Copyright
©The Author(s) 2025.
World J Hepatol. May 27, 2025; 17(5): 104519
Published online May 27, 2025. doi: 10.4254/wjh.v17.i5.104519
Published online May 27, 2025. doi: 10.4254/wjh.v17.i5.104519
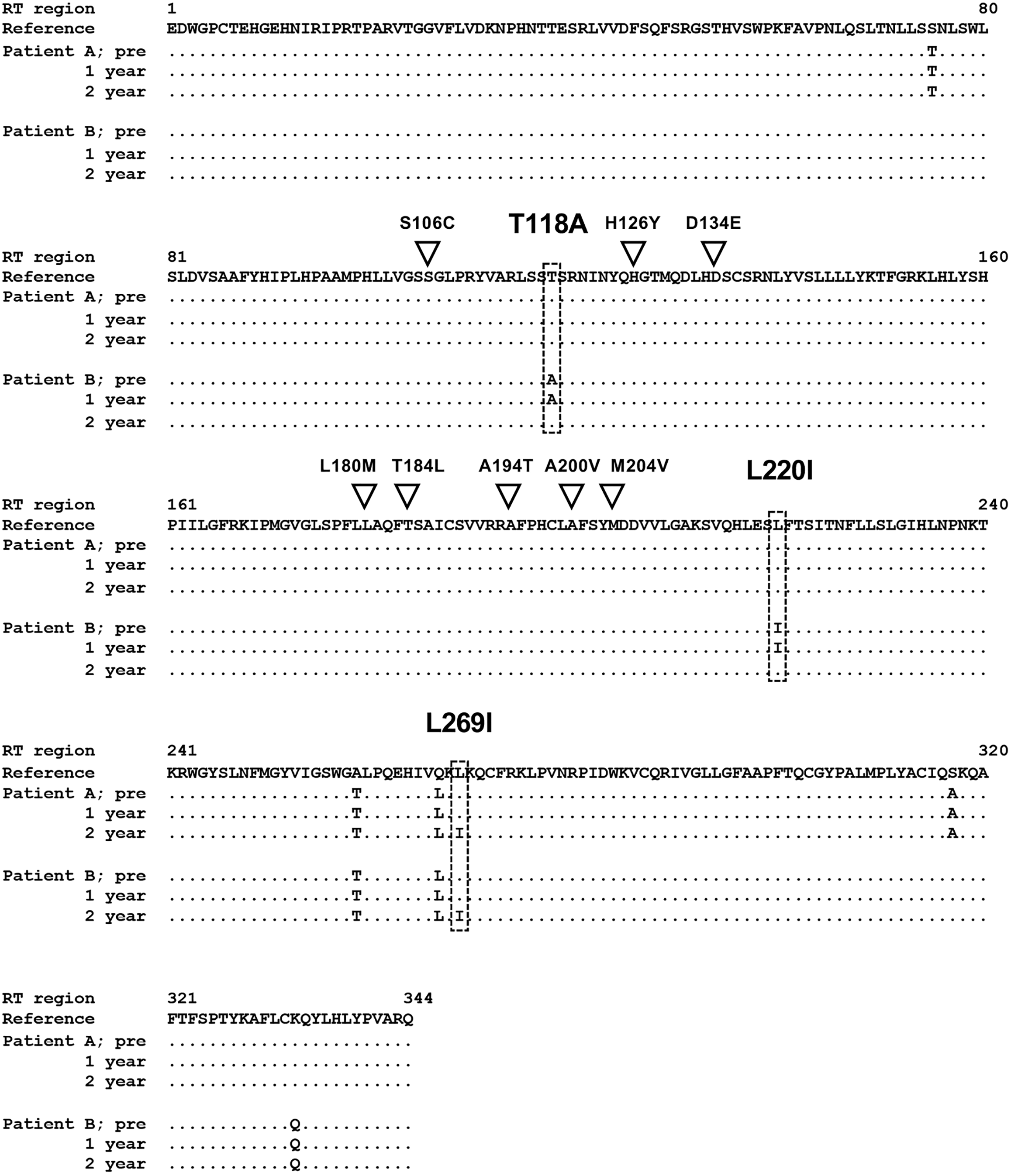
Figure 2 Alignment of reverse transcriptase regions of hepatitis B virus strains detected in tenofovir alafenamide fumarate-incomplete responders.
The reverse transcriptase regions of the hepatitis B virus strains detected in tenofovir alafenamide fumarate (TAF)-incomplete responders before treatment (pre), 1 year after TAF treatment (1 year), and 2 years after TAF treatment (2 year) were directly sequenced, and deduced amino acids were aligned with the reference sequence of genotype C (accession number: GQ872210). The inverted triangle indicates the positions of the reported resistance-associated mutations to tenofovir disoproxil fumarate and TAF. A box with a dotted line indicates the positions of the mutations detected in this study. RT: Reverse transcriptase.
- Citation: Yamada N, Igarashi H, Murayama A, Suzuki M, Yasuda K, Saito M, Isogawa M, Kato T. Deep sequencing analysis of hepatitis B virus in patients with incomplete response to tenofovir alafenamide fumarate treatment. World J Hepatol 2025; 17(5): 104519
- URL: https://www.wjgnet.com/1948-5182/full/v17/i5/104519.htm
- DOI: https://dx.doi.org/10.4254/wjh.v17.i5.104519