Published online Oct 27, 2018. doi: 10.4254/wjh.v10.i10.645
Peer-review started: April 21, 2018
First decision: May 8, 2018
Revised: June 8, 2018
Accepted: June 27, 2018
Article in press: June 28, 2018
Published online: October 27, 2018
Hepatocellular carcinoma (HCC) is one of the most prevalent malignancies worldwide and the second leading cause of death among all cancer types. Deregulation of the networks of tissue-specific transcription factors (TFs) observed in HCC leads to profound changes in the hepatic transcriptional program that facilitates tumor progression. In addition, recent reports suggest that substantial aberrations in the production of TF isoforms occur in HCC. In vitro experiments have identified distinct isoform-specific regulatory functions and related biological effects of liver-specific TFs that are implicated in carcinogenesis, which may be relevant for tumor progression and clinical outcome. This study reviews available data on the expression of isoforms of liver-specific and ubiquitous TFs in the liver and HCC and their effects, including HNF4α, C/EBPs, p73 and TCF7L2, and indicates that assessment of the ratio of isoforms and targeting specific TF variants may be beneficial for the prognosis and treatment of HCC.
Core tip: This paper aims to analyze existing data on the spectrum of isoforms of liver-specific transcription factors produced in the liver and hepatocellular carcinoma (HCC) and implicated in carcinogenesis, their distinct regulatory functions and subsequent isoform-dependent biological effects which may be relevant for tumor progression and clinical outcomes in HCC patients.
- Citation: Krivtsova O, Makarova A, Lazarevich N. Aberrant expression of alternative isoforms of transcription factors in hepatocellular carcinoma. World J Hepatol 2018; 10(10): 645-661
- URL: https://www.wjgnet.com/1948-5182/full/v10/i10/645.htm
- DOI: https://dx.doi.org/10.4254/wjh.v10.i10.645
According to the current estimations of The Human Genome Sequencing Consortium, the human genome is predicted to comprise less than 20000 protein-coding genes[1]. Due to concerted efforts of The Human Proteome Project, 85% of the predicted proteins have already been identified[2]. Multiple sources of evidence suggest that this amount may be underestimated due to the existence of protein isoforms arising from the usage of alternative promoters or translation start sites (TSSs), alternative splicing regulated by ubiquitous and tissue-specific splicing factors, which affects the transcripts of 92%-94% of genes, and alternative cleavage and polyadenylation[3-5]. Interestingly, the occurrence of both alternative promoters and multiple conserved TSSs positively correlates with alternative splicing events[4,6,7]. However, although most multi-exon genes produce several alternative isoforms, 85% of the transcriptome is generally represented by a single major gene transcript[8].
Deregulation of gene expression[9] and the generation of aberrant alternative isoforms are commonly observed in multiple cancer types[10,11]. Mechanisms underlying the misregulation of the production of alternative isoforms in cancer have been thoroughly described elsewhere[12-15] and are beyond the scope of this review.
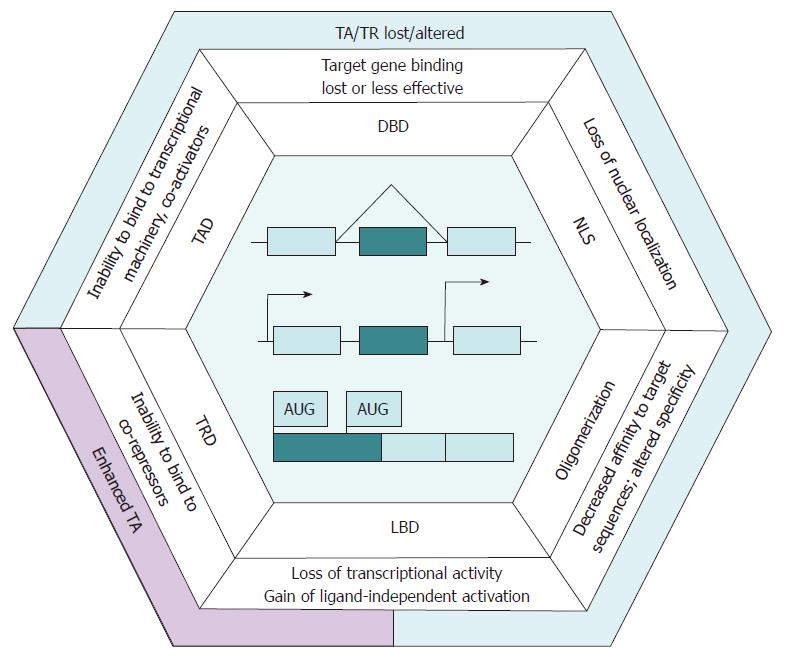
Distinct transcriptional programs of particular cell types are principally controlled by tissue-specific transcription regulators[16]. Due to alternative promoter usage, transcription factors (TFs) produce a tissue-specific pattern of alternative isoform expression that may be altered in carcinogenesis[12], and the imbalance of isoform production in tumors contributes to a flexible contextdependent diversification of cell phenotypes[17,18]. Somatic mutations and abnormal activation of signaling pathways in cancer may cause a differential regulation of the abundance and activity of splicing factors and lead to an increase in the production of alternative transcripts, including those for TFs[14]. Additionally, the transcripts of genes encoding TFs tend to contain multiple conserved TSSs, which may contribute to the production of protein isoforms. In the case of TFs, these complex regulatory mechanisms specifying alternative isoform production frequently affect functional domains[19], and their disruption in cancer may result in the misregulation of networks of their target genes (Figure 1).
Hepatocellular carcinoma (HCC) is the most frequent type of liver cancer and the second leading cause of death among all cancer types. Although recurrent driver genes and somatic mutations have been identified in HCC, most of them cannot yet be considered as druggable targets for therapy[20]. Massive deregulation of the liver-specific TF network[16] and aberrant alternative splicing[14,21,22] have been reported in HCC. The latter arises from abnormal expression, altered transcript splicing and a high mutation rate of genes encoding splicing factors that exert an effect in hepatocarcinogenesis and result in profound changes in the isoform balance compared with the normal liver. Importantly, up to 9% of differentially spliced transcripts in HCC originate from TF-coding genes[23,24]. These profound changes in the ratios of TF isoforms should likely result in a substantial modification of gene expression programs and therefore produce tumor phenotype alterations with a distinct clinical impact. Thus, data on the induction of expression of tumor-specific TF variants may be useful for the prognosis and development of approaches to isoform-specific therapy for HCC.
Nevertheless, current data concerning the regulation of the TF network in hepatocarcinogenesis by alteration of their isoform balance are rather diverse. In this review, we consider structural properties, biological effects and the clinical impact of the most investigated TF isoforms specific to liver and HCC. Essential features of these TFs and their isoforms are summarized in Table 1.
| TF | No. of isoforms expressed in the liver | Types of isoforms | Regulated processes (canonical isoform) | Upstream signaling pathways | Target genes expressed in the liver | Isoforms that presumably promote hepatocarcinogenesis | Isoforms that exhibit tumor-suppressive effects | Ref. |
| C/EBPβ | 3 | TA, DN | Metabolism POS: apoptosis, inflammatory response NEG: proliferation | TNF | UP(LAP1): cytochrome p450 genes UP(LIP): GADD45B DOWN (LAP1): CCNA, CCNE, CDK2, PCNA DOWN (LIP): CLU, NUMB | C/EBPβ-LIP | C/EBPβ-LAP1, C/EBPβ-LAP2 | [59,65,74,76,77, 78,139] |
| ERα | 4 | DN | POS: proliferation | Estrogen signaling | UP: CCND1, HNRNPH2, MYC, RET, WWC1 | ERα-36, ERα-46, ER-αΔ5 | ERα-66 | [41,43,44,47,139,140] |
| HIF1α | 3 | DN | NEG: apoptosis, inflammatory response | PI3K/Akt, mTOR | UP: VEGFA | HIF1α1.1 | HIF1α516, HIF1α736 | [125,131,133,134,139] |
| HNF4α | 9-12 | TA | POS: angiogenesis | AMPK, Hippo, TGFβ | UP: CDH1, CDKN1A, HNF1a, PERP DOWN: BMP7, HMGA2, MYC, SNAI1, SNAI2 | HNF4α7-α9 | HNF4α1-α3 | [28,29,37,139,141,142] |
| KLF6 | 3 | TA, DN | POS: differentiation, morphogenesis, apoptosis | TGFβ | UP(wtKLF6): ССND1, CDH1, CDK4 UP(SV2): CDKN1A DOWN(wtKLF6): MDM2 DOWN(SV1): CDKN1A | SV1 | wtKLF6, SV2 | [105,108-114,143] |
| p73 | 3 | TA, DN | NEG: proliferation, invasion, metastases | p53, Hippo, mTOR | UP: BCL2 family genes, caspases, CD95, TNF-R, TRAIL-R | ΔN-p73, ΔEx2p73, ΔEx2/3p73 | TA-p73 | [87-91,139,144] |
| PGC1α | 3 | TA, DN | POS: proliferation, differentiation, apoptosis | AMPK, Insulin signaling | UP: GLUT4, PDK4, PEPCK, PPARA, PPARG | PGC1α1-a, LPGC1α, NT-Pgc 1α-a* (*promote replication and assembly of in HBV and HCV) | [49,52,54-56,139] | |
| TCF4 (TCF7L2) | 17 | TA | POS: apoptosis | Wnt, Hippo | UP: AXIN2, CCND1, IRS1, JUN, MMP7, WISP1 | TCF-4J, TCF-4C | TCF-4B, TCF-4K | [98-102,139,145] |
| WT1 | 4 | DN | NEG: cell cycle progression | UP: BCL2 family genes, cFLIP DOWN: FADD, HNF4A, NF-κB | WT1(+/) 17АА(+)KTS | WT1(-)KTS | [119-122,146] | |
| ZIP (ZGPAT) | 2 | DN | POS: gluconeogenesis | DOWN: CDC25A, EGFR, FGF5, FGF14, PDGFB, PTEN, RGS3, TBPL1, VCAM1 | sZIP | ZIP(fl) | [135-137] | |
Nuclear receptor HNF4α (NR2A1) is a key regulator of hepatocyte differentiation. Bound to DNA as a homodimer, it modulates the expression of nearly 42% of the genes expressed in hepatocytes, including a wide spectrum of hepato-specific genes, either directly or through activation of other liver-enriched TFs[25,26]. Apart from the regulation of differentiation and morphogenesis, HNF4α acts as a tumor suppressor in the liver. It has been shown to inhibit proliferation through the induction of CDKN1A and repression of BMP7 and MYC, to regulate the expression of the p53/p63-dependent apoptotic effector PERP and to interfere with epithelial-mesenchymal transition (EMT) via repression of SNAI1, SNAI2 and HMGA2 and the upregulation of CDH1[27,28].
