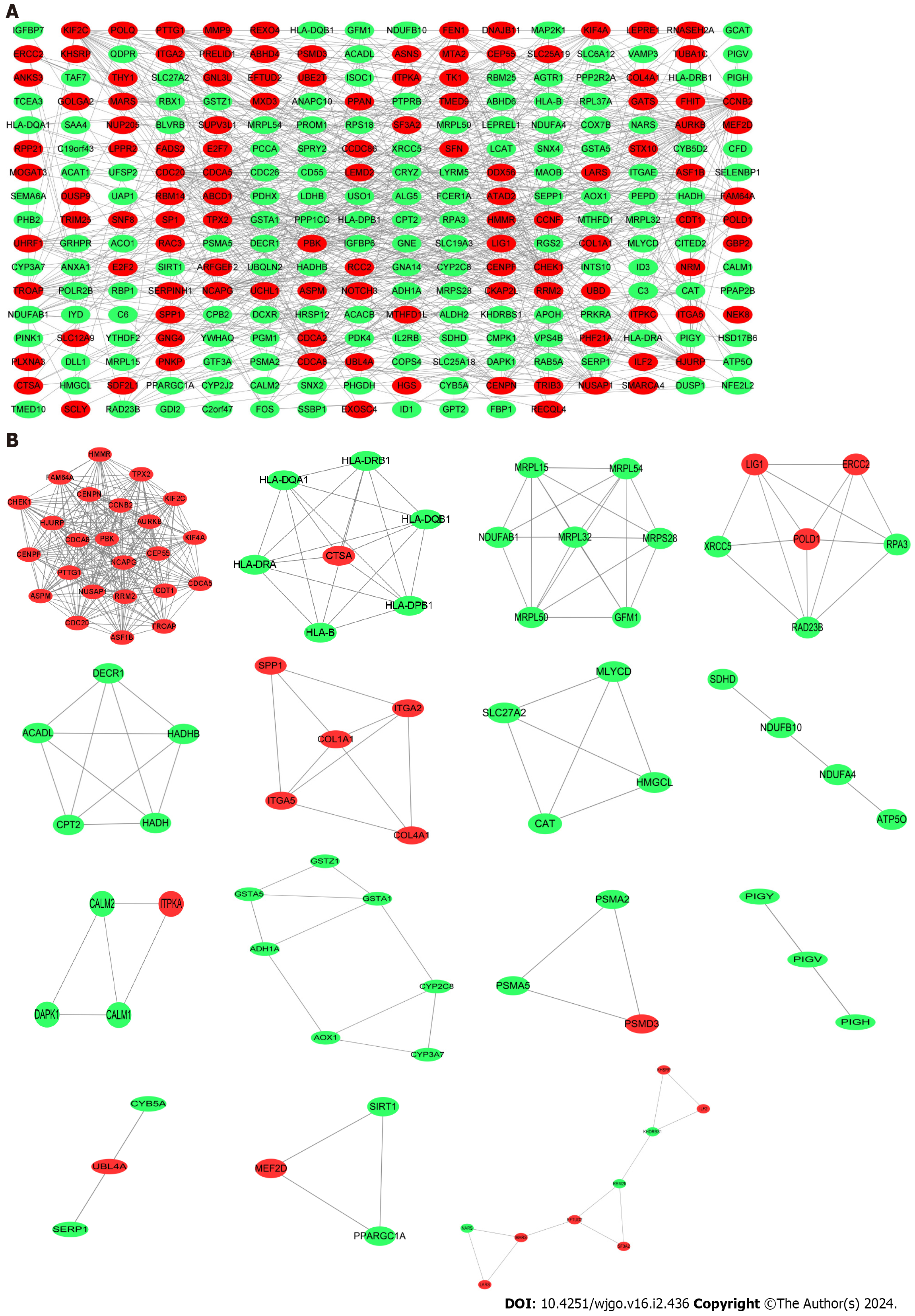
Figure 4 Protein–protein interaction network construction.
