Published online Mar 26, 2015. doi: 10.4252/wjsc.v7.i2.461
Peer-review started: August 15, 2014
First decision: September 28, 2014
Revised: October 29, 2014
Accepted: October 31, 2014
Article in press: November 3, 2014
Published online: March 26, 2015
Processing time: 217 Days and 15.3 Hours
Cancer is a highly heterogeneous group of diseases that despite improved treatments remain prevalent accounting for over 14 million new cases and 8.2 million deaths per year. Studies into the process of carcinogenesis are limited by lack of appropriate models for the development and pathogenesis of the disease based on human tissues. Primary culture of patient samples can help but is difficult to grow for a number of tissues. A potential opportunity to overcome these barriers is based on the landmark study by Yamanaka which demonstrated the ability of four factors; Oct4, Sox2, Klf4, and c-Myc to reprogram human somatic cells in to pluripotency. These cells were termed induced pluripotent stem cells (iPSCs) and display characteristic properties of embryonic stem cells. This technique has a wide range of potential uses including disease modelling, drug testing and transplantation studies. Interestingly iPSCs also share a number of characteristics with cancer cells including self-renewal and proliferation, expression of stem cell markers and altered metabolism. Recently, iPSCs have been generated from a number of human cancer cell lines and primary tumour samples from a range of cancers in an attempt to recapitulate the development of cancer and interrogate the underlying mechanisms involved. This review will outline the similarities between the reprogramming process and carcinogenesis, and how these similarities have been exploited to generate iPSC models for a number of cancers.
Core tip: Human induced pluripotent stem cells (iPSCs) represent a novel method for studying the mechanisms of cancer development and progression. Recently, a number of studies have generated iPSCs from human cancer cells and cell lines, which can then be used as a model for carcinogenesis. This review outlines the similarities that exist between pluripotent and malignant cells and summarizes available studies that have generated iPSC models of cancer.
- Citation: Curry EL, Moad M, Robson CN, Heer R. Using induced pluripotent stem cells as a tool for modelling carcinogenesis. World J Stem Cells 2015; 7(2): 461-469
- URL: https://www.wjgnet.com/1948-0210/full/v7/i2/461.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v7.i2.461
Cancer is a heterogeneous group of diseases that accounts for an estimated 14.1 million new cases and 8.2 million deaths annually, with the number of cases expected to increase to 24 million annually by 2035[1]. Surgical removal, chemotherapy and/or radiotherapy have become the mainstay treatment for cancer. In recent decades research has focused on improving current drugs and developing targeted therapies specific to the defining biological properties of tumours. Despite the development of these therapeutic interventions, the death rate from cancer remains high. One major reason for this is the limitations of current representative pre-clinical models for human carcinogenesis, including the difficulty in growing and expanding primary cultures. Induced pluripotent stem cells (iPSCs) offer a relevant and unlimited system to study the development and progression of cancer. This review will discuss the current models for carcinogenesis, similarities between cancer and stem cells, and recent iPSC models of human cancers.
Cell lines developed from human tumours are often used to study carcinogenesis. These are inexpensive and can be maintained over lengthy periods; however prolonged culture can alter the characteristics of cells resulting in them becoming less representative of primary tumours[2]. Alterations in gene expression with prolonged in vitro culture are also associated with the use of cell lines, with studies using microarray data from snap-frozen normal human tissue, primary tumour biopsy tissue and tumour-derived cell lines identifying that only 2% of tissue specific and 5% of tumour specific genes were expressed when compared to their equivalent tissue or tumour[3]. Primary cell cultures better represent inter-patient heterogeneity which exists due to differences in tumorigenic cell properties and numbers, variation in cell of origin and frequency of mutations[4,5], however cultures have a limited life span and are difficult to obtain, maintain and expand. A number of animal models for carcinogenesis exist and have greatly increased our knowledge of cancer. However, animal models are not fully representative of carcinogenesis in the human setting due to inherent species differences including organism size and longevity as well as cancer susceptibility[6].
Stem cells are defined by their ability for self-renewal and differentiation into a range of cell types. Their ability to replicate indefinitely overcomes the limitations of current human tissue models as they are able to generate a limitless supply of human cells. Somatic stem cells are present within many organs and are defined by their ability for both self-renewal and differentiation to maintain homeostasis[7]. These cells could be used to model development and disease; however adult stem cells comprise rare populations that are not easily identifiable.