HNF4A is transcribed from one of its alternative promoters, P1 or P2, which are regulated in a tissue- and developmental stage-specific manner. Additional exon inclusion and alternative splicing result in the generation of up to 12 HNF4α variants sharing common DNA-binding (DBD) and ligand-binding domains (LBD) and differing in the trans-activation domain (TAD) and repressor F domain[29]. The isoforms transcribed from P2 (HNF4α7- α12) are devoid of the TAD activation function (AF)-1 region involved in the interaction with co-activators, while demonstrating a significant decrease in trans-activation (TA) properties[30]. The full-length F domain of HNF4α1 and HNF4α7 variants impair their TA potential because it masks the AF-2-independent co-factor binding site. In contrast, the 10-amino-acid (AA) insert in the proline-rich F domain region of HNF4α2/α8 isoforms prevents masking, which increases the effectiveness of target gene activation[31]. Thus, the structural diversity of HNF4α variants determines a lower transcriptional activity of Р2-derived isoforms (HNF4α7-α12) that lack AF-1, compared to Р1-derived variants (HNF4α1-α6), and a higher transcriptional activity of isoforms that are devoid of the F domain, compared to HNF4α1/α7.
HNF4α3/α9 isoforms derive from skipping a splice site in exon 8 and contain an extended reading frame with an alternative termination site, thus encoding a protein with a completely different C-terminal F domain with undetermined function[32]. The existence of HNF4α4-α6 variants containing additional exons 1B and 1C has not been clearly proven[32]. HNF4α10-α12 isoforms bearing extended TAD due to exon 1E translation are expressed in various hepatoma cell lines, but their functions remain unclear[33].
P1 variants are predominant in the normal liver and regulate the expression of hepatocyte differentiation markers[32,34]. HNF4αP2 isoforms are prevalent at the early stages of embryogenesis; they preferentially activate promoters of early hepatic genes[35]. In the postnatal liver, the P2 promoter is repressed due to the binding of HNF4αP1 isoforms.
Deregulation of the expression of HNF4α isoforms is a frequent event in hepatocarcinogenesis. At the early stages of hepatocarcinogenesis, moderately differentiated HCC cells are characterized by activation of the expression of embryonic HNF4aP2 isoforms, which is associated with vascular invasion and poor overall survival in HCC patients[36]. In advanced dedifferentiated HCCs, which lose epithelial morphology, the expression of both P1 and P2 variants is repressed[34,37,38].
Exogenous expression of HNF4α1 in dedifferentiated HCC cells results in the restoration of epithelial morphology via the upregulation of epithelial markers (E-cadherin, connexin32, ZO-1), decrease in proliferation and reactivation of genes specific to differentiated hepatocytes, and decrease in in vivo tumor growth and metastatic potential[37,39,40]. Overall, these data indicate that HNF4αP1 reactivation might be beneficial as a therapeutic approach for HCC treatment. Further experiments are required to elucidate the role and clinical impact of particular HNF4α isoforms and to implement these findings into the development of therapeutic and prognostic approaches to HCC treatment.
Nuclear receptor ERα encoded by the ESR1 gene induces proliferation and exhibits anti-apoptotic and anti-inflammatory activities[41] via estrogen-dependent binding to estrogen response elements of target genes. Non-canonical activation of ERα target genes via secondary messengers, activation of membrane-bound ERα isoforms or protein-protein interactions have also been described[42].
Canonical 66-kDa ERα (ERα-66) matches the conventional structure of nuclear receptors. Additionally, numerous ERα splice variants that differ in promoter usage and the presence of exons encoding TAD, DBD, LBD and the nuclear localization signal (NLS) region have been reported in normal and tumor tissues[42]. Apart from ERα-66, the expression of 3 dominant-negative (DN) isoforms has been reported in the liver, namely, ERα-46 lacking AF-1 TAD, ERαΔ5 harboring an incomplete LBD, and ERα-36, which is transcribed from an alternative promoter and is deficient in both AF-1 and LBD[41].
Investigation of the properties of truncated isoforms in various cell lines has demonstrated that the dimerization of a shorter isoform with the full-length one represses its transcriptional activity. Membrane-bound fractions of ERα-46 and ERα-36 are believed to interfere with the function of ERα-66. Moreover, ERα-36 lacks the NLS but is able to activate the MAPK/ERK cascade upon treatment with estrogens or tamoxifen[41-45]. A constitutively active ER-αΔ5 variant exhibits approximately 10%-15% of the ligand-bound ERα-66 activity[46].
ERα-66 is highly expressed in the normal liver and, to a lesser extent, in cirrhotic tissue, while its transcription in HCC is substantially or completely suppressed. Higher ERα-66 expression is associated with a better overall and recurrence-free survival of HCC patients and is negatively correlated with the tumor size and extra-hepatic metastasis. The mRNA of DN ERα-46 is detected in non-tumor, cirrhotic and tumor liver tissues. Upregulation of ERαΔ5 in HCC causes repression of ERα-66 transcriptional activity through heterodimerization. An increase in ERα-36 isoform production has been reported in HCC and hepatoma cell lines; it correlates with the downregulation of the full-length isoform[43,44,47] and may arise from ESR1 promoter hypermethylation, which is frequently observed in HCC[48]. Thus, the shift in balance of ERα variants towards truncated isoforms with DN properties can essentially modulate the impact of the estrogen receptor signaling pathway in hepatocarcinogenesis and is likely associated with an unfavorable HCC prognosis.
PGC-1α (PPARGC1A) is an inducible adaptor of the PPARγ co-activator 1 family. It has been shown to regulate adaptive reactions of energy metabolism in brain, cardiac and skeletal muscles, adipose tissue and liver[49]. PGC-1α is a key inducer of gluconeogenesis in the fasted liver[50]. A significant decrease in PGC-1α expression has been reported in primary human HCCs[51].
The N-terminus of PGC-1α protein contains a TAD with co-activator and nuclear receptor (HNF4α, PPARα, PPARγ, LXRα) binding sites[52] followed by phosphorylation and acetylation sites that modulate PGC-1α activity[53]. The NLS region, an arginine-serine-rich domain that enables binding to TFs and RNA-recognition motifs, is located in the C-terminal part of PGC-1α. C-terminal domains are involved in mRNA processing and splicing[53].
PGC-1α transcription is controlled by several tissue-specific promoters and is often coupled with alternative splicing. Processed alternative transcripts give rise to a set of TA-competent isoforms and DN variants lacking C-terminal domains or with truncations in their co-activator and co-repressor regions[53]. In human tissues with intense energy metabolism, the full-size PGC-1α1-a is a prevalent isoform[52]. In murine liver, mRNAs encoding Pgc1α1a and NT-Pgc1αa are transcribed from the proximal promoter. The NT-Pgc1αa variant preserves the co-activator binding domain and a portion of the co-repressor binding domain, but it lacks the C-terminal part[54]. Although less effective, NT-Pgc1αa is sufficient to stimulate gluconeogenesis in PGC-1α1-a-/- hepatocytes[55]. The hepato-specific promoter located downstream of the proximal one is active in human fasted liver[52] or in response to hepatitis C virus (HCV) induced endoplasmic reticulum (ER) stress. Its activation stimulates the expression of a liver-specific TA-isoform, L-PGC-1α, which is deficient in the first 127 aa but is able to co-activate most PGC-1α-interacting nuclear receptors except liver X receptor α (LXRα)[56]. LXRα has been shown to repress the FOXM1 transcriptional regulator and its target genes CCNB1 and CCND1 in HCC cells[57]. Thus, the reduced LXRα activity may stimulate proliferation in hepatoma cell lines. The reduced LXRα activity also causes the deregulation of cholesterol metabolism, resulting in liver damage and inflammation, which facilitates HCC progression[58]. Hence, the elevated L-PGC-1α level is likely to be implicated in hepatocarcinogenesis. However, PGC-1α1-a induction occurs along with L-PGC-1α upregulation in the fasted liver and during HCV-induced ER stress. As the result, a higher PGC-1α1-a/L-PGC-1α ratio abolishes the effects of the alternative isoform. Currently, the expression of PGC-1α isoforms has been investigated in HCC cell lines but not in HCC clinical samples. Further research is required to determine a possible impact of the imbalance of PGC-1α isoforms in hepatocarcinogenesis.
CCAAT enhancer binding proteins (C/EBPs) belong to a liver-enriched bZIP TF family; they regulate the expression of genes involved in proliferation, differentiation, inflammatory response and metabolism. C/EBPs bind to DNA as homo- and heterodimers, and the heterodimerization of C/EBPs can alter target site recognition[59].
Two C/EBP family members are predominant in normal liver. CEBPA is highly expressed in adult liver under normal conditions, whereas the expression of CEBPB is low in hepatocytes and can be induced by pro-inflammatory cytokines, hormones, cAMP and other agents[59,60]. The induction of hepatocyte proliferation is accompanied by C/EBPα downregulation and increases C/EBPβ levels[61]. Inactivation of the CEBPB gene significantly reduces the regenerative response in mice after a partial hepatectomy, whereas CEBPA-deficient murine hepatocytes demonstrate a high proliferative activity[59]. While CEBPA is mainly considered as a tumor suppressor that is downregulated in HCC[62], Lu et al[63,64] reported its upregulation in liver cancer, which was associated with escape from starvation-induced cell death and a poor prognosis. CEBPB expression is not altered in HCC, albeit the C/EBPβ protein level is decreased[65].
The N-termini of C/EBPα and C/EBPβ proteins contain a number of activation domains and negative regulatory regions that significantly differ between these TFs, while their C-termini contain a highly homologous basic sequence-specific DNA recognition region and leucine zipper dimerization domain. Both CEBPA and CEBPB are intronless genes. Alternative protein isoforms arise due to the alternative usage of TSSs or regulated proteolysis[59].
The 42-kDa and 30-kDa C/EBPα isoforms differ in their amino termini with the p30 isoform possessing a lower activation potential than the predominant full-length variant due to the absence of two activation domains[59,62]. Apart from the DN activity, p30 has been proposed to bind and regulate a distinct set of target genes[66]. While both the p42 and p30 variants have been detected in liver, to date, the changes in the isoform ratio in HCC have not been quantitatively investigated, and reports of CEBPA upregulation leading to poor outcomes in НСС patients might arise from an isoform imbalance. Indeed, the induction of CEBPA expression in various models of liver disease leads to disease reversal and results in lower levels of liver damage markers and reduced tumor burden in rodents with cirrhotic HCC[67].
C/EBPβ isoforms are subdivided into 2 classes. Liver-enriched activating protein isoforms are able to induce the expression of target genes. C/EBPβ-LAP1 (LAP*) exhibits a stronger TA potential than C/EBPβ-LAP2 (LAP), which is deficient in the short N-terminal portion of the activation domain. C/EBPβ-LIP, a liver-enriched inhibitory protein, lacks the TAD but preserves a part of the negative regulation domain, and bZIP and acts as a DN regulator of transcription[59]. LAP1 is the major C/EBPβ variant expressed in hepatocytes[65]. According to Fang et al[65], LAP1 is significantly downregulated in HCC, whereas the LAP2 level is unaltered, and LIP is underrepresented. These observations are partly in line with reports on LAP1 isoform downregulation in squamous cell carcinoma[68] and breast cancer[69]. Albeit LAP1 variant expression is reduced in HCC, its expression is even weaker in liver cancer stem cells[70]. Conversely, LAP2 and LIP are induced in these tumor types, and a high LIP/LAP ratio is indicative of an advanced stage and poor prognosis in breast cancer[71].