A: Protein–protein interaction network of shared differentially expressed genes (DEGs) for coronavirus disease 2019 and liver cancer; B: Subnetworks of shared DEGs. Highlighted red nodes indicate up-regulated DEGs and green nodes indicate down-regulated DEGs.
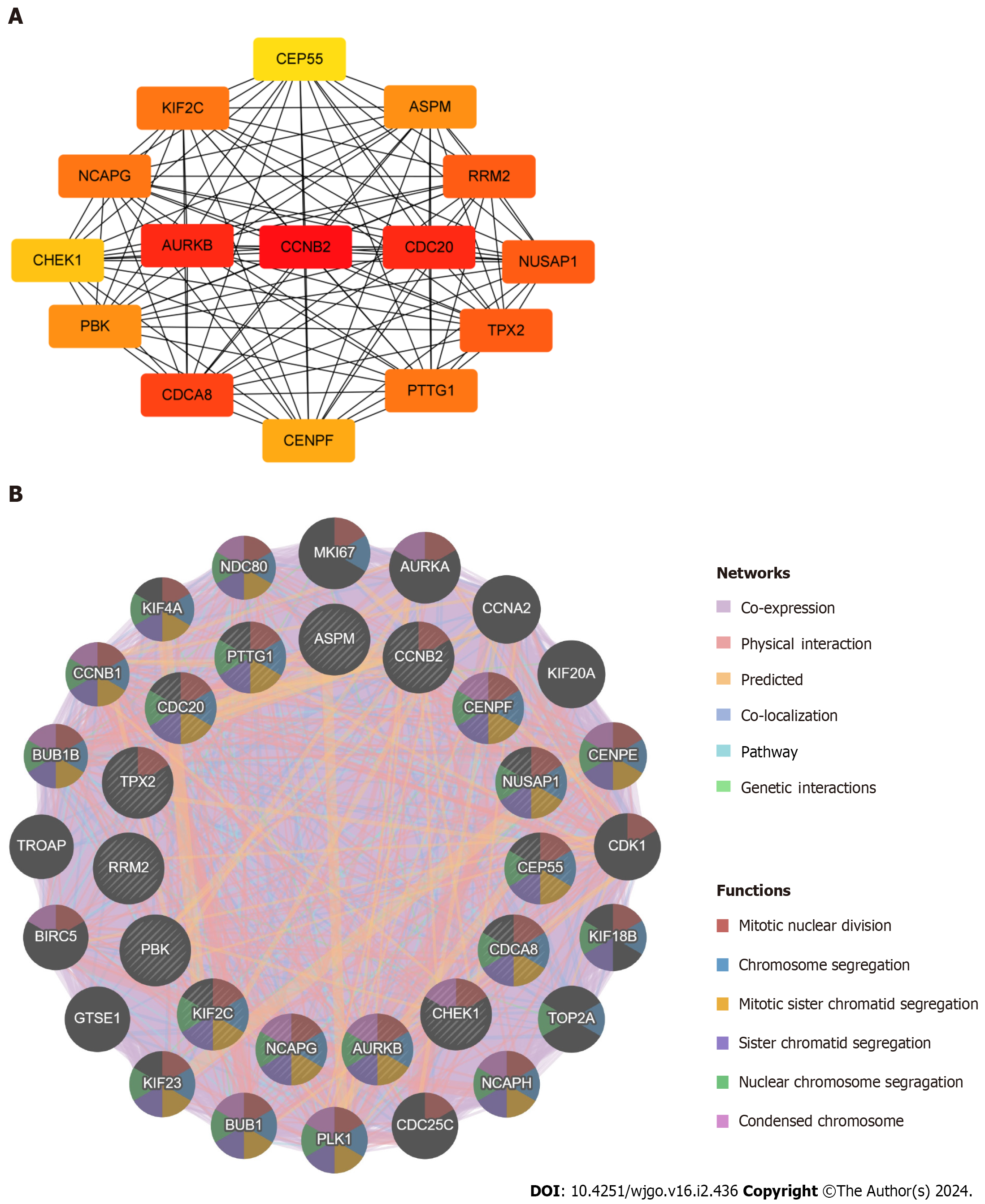
Figure 5 Identification and analysis of the hub genes.