The first human embryonic stem cell line was derived from human blastocysts in 1998 by Thomson et al[8]. Stem cells have two defining features; self-renewal and indefinite proliferation, meaning a limitless supply of tissue can be derived from these cells. Due to these properties, it is hoped that stem cells can be used as a system for disease modelling and drug discovery. However, research using human embryonic stem cells (hESCs) is hampered due to the ethical issues surrounding ESCs and the stringent restrictions enforced as a result.
An alternative to hESCs and adult stem cells are induced pluripotent stem cells (iPSCs), generated in a landmark study by Takahashi et al[9]. From a screen of 24 candidates, 4 factors were identified which were able to reprogram mouse somatic cells to pluripotency; Oct3/4, Sox2, c-Myc and Klf4. These cells showed characteristics of ESCs including morphology, marker expression and the ability to form all three embryonic germ layers[9]. Subsequently, the same four factors were shown to also have the ability to reprogram human adult dermal fibroblasts to iPSCs[10]. An alternative cocktail of factors consisting of Oct4, Sox2, Nanog and Lin28 was also shown to generate iPSCs from human fibroblasts[11]. Human iPSCs possess a number of features that are typical of ESCs including self-renewal and expression of ESC marker genes. Importantly, as for hESCs, iPSCs have the ability to differentiate both in vitro, via the formation of embryoid bodies (EBs) comprising all three germ layers and in vivo, as demonstrated by the formation of teratomas[10]. The use of iPSCs also resolves the ethical issues associated with hESCs, a controversial topic due to the use of blastocyst stage human embryos to derive the cells[12]. As iPSCs can be generated from somatic cells, they also show great potential for developing patient specific models of diseases, which can be used to study the underlying mechanisms of disease development and the efficacy of treatments[13].
A number of similarities exist between the processes of reprogramming and carcinogenesis. Cancer cells have a number of defined characteristics including sustained proliferative signaling and replicative immortality[14]. Stem cells also possess this intrinsic ability for both self-renewal and proliferation, highlighting their similarity to cancer cells.
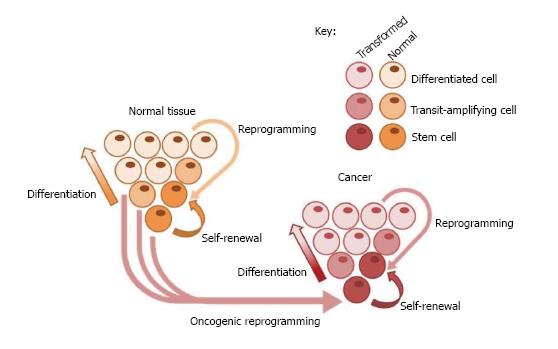
Tumours, like normal tissue, are heterogeneous populations of cells, varying in phenotype, function and gene expression[15]. Furthermore, studies from a number of cancers have shown that not all cells can regenerate tumours upon injection into immunodeficient mice, a functional assay which is now used to identify cells termed cancer stem cells (CSCs)[16]. CSCs can be defined as tumour cells which have the ability for both self-renewal to maintain the stem cell pool, and differentiation to the heterogeneous cell types which maintain the tumour[17], and therefore share the quintessential properties of normal stem cells (Figure 1). Importantly, due to these properties, CSCs which survive chemotherapy are able to re-establish tumours[18]. Whilst the origin of CSCs is not fully known, it has been suggested that CSCs could arise as a result of mutations in stem, transit-amplifying, differentiated or cancer cells leading to acquisition of malignant and/or stem cell properties[19]. Understanding the process by which CSCs are generated and how they maintain tumours is critical to develop therapies which target this cell population and therefore prevent tumour recurrence.
CSCs were originally identified in acute myeloid leukemia, however putative CSCs have now been identified in solid cancers including breast, colon, brain, prostate, and lung cancer[20]. These cells could be used to study tumours however they are difficult to isolate, particularly in solid tumours which are very heterogeneous and therefore are not fully represented by tissue samples taken from a single site within the tumour[21]. Reprogramming of primary cells of interest derived from patients into iPSCs could allow generation of disease- and patient-specific models to study carcinogenesis and to further interrogate the CSC model. An interesting addition to the CSC model is the concept of tumour cell plasticity. A number of studies have shown that non-CSCs can dedifferentiate and acquire properties of stem cells, converting them to CSCs which can then maintain the tumour. In colorectal cancer, Wnt signaling induced by the tumour microenvironment activates CSC properties in differentiated cancer cells[22], whilst transformed human mammary epithelial cells can spontaneously convert to cells with a CSC phenotype both in vitro and in vivo[23].