In murine liver regeneration models, C/EBPβ regulates the proliferation of hepatocytes via activation of the transcription of E2F-regulated genes that are essential for DNA replication (Mcm3, Cdc6) and reparation (Msh2, Msh5) and Cebpa repression[72,73]. C/EBPβ-LAP expression leads to a delay of the G1/S transition, which results in the synchronization of hepatocytes after a partial hepatectomy due to the downregulation of CCNA, CCNE, PCNA and CDK2, whereas the expression of C/EBPβ-LIP induces proliferation. Intriguingly, overexpression of LAP leads to an increase in the C/EBPα p30/p42 ratio, while expression of the LIP variant upregulates both C/EBPα isoforms[74]. Exogenous expression of C/EBPβ-LAP (but not the –LIP variant) arrests cell cycle progression in HepG2 and Hep3B hepatoma cells[75,76]. C/EBPβ-LIP contributes to the survival of tumor cells. Drug-induced apoptosis is significantly reduced in Hep3B cells overexpressing the C/EBPβ-LIP but not the C/EBPβ-LAP variant[76].
Emerging data also link C/EBPβ expression to the metastatic potential of tumor cells. LAP1/2 overexpression represses Huh7 migration in vitro and metastasis in vivo via the induction of orsomucoid 2 (ORM2) gene expression, whereas LIP does not affect ORM2 levels[65]. In vivo experiments also demonstrate that LAP1-transfected hepatoma cells possess a reduced ability to form subcutaneous tumors in nude mice and that the expression of Ki-67 and cancer stem cell markers is reduced in tumors originating from these cells[70].
The complex impact of CEBP isoform variants on the fine regulation of liver-specific gene expression can be illustrated by the regulation of the CYP3A4 gene encoding the most abundant type of cytochrome P450 in the liver. CYP3A4 expression is modulated by liver-specific TFs, including C/EBPα and C/EBPβ, which have multiple binding sites in their regulatory region[77,78]. While TA-competent isoforms induce the expression of CYP3A4, the DN variant LIP inhibits it, and an increase in the LIP/LAP ratio results in the repression of CYP3A4 in HepG2 cells[78]. As both TFs, including their DN variants, are able to heterodimerize, not only the levels of C/EBP proteins themselves but also the interactions between their isoforms, may affect the regulation of the metabolism of xenobiotics.
Taken together, the data on the abundance of C/EBPα and C/EBPβ variants in HCC remain limited and contradictory. CEBPA has previously been identified as a tumor suppressor that tends to be downregulated in HCC. Although there are limited data on the ratio of C/EBPα isoforms and their distinct effects in HCC, the capability of TA-competent and DN C/EBPα variants to form dimers with C/EBPβ isoforms increases the complexity of the network of target gene regulation and should be further investigated. In contrast, growing evidence of a variety of diverse effects arising from the regulation of expression by individual C/EBPβ isoforms indicates that evaluation of the ratio of LAP/LIP isoforms and their functions may be of value for HCC prognosis and treatment.
p53, p63 and p73 TFs of the p53 family regulate cell cycle progression and induce programmed cell death. Proteins of the p53 family have a similar domain structure, with TAD in their N-terminus followed by a proline-rich region, DBD and oligomerization domain. p63 and p73 possess a longer C-terminus encompassing the sterile α motif[79]. Since DBD shares substantial homology among the p53 family members, these TFs are able to regulate the expression of common target genes, including the regulation of each other’s expression, although each transcription factor has specific target genes[80].
The p53 family proteins bind DNA as tetramers. The wild-type p53 forms tetramers only with p53 variants, while mutant p53 variants can oligomerize with p63 and p73, thus reducing the activation of their target genes[81]. p63 and p73 can form mixed tetramers with varying transcriptional activity[82].
Alternative transcription and translation start sites and alternative splicing of the first exons encoding TAD result in the generation of a wide spectrum of N-terminally truncated (ΔN-) p53 family proteins that generally exert a DN effect over the full-length isoform activity. Additionally, the resultant proteins vary in their C-terminal domains due to alternative splicing[80]. p53 is frequently mutated or inactivated in cancer[80]. While p53 dysfunction is clearly associated with HCC progression, the only isoform proposed to play a significant role in hepatocarcinogenesis is the Δ40p53α variant translated from the second in-frame AUG of TP53 mRNA and lacking 39 AA of TAD. Δ40p53α has been demonstrated to be induced after doxorubicin treatment. Its exogenous expression suppresses proliferation, colony formation and induced senescence in hepatoma cells[83]. p63 is not expressed in the normal liver; however, the expression of ΔN isoforms is detected in p53-null hepatocytes. A trans-activating p63 variant (TAp63) is expressed in hepatoma cell lines irrespective of p53 status[84]. The TAp63 knockdown in HepG2 and Hep3B hepatoma cells leads to increased proliferation and colony formation. TAp63 expression negatively correlates with tumor size, intrahepatic metastasis, and distant metastasis and, according to the results of a retrospective analysis, a low TAp63 level is associated with poor survival in HCC patients[85,86].
The activation-competent TAp73 is absent in normal hepatocytes but is widely expressed in hepatoma cell cultures and human HCC samples[87-89]. Intriguingly, alternative splicing of full-length TAD-encoding transcripts originating from the first promoter (TA-promoter) is the main source of TAD-deficient pro-oncogenic p73 isoforms in HCC. The TA-promoter-driven and aberrantly spliced DN isoforms Δex2p73 and Δex2/3p73 that lack TAD, and ΔN’-p73 composed solely of TAD, are the most abundant p73 variants in HCC cells[88]. Both TAp73 and ΔTA variants including Δex2/3p73 are upregulated in hepatitis B virus-associated HCC[90]. ΔN-p73, an internal promoter-derived variant devoid of TAD, is detected both in hepatocytes and HCC and is overexpressed in 37% of tumors[87,89].
Normally, the induction of TAp73 expression inhibits cell proliferation; however, hepatoma cells tolerate its effects, presumably due to the co-expression of ΔTA variants[87]. The liver-specific expression of ΔNp73 in transgenic mice causes the formation of hepatic adenomas and subsequently to development of HCC in most animals[91]. Overexpression of the DN ΔN-p73 variant is associated with poor survival in HCC patients[89].
p63 and p73 initiate programmed cell death via a common mechanism. DNA damage induces TAp63 or TAp73-dependent TA of genes encoding the death receptors CD95, TNF-R and TRAIL-R, caspases and pro-apoptotic Bcl-2 proteins[91]. The expression of DN p63 and p73 variants abolishes the induction of pro-apoptotic genes and may cause resistance to chemotherapy[89,92]. Thus, since pro-oncogenic p53-family TF isoforms are generally expressed at low levels or are absent in non-transformed hepatocytes, targeting these variants may provide an option to counteract their DN effects on pro-apoptotic p53, p63 and p73 isoforms to improve the efficiency of drug therapy in HCC patients.
TCF-4 (TCF7L2) is a basic helix-loop-helix (bHLH) TF of the LEF/TCF family that modulates the expression of Wnt signaling pathway target genes involved in proliferation, differentiation, apoptosis, cell polarity and motility upon β-catenin binding[93]. The conserved domain structure of TCF-4 is characteristic of the LEF/TCF family; it comprises the N-terminal β-catenin-binding domain (BCBD), followed by the context-dependent regulatory domain, high-mobility group DBD, and E-tail, which includes the second sequence-specific DBD, C-clamp and binding region for the transcriptional repressor CtBP[94].
The context-dependent regulatory domain harbors several regulatory motifs: LVPQ and SxxSS, implicated in the repression of β-catenin-mediated TA, and the Groucho/TLE co-repressor binding site. The C-clamp contains a 30 AA motif that is required for binding to weak Wnt response elements[93,95]. Approximately 20 TCF-4 isoforms generated by alternative splicing and alternative transcription start site usage have been reported[96]. Variants transcribed from the internal transcription start sites 2 and 3 lack BCBD and act as transcriptional repressors of β-catenin target genes[97]. Additionally, alternatively spliced isoforms differ by the presence of exon 4, which confers transcription repressor function, LPQV- and SxxSS motifs, and by the splicing pattern of exons 13-17 encoding a part of the C-clamp[98,99].
TCF-4 is highly expressed in hepatocytes. The pattern of expression of TCF-4 variants depends on the degree of cell differentiation[98]. The isoform TCF-4B that is predominant in normal liver lacks exon 4, the SxxSS, C-clamp and CtBP-binding region, which makes it a potent β-catenin-mediated trans-activator[98]. TCF-4B expression is maintained in HCCs and in differentiated hepatoma cell lines[98,100].
LPQV and SxxSS motif-containing variants expressed in normal liver are not substantially altered in liver tumors[100], except for a TCF-4K isoform. TCF-4K retains a portion of the C-clamp and the full CtBP-binding site[98]; it is significantly downregulated in HCC[100]. Conversely, TCF-4C and -4J are upregulated in HCC, specifically in poorly differentiated tumors. Along with TCF-4B, TCF-4C demonstrates the highest transcriptional activity among the TCF-4 isoforms due to the lack of any repressor motifs and domains. TCF-4J, that is deficiency in most repressor units, preserves the LVPQ motif and CtBP-binding region, which results in a reduced TA ability[98,99,101].
The exogenous expression of TCF-4J in hepatoma cell cultures induces the expression of genes that are overexpressed in poorly differentiated HCCs and are distinct from SxxSS-containing TCF-4K activated targets. Additionally, TCF-4J has been demonstrated to interact with the liver-specific master regulator HNF4α, affecting its target gene expression[96]. Thus, hepatic differentiation may be affected by the balance of certain TCF-4 isoforms[102]. Alternatively, it was proposed that HNF4α competes with TCF-4 for its binding sites in vitro in human colorectal cancer cells in an isoform-dependent manner, where the HNF4α2, but not the HNF4α8, P2-driven variant, might displace TCF-4 in the AP-1 transcriptional complex[103]. Although there are no data on their interplay in the liver, this possible interaction may also be important for the indirect regulation of gene expression mediated by the Wnt signaling pathway and relevant for hepatocarcinogenesis.
The exogenous expression of SxxSS-deficient TCF-4B, -4C and -4J isoforms in hepatoma cell lines results in the upregulation of Wnt-responsive genes, thus promoting cell proliferation. TCF-4C and -4J overexpression also induces colony formation and promotes cell migration in hepatoma cell lines. In contrast, a SxxSS-containing TCF-4K variant reduces the proliferation rate and colony formation but stimulates cell migration[98,99,101,102].