A: Protein–protein interaction network of hub genes; B: Biological function analysis of hub genes.
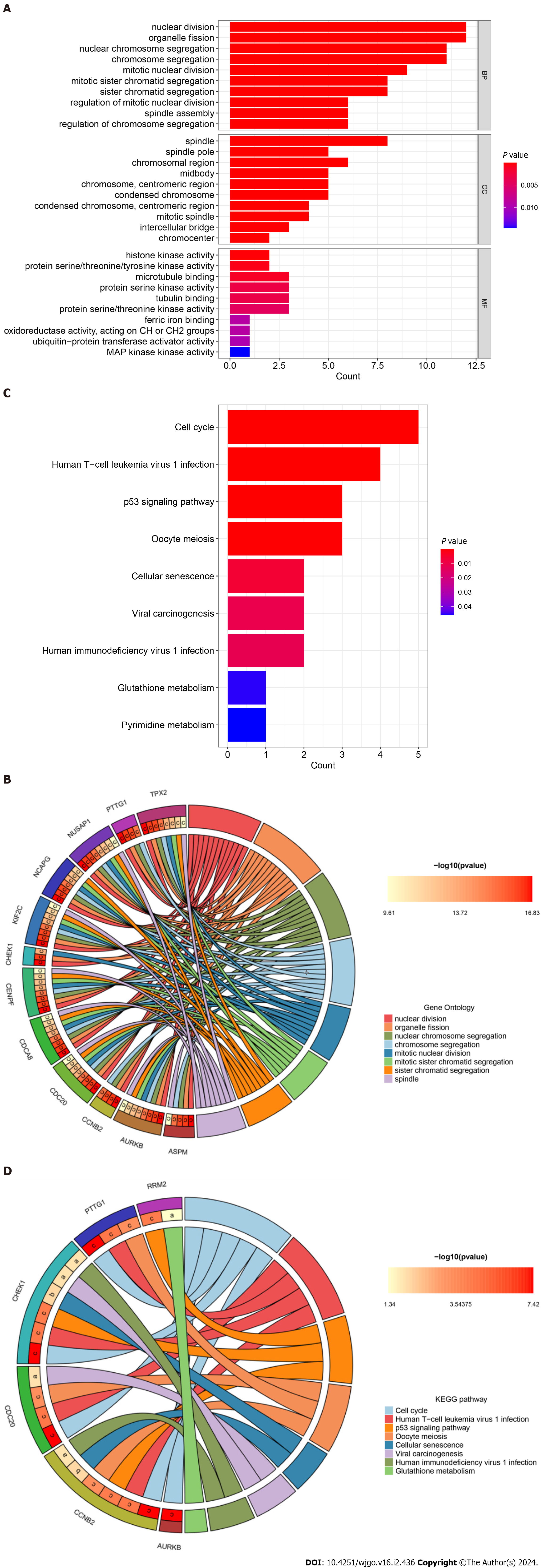
Figure 6 Gene Ontology annotation and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of hub genes.
A: Bar plot of Gene Ontology (GO) function annotation of hub genes; B: Circle plot of GO function annotation of hub genes; C: Bar plot of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of hub genes; D: Circle plot of KEGG pathway enrichment analysis of hub genes. KEGG: Kyoto Encyclopedia of Genes and Genomes.