Carcinogenesis is a multi-step process consisting of initiation, promotion, conversion to malignancy and progression[24]. Cancer is known to undergo clonal expansion throughout this process, resulting from an initial change or mutation in a single cell which confers a growth advantage allowing for enhanced proliferation and expansion of this population. Further mutations then occur within cells forming subpopulations which expand and continue to accumulate changes and mutations which further alter proliferation, cause loss of differentiation and enhance invasive potential, resulting in a malignancy which is genetically and phenotypically divergent from the normal tissue counterpart[25]. Clonality is also a defining feature of stem cells, and is often used to measure the ability of such cells to self-renew[26].
Epithelial-mesenchymal transition (EMT) is defined as a conversion of cells from an epithelial to mesenchymal phenotype with loss of epithelial properties such as apical-basal polarization and cell-cell adhesion and gain of mesenchymal properties such as motility, degradation of the extracellular matrix and resulting invasiveness[27,28]. This process is critical for normal development- during gastrulation for formation of the mesoderm, and also at subsequent stages such as neural crest formation and heart development[29]. Similarly, EMT occurs during differentiation of ESCs[30], whilst its opposing process mesenchymal-epithelial transition is necessary to reprogram cells to pluripotency[31]. EMT has also been implicated in cancer with roles in a number of cancer hallmarks. The EMT inducers SNAIL1 and SNAIL2 correlate with relapse and survival in breast, ovarian and colorectal cancers, suggesting that EMT is associated with poor outcome[28]. Expression of EMT genes is also associated with cancer progression as these expression profiles are identified at the invasive front of a number of cancers including colon carcinoma, papillary thyroid carcinoma and some breast carcinomas[28]. Furthermore, an EMT expression profile was shown to associate with development of metastasis in cutaneous malignant melanoma[32].
TGFβ, an inducer of EMT, has a dual role in cancer, acting as a tumour suppressor and activator of apoptosis whilst also promoting immune tolerance, invasion and metastasis[33]. TWIST1/2 proteins, which also act as EMT inducers, are increased in a number of human cancers and prevent senescence in cancer cells by inhibition of the p53 and Rb pathways[34]. Finally, EMT has also been implicated in resistance to chemotherapies, with evidence that a number of chemoresistant cell lines are induced to undergo EMT[35,36].
Transcription factors which play a critical role in reprogramming cells to pluripotency have also been identified in human cancers. Oct4 is a transcription factor with roles in embryogenesis[37] which is expressed in all testicular germ cell tumours and also pre-malignant carcinoma in situ lesions[38], as well as in breast, pancreas and colon cancer cDNA panels[39]. Furthermore, induced expression of Oct4 in the somatic cells of mice results in epithelial dysplasia, providing further evidence of a role for stem cell genes in carcinogenesis[40].
Immunohistochemical staining of a panel of oral squamous cell carcinoma (OSCC) patients identified an increased incidence of Oct4 expression with advanced stages of the disease, with enhanced nuclear staining in higher grade oral cancers[41]. Transduction of primary human breast epithelial cultures with Oct4 generated colonies with self-renewal capacity, which similarly to oral squamous cells with overexpressed Oct4 showed enhanced expression of mesenchymal markers and a loss of epithelial markers. These cells were also highly tumorigenic, forming poorly differentiated, high grade tumours after injection into nude mice[42]. Oct4 has also been implicated in cervical cancer with increasing numbers of Oct4 expressing cells in both carcinoma in situ and invasive cervical cancer compared to normal cervical tissue, suggesting a role for Oct4 in the development of cervical cancer. Overexpression of Oct4 in cervical cancer cell lines resulted in increased tumour volume and weight upon injection into mice, and decreased apoptosis both in vitro and in vivo compared to controls[43].
MYC encodes the Myc transcription factor which regulates genes involved in cell growth and proliferation[44]. Myc is a known oncogene and one of the most commonly altered genes in human cancers with copy number alterations in 14% of cancer samples[45]. Translocation of c-Myc from chromosome 8 to chromosome 2, 14 or 22 in Burkitt lymphoma cells was the first evidence for the role of Myc in cancer development[46]. Myc translocation is also common in multiple myeloma where the oncogene is fused to an IgH or IgL locus in early carcinogenesis[47]. Abnormalities at the c-Myc or l-Myc locus have been identified in 19/20 multiple myeloma cell lines and 50% of advanced primary cancers[48]. Finally, Myc has also been implicated as a downstream target of deregulated Notch signalling which is apparent in T cell leukaemia, with the two molecules concurrently regulating leukaemic cell growth and proliferation[49].