TCF-4J-expressing hepatoma cells demonstrate higher tumorigenicity than TCF-4K-overexpressing ones. Tumors derived from TCF-4J-overexpressing cells also express high levels of HIF-2a and EGFR, which confer hypoxia resistance characteristic of an aggressive tumor phenotype[100]. Overall, these data indicate that the imbalance of TCF-4 isoforms observed in HCC contributes to multiple processes including dedifferentiation, increased proliferation, survival and metastatic potential of tumor cells and thus confers an aggressive tumor phenotype.
The tumor suppressor KLF6 belongs to the C2H2 zinc finger domain Sp1/KLF family of TFs, which are essential regulators of proliferation, differentiation and migration. Biological functions of KLFs are disrupted in a wide variety of tumors[104]. In HCC, KLF6 is frequently inactivated due to the loss of heterozygosity or point mutations[105].
The KLF6 molecule contains N-terminal TAD region, an NLS and three zinc fingers located in the C-terminus that allow DNA binding. Several KLF6 isoforms are produced in vivo due to the activation of cryptic splice sites[104]. In the liver, the full-length wtKLF6 isoform is prevalent, while truncated SV1 and SV2 variants are also expressed[106]. In contrast to wtKLF6, which is accumulated in the nucleus, SV1 and SV2 proteins lacking the NLS are mainly localized in the cytoplasm[107]. The SV1 variant is also defective in DNA-binding due to the absence of all zinc fingers[104]. The downregulation of wtKLF6 in dysplastic nodules is an early event in hepatocarcinogenesis, while its further decrease is observed in highly malignant HCCs and is associated with a poor prognosis[108,109]. The downregulation of wtKLF6 in HCC is often accompanied by a decrease in SV2 production and upregulation of the SV1 isoform, while higher SV1/wtKLF6 ratios correlate with an advanced tumor stage[106,108,110].
Several mechanisms regulating the balance of SV1 and wtKLF6 in HCC have been proposed. HGF- or Ras-dependent activation of the PI3K/AKT signaling pathway induces alterations of KLF6 splicing, which leads to enhanced generation of the SV1 variant[111,112]. In addition, miRNA-1301, which is abundant in HCC samples, and miRNA-210 have been found to specifically target wtKLF6 but not the SV1 variant[113].
The SV1 isoform counteracts wtKLF6 and SV2 and demonstrates obvious oncogenic properties. It has been proposed to carry out DN functions via the cytoplasmic sequestration of wtKLF6, which leads to its subsequent proteasomal degradation[110]. Both wtKLF6 depletion and SV1 overexpression significantly accelerate tumorigenesis in diethylnitrosamine-treated mice[110]. In human hepatoma cell lines, wtKLF6 depletion and SV1 exogenous expression enhance proliferation via the downregulation of CDKN1A and CCNB1 target genes[110-112], whereas SV2 expression has an opposite effect[106].
KLF6 variants are implicated in the regulation of apoptosis. The exogenously expressed wtKLF6 induces transcriptional repression of the MDM2 gene, thus securing the stabilization of the p53 level[109]. SV2 overexpression results in p53-dependent upregulation of the apoptosis inducers Bax and PUMA[106].
SV1 promotes the migration of hepatoma cell lines; it is proposed to favor metastasis via inhibition of the function of wtKLF6 implicated in the regulation of Rho family GTPase activity[114]. Overexpression of SV1 also upregulates the expression of mesenchymal marker genes, whereas wtKLF6 is essential for the maintenance of E-cadherin expression but does not affect other EMT-associated markers[113,114]. Thus, since the DN SV1 isoform of the KLF6 tumor suppressor performs obvious pro-oncogenic functions and is upregulated in HCC, it can be considered as a prognostic factor and candidate target for siRNA-mediated therapeutic inactivation in liver tumors. Alternatively, targeting miRNAs implicated in wtKLF6 downregulation may also result in the restoration of the KLF6 isoform balance to reduce the pro-oncogenic effects of the SV1 TF variant.
WT1, a TF of the C2H2-type zinc-finger protein family, is involved in the regulation of cell differentiation, proliferation and apoptosis. Depending on the tissue-specific context, WT1 exhibits either tumor-suppressive or oncogenic properties[115], but in most solid tumors, including HCC, the upregulation of WT1 is associated with a poor prognosis[116,117]. The N-terminal proline-rich region of WT1 contains a homodimerization domain and trans-repression and trans-activation domains that are responsible for co-factor binding. The C-terminus comprises DBD with 4 C2H2-type Zn-fingers[115].
Although up to 36 alternative isoforms of WT1 produced by a combination of alternative start codon usage, alternative splicing and RNA editing have been predicted, 4 major isoforms that vary in exon 5 (17-AA insertion - “17AA”) and/or exon 9 (3 AA insertion - “KTS”) splicing have mostly been investigated[115,118]. The 17AA insertion located between the proline-rich region and the first zinc finger and the KTS triplet between the third and fourth Zn-fingers have been demonstrated to change the DBD conformation and diminish WT1 DNA-binding[115]. Therefore, the WT1 isoforms harboring these insertions have been suggested to be less transcriptionally active[119]. While WT1 expression in the normal liver is very low, in the cirrhotic liver and HCC tissue, all 4 major WT1 variants are upregulated[116,120,121].
The functional differences in the KTS-variable WT1 isoforms have been investigated in HCC model systems. In HepG2 and Hep3B hepatomas, exogenously expressed WT1 KTS(-) isoforms, but not KTS(+), act as tumor suppressors, triggering a p53-independent apoptotic program[119]. Conversely, in Huh7 and HLE cells, the major 17AA(+) KTS(+) isoform inhibits apoptosis via transcriptional activation of the cFLIP apoptotic inhibitor and repression of FADD, a caspase cascade activator[122]. These data imply that cell fate decisions may be dependent on the ratio of KTS(+)/KTS(-) isoforms in HCC cells.
The WT1 knockdown in PLC/PRF/5 HCC cells stimulates differentiation through upregulation of the key hepato-specific regulator HNF4α and leads to the loss of resistance to apoptosis[121]. In contrast, ectopic equimolar expression of 4 major WT1 variants in a primary rat hepatocytes culture induces a decrease in HNF4α expression and dedifferentiation[120]. Overall, the existing ambiguity regarding the impact of WT1 on different tumors could be explained not only by the tissue-specific context but also by a different spectrum of expressed WT1 variants. Importantly, the expression of minor WT1 isoforms, in addition to the major ones, and their impact on HCC development, progression and prognosis has not been thoroughly investigated. Thus, further investigation of WT1 isoform properties and the development of approaches for WT1 silencing or shifting the balance towards DN variants that counteract its pro-oncogenic and apoptosis resistance functions may benefit HCC prognosis and treatment. Apart from gene silencing, enforced expression of DN WT1 variants may be considered a therapeutic option for HCC to oppose the dedifferentiation and acquisition of resistance to apoptosis conferred by extrinsically expressed WT1.
bHLH-PAS protein HIF1α promotes tumor progression through the regulation of the adaptive response to hypoxia[123]. HIF1α has been proven to be the most potent inducer of the expression of vascular endothelial growth factors and other hypoxia-responsible element-containing genes in various cell type-dependent contexts[124-126]. HIF1α is not detected in hepatocytes under normoxia; its expression is triggered by low oxygen levels in the murine liver[127]. HIF1α expression is induced in dysplastic liver nodules during malignization and is further increased in HCC[128], where its upregulation correlates with vascular invasion and a poor prognosis[129]. In Hep3B hepatoma cells, HIF1α modulates the expression and alternative splicing of its target genes to facilitate tumor cell metabolism adaptation to hypoxia[130].
The amino-terminal region of HIF1α harbors a bHLH DBD followed by the PAS domain, which enables its heterodimerization with HIF1β, a component of the TA-competent HIF1 complex, and by the oxygen-dependent degradation domain (ODDD). ODDD contains proline residues that are modified under normoxia and potentiate binding to VHL ubiquitin ligase to provide ubiquitin-dependent degradation of the TF. The C-terminal part of HIF1α contains 2 TADs and the NLS region[125,131].
HIF1α transcription is regulated by the universal I.1 promoter that drives expression of the major HIF1α1.1 isoform or by two tissue-specific promoters, I.2 and I.3, which give rise to minor HIF1α1.2 and HIF1α1.3 variants[132]. Alternative splicing of the HIF1α1.1 transcript leads to frameshifts and generates isoforms with impaired activity. DN cytoplasm-localized HIF1α516 and HIF1α557 variants are devoid of the NLS and both TADs as a result of the exclusion of exons 11/12; thus, they escape hypoxia-induced nuclear translocation and lack TA properties. The HIF1α736 variant lacks the C-terminal TAD due to the exclusion of exon 14 and demonstrates weaker TA properties compared with HIF1α1.1[123,133,134].
HIF1α1.1 is a predominant isoform generated under hypoxic conditions in hepatoma cell lines. The expression of minor fractions of HIF1α516 and HIF1α736 that inhibit the activity of HIF1α1.1 through competitive binding to HIF1β and thereby reduce the expression of HIF1 target genes has also been identified in hepatoma cells[123,134]. Although the biological role of these variants in hepatocarcinogenesis is underexplored and no survival analysis addressing their impact has been carried out to date, further investigation of DN HIF1α variants is required to define the possible utility of distinct isoforms in prognosis and therapy.
ZIP (ZGPAT) is a DNA-binding transcriptional repressor that contains a C3H1-type zinc finger, TUDOR, G-patch, coiled-coil domains. ZIP-mediated transcriptional repression of target genes is achieved through the recruitment of the nucleosome remodeling and deacetylase (NuRD) complex[135]. Dimerization of ZIP is essential for its DNA binding ability[136]. ZIP target genes encode growth factors and their receptors, cell cycle regulators, components of the MAPK signaling pathway, actin cytoskeleton, and tight and gap junctions, particularly EGFR, PTEN, CDC25A, FGF5, FGF14, PDGFB, RGS3, and VCAM1[135].
Two ZIP isoforms have been identified. The full-length one acts as a transcriptional repressor, whereas a truncated sZIP variant deficient in DBD but capable of competitive binding with NuRD and, presumably, of dimerizing with ZIP decreases the inhibitory effect of ZIP[136,137]. Unlike most cell types, hepatocytes express both truncated and full-length ZIP variants[135]. Induced ZIP overexpression in HepG2 cells result in EGFR transcriptional downregulation, growth inhibition and a reduced clonogenic potential. In contrast, sZIP overexpression or ZIP/sZIP co-expression induce clearly opposite effects, indicating that the shortened isoform can counteract the tumor-suppressing activity of ZIP[137].