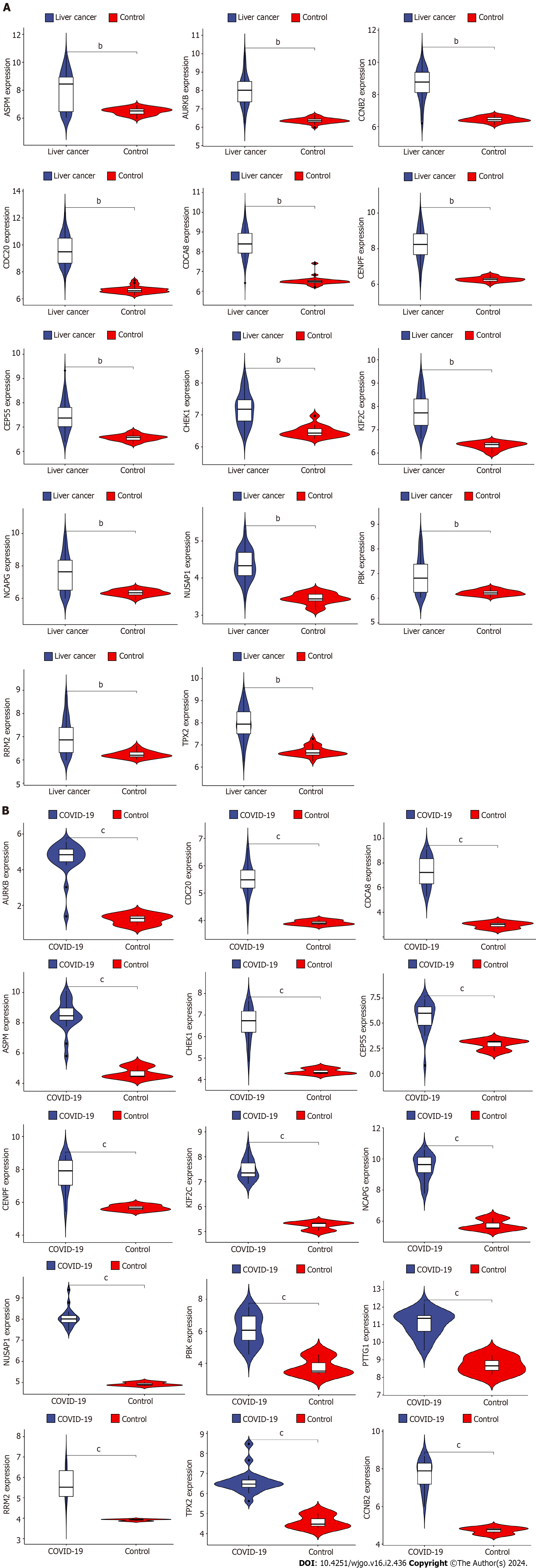
Figure 7 Validation of differential expression of hub genes in liver cancer and coronavirus disease 2019.
A: Differential expression analysis of hub genes in liver cancer dataset; B: Differential expression analysis of hub genes in coronavirus disease 2019 dataset. COVID-19: Coronavirus disease 2019. bP < 0.01, cP < 0.001.