Myc has also been implicated in solid tumours. c-Myc amplification occurs in 25% of primary breast carcinomas with protein overexpression in 45% of breast tumours, and was significantly correlated with poorly differentiated and highly proliferative tumours[50]. Overexpression of c-Myc mRNA is also thought to occur in 60%-80% of human colorectal adenocarcinomas[51] and has been shown to enhance colon cancer cell angiogenesis via inhibition of HIF-1α degradation and promotion of VEGF expression[52].
Myc is overexpressed or activated in over 50% of human cancers, but in most human cells cannot independently induce tumourigenesis, instead cooperating with other events such as loss of p53 or overexpression of Bcl2 to bypass normal cell checkpoints and initiate carcinogenesis[53]. Interestingly, the level of Myc expression has been shown to produce varying effects, with low level deregulated Myc resulting in proliferation alone whilst high levels of Myc overexpression are required to activate tumour suppressor mechanisms such as ARF induction leading to cellular apoptosis[54].
As is the case with Oct4, Nanog expression correlates with poor survival in OSCC and also in nasopharyngeal carcinoma if co-expressed with Oct4[41,55]. High levels of Nanog expression are also present in breast, lung, ovarian and colon cancer cell lines, and furthermore in cervical cancer Nanog expression increases with progression from cervical intraepithelial neoplasia to invasive cervical carcinoma and is associated with poorer prognosis. Inhibition of Nanog by siRNA decreased proliferation in a mouse model of colon cancer, implicating Nanog as a potential therapeutic target in the disease[56].
Sox2 is contained within the most significantly amplified peak in lung and esophageal squamous cell carcinoma (SSC), identifying it as an oncogene for these cancers[57]. Sox2 is also expressed in both mouse and human pre-neoplastic skin lesions and squamous cell carcinomas, but not in normal epidermis. Deletion of Sox2 in melanoma or SSC caused regression of tumours, and identified a number of genes involved in proliferation, stemness and cell survival which are regulated by Sox2, providing further evidence of a role for Sox2 in carcinogenesis[58].
A number of studies have suggested a role for typical stem cell genes in carcinogenesis. An 11-gene stem cell-like signature found in primary prostate tumours can be used as a predictive tool for prostate, breast and lung cancer patients, amongst other cancer types[59]. Construction of a gene module map by Chang et al[60] identified modules specific to adult and murine ESCs and found that the ESC module was activated in human epithelial cancers. Activation of these genes was associated with poorly differentiated breast tumours with an increased risk of metastasis and death, and was also associated with an increased risk of death in lung adenocarcinomas. The c-Myc oncogene was subsequently shown to induce the ESC-like module both in vitro and in vivo[60], consistent with the well-described role of c-Myc in stem cell self-renewal[61]. An ESC signature developed by Weinberg’s group showed enrichment of stem cell genes in poorly differentiated and larger breast tumours and again this was associated with a poorer survival. The ESC signature was also associated with high grade gliomas and bladder carcinomas[62].
Altered metabolism has been implicated in cancer due to the “Warburg effect” whereby cancer cells use glycolysis for energy production rather than mitochondrial oxidative phosphorylation[63]. Aerobic glycolysis occurs in cultured cancer cells and pluripotent stem cells[64]. This is mediated by uncoupling proteins (UCPs) which uncouple oxidative phosphorylation from glycolysis, including UCP2 which is thought to be important for pluripotency[65] and is also increased in most human color cancers[66] as well as showing high expression in 90% of ovarian carcinomas and 94% of breast carcinomas[67]. Recent studies have found that during induction of pluripotency in somatic cells, genes involved in glycolysis are upregulated whilst those involved in oxidative pathways are downregulated, mediating a switch from oxidative phosphorylation to glycolysis[68] in a similar manner to that seen in carcinogenesis.
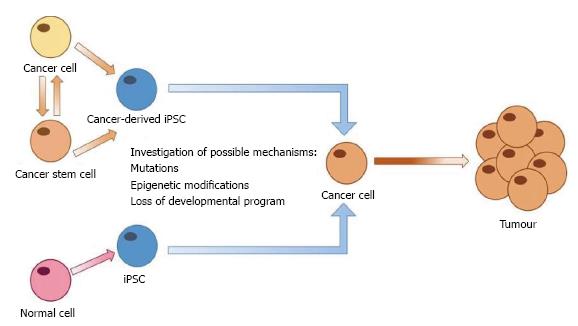
The generation of iPSCs from human somatic cells heralds a new era in disease modelling, allowing the development of patient specific models. As previously mentioned, despite improvements in cancer treatment the disease is still a major cause of morbidity and mortality. The lack of a relevant model to study the development of cancers and their progression has limited research which is suitable for translation to the clinical setting. Generation of iPSCs from human cancer cells represents an opportunity to develop in vitro models of carcinogenesis for specific cancer types (Figure 2).
