Published online Oct 26, 2011. doi: 10.4252/wjsc.v3.i10.89
Revised: September 14, 2011
Accepted: September 21, 2011
Published online: October 26, 2011
Recent advances in reprograming somatic cells from normal and diseased tissues into induced pluripotent stem cells (iPSCs) provide exciting possibilities for generating renewed tissues for disease modeling and therapy. However, questions remain on whether iPSCs still retain certain markers (e.g. aging) of the original somatic cells that could limit their replicative potential and utility. A reliable biological marker for measuring cellular aging is telomere length, which is maintained by a specialized form of cellular polymerase known as telomerase. Telomerase is composed of the cellular reverse transcriptase protein, its integral RNA component, and other cellular proteins (e.g. dyskerin). Mutations in any of these components of telomerase can lead to a severe form of marrow deficiency known as dyskeratosis congenita (DC). This review summarizes recent findings on the effect of cellular reprograming via iPS of normal or DC patient-derived tissues on telomerase function and consequently on telomere length maintenance and cellular aging. The potentials and challenges of using iPSCs in a clinical setting will also be discussed.
- Citation: Ly H. Telomere dynamics in induced pluripotent stem cells: Potentials for human disease modeling. World J Stem Cells 2011; 3(10): 89-95
- URL: https://www.wjgnet.com/1948-0210/full/v3/i10/89.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v3.i10.89
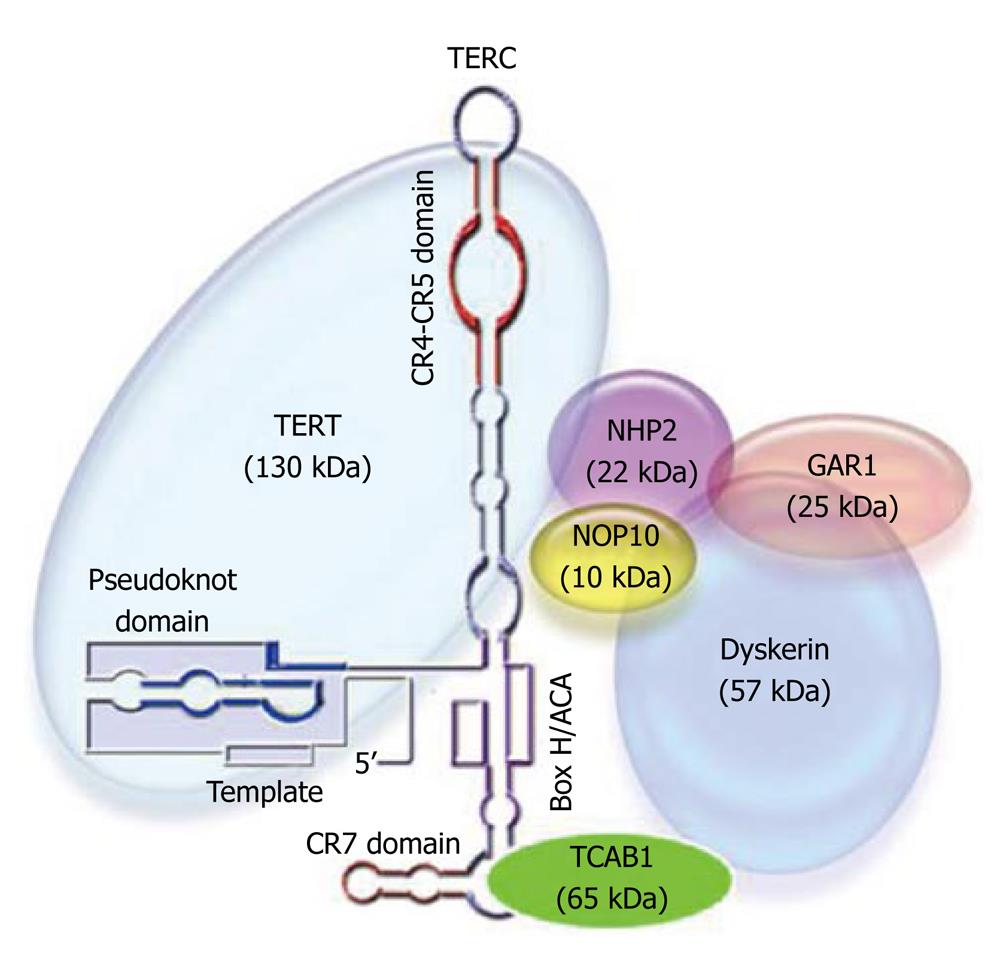
Telomerase is a ribonucleoprotein complex whose main function is to elongate chromosomal 3’ end sequences known as telomeres. The function of this unique enzyme in terminal DNA elongation is necessary in order to overcome the “end-replication problem” whereby conventional DNA polymerases cannot fully replicate linear DNAs[1,2]. Human telomerase is composed of a specialized reverse transcriptase protein (hTERT) and its intrinsic RNA template (hTERC), as well as the associated proteins dyskerin, NOP10, NHP2, GAR1, and TCAB1 (Figure 1). Telomere erosion (by 50-100 bp per cellular division) limits the replicative capacity of the majority of somatic cells, which do not express active telomerase[3,4]. Cells whose telomeres shorten to a “critical length” enter cellular crisis, which is characterized by replicative senescence or apoptosis, meaning cells either stop dividing or commit suicide[5,6]. Stem cells, germ cells, and certain types of somatic cells (e.g. lymphocytes) express the telomerase enzyme, allowing them to maintain telomere length and escape cellular crisis.