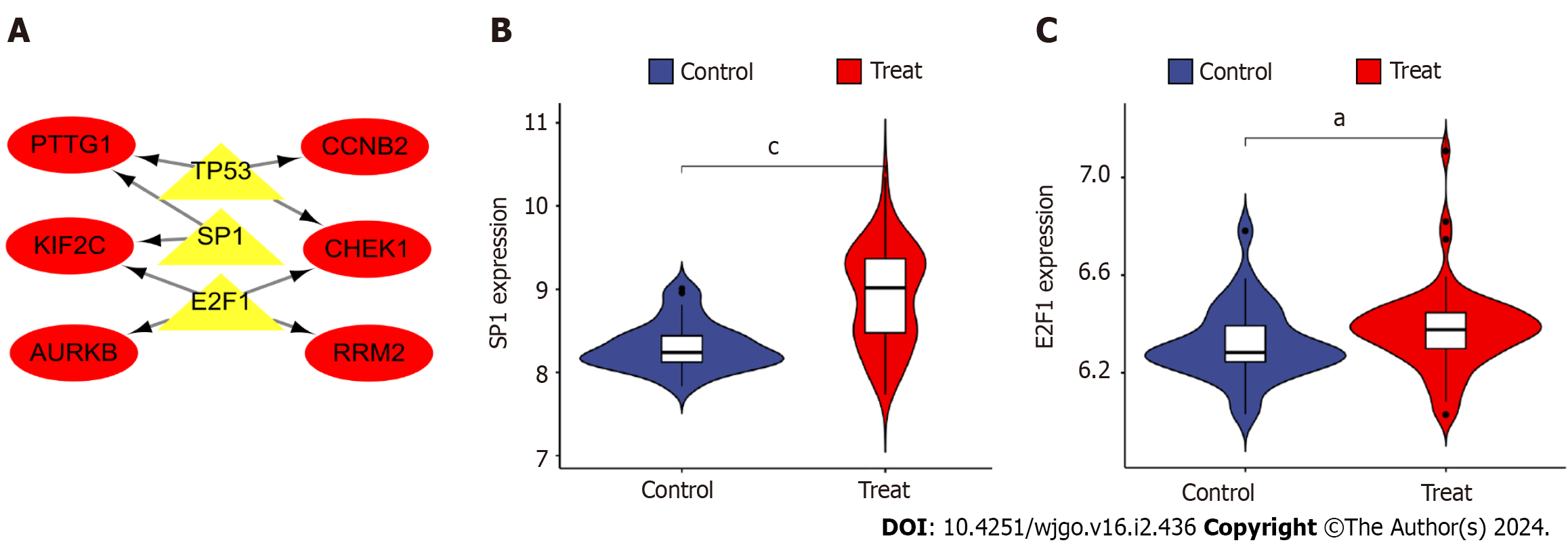
Figure 8 Construction and analysis of the regulatory network of transcription factor genes and hub genes.
A: Transcription factor (TF)-hub gene regulatory network, highlighted red nodes indicate hub genes and yellow nodes indicate TF genes; B: Analysis of differential expression of TF gene e2f transcription factor 1 in disease; C: Differential analysis of TF gene sp1 transcription factor expression in disease. aP < 0.05, cP < 0.001 E2F1: e2f transcription factor 1; SP1: sp1 transcription factor; AURKB: Aurora kinase B; CCNB2: Cyclin B2; CHEK1: Checkpoint kinase 1.
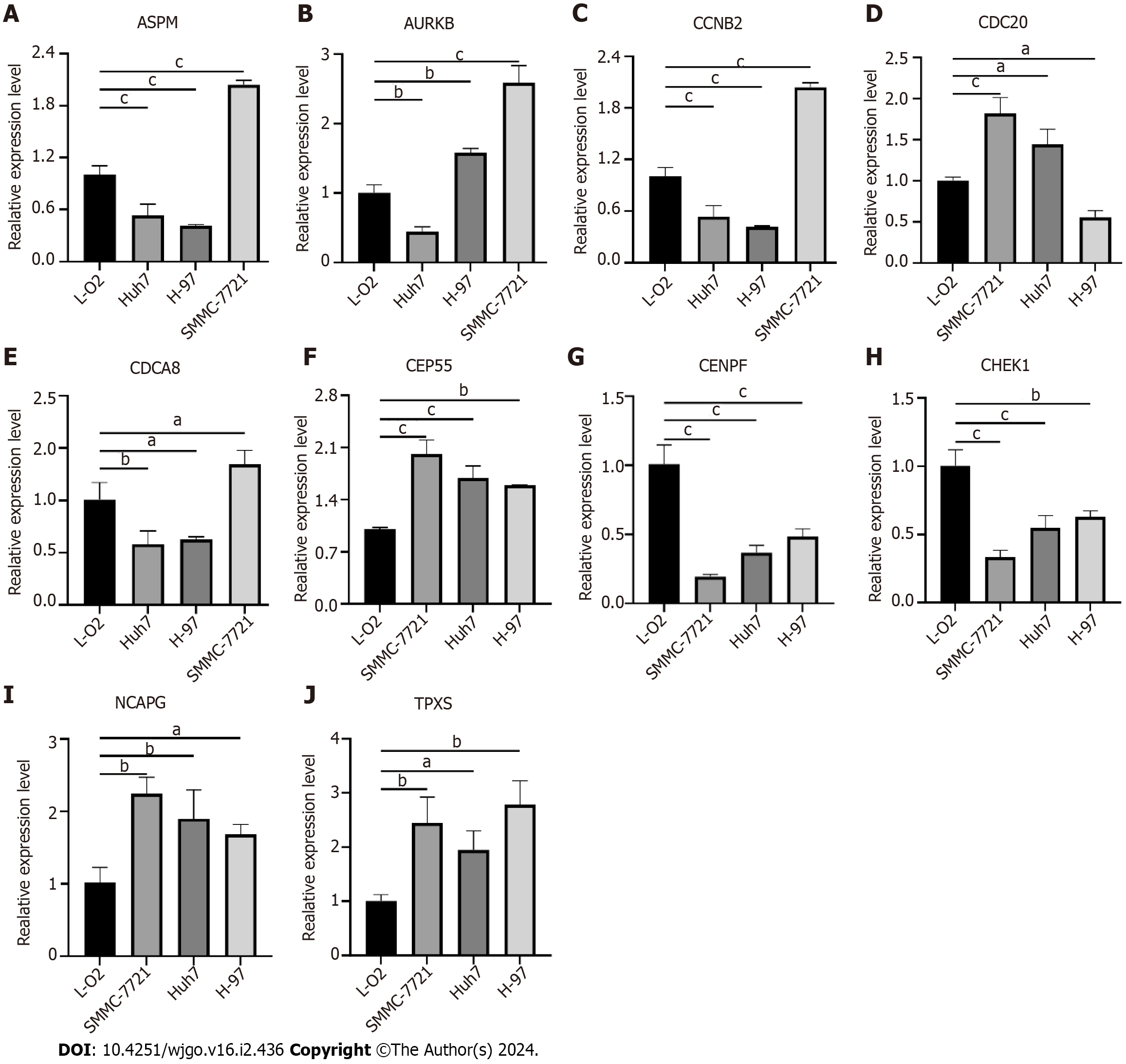
Figure 9 Validation of hub genes expression in liver cancer cells.
A: Assembly factor for spindle microtubules (ASPM); B: Aurora kinase B (AURKB); C: Cyclin B2 (CCNB2); D: Cell division cycle 20 (CDC20); E: Cell division cycle associated 8 (CDCA8); F: Centrosomal protein 55 (CEP55); G: Centromere protein F (CENPF); H: Checkpoint kinase 1 (CHEK1); I: Non-SMC condensin I complex subunit G (NCAPG); J: Targeting protein for xklp2 (TPX2). aP < 0.05, bP < 0.01, cP < 0.001 ASPM: Assembly factor for spindle microtubules; AURKB: Aurora kinase B; CCNB2: Cyclin B2; CDC20: Cell division cycle 20; CDCA8: Cell division cycle associated 8; CEP55: Centrosomal protein 55; CENPF: Centromere protein F; CHEK1: Checkpoint kinase 1; NCAPG: Non-SMC condensin I complex subunit G; TPX2: Targeting protein for xklp2.
- Citation: Rong Y, Tang MZ, Liu SH, Li XF, Cai H. Comprehensive analysis of the potential pathogenesis of COVID-19 infection and liver cancer. World J Gastrointest Oncol 2024; 16(2): 436-457
- URL: https://www.wjgnet.com/1948-5204/full/v16/i2/436.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v16.i2.436