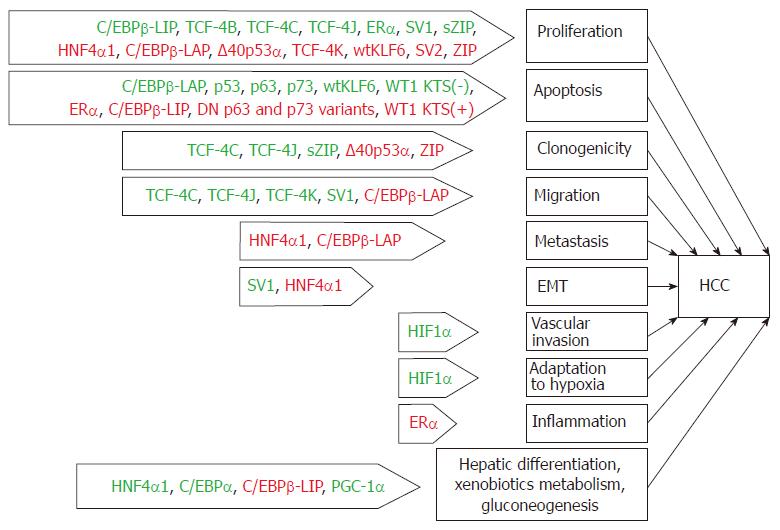
Aberrant alternative splicing has recently been proposed to be an additional hallmark of cancer[138]. We believe it reasonable to consider this hallmark in a broad sense, i.e., as the generation of transcripts and protein variants that are not characteristic of a particular cell type. Such an interpretation stems from the observation that the mechanism of isoform generation is not limited to alternative splicing and can be based on additional mechanisms that are listed above or, frequently, on their combination. The production of aberrant isoforms is certainly not only a hallmark of cancer but also the cause of the development of other distinguishing characteristics of tumor cells. This statement is particularly relevant in regard to TF isoforms since TFs act as hubs that convert various incoming signals to a wide variety of target genes to modulate their expression, and the processes presumably regulated by these atypical isoforms might also be relevant for hepatocarcinogenesis (Figure 2).
The existence of TF isoform variants enables their functional diversification for the tight tissue- and condition-specific regulation of target genes. In contrast, aberrant production of TF variants with different TA properties can result in a significant alteration of the transcriptional program of the cell and, eventually, in its malignization. For instance, the overproduction of DN isoforms of TFs that are able to dimerize with functionally active variants of the corresponding TFs or compete for co-factors and/or binding to target gene promoters causes a reduction or inhibition of the functional activity of such TFs. The abnormal isoform production may result in mRNA degradation or structural changes that cause cytoplasmic localization, constitutive activation of TFs or enhanced TA ability and other alterations that eventually affect the expression of target genes.
The rapid development of transcriptome sequencing and proteomic technologies facilitates the detection, quantitative assessment and acquisition of statistically significant data on alterations of TF isoform ratios. Recently, global profiling of alternative splicing in hepatitis-associated and virus-free HCCs using a TCGA-LIHC dataset revealed deep perturbations in RNA splicing concomitant with altered expression levels and isoform patterns of splicing factors. Among the spectrum of alternatively spliced transcripts, those that are common for HCC or those that change depending on hepatitis infection status have been identified[24]. Notably, 7.6% to 9.0% of differentially spliced transcripts in HCCs originate from TF-encoding genes, including those involved in the regulation of the specification, differentiation or malignant transformation of hepatocytes (HNF1B, FOXM1, PPARA, NR1I3/CAR, NR3C1/GR)[24]. However, very few of the identified TFs have been previously investigated with regard to the biological effects or clinical implications of their isoforms. The available data on TF isoforms that are produced in the liver and HCC are summarized in Table 1.
The application of methods that allow the differential detection of TF isoforms appears to be crucial for the identification of clinically relevant alterations. Data based on the evaluation of the total level of gene expression or protein synthesis may be ambiguous, as such approaches mask the contribution of individual TF isoforms possessing distinct transcriptional activities to the overall pool of the corresponding TF. For example, several oncogenic TCF4 isoforms implicated in hepatocarcinogenesis are substantially upregulated in HCC, whereas the expression of other variants is usually decreased or unaltered.
Additionally, the ratio and functional impact of TF isoforms may significantly depend on the tissue-specific context. In the liver, HNF4a isoforms that are translated from mRNA transcribed from the P1 promoter are predominant and clearly demonstrate tumor-suppressive properties[37]. In the colon epithelium, HNF4A isoforms transcribed from both promoters are normally expressed. Unlike HCC cells, non-isoform-specific HNF4A knockdown in colorectal carcinoma cell lines inhibits proliferation[147], while HNF4αP1 downregulation in vivo is associated with a higher metastatic potential in CRC, raising the possibility that this isoform group nevertheless implements a tumor suppressive function[148]. Thus, the isoform-discerning approach may resolve the existing contradictions in understanding the roles of certain TFs that are believed to exert opposing effects in different types of tumors.
Overall, knowledge of the effects of particular isoforms, their tissue-specific expression profiles and shifts in their ratios in disease may be of practical value for HCC prognosis and outlining potential therapeutic targets. Unfortunately, the available data on TF isoforms are mostly based on in vitro studies, the results of which are not to be directly extrapolated since gene expression, splicing and the effects of those variants may differ in vivo[149,150]. Few of the isoforms discussed above have been investigated in terms of their prognostic significance in HCC, although their aberrant production is associated with clinical features and outcomes in other types of cancer. To date, only the overproduction of HNF4αP2, ΔN-p73 and the downregulation of the TAp63 and ERα-66 isoforms have been reported to be significantly associated with the poor survival in HCC patients. A comprehensive understanding of the functions and regulation of particular isoforms may be further applied to the development of new therapeutic approaches. For instance, the upregulation of the anti-apoptotic Bcl-x(L) isoform of Bcl-2-like protein 1 is frequently observed in HCC. Its knockdown in hepatoma cell lines through RNA-interference has been demonstrated to induce apoptosis after staurosporine treatment in originally resistant cells[151]. A similar approach may be applied to inhibit isoforms that exert definite oncogenic effects, such as SxxSS-deficient TCF4 variants, which are overexpressed in HCC and possess high transcriptional activity due to the absence of co-repressor binding or posttranslational modifications that normally decrease TCF4 activity.
Identification and targeting of the regulators that control the production of particular TF variants would also be beneficial. For instance, tumor-suppressive HNF4α and KLF6 isoforms are frequently downregulated in HCC and can induce at least partial reversion of the malignant phenotype. In contrast, c-MET- and EGFR-mediated activation of signaling pathways that are essential for hepatocarcinogenesis has been demonstrated to drastically alter the expression of multiple splicing factors and induce the production of DN KLF6 and p73 isoforms[14]. Thus, targeted inhibition of the indicated signaling pathways may be considered as an additional strategy to prevent aberrant isoform synthesis. Since the expression of genes encoding splicing machinery components is altered in HCC, their inhibition may be considered as an additional way to reduce aberrant splicing. Several small molecule splicing modulators targeting basal splicing machinery or splicing factors and their regulators have been identified. Such modulators effectively inhibit tumor growth in vivo, but they lack specificity; in addition, they have been demonstrated to be toxic in xenograft experiments and clinical trials[152]. Alternatively, whereas some components of the splicing regulatory network are recurrently mutated in HCC[23], targeting these mutant proteins with specific small molecules or antibodies may be relevant. Thus, further expansion of knowledge on the functions of TF variants, mechanisms of their generation and switching in HCC should shed light on additional mechanisms of hepatocarcinogenesis. Hopefully, it will also facilitate the discovery of new clinically relevant prognostic markers and therapeutic targets and contribute to the development of novel approaches to cancer treatment.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Russia
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Luo GH, Roohvand F, Sukocheva OA S- Editor: Ji FF L- Editor: A E- Editor: Bian YN
| 1. | Ezkurdia I, Juan D, Rodriguez JM, Frankish A, Diekhans M, Harrow J, Vazquez J, Valencia A, Tress ML. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum Mol Genet. 2014;23:5866-5878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 384] [Cited by in F6Publishing: 320] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 2. | Paik YK, Overall CM, Deutsch EW, Hancock WS, Omenn GS. Progress in the Chromosome-Centric Human Proteome Project as Highlighted in the Annual Special Issue IV. J Proteome Res. 2016;15:3945-3950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3539] [Cited by in F6Publishing: 3653] [Article Influence: 228.3] [Reference Citation Analysis (0)] |
| 4. | Bazykin GA, Kochetov AV. Alternative translation start sites are conserved in eukaryotic genomes. Nucleic Acids Res. 2011;39:567-577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 5. | Jacox E, Gotea V, Ovcharenko I, Elnitski L. Tissue-specific and ubiquitous expression patterns from alternative promoters of human genes. PLoS One. 2010;5:e12274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823-4832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 132] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 7. | Xin D, Hu L, Kong X. Alternative promoters influence alternative splicing at the genomic level. PLoS One. 2008;3:e2377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Gonzàlez-Porta M, Frankish A, Rung J, Harrow J, Brazma A. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14:R70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 183] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 9. | Peng L, Bian XW, Li DK, Xu C, Wang GM, Xia QY, Xiong Q. Large-scale RNA-Seq Transcriptome Analysis of 4043 Cancers and 548 Normal Tissue Controls across 12 TCGA Cancer Types. Sci Rep. 2015;5:13413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 82] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 10. | Miura K, Fujibuchi W, Sasaki I. Alternative pre-mRNA splicing in digestive tract malignancy. Cancer Sci. 2011;102:309-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 11. | Shkreta L, Bell B, Revil T, Venables JP, Prinos P, Elela SA, Chabot B. Cancer-Associated Perturbations in Alternative Pre-messenger RNA Splicing. Cancer Treat Res. 2013;158:41-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 238] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 13. | Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 850] [Cited by in F6Publishing: 886] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 14. | Berasain C, Elizalde M, Urtasun R, Castillo J, García-Irigoyen O, Uriarte I, Latasa MU, Prieto J, Avila MA. Alterations in the expression and activity of pre-mRNA splicing factors in hepatocarcinogenesis. Hepat Oncol. 2014;1:241-252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 15. | Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H. Translation from unconventional 5’ start sites drives tumour initiation. Nature. 2017;541:494-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 225] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 16. | Lazarevich NL, Fleishman DI. Tissue-specific transcription factors in progression of epithelial tumors. Biochemistry (Mosc). 2008;73:573-591. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 17. | Bonomi S, Gallo S, Catillo M, Pignataro D, Biamonti G, Ghigna C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol. 2013;2013:962038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 18. | Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34:1-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 19. | Talavera D, Orozco M, de la Cruz X. Alternative splicing of transcription factors’ genes: beyond the increase of proteome diversity. Comp Funct Genomics. 2009;905894. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, Gores G. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1401] [Cited by in F6Publishing: 1630] [Article Influence: 203.8] [Reference Citation Analysis (2)] |