Human telomerase reverse transcriptase (hTERT) has been extensively characterized[7]. The protein is defined by the catalytic domain, which contains seven conserved reverse transcriptase motifs essential for enzymatic activity. Functionally important regions have also been defined in the human RNA template (hTERC) (Figure 1)[8]. The template region is absolutely required for the hTERT protein to reverse transcribe it into telomeric DNA repeats. Other regions include the pseudoknot domain, required for hTERT binding and enzymatic activity, and the Box H/ACA domain, which is required for hTERC RNA processing and stability. Proper assembly and function of the telomerase complex also require other cellular proteins, such as dyskerin, NOP10, NHP2, and GAR1 (Figure 1).
Dyskeratosis congenita (DC) is a rare inherited disorder characterized by mucosal leukoplakia, nail dystrophy, and abnormal skin pigmentation[9]. The majority of the cases occur in children, who, in addition to the aforementioned physical anomalies, suffer from bone-marrow failure syndromes and sometimes other symptoms indicative of premature aging, such as dental abnormalities, esophagostenosis, alopecia, and pulmonary disorders. Hematopoietic malignancies (e.g. MDS, Hodgkin’s and acute myelogenous leukemias) and/or solid tumors of the GI tract, nasopharynx and skin have also been observed in some DC patients[10]. Since this disease affects rapidly renewing tissues, it has been speculated that DC is a telomerase disease. In support of this theory, most DC patients have short telomeres[11,12] and carry mutations in one of the three main components of the telomerase holoenzyme complex, dyskerin (DKC), hTERT protein, and hTERC RNA[13].
There are three different patterns of DC disease inheritance: X-linked recessive, autosomal dominant, and autosomal recessive. The X-linked form of the disease is most severe and is caused by mutations in the DKC1 gene on chromosome Xq28 that encodes dyskerin. Dyskerin is a nucleolar protein that is predicted to function in ribosomal RNA processing, in addition to a role in the biogenesis of the telomerase enzyme complex. Indeed, primary fibroblasts and lymphoblasts from X-linked DC patients have a lower level of hTERC RNA, which corresponds to lower levels of telomerase enzymatic activity and shorter telomere lengths than matched normal cells[14,15]. Interestingly, most Dkc1 mutations are missense mutations and one contains a 3’ deletion, indicating that frameshift and null mutations are possibly incompatible with life[16-26]. Indeed, a DKC1-null mouse is embryonic lethal[27]. In humans, one mutation (A353V) accounts for approximately 30% of all X-linked DC cases and is also seen quite frequently in a severe form of a disease known as Hoyeraal-Hreidarsson syndrome.