Recently, a number of studies have generated iPSCs from cancer cells with the hope of developing such a model (summarized in Table 1). One of the earliest attempts to generate iPSCs from malignant cells used retroviral transduction of Oct3/4, Sox2, Klf4 and c-Myc into cancer cell lines from pancreatic, liver, stomach and colorectal cancers. These cancer-derived iPSCs had slower proliferation and increased sensitivity to chemotherapeutic agents in comparison to their parental cells[69].
| Malignancy | Study | Ref. |
| PDAC | Model of early PDAC and progression | Kim et al[73], 2013 |
| CML | iPSCs from CML cell lines and primary patient samples | Carette et al[70], 2010 |
| Kumano et al[72], 2012 | ||
| JML | Derived iPSCs from 2 JML patients | Gandre-Babbe et al[75], 2013 |
| Gastrointestinal cancer | Generated iPSCs from multiple GI cancer cell lines | Miyoshi et al[69], 2010 |
| Glioblastoma | iPSCs generated from glioblastoma-derived neural stem cells | Stricker et al[76], 2014; Stricker et al[77], 2013 |
Carette et al[70] generated iPSCs from the KBM7 chronic myeloid leukemia (CML) cell line. Interestingly, despite the sensitivity of the KBM7 cell line to imatinib, the iPSCs generated lost their BCR-ABL dependence and became resistant to imatinib[70], the tyrosine kinase inhibitor which targets the BCR-ABL protein which defines CML[71]. In a further study, Kumano et al[72] derived iPSCs from primary cultures from two CML patients and showed stem cell morphology and markers along with the ability to differentiate into haematopoietic progenitors which expressed the BCR-ABL fusion protein. Again, these iPSCs were generated from imatinib-sensitive patients but became resistant. Once differentiated, immature cells (CD34+38-90+45+) were identified which were resistant to imatinib and demonstrated phenotypic similarities to CML stem cells. This cell population may therefore be useful to interrogate the role of stem cells in CML and the mechanisms underlying the development of resistance[72].
iPSCs have also been used to study pancreatic ductal adenocarcinoma (PDAC), which currently has no suitable model. iPSC lines were generated from human tumours and injected into immunodeficient mice. After 3 mo, pancreatic intraepithelial neoplasia-like structures could be identified in 9 out of 10 teratomas, and by 9 mo solid tumours were present, suggesting that PDAC derived iPSCs can capture the process of carcinogenesis from pre-malignant lesions to the malignant phenotype. In vitro 3D culture of cells harvested from mice at 3 mo identified 25 proteins which were secreted by all teratomas, of which 8 have been previously reported in PDAC, pancreatic epithelial neoplasia or intraductal papillary mucinous neoplasms[73]. However, although 9 patient samples were used in the study, only 1 iPSC line with a cancer genotype was generated from a single patient, which is consistent with other studies suggesting the difficulty of reprogramming malignant cells[74].
A model of juvenile myelomonocytic leukemia using iPSCs derived from two patients has also recently been generated. Cells were able to differentiate to myeloid cells which showed phenotypic similarities to the primary tumours including enhanced proliferation, suggesting they could be a useful resource to model the disease[75].
iPSCs have also been generated from a number of gastrointestinal cell lines using retroviral transduction. These cancer-derived iPSCs showed reduced tumourigenic potential in vivo as well as increased sensitivity to anti-cancer drugs and decreased proliferative rate[69]. This decrease in tumour forming ability is a concern for the use of cancer-derived iPSCs as it is thought that reprogramming cells removes epigenetic marks which are important for lineage identity and malignancy. However, lineage specificity may remain due to incomplete reprogramming and can also be induced by differentiation. For example, in iPSCs derived from glioblastomas both lineage and cancer associated methylation marks were reset however during differentiation along the neural lineage cells maintained their malignant phenotype[76,77].