| 21. | Berasain C, Goñi S, Castillo J, Latasa MU, Prieto J, Avila MA. Impairment of pre-mRNA splicing in liver disease: mechanisms and consequences. World J Gastroenterol. 2010;16:3091-3102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 32] [Cited by in F6Publishing: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Liu L, Xie S, Zhang C, Zhu F. Aberrant regulation of alternative pre-mRNA splicing in hepatocellular carcinoma. Crit Rev Eukaryot Gene Expr. 2014;24:133-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 23. | Cancer Genome Atlas Research Network. Electronic address: wheeler@bcm.edu; Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1175] [Cited by in F6Publishing: 1608] [Article Influence: 229.7] [Reference Citation Analysis (1)] |
| 24. | Tremblay MP, Armero VE, Allaire A, Boudreault S, Martenon-Brodeur C, Durand M, Lapointe E, Thibault P, Tremblay-Létourneau M, Perreault JP. Global profiling of alternative RNA splicing events provides insights into molecular differences between various types of hepatocellular carcinoma. BMC Genomics. 2016;17:683. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 25. | Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378-1381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1027] [Cited by in F6Publishing: 998] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 26. | Lazarevich NL, Al’pern DV. Hepatocyte nuclear factor 4 (HNF4) in epithelial development and carcinogenesis. Mol Biol (Mosk). 2008;42:786-797. [PubMed] [Cited in This Article: ] |
| 27. | Cicchini C, Amicone L, Alonzi T, Marchetti A, Mancone C, Tripodi M. Molecular mechanisms controlling the phenotype and the EMT/MET dynamics of hepatocyte. Liver Int. 2015;35:302-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Bonzo JA, Ferry CH, Matsubara T, Kim JH, Gonzalez FJ. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem. 2012;287:7345-7356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 145] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 29. | Yusuf D, Butland SL, Swanson MI, Bolotin E, Ticoll A, Cheung WA, Zhang XY, Dickman CT, Fulton DL, Lim JS. The transcription factor encyclopedia. Genome Biol. 2012;13:R24. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 30. | Green VJ, Kokkotou E, Ladias JA. Critical structural elements and multitarget protein interactions of the transcriptional activator AF-1 of hepatocyte nuclear factor 4. J Biol Chem. 1998;273:29950-29957. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 31. | Sladek FM, Ruse MD Jr, Nepomuceno L, Huang SM, Stallcup MR. Modulation of transcriptional activation and coactivator interaction by a splicing variation in the F domain of nuclear receptor hepatocyte nuclear factor 4alpha1. Mol Cell Biol. 1999;19:6509-6522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 103] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 32. | Huang J, Karakucuk V, Levitsky LL, Rhoads DB. Expression of HNF4alpha variants in pancreatic islets and Ins-1 beta cells. Diabetes Metab Res Rev. 2008;24:533-543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Huang J, Levitsky LL, Rhoads DB. Novel P2 promoter-derived HNF4alpha isoforms with different N-terminus generated by alternate exon insertion. Exp Cell Res. 2009;315:1200-1211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Tanaka T, Jiang S, Hotta H, Takano K, Iwanari H, Sumi K, Daigo K, Ohashi R, Sugai M, Ikegame C. Dysregulated expression of P1 and P2 promoter-driven hepatocyte nuclear factor-4alpha in the pathogenesis of human cancer. J Pathol. 2006;208:662-672. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 137] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 35. | Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 2006;20:2293-2305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 213] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 36. | Cai SH, Lu SX, Liu LL, Zhang CZ, Yun JP. Increased expression of hepatocyte nuclear factor 4 alpha transcribed by promoter 2 indicates a poor prognosis in hepatocellular carcinoma. Therap Adv Gastroenterol. 2017;10:761-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 37. | Lazarevich NL, Cheremnova OA, Varga EV, Ovchinnikov DA, Kudrjavtseva EI, Morozova OV, Fleishman DI, Engelhardt NV, Duncan SA. Progression of HCC in mice is associated with a downregulation in the expression of hepatocyte nuclear factors. Hepatology. 2004;39:1038-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 175] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 38. | Lazarevich NL, Shavochkina DA, Fleishman DI, Kustova IF, Morozova OV, Chuchuev ES, Patyutko YI. Deregulation of hepatocyte nuclear factor 4 (HNF4)as a marker of epithelial tumors progression. Exp Oncol. 2010;32:167-171. [PubMed] [Cited in This Article: ] |
| 39. | Späth GF, Weiss MC. Hepatocyte nuclear factor 4 provokes expression of epithelial marker genes, acting as a morphogen in dedifferentiated hepatoma cells. J Cell Biol. 1998;140:935-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 121] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 40. | Yin C, Lin Y, Zhang X, Chen YX, Zeng X, Yue HY, Hou JL, Deng X, Zhang JP, Han ZG. Differentiation therapy of hepatocellular carcinoma in mice with recombinant adenovirus carrying hepatocyte nuclear factor-4alpha gene. Hepatology. 2008;48:1528-1539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 41. | Shi L, Feng Y, Lin H, Ma R, Cai X. Role of estrogen in hepatocellular carcinoma: is inflammation the key? J Transl Med. 2014;12:93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 42. | Barone I, Brusco L, Fuqua SA. Estrogen receptor mutations and changes in downstream gene expression and signaling. Clin Cancer Res. 2010;16:2702-2708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 43. | Han J, Ding L, Yuan B, Yang X, Wang X, Li J, Lu Q, Huang C, Ye Q. Hepatitis B virus X protein and the estrogen receptor variant lacking exon 5 inhibit estrogen receptor signaling in hepatoma cells. Nucleic Acids Res. 2006;34:3095-3106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Miceli V, Cocciadiferro L, Fregapane M, Zarcone M, Montalto G, Polito LM, Agostara B, Granata OM, Carruba G. Expression of wild-type and variant estrogen receptor alpha in liver carcinogenesis and tumor progression. OMICS. 2011;15:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 45. | Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc Natl Acad Sci USA. 2006;103:9063-9068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 305] [Cited by in F6Publishing: 298] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 46. | Fuqua SA, Fitzgerald SD, Chamness GC, Tandon AK, McDonnell DP, Nawaz Z, O’Malley BW, McGuire WL. Variant human breast tumor estrogen receptor with constitutive transcriptional activity. Cancer Res. 1991;51:105-109. [PubMed] [Cited in This Article: ] |
| 47. | Zhang J, Ren J, Wei J, Chong CC, Yang D, He Y, Chen GG, Lai PB. Alternative splicing of estrogen receptor alpha in hepatocellular carcinoma. BMC Cancer. 2016;16:926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 48. | Hishida M, Nomoto S, Inokawa Y, Hayashi M, Kanda M, Okamura Y, Nishikawa Y, Tanaka C, Kobayashi D, Yamada S. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Oncol. 2013;43:88-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 49. | Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 997] [Cited by in F6Publishing: 1048] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 50. | Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1422] [Cited by in F6Publishing: 1409] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 51. | Wang B, Hsu SH, Frankel W, Ghoshal K, Jacob ST. Stat3-mediated activation of microRNA-23a suppresses gluconeogenesis in hepatocellular carcinoma by down-regulating glucose-6-phosphatase and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha. Hepatology. 2012;56:186-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 52. | Felder TK, Soyal SM, Oberkofler H, Hahne P, Auer S, Weiss R, Gadermaier G, Miller K, Krempler F, Esterbauer H. Characterization of novel peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) isoform in human liver. J Biol Chem. 2011;286:42923-42936. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 53. | Martínez-Redondo V, Pettersson AT, Ruas JL. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia. 2015;58:1969-1977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 134] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 54. | Zhang Y, Huypens P, Adamson AW, Chang JS, Henagan TM, Boudreau A, Lenard NR, Burk D, Klein J, Perwitz N. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J Biol Chem. 2009;284:32813-32826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 55. | Chang JS, Jun HJ, Park M. Transcriptional coactivator NT-PGC-1α promotes gluconeogenic gene expression and enhances hepatic gluconeogenesis. Physiol Rep. 2016;4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Yao W, Cai H, Li X, Li T, Hu L, Peng T. Endoplasmic reticulum stress links hepatitis C virus RNA replication to wild-type PGC-1α/liver-specific PGC-1α upregulation. J Virol. 2014;88:8361-8374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 57. | Hu C, Liu D, Zhang Y, Lou G, Huang G, Chen B, Shen X, Gao M, Gong W, Zhou P. LXRα-mediated downregulation of FOXM1 suppresses the proliferation of hepatocellular carcinoma cells. Oncogene. 2014;33:2888-2897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 58. | Bakiri L, Hamacher R, Graña O, Guío-Carrión A, Campos-Olivas R, Martinez L, Dienes HP, Thomsen MK, Hasenfuss SC, Wagner EF. Liver carcinogenesis by FOS-dependent inflammation and cholesterol dysregulation. J Exp Med. 2017;214:1387-1409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 72] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 59. | Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561-575. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1019] [Cited by in F6Publishing: 1043] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 60. | Takiguchi M. The C/EBP family of transcription factors in the liver and other organs. Int J Exp Pathol. 1998;79:369-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 86] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 61. | Rana B, Xie Y, Mischoulon D, Bucher NL, Farmer SR. The DNA binding activity of C/EBP transcription factor is regulated in the G1 phase of the hepatocyte cell cycle. J Biol Chem. 1995;270:18123-18132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 128] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 62. | Lourenço AR, Coffer PJ. A tumor suppressor role for C/EBPα in solid tumors: more than fat and blood. Oncogene. 2017;36:5221-5230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 63. | Lu GD, Leung CH, Yan B, Tan CM, Low SY, Aung MO, Salto-Tellez M, Lim SG, Hooi SC. C/EBPalpha is up-regulated in a subset of hepatocellular carcinomas and plays a role in cell growth and proliferation. Gastroenterology. 2010;139:632-643, 643.e1-643.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 64. | Lu GD, Ang YH, Zhou J, Tamilarasi J, Yan B, Lim YC, Srivastava S, Salto-Tellez M, Hui KM, Shen HM. CCAAT/enhancer binding protein α predicts poorer prognosis and prevents energy starvation-induced cell death in hepatocellular carcinoma. Hepatology. 2015;61:965-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 57] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 65. | Fang T, Cui M, Sun J, Ge C, Zhao F, Zhang L, Tian H, Zhang L, Chen T, Jiang G. Orosomucoid 2 inhibits tumor metastasis and is upregulated by CCAAT/enhancer binding protein β in hepatocellular carcinomas. Oncotarget. 