The autosomal dominant form of DC (AD-DC) is much less severe and less common than the X-linked form. Mutations in hTERT protein and hTERC RNA, as well as in the telomere binding protein TIN2, have been associated with AD-DC[28]. The vast majority of these mutations are heterozygous, resulting in a haploinsufficiency effect on telomerase function that accounts for the observed telomere shortening. In AD-DC families, the genetic lesion does not change, yet the onset of disease features occurs, on average, 20 years earlier in the children than in their parents. Telomere length appears to play a role in this accelerated disease presentation in later generations, as telomeres are significantly shorter in the later generations of affected families than in the earlier ones, leading to the “disease anticipation” idea based on telomere length measurement[29].
The causal gene(s) for the autosomal recessive form of DC remain somewhat elusive. A homozygous mutation (R34W) in the telomerase-associated NOP10 protein was found in all 3 affected members of a single family and appears to segregate with the disease, as unaffected family members are heterozygous. Patients and unaffected carriers do in fact have significantly shorter telomeres than controls. However, this mutation was not identified in any of the other 15 families screened, suggesting that it may be a very rare genetic risk factor[30]. A recent screen of another small cohort of DC patients identified two out of nine unrelated patients with unique compound heterozygous missense mutations in the TCAB1 locus (gene names WDR79 and WRAP53)[31]. TCAB1 is a WD40-repeat containing protein that binds the CAB box sequence within TERC[32]. It is a constituent of the active telomerase holoenzyme and inhibition of TCAB1 prevents telomerase from localizing to Cajal bodies where RNA-protein complexes are assembled and modified[33]. The proband from one of the families has mutations in exon 2 (F164L) and exon 8 (R398W) of the gene, and the proband of the second family has mutations in exon 7 (H376Y) and exon 9 (G435R). They have classic DC symptoms and much shorter telomeres than healthy age-matched controls. The healthy parents and siblings of each proband carry only a single mutant TCAB1 allele, which is consistent with autosomal recessive inheritance. The mutations were not detected in 380 control individuals, again suggesting that these are rare mutations[31].
The successful cloning of an entirely new animal (e.g. Dolly the sheep) from a single cell via somatic cell nuclear transfer (SCNT) technology heralded humankind into a brave new era of genetic engineering[34]. For the first time, it is possible to reprogram a somatic cell to behave “young” again; to coax it into behaving like an embryonic stem cell that can then differentiate into cells of a variety of different lineages, which is the hallmark of pluripotency. By all measures, this is an intrepid undertaking with an outcome that is beyond anyone’s expectations. SCNT technology involves transferring a somatic cell nucleus into an enucleated donor oocyte and stimulating this chimeric cell to divide and differentiate into cells of different lineages, the exact mechanisms of which are unknown[34]. Factors that can allow the cloned cell to achieve pluripotency remained largely unknown until a seminal discovery made by Takahashi and colleagues reported in 2006 that only four transcriptional factors (Sox2, c-Myc, Oct4, and Klf4) were needed to reprogram mouse fibroblasts to pluripotency[35]. In other words, it takes only four cellular factors to reprogram somatic cells to induced pluripotent stem cells (iPSCs), albeit at relatively low frequencies that vary by the age and tissue origin of the cells[35-39]. Other researchers have shown that different cellular factors (i.e., Nanog and Lin28) can also be used to achieve a similar outcome[40]. However, it was noted that many SCNT-transduced cells failed to divide and possibly entered a stage of cellular senescence or apoptosis[41], which might involve some of the known stress-activated senescence genes (e.g. p53, p21WAF/CIP and p16INK4a) of the p53 and pRb stress response pathways[41-45].