An alternative method to study carcinogenesis using iPSCs is to reprogram normal cells to pluripotency and follow their development to interrogate the processes which contribute to cancer development (Figure 2). iPSCs are able to differentiate into all adult cell types and can therefore be used to model development of human tissues[78]. iPSC models of organ and tissue development can then be monitored in real-time to identify any changes which may induce the onset of carcinogenesis. Recently, an in vitro model of skin was developed using iPSCs differentiated to keratinocytes and fibroblasts, providing an iPSC-generated in vitro model of a human organ[79].
Alternatively, iPSCs from normal somatic cells could also be manipulated to study carcinogenesis through overexpression or silencing of oncogenes, tumour suppressor genes and other factors thought to play a role in carcinogenesis, or alteration of the microenvironment. The response of cells to these changes can then be studied to determine the roles of such factors in cancer initiation and progression.
Pluripotent cells and cancer cells share a number of characteristics including the ability for continual proliferation and self-renewal. iPSCs offer an opportunity to develop disease-specific models for carcinogenesis by reprogramming malignant cells. A number of studies have successfully generated iPSCs from human tumours and cancer cell lines and used them to study the underlying mechanisms of cancer. Generation of iPSCs from other cancer types is necessary to develop relevant in vitro models for carcinogenesis.
P- Reviewer: Alsanie WF, Czyz J, Yasuhara T S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0,Cancer incidence and mortality worldwide: IARC CancerBase No. 11[Internet]. Lyon, France: International Agency for Research on Cancer 2013; Available from: http: //globocan.iarc.fr. |
| 2. | Kaur G, Dufour JM. Cell lines: Valuable tools or useless artifacts. Spermatogenesis. 2012;2:1-5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 321] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 3. | Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). Proc Natl Acad Sci USA. 2005;102:2052-2057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 137] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 4. | Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214-218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3848] [Cited by in RCA: 4122] [Article Influence: 343.5] [Reference Citation Analysis (0)] |
| 5. | Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1549] [Cited by in RCA: 1801] [Article Influence: 150.1] [Reference Citation Analysis (0)] |
| 6. | Rangarajan A, Weinberg RA. Opinion: Comparative biology of mouse versus human cells: modelling human cancer in mice. Nat Rev Cancer. 2003;3:952-959. [PubMed] |
| 7. | Collins AT, Habib FK, Maitland NJ, Neal DE. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J Cell Sci. 2001;114:3865-3872. [PubMed] |
| 8. | Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145-1147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11399] [Cited by in RCA: 10426] [Article Influence: 386.1] [Reference Citation Analysis (0)] |
| 9. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18176] [Article Influence: 956.6] [Reference Citation Analysis (0)] |
| 10. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14305] [Article Influence: 841.5] [Reference Citation Analysis (0)] |
| 11. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7241] [Article Influence: 402.3] [Reference Citation Analysis (0)] |
| 12. | de Wert G, Mummery C. Human embryonic stem cells: research, ethics and policy. Hum Reprod. 2003;18:672-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 149] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 13. | Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 657] [Cited by in RCA: 651] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 14. | Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19834] [Cited by in RCA: 19494] [Article Influence: 779.8] [Reference Citation Analysis (0)] |
| 15. | Marte B. Tumour heterogeneity. Nature. 2013;501:327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 16. | Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 595] [Cited by in RCA: 650] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 17. | Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339-9344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2176] [Cited by in RCA: 2199] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 18. | Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic implications of cancer stem cells. Curr Opin Genet Dev. 2004;14:43-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 403] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 19. | Wu XZ. Origin of cancer stem cells: the role of self-renewal and differentiation. Ann Surg Oncol. 2008;15:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 20. | Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2552] [Cited by in RCA: 2624] [Article Influence: 154.4] [Reference Citation Analysis (0)] |
| 21. | Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1460] [Cited by in RCA: 1678] [Article Influence: 152.5] [Reference Citation Analysis (0)] |
| 22. | Vermeulen L, De Sousa E Melo F, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1301] [Cited by in RCA: 1445] [Article Influence: 96.3] [Reference Citation Analysis (1)] |
| 23. | Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108:7950-7955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 883] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 24. | Vincent TL, Gatenby RA. An evolutionary model for initiation, promotion, and progression in carcinogenesis. Int J Oncol. 2008;32:729-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 25. | Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4428] [Cited by in RCA: 4182] [Article Influence: 85.3] [Reference Citation Analysis (1)] |
