Copyright
©The Author(s) 2025.
World J Hepatol. Jun 27, 2025; 17(6): 107329
Published online Jun 27, 2025. doi: 10.4254/wjh.v17.i6.107329
Published online Jun 27, 2025. doi: 10.4254/wjh.v17.i6.107329
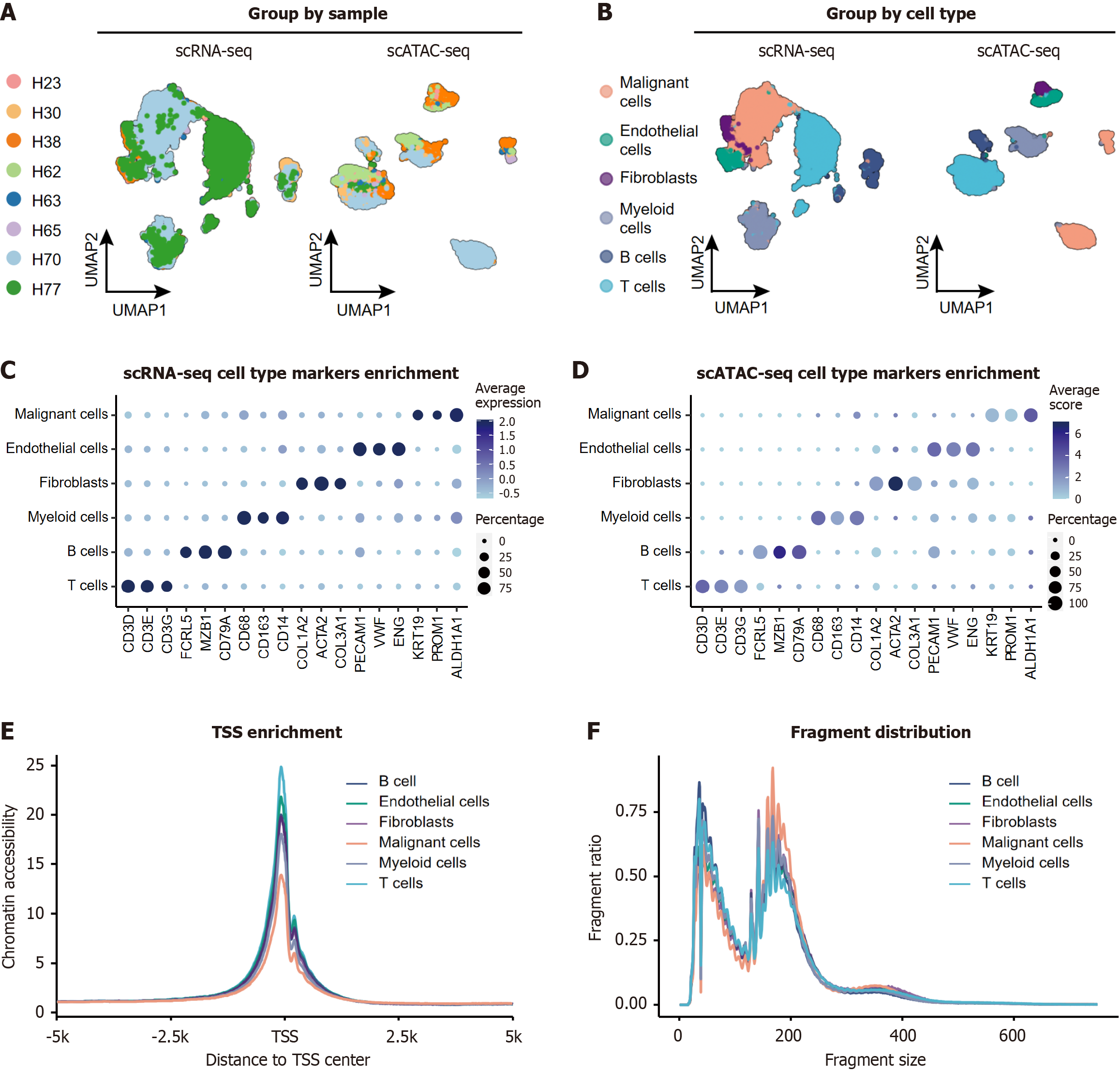
Figure 2 Integrated single-cell multi-omics characterization of hepatocellular carcinoma tumor from the same patients.
A: Uniform manifold approximation and projection plot of 15660 single-cell RNA-sequencing (scRNA-seq) cells (left) and UMAP plot of 10201 single-cell assay for transposase-accessible chromatin with sequencing (scATAC-seq) cells (right) color-coded by patient origin; B: UMAP plot of 15660 scRNA-seq cells (left) and UMAP plot of 10201 scATAC-seq cells (right) color-coded by annotated cell types; C: Dot plot showing canonical marker gene expression (rows) across cell types (columns) in scRNA-seq. Dot size: Fraction of expressing cells; color: Mean normalized expression; D: Dot plot showing chromatin accessibility of marker gene promoters (rows) across cell types (columns) scATAC-seq. Dot size: Fraction of expressing cells; color: Mean normalized expression; E: Transcription start site enrichment profiles stratified by major cell types. Malignant hepatocytes exhibit reduced transcription start site signal compared to non-malignant cells; F: Fragment size distributions across cell types. Malignant cells display longer nucleosome-free fragments. UMAP: Uniform manifold approximation and projection; scRNA-seq: Single-cell RNA-sequencing; ATAC-seq: Assay for transposase-accessible chromatin using sequencing; scATAC-seq: Single-cell assay for transposase-accessible chromatin with sequencing; TSS: Transcription start site.
- Citation: Xi XL, Yang YD, Liu HL, Jiang J, Wu B. Chromatin accessibility module identified by single-cell sequencing underlies the diagnosis and prognosis of hepatocellular carcinoma. World J Hepatol 2025; 17(6): 107329
- URL: https://www.wjgnet.com/1948-5182/full/v17/i6/107329.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i6.107329