It was shown that telomeres of cells collected from the SCNT cloned sheep were shorter than those from age-matched control animals[46,47], raising a concern that the cloned cells might inherit telomeres of similar lengths as those from the donor nucleus. However, subsequent analysis of telomere lengths from other cloned animals (e.g. cattles) have shown that telomeres were elongated during the cloning process[48] due possibly to reactivation of telomerase in blastocyst stage embryos in the cloned nucleus[49].
It was demonstrated that iPSCs could not be generated from somatic cells from late generations (G3) of telomerase-null mice, possibly due to the high degrees of genomic instability as a result of telomere fusions, and that reintroduction of telomerase could restore the efficiency of generating iPSCs[50]. Interestingly, iPSCs derived from normal human and mouse cells show progressive telomere elongation with passaging in cultures, indicating that telomere elongation can occur post-programming[50-53]. Studies have also shown that telomerase enzymatic activity is significantly activated upon iPS manipulation[38-40,50,52,54] as a result of upregulated expression of the TERT protein and TERC RNA, as well as of the associated protein dyskerin[52,54]. The level of increase in TERT transcript and telomerase function, however, differs between human and mouse cells, about 100 fold in human iPSCs and a modest 2-3 fold in mouse iPSCs, possibly reflecting mechanistic differences in telomerase regulation in different organisms. During normal embryonic development, telomerase function is down regulated upon differentiation of iPSCs into different lineages, resulting in a telomere shorting effect. Several factors, including but not necessarily limited to chromatin structure, can influence the dynamics of telomere maintenance during and/or post-programming of the cells.
Telomere chromatin structure has been shown to be altered during the reprogramming process. High levels of trimethylation of histone at lysine 9 (H3K9) and of histone H4 at lysine 20 (H4K20) are normally observed at telomeric regions, whereas lower levels, as in embryonic stem cells, are detected upon cellular reprograming[50]. The subtelomeric DNA regions in human iPSCs have also been shown to be hypermethylated, as compared to those in the original somatic cells, and to contain high degrees of heterogeneity in their methylation patterns[51]. In contrast to human iPSCs, no obvious changes were observed in the subtelomeric regions of mouse fibroblasts upon reprogramming[50]. It is possible that the reprogramming-induced changes in methylation at or near the subtelomeric regions may alter the expression of genes in the vicinity. For example, it has recently been shown that mouse and human iPSCs upregulate the expression of TERRA RNA[50,51], which is known to regulate both telomere length and its chromatin structure[55]. Deng and colleagues have shown that the association of TERRA RNA with a telomeric DNA binding protein TRF2 can facilitate heterochromatin structural formation via its association with the histone HP1α and trimethylated H3K9[56]. While heterogeneity in levels of telomere-specific gene expression may exist in human vs mouse iPSCs, a general consensus is that the chromatin structures at subtelomeric and/or telomeric regions can change upon reprogramming and can revert back as in the original somatic cells[57-59].
Recent studies (described above) suggest that reprogramming of somatic cells via iPS technology can hold great promise for correcting defects in telomerase function and/or telomere attrition observed in patients with DC, a disease with short telomeres leading to limited bone-marrow stem cell reserve and renewal capacity[60]. The question is whether iPS reprogramming of somatic cells collected from DC patients can reactivate telomerase enzymatic activity to elongate telomeres. To address this question, Agarwal and colleagues[54] attempted to reprogram fibroblasts collected from either an X-linked DC patient with the del37L mutation in the dyskerin gene[14,26] or an autosomal dominant DC patient who is heterozygous for a truncated form of the hTERC gene (del378-451) due to an 821 bp deletion on chromosome 3q26.2-3[60]. In both cases, the mutations greatly reduced the levels of hTERC RNA expression in the cells[15,60]. The patients’ primary fibroblasts were retrovirally transduced with Oct4, Sox2, Klf4, and c-Myc genes, the pluripotent phenotype of the iPSCs was monitored by conventional iPSC assays[61], and the mean telomere lengths were measured by Southern blot. Significant upregulation of telomerase activity was observed in DC patient cells-derived iPSCs, which correspondingly showed telomere elongation upon cellular passaging. More importantly, differentiated cells showed down-regulated telomerase activity and accelerated telomere attrition. Given that the hTERC gene expression in the original fibroblast cells has been shown to be suboptimal[15,60], it is quite unexpected that telomere lengths in their iPSCs can be elongated to a degree similar to control fibroblasts. A possible explanation is that the hTERC gene expression in the iPSCs was found to be significantly upregulated (by 3-8 fold); this the authors attributed to a unique feature of pluripotency as several of the telomerase-associated genes could be targeted by pluripotency-associated transcription factors[54].
One exciting potential of iPSCs is to use them to model the pathogenesis of human disease in vitro. Toward this end, Batista et al[62] have recently derived iPSCs from fibroblasts collected from: autosomal dominant DC patients with mutations in hTERT (P704S and R979); X-linked DC patients with dyskerin mutations (L54V and del37L); and autosomal recessive DC patient with the recently identified disease-associated mutations in TCAB1 gene (H376Y/G435R)[31], using either retrovirus or lentivirus expressing the four required transcription factors (Sox2, c-Myc, Klf4 and Oct4). The authors found that, even in the undifferentiated state, iPSCs derived from these DC patients exhibit the precise features of each form of the disease. Unlike an earlier study[54], profound defects in telomere maintenance were observed[62]; the reasons for the discrepancy between the studies are unclear but is likely due to possible differences in experimental conditions or statistical variations among the iPS clones[63]. In the Batista study[62], iPSCs derived from the hTERT-mutated cells with telomerase haploinsufficiency exhibited blunted telomere elongation effect during reprogramming. In iPSCs from X-linked DC patients, dyskerin mutations severely impaired telomerase assembly and function, and hence disrupted telomere synthesis during reprogramming. In iPSCs derived from cells with the TCAB1 mutations, which led to the mis-localization of the telomerase enzyme from Cajal bodies to nucleoli, proper telomere synthesis was abrogated during reprogramming. Prolonged passaging of some of the undifferentiated iPSC cultures could lead to progressive telomere attrition and eventual loss of self-renewal capacity of the cultures, closely mimicking processes that might occur to the tissue stem cells of the patients. These findings suggest that iPSCs can serve as a good cell-culture-based system for disease modeling and for developing therapeutic strategies (e.g. drug screening).
While recent studies have shown great potential for iPSCs in disease modeling, several obstacles still exist before contemplating the clinical application of iPSCs to treat human diseases. First and foremost, since most successful iPS studies involve the use of retroviral or lentiviral transduction, safety is a principle and valid concern. Several non-viral techniques to deliver the transgenes have recently been developed that should lessen the concern of possible turmorigenesis[64]. Despite recent advances in iPS technology development, the efficiency of reprogramming somatic cells still remains an issue. Recent observations have also indicated that the epigenetic changes at telomeres and elsewhere in the genome of iPSCs are not necessarily identical to those found in embryonic stem cells[65]. This line of investigation clearly deserves more attention as any aberrance in chromatin structure and function potentially renders the iPSCs useless, or worst yet, prone to chromosome instability.This would lead to acquisition of undesired mutations[66] and/or increase in chromosome copy number variations[67-69] that cause enhanced susceptibility to cellular transformation. It is not entirely clear either how to differentiate iPSCs into various cellular lineages in order to generate tissue-specific stem cells for clinical utility.
While the therapeutic usage of iPSCs in clinics appears to be beyond the reach of current technologies[70], several recent studies have provided exciting proof of concepts. A number of human and mouse fibroblast-derived iPSCs have been successfully differentiated into a variety of tissue/cell types, such as cardiomyocytes[71,72], hematopoietic cells[73,74], endothelial-like cells[75], insulin-secreting islet-like cells[76], retinal pigment epithelial cells[77,78], and neurons[79,80]. Using a humanized mouse model of sickle cell