| 26. | Li L, Wang BH, Wang S, Moalim-Nour L, Mohib K, Lohnes D, Wang L. Individual cell movement, asymmetric colony expansion, rho-associated kinase, and E-cadherin impact the clonogenicity of human embryonic stem cells. Biophys J. 2010;98:2442-2451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 27. | Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4715] [Cited by in RCA: 6214] [Article Influence: 564.9] [Reference Citation Analysis (0)] |
| 28. | Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6805] [Cited by in RCA: 7739] [Article Influence: 483.7] [Reference Citation Analysis (0)] |
| 29. | Larue L, Bellacosa A. Epithelial-mesenchymal transition in development and cancer: role of phosphatidylinositol 3’ kinase/AKT pathways. Oncogene. 2005;24:7443-7454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 884] [Cited by in RCA: 952] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 30. | Kim YS, Yi BR, Kim NH, Choi KC. Role of the epithelial-mesenchymal transition and its effects on embryonic stem cells. Exp Mol Med. 2014;46:e108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 31. | Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 895] [Cited by in RCA: 924] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 32. | Alonso SR, Tracey L, Ortiz P, Pérez-Gómez B, Palacios J, Pollán M, Linares J, Serrano S, Sáez-Castillo AI, Sánchez L. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450-3460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 252] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 33. | Massagué J. TGFbeta in Cancer. Cell. 2008;134:215-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2725] [Cited by in RCA: 3131] [Article Influence: 184.2] [Reference Citation Analysis (0)] |
| 34. | Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, Fauvet F, Puisieux I, Doglioni C, Piccinin S. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 541] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 35. | Kajiyama H, Shibata K, Terauchi M, Yamashita M, Ino K, Nawa A, Kikkawa F. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int J Oncol. 2007;31:277-283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 30] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 36. | Yang AD, Fan F, Camp ER, van Buren G, Liu W, Somcio R, Gray MJ, Cheng H, Hoff PM, Ellis LM. Chronic oxaliplatin resistance induces epithelial-to-mesenchymal transition in colorectal cancer cell lines. Clin Cancer Res. 2006;12:4147-4153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 433] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 37. | Jerabek S, Merino F, Schöler HR, Cojocaru V. OCT4: dynamic DNA binding pioneers stem cell pluripotency. Biochim Biophys Acta. 2014;1839:138-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 38. | Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 339] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 39. | Monk M, Holding C. Human embryonic genes re-expressed in cancer cells. Oncogene. 2001;20:8085-8091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 267] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 40. | Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 651] [Cited by in RCA: 659] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 41. | Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, Chou SH, Chien CS, Ku HH, Lo JF. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085-4095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 513] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 42. | Beltran AS, Rivenbark AG, Richardson BT, Yuan X, Quian H, Hunt JP, Zimmerman E, Graves LM, Blancafort P. Generation of tumor-initiating cells by exogenous delivery of OCT4 transcription factor. Breast Cancer Res. 2011;13:R94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 43. | Wang YD, Cai N, Wu XL, Cao HZ, Xie LL, Zheng PS. OCT4 promotes tumorigenesis and inhibits apoptosis of cervical cancer cells by miR-125b/BAK1 pathway. Cell Death Dis. 2013;4:e760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 44. | Dang CV. MYC on the path to cancer. Cell. 2012;149:22-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1984] [Cited by in RCA: 2585] [Article Influence: 198.8] [Reference Citation Analysis (0)] |
| 45. | Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899-905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3271] [Cited by in RCA: 3053] [Article Influence: 203.5] [Reference Citation Analysis (0)] |
| 46. | Dalla-Favera R, Bregni M, Erikson J, Patterson D, Gallo RC, Croce CM. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc Natl Acad Sci USA. 1982;79:7824-7827. [PubMed] |
| 47. | Wiener F, Potter , M . In: Kirsch IR, editor. The causes and consequences of chromosomal aberrations. New York: CRC Press 1993; 91-124. |
| 48. | Shou Y, Martelli ML, Gabrea A, Qi Y, Brents LA, Roschke A, Dewald G, Kirsch IR, Bergsagel PL, Kuehl WM. Diverse karyotypic abnormalities of the c-myc locus associated with c-myc dysregulation and tumor progression in multiple myeloma. Proc Natl Acad Sci USA. 2000;97:228-233. [PubMed] |
| 49. | Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O’Neil J, Neuberg D, Weng AP. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261-18266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 679] [Cited by in RCA: 661] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 50. | Naidu R, Wahab NA, Yadav M, Kutty MK. Protein expression and molecular analysis of c-myc gene in primary breast carcinomas using immunohistochemistry and differential polymerase chain reaction. Int J Mol Med. 2002;9:189-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 51. | Smith DR, Goh HS. Overexpression of the c-myc proto-oncogene in colorectal carcinoma is associated with a reduced mortality that is abrogated by point mutation of the p53 tumor suppressor gene. Clin Cancer Res. 1996;2:1049-1053. [PubMed] |
| 52. | Chen C, Cai S, Wang G, Cao X, Yang X, Luo X, Feng Y, Hu J. c-Myc enhances colon cancer cell-mediated angiogenesis through the regulation of HIF-1α. Biochem Biophys Res Commun. 2013;430:505-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 53. | Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 434] [Cited by in RCA: 627] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 54. | Murphy DJ, Junttila MR, Pouyet L, Karnezis A, Shchors K, Bui DA, Brown-Swigart L, Johnson L, Evan GI. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell. 2008;14:447-457. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 376] [Cited by in RCA: 362] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 55. | Luo W, Li S, Peng B, Ye Y, Deng X, Yao K. Embryonic stem cells markers SOX2, OCT4 and Nanog expression and their correlations with epithelial-mesenchymal transition in nasopharyngeal carcinoma. PLoS One. 2013;8:e56324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 148] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 56. | Noh KH, Kim BW, Song KH, Cho H, Lee YH, Kim JH, Chung JY, Kim JH, Hewitt SM, Seong SY. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J Clin Invest. 2012;122:4077-4093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 57. | Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238-1242. [PubMed] |
| 58. | Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, Delatte B, Caauwe A, Lenglez S, Nkusi E. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 513] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 59. | Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503-1521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 683] [Cited by in RCA: 715] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 60. | Wong DJ, Liu H, Ridky TW, Cassarino D, Segal E, Chang HY. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333-344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 606] [Cited by in RCA: 575] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 61. | Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747-2763. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 613] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 62. | Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499-507. [PubMed] |
| 63. | Warburg O. On the origin of cancer cells. Science. 1956;123:309-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9117] [Cited by in RCA: 9914] [Article Influence: 143.7] [Reference Citation Analysis (0)] |
| 64. | Zhang J, Nuebel E, Daley GQ, Koehler CM, Teitell MA. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell. 2012;11:589-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 371] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 65. | Shyh-Chang N, Zheng Y, Locasale JW, Cantley LC. Human pluripotent stem cells decouple respiration from energy production. EMBO J. 2011;30:4851-4852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 30] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Horimoto M, Resnick MB, Konkin TA, Routhier J, Wands JR, Baffy G. Expression of uncoupling protein-2 in human colon cancer. Clin Cancer Res. 2004;10:6203-6207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 67. | Ayyasamy V, Owens KM, Desouki MM, Liang P, Bakin A, Thangaraj K, Buchsbaum DJ, LoBuglio AF, Singh KK. Cellular model of Warburg effect identifies tumor promoting function of UCP2 in breast cancer and its suppression by genipin. PLoS One. 2011;6:e24792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 68. | Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerías A, Batchelder EM, Plongthongkum N, Lutz M. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 402] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 69. | Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, Nagano H, Sekimoto M, Doki Y, Mori M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA. 2010;107:40-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 70. | Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, Brummelkamp TR. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039-4042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 71. | Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85-109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (1)] |
| 72. | Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, Nagae G, Ueda K, Nakazaki K, Kamikubo Y. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234-6242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 73. | Kim J, Hoffman JP, Alpaugh RK, Rhim AD, Reichert M, Stanger BZ, Furth EE, Sepulveda AR, Yuan CX, Won KJ. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell Rep. 2013;3:2088-2099. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 143] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 74. | Ramos-Mejia V, Fraga MF, Menendez P. iPSCs from cancer cells: challenges and opportunities. Trends Mol Med. 2012;18:245-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 75. | Gandre-Babbe S, Paluru P, Aribeana C, Chou ST, Bresolin S, Lu L, Sullivan SK, Tasian SK, Weng J, Favre H. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood. 2013;121:4925-4929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 76. | Stricker S, Pollard S. Reprogramming cancer cells to pluripotency: an experimental tool for exploring cancer epigenetics. Epigenetics. 2014;9:798-802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 77. | Stricker SH, Feber A, Engström PG, Carén H, Kurian KM, Takashima Y, Watts C, Way M, Dirks P, Bertone P. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654-669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 78. | Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nat Rev Drug Discov. 2011;10:915-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 355] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 79. | Itoh M, Umegaki-Arao N, Guo Z, Liu L, Higgins CA, Christiano AM. Generation of 3D skin equivalents fully reconstituted from human induced pluripotent stem cells (iPSCs). PLoS One. 2013;8:e77673. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 161] [Article Influence: 13.4] [Reference Citation Analysis (0)] |