Copyright
©The Author(s) 2015.
World J Stem Cells. May 26, 2015; 7(4): 700-710
Published online May 26, 2015. doi: 10.4252/wjsc.v7.i4.700
Published online May 26, 2015. doi: 10.4252/wjsc.v7.i4.700
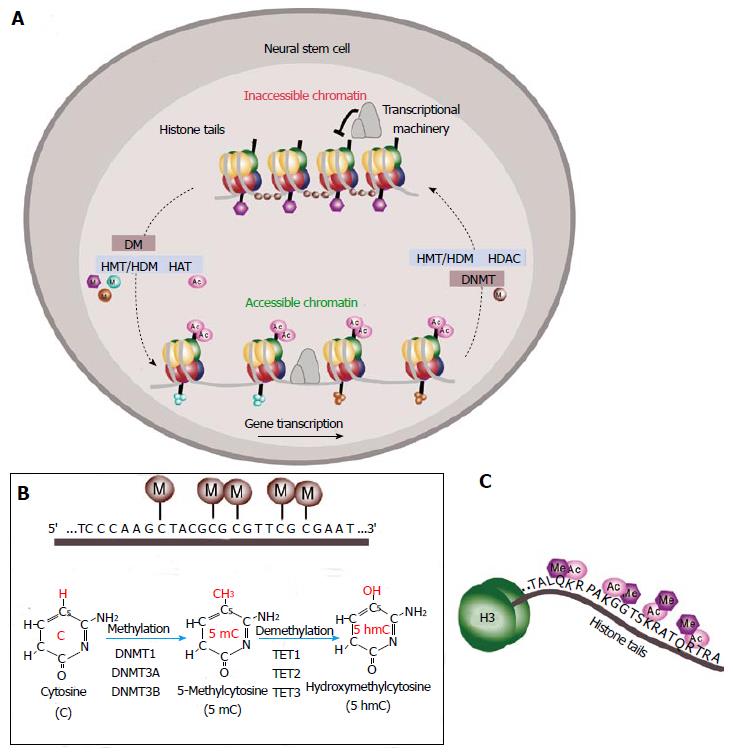
Figure 2 Epigenetic regulation of gene expression.
A: Schematic of DNA methylation and histone modifications in neural stem cells (NSCs). DNA is compressed through interactions with histones and methyl groups (M) are added to cytosine-guanine (CpGs) dinucleotides in regulatory regions. Histone methylation reactions are catalyzed by histone methyltransferases (HMTs) and the reverse process is mediated by histone demethylases (HDMs). Histone acetylation is mediated by histone acetyltransferases (HATs) that leads to chromatin decondensation (accessible chromatin) and transcription activation. Histone deacetylases (HDACs) catalyze the reverse process inducing inactivation of transcription (inaccessible chromatin); B: Schematic of DNA methylation at the cytosine-guanine dinucleotides in gene regulatory regions. Methylation reactions are mediated by DNA methyltransferases (DNMTs) that transfer methyl groups (M) to the fifth position of the pyrimidine ring. This is a reversible process mediated by the ten-eleven translocation (TET) family of enzymes TET1, TET2 and TET3 dioxygenases that catalyze the conversion of the modified genomic base 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) playing a key role in active DNA demethylation; C: Schematic of the histone tail showing multiple sites for epigenetic modifications as acetylation (Ac) or methylation (Me).
- Citation: Montalbán-Loro R, Domingo-Muelas A, Bizy A, Ferrón SR. Epigenetic regulation of stemness maintenance in the neurogenic niches. World J Stem Cells 2015; 7(4): 700-710
- URL: https://www.wjgnet.com/1948-0210/full/v7/i4/700.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i4.700