2015;6:16106-16119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 66. | Wang C, Chen X, Wang Y, Gong J, Hu G. C/EBPalphap30 plays transcriptional regulatory roles distinct from C/EBPalphap42. Cell Res. 2007;17:374-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 67. | Reebye V, Huang KW, Lin V, Jarvis S, Cutilas P, Dorman S, Ciriello S, Andrikakou P, Voutila J, Saetrom P. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. Oncogene. 2018;37:3216-3228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 68. | Anand S, Ebner J, Warren CB, Raam MS, Piliang M, Billings SD, Maytin EV. C/EBP transcription factors in human squamous cell carcinoma: selective changes in expression of isoforms correlate with the neoplastic state. PLoS One. 2014;9:e112073. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 69. | Zahnow CA. CCAAT/enhancer-binding protein beta: its role in breast cancer and associations with receptor tyrosine kinases. Expert Rev Mol Med. 2009;11:e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 70. | Yang LH, Wang Y, Qiao S, Wang MJ, Chen F, Zi XY, Li JX, Zhang HB, Yu B, Hu YP. Liver-enriched activator protein 1 as an isoform of CCAAT/enhancer-binding protein beta suppresses stem cell features of hepatocellular carcinoma. Cancer Manag Res. 2018;10:873-885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 71. | Milde-Langosch K, Löning T, Bamberger AM. Expression of the CCAAT/enhancer-binding proteins C/EBPalpha, C/EBPbeta and C/EBPdelta in breast cancer: correlations with clinicopathologic parameters and cell-cycle regulatory proteins. Breast Cancer Res Treat. 2003;79:175-185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 72. | Wang H, Larris B, Peiris TH, Zhang L, Le Lay J, Gao Y, Greenbaum LE. C/EBPbeta activates E2F-regulated genes in vivo via recruitment of the coactivator CREB-binding protein/P300. J Biol Chem. 2007;282:24679-24688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 73. | Wang GL, Salisbury E, Shi X, Timchenko L, Medrano EE, Timchenko NA. HDAC1 promotes liver proliferation in young mice via interactions with C/EBPbeta. J Biol Chem. 2008;283:26179-26187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 74. | Luedde T, Duderstadt M, Streetz KL, Tacke F, Kubicka S, Manns MP, Trautwein C. C/EBP beta isoforms LIP and LAP modulate progression of the cell cycle in the regenerating mouse liver. Hepatology. 2004;40:356-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 75. | Buck M, Turler H, Chojkier M. LAP (NF-IL-6), a tissue-specific transcriptional activator, is an inhibitor of hepatoma cell proliferation. EMBO J. 1994;13:851-860. [PubMed] [Cited in This Article: ] |
| 76. | Saint-Auret G, Danan JL, Hiron M, Blache C, Sulpice E, Tendil S, Daveau M, Gidrol X, Salier JP. Characterization of the transcriptional signature of C/EBPbeta isoforms (LAP/LIP) in Hep3B cells: implication of LIP in pro-survival functions. J Hepatol. 2011;54:1185-1194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 77. | Rodríguez-Antona C, Bort R, Jover R, Tindberg N, Ingelman-Sundberg M, Gómez-Lechón MJ, Castell JV. Transcriptional regulation of human CYP3A4 basal expression by CCAAT enhancer-binding protein alpha and hepatocyte nuclear factor-3 gamma. Mol Pharmacol. 2003;63:1180-1189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 80] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 78. | Martínez-Jiménez CP, Gómez-Lechón MJ, Castell JV, Jover R. Transcriptional regulation of the human hepatic CYP3A4: identification of a new distal enhancer region responsive to CCAAT/enhancer-binding protein beta isoforms (liver activating protein and liver inhibitory protein). Mol Pharmacol. 2005;67:2088-2101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 79. | Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 2010;17:901-911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 80. | Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13:962-972. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 367] [Cited by in F6Publishing: 389] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 81. | Ferraiuolo M, Di Agostino S, Blandino G, Strano S. Oncogenic Intra-p53 Family Member Interactions in Human Cancers. Front Oncol. 2016;6:77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 82. | Natan E, Joerger AC. Structure and kinetic stability of the p63 tetramerization domain. J Mol Biol. 2012;415:503-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 83. | Ota A, Nakao H, Sawada Y, Karnan S, Wahiduzzaman M, Inoue T, Kobayashi Y, Yamamoto T, Ishii N, Ohashi T. Δ40p53α suppresses tumor cell proliferation and induces cellular senescence in hepatocellular carcinoma cells. J Cell Sci. 2017;130:614-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 84. | Petitjean A, Cavard C, Shi H, Tribollet V, Hainaut P, Caron de Fromentel C. The expression of TA and DeltaNp63 are regulated by different mechanisms in liver cells. Oncogene. 2005;24:512-519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 85. | Yao JY, Pao CC, Chen JK. Transcriptional activity of TAp63 promoter is regulated by c-jun. J Cell Physiol. 2010;225:898-904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 86. | Gu H, Fang Z, Cai X, Song R, Lin M, Ye J, Ding X, Ke Q, Chen H, Gong C. Highly expressed histone deacetylase 5 promotes the growth of hepatocellular carcinoma cells by inhibiting the TAp63-maspin pathway. Am J Cancer Res. 2018;8:462-475. [PubMed] [Cited in This Article: ] |
| 87. | Sayan AE, Sayan BS, Findikli N, Ozturk M. Acquired expression of transcriptionally active p73 in hepatocellular carcinoma cells. Oncogene. 2001;20:5111-5117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 51] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 88. | Stiewe T, Tuve S, Peter M, Tannapfel A, Elmaagacli AH, Pützer BM. Quantitative TP73 transcript analysis in hepatocellular carcinomas. Clin Cancer Res. 2004;10:626-633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 89. | Müller M, Schilling T, Sayan AE, Kairat A, Lorenz K, Schulze-Bergkamen H, Oren M, Koch A, Tannapfel A, Stremmel W. TAp73/Delta Np73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564-1577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 143] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 90. | Knoll S, Fürst K, Thomas S, Villanueva Baselga S, Stoll A, Schaefer S, Pützer BM. Dissection of cell context-dependent interactions between HBx and p53 family members in regulation of apoptosis: a role for HBV-induced HCC. Cell Cycle. 2011;10:3554-3565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 91. | Tannapfel A, John K, Mise N, Schmidt A, Buhlmann S, Ibrahim SM, Pützer BM. Autonomous growth and hepatocarcinogenesis in transgenic mice expressing the p53 family inhibitor DNp73. Carcinogenesis. 2008;29:211-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 92. | Schuster A, Schilling T, De Laurenzi V, Koch AF, Seitz S, Staib F, Teufel A, Thorgeirsson SS, Galle P, Melino G. ΔNp73β is oncogenic in hepatocellular carcinoma by blocking apoptosis signaling via death receptors and mitochondria. Cell Cycle. 2010;9:2629-2639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 93. | Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25:7492-7504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 310] [Cited by in F6Publishing: 317] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 94. | Cadigan KM, Waterman ML. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol. 2012;4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 443] [Cited by in F6Publishing: 494] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 95. | Atcha FA, Syed A, Wu B, Hoverter NP, Yokoyama NN, Ting JH, Munguia JE, Mangalam HJ, Marsh JL, Waterman ML. A unique DNA binding domain converts T-cell factors into strong Wnt effectors. Mol Cell Biol. 2007;27:8352-8363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 96. | Neve B, Le Bacquer O, Caron S, Huyvaert M, Leloire A, Poulain-Godefroy O, Lecoeur C, Pattou F, Staels B, Froguel P. Alternative human liver transcripts of TCF7L2 bind to the gluconeogenesis regulator HNF4α at the protein level. Diabetologia. 2014;57:785-796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 97. | Prokunina-Olsson L, Welch C, Hansson O, Adhikari N, Scott LJ, Usher N, Tong M, Sprau A, Swift A, Bonnycastle LL. Tissue-specific alternative splicing of TCF7L2. Hum Mol Genet. 2009;18:3795-3804. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 86] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 98. | Tsedensodnom O, Koga H, Rosenberg SA, Nambotin SB, Carroll JJ, Wands JR, Kim M. Identification of T-cell factor-4 isoforms that contribute to the malignant phenotype of hepatocellular carcinoma cells. Exp Cell Res. 2011;317:920-931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 99. | Tomimaru Y, Xu CQ, Nambotin SB, Yan T, Wands JR, Kim M. Loss of exon 4 in a human T-cell factor-4 isoform promotes hepatic tumourigenicity. Liver Int. 2013;33:1536-1548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 100. | Koga H, Tsedensodnom O, Tomimaru Y, Walker EJ, Lee HC, Kim KM, Yano H, Wands JR, Kim M. Loss of the SxxSS motif in a human T-cell factor-4 isoform confers hypoxia resistance to liver cancer: an oncogenic switch in Wnt signaling. PLoS One. 2012;7:e39981. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 101. | Tomimaru Y, Koga H, Shin TH, Xu CQ, Wands JR, Kim M. The SxxSS motif of T-cell factor-4 isoforms modulates Wnt/β-catenin signal activation in hepatocellular carcinoma cells. Cancer Lett. 2013;336:359-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 102. | Tomimaru Y, Koga H, Yano H, de la Monte S, Wands JR, Kim M. Upregulation of T-cell factor-4 isoform-responsive target genes in hepatocellular carcinoma. Liver Int. 2013;33:1100-1112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 103. | Vuong LM, Chellappa K, Dhahbi JM, Deans JR, Fang B, Bolotin E, Titova NV, Hoverter NP, Spindler SR, Waterman ML. Differential Effects of Hepatocyte Nuclear Factor 4α Isoforms on Tumor Growth and T-Cell Factor 4/AP-1 Interactions in Human Colorectal Cancer Cells. Mol Cell Biol. 2015;35:3471-3490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 104. | DiFeo A, Martignetti JA, Narla G. The role of KLF6 and its splice variants in cancer therapy. Drug Resist Updat. 2009;12:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 103] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 105. | Kremer-Tal S, Reeves HL, Narla G, Thung SN, Schwartz M, Difeo A, Katz A, Bruix J, Bioulac-Sage P, Martignetti JA. Frequent inactivation of the tumor suppressor Kruppel-like factor 6 (KLF6) in hepatocellular carcinoma. Hepatology. 2004;40:1047-1052. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 106. | Hanoun N, Bureau C, Diab T, Gayet O, Dusetti N, Selves J, Vinel JP, Buscail L, Cordelier P, Torrisani J. The SV2 variant of KLF6 is down-regulated in hepatocellular carcinoma and displays anti-proliferative and pro-apoptotic functions. J Hepatol. 2010;53:880-888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 107. | Narla G, Difeo A, Reeves HL, Schaid DJ, Hirshfeld J, Hod E, Katz A, Isaacs WB, Hebbring S, Komiya A. A germline DNA polymorphism enhances alternative splicing of the KLF6 tumor suppressor gene and is associated with increased prostate cancer risk. Cancer Res. 2005;65:1213-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 180] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 108. | Kremer-Tal S, Narla G, Chen Y, Hod E, DiFeo A, Yea S, Lee JS, Schwartz M, Thung SN, Fiel IM. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J Hepatol. 2007;46:645-654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 109. | Tarocchi M, Hannivoort R, Hoshida Y, Lee UE, Vetter D, Narla G, Villanueva A, Oren M, Llovet JM, Friedman SL. Carcinogen-induced hepatic tumors in KLF6+/- mice recapitulate aggressive human hepatocellular carcinoma associated with p53 pathway deregulation. Hepatology. 2011;54:522-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 110. | Vetter D, Cohen-Naftaly M, Villanueva A, Lee YA, Kocabayoglu P, Hannivoort R, Narla G, M Llovet J, Thung SN, Friedman SL. Enhanced hepatocarcinogenesis in mouse models and human hepatocellular carcinoma by coordinate KLF6 depletion and increased messenger RNA splicing. Hepatology. 2012;56:1361-1370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 111. | Yea S, Narla G, Zhao X, Garg R, Tal-Kremer S, Hod E, Villanueva A, Loke J, Tarocchi M, Akita K. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology. 2008;134:1521-1531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 112. | Muñoz Ú, Puche JE, Hannivoort R, Lang UE, Cohen-Naftaly M, Friedman SL. Hepatocyte growth factor enhances alternative splicing of the Kruppel-like factor 6 (KLF6) tumor suppressor to promote growth through SRSF1. Mol Cancer Res. 2012;10:1216-1227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 113. | Liang WC, Wang Y, Xiao LJ, Wang YB, Fu WM, Wang WM, Jiang HQ, Qi W, Wan DC, Zhang JF. Identification of miRNAs that specifically target tumor suppressive KLF6-FL rather than oncogenic KLF6-SV1 isoform. RNA Biol. 2014;11:845-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 114. | Ahronian LG, Zhu LJ, Chen YW, Chu HC, Klimstra DS, Lewis BC. A novel KLF6-Rho GTPase axis regulates hepatocellular carcinoma cell migration and dissemination. Oncogene. 2016;35:4653-4662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 115. | Hohenstein P, Hastie ND. The many facets of the Wilms’ tumour gene, WT1. Hum Mol Genet. 2006;15 Spec No 2:R196-R201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 292] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 116. | Sera T, Hiasa Y, Mashiba T, Tokumoto Y, Hirooka M, Konishi I, Matsuura B, Michitaka K, Udaka K, Onji M. Wilms’ tumour 1 gene expression is increased in hepatocellular carcinoma and associated with poor prognosis. Eur J Cancer. 2008;44:600-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 117. | Qi XW, Zhang F, Wu H, Liu JL, Zong BG, Xu C, Jiang J. Wilms’ tumor 1 (WT1) expression and prognosis in solid cancer patients: a systematic review and meta-analysis. Sci Rep. 2015;5:8924. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 118. | Tatsumi N, Hojo N, Sakamoto H, Inaba R, Moriguchi N, Matsuno K, Fukuda M, Matsumura A, Hayashi S, Morimoto S. Identification of a Novel C-Terminal Truncated WT1 Isoform with Antagonistic Effects against Major WT1 Isoforms. PLoS One. 2015;10:e0130578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 119. | Menke AL, Shvarts A, Riteco N, van Ham RC, van der Eb AJ, Jochemsen AG. Wilms’ tumor 1-KTS isoforms induce p53-independent apoptosis that can be partially rescued by expression of the epidermal growth factor receptor or the insulin receptor. Cancer Res. 1997;57:1353-1363. [PubMed] [Cited in This Article: ] |
| 120. | Berasain C, Herrero JI, García-Trevijano ER, Avila MA, Esteban JI, Mato JM, Prieto J. Expression of Wilms’ tumor suppressor in the liver with cirrhosis: relation to hepatocyte nuclear factor 4 and hepatocellular function. Hepatology. 2003;38:148-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 121. | Perugorria MJ, Castillo J, Latasa MU, Goñi S, Segura V, Sangro B, Prieto J, Avila MA, Berasain C. Wilms’ tumor 1 gene expression in hepatocellular carcinoma promotes cell dedifferentiation and resistance to chemotherapy. Cancer Res. 2009;69:1358-1367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 122. | Uesugi K, Hiasa Y, Tokumoto Y, Mashiba T, Koizumi Y, Hirooka M, Abe M, Matsuura B, Onji M. Wilms’ tumor 1 gene modulates Fas-related death signals and anti-apoptotic functions in hepatocellular carcinoma. J Gastroenterol. 2013;48:1069-1080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 123. | Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12:9-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1159] [Cited by in F6Publishing: 1283] [Article Influence: 98.7] [Reference Citation Analysis (0)] |
| 124. | Shinojima T, Oya M, Takayanagi A, Mizuno R, Shimizu N, Murai M. Renal cancer cells lacking hypoxia inducible factor (HIF)-1alpha expression maintain vascular endothelial growth factor expression through HIF-2alpha. Carcinogenesis. 2007;28:529-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 129] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 125. | Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130-6134. [PubMed] [Cited in This Article: ] |
| 126. | Warnecke C, Zaborowska Z, Kurreck J, Erdmann VA, Frei U, Wiesener M, Eckardt KU. Differentiating the functional role of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2alpha target gene in Hep3B and Kelly cells. FASEB J. 2004;18:1462-1464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 300] [Cited by in F6Publishing: 310] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 127. | Stroka DM, Burkhardt T, Desbaillets I, Wenger RH, Neil DA, Bauer C, Gassmann M, Candinas D. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J. 2001;15:2445-2453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 542] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 128. | Nakamura K, Zen Y, Sato Y, Kozaka K, Matsui O, Harada K, Nakanuma Y. Vascular endothelial growth factor, its receptor Flk-1, and hypoxia inducible factor-1alpha are involved in malignant transformation in dysplastic nodules of the liver. Hum Pathol. 2007;38:1532-1546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 129. | Cao S, Yang S, Wu C, Wang Y, Jiang J, Lu Z. Protein expression of hypoxia-inducible factor-1 alpha and hepatocellular carcinoma: a systematic review with meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38:598-603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 130. | Sena JA, Wang L, Heasley LE, Hu CJ. Hypoxia regulates alternative splicing of HIF and non-HIF target genes. Mol Cancer Res. 2014;12:1233-1243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 131. | Gothié E, Richard DE, Berra E, Pagès G, Pouysségur J. Identification of alternative spliced variants of human hypoxia-inducible factor-1alpha. J Biol Chem. 2000;275:6922-6927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 80] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 132. | Monsef N, Soller M, Panagopoulos I, Abrahamsson PA. HIF1alpha isoforms in benign and malignant prostate tissue and their correlation to neuroendocrine differentiation. BMC Cancer. 2010;10:385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 133. | Chun YS, Choi E, Yeo EJ, Lee JH, Kim MS, Park JW. A new HIF-1 alpha variant induced by zinc ion suppresses HIF-1-mediated hypoxic responses. J Cell Sci. 2001;114:4051-4061. [PubMed] [Cited in This Article: ] |
| 134. | Chun YS, Choi E, Kim TY, Kim MS, Park JW. A dominant-negative isoform lacking exons 11 and 12 of the human hypoxia-inducible factor-1alpha gene. Biochem J. 2002;362:71-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 135. | Li R, Zhang H, Yu W, Chen Y, Gui B, Liang J, Wang Y, Sun L, Yang X, Zhang Y. ZIP: a novel transcription repressor, represses EGFR oncogene and suppresses breast carcinogenesis. EMBO J. 2009;28:2763-2776. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 136. | Gui B, Han X, Zhang Y, Liang J, Wang D, Xuan C, Yu Z, Shang Y. Dimerization of ZIP promotes its transcriptional repressive function and biological activity. Int J Biochem Cell Biol. 2012;44:886-895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 137. | Yu W, Li R, Gui B, Shang Y. sZIP, an alternative splice variant of ZIP, antagonizes transcription repression and growth inhibition by ZIP. J Biol Chem. 2010;285:14301-14307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 138. | Ladomery M. Aberrant alternative splicing is another hallmark of cancer. Int J Cell Biol. 2013;2013:463786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 139. | Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353-D361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4407] [Cited by in F6Publishing: 4935] [Article Influence: 616.9] [Reference Citation Analysis (0)] |
| 140. | Yamaga R, Ikeda K, Horie-Inoue K, Ouchi Y, Suzuki Y, Inoue S. RNA sequencing of MCF-7 breast cancer cells identifies novel estrogen-responsive genes with functional estrogen receptor-binding sites in the vicinity of their transcription start sites. Horm Cancer. 2013;4:222-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 141. | Alder O, Cullum R, Lee S, Kan AC, Wei W, Yi Y, Garside VC, Bilenky M, Griffith M, Morrissy AS. Hippo signaling influences HNF4A and FOXA2 enhancer switching during hepatocyte differentiation. Cell Rep. 2014;9:261-271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 142. | Lu H. Crosstalk of HNF4α with extracellular and intracellular signaling pathways in the regulation of hepatic metabolism of drugs and lipids. Acta Pharm Sin B. 2016;6:393-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 143. | Botella LM, Sanz-Rodriguez F, Komi Y, Fernandez-L A, Varela E, Garrido-Martin EM, Narla G, Friedman SL, Kojima S. TGF-beta regulates the expression of transcription factor KLF6 and its splice variants and promotes co-operative transactivation of common target genes through a Smad3-Sp1-KLF6 interaction. Biochem J. 2009;419:485-495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 144. | Rosenbluth JM, Mays DJ, Pino MF, Tang LJ, Pietenpol JA. A gene signature-based approach identifies mTOR as a regulator of p73. Mol Cell Biol. 2008;28:5951-5964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 145. | Wands JR, Kim M. Signaling Pathways in Viral Related Pre-neoplastic Liver Disease and Hepatocellular Carcinoma. In: Wang XW, Grisham JW, Thorgeirsson SS, editors. Molecular Genetics of Liver Neoplasia. 2011;103-127. [Cited in This Article: ] |
| 146. | Wagner KD, Cherfils-Vicini J, Hosen N, Hohenstein P, Gilson E, Hastie ND, Michiels JF, Wagner N. The Wilms’ tumour suppressor Wt1 is a major regulator of tumour angiogenesis and progression. Nat Commun. 2014;5:5852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 147. | Algamas-Dimantov A, Yehuda-Shnaidman E, Peri I, Schwartz B. Epigenetic control of HNF-4α in colon carcinoma cells affects MUC4 expression and malignancy. Cell Oncol (Dordr). 2013;36:155-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 148. | Oshima T, Kawasaki T, Ohashi R, Hasegawa G, Jiang S, Umezu H, Aoyagi Y, Iwanari H, Tanaka T, Hamakubo T. Downregulated P1 promoter-driven hepatocyte nuclear factor-4alpha expression in human colorectal carcinoma is a new prognostic factor against liver metastasis. Pathol Int. 2007;57:82-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 149. | Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 372] [Cited by in F6Publishing: 380] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 150. | Katt ME, Placone AL, Wong AD, Xu ZS, Searson PC. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front Bioeng Biotechnol. 2016;4:12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 371] [Cited by in F6Publishing: 434] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 151. | Takehara T, Liu X, Fujimoto J, Friedman SL, Takahashi H. Expression and role of Bcl-xL in human hepatocellular carcinomas. Hepatology. 2001;34:55-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 203] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 152. | Salton M, Misteli T. Small Molecule Modulators of Pre-mRNA Splicing in Cancer Therapy. Trends Mol Med. 2016;22:28-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 111] [Article Influence: 12.3] [Reference Citation Analysis (0)] |