Copyright
©The Author(s) 2025.
World J Hepatol. Jun 27, 2025; 17(6): 107329
Published online Jun 27, 2025. doi: 10.4254/wjh.v17.i6.107329
Published online Jun 27, 2025. doi: 10.4254/wjh.v17.i6.107329
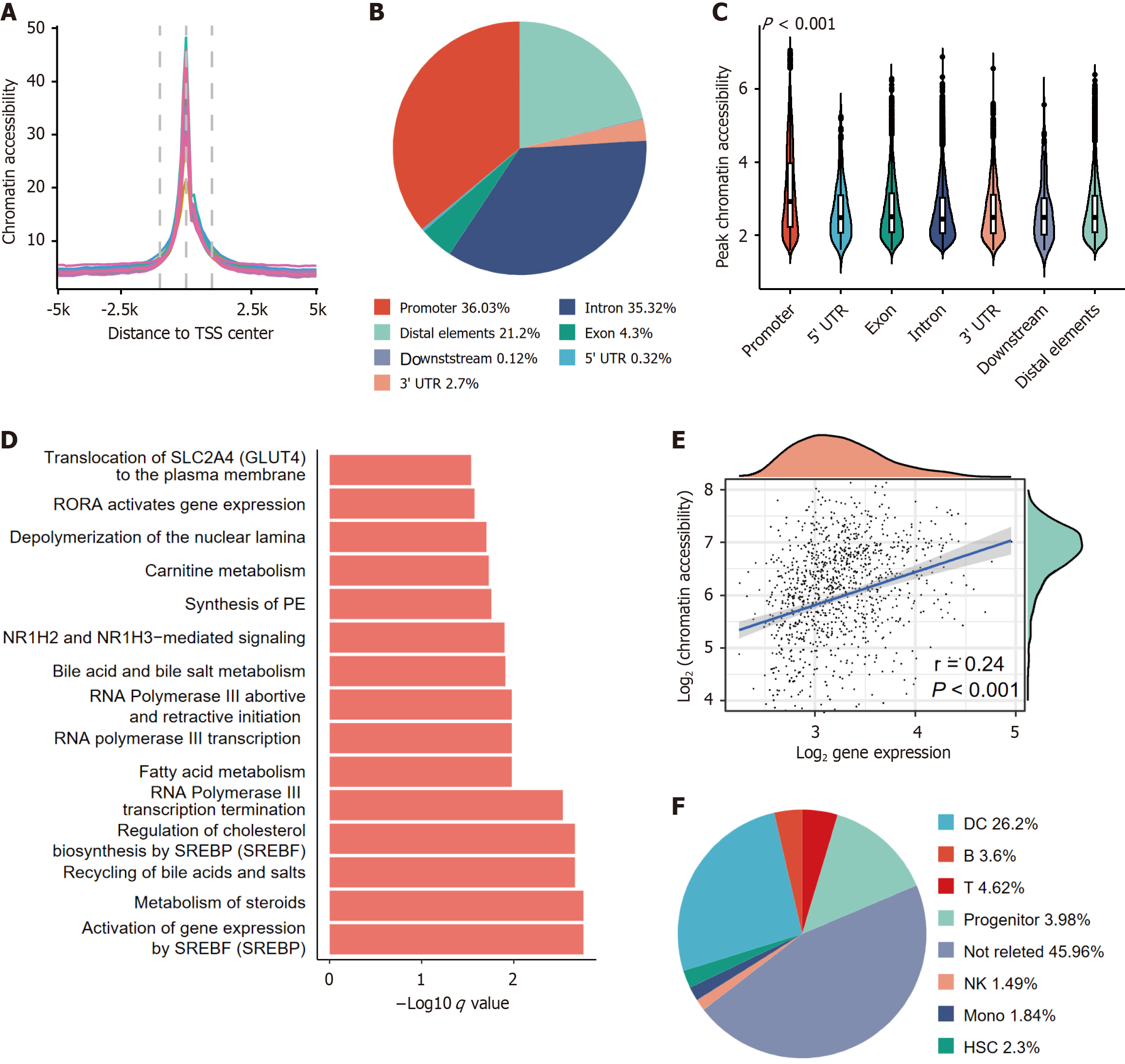
Figure 1 Bulk the assay for transposase-accessible chromatin with sequencing dataset identifies chromatin accessibility dynamics and tumor-associated pathways in hepatocellular carcinoma.
A: Transcription start site enrichment profiles (± 5 kb from transcription start site) validating sequencing quality across hepatocellular carcinoma (HCC) and paired normal samples; B: Genomic distribution (proportions of promoter, intronic, and distal regions) of chromatin accessibility peaks unique to HCC; C: Chromatin accessibility scores across genomic compartments. Promoter regions show the highest accessibility (Wilcoxon test, P < 0.001); D: Top enriched REACTOM pathways for genes with recurrent promoter-accessible peaks (adjusted P < 0.05); E: Weak but significant correlation between promoter accessibility and corresponding gene expression (Pearson r = 0.24, P < 0.001); F: Immune infiltration-associated chromatin features across HCC-specific peaks. Criteria for immune-associated peaks: Enhanced accessibility in immune cells; correlation with cytolytic activity. TSS: Transcription start site; UTR: Untranslated region; PE: Phosphatidyl ethanolamine; DC: Dendritic cells; NK: Natural killer cells; HSC: Hematopoietic stem cells.
- Citation: Xi XL, Yang YD, Liu HL, Jiang J, Wu B. Chromatin accessibility module identified by single-cell sequencing underlies the diagnosis and prognosis of hepatocellular carcinoma. World J Hepatol 2025; 17(6): 107329
- URL: https://www.wjgnet.com/1948-5182/full/v17/i6/107329.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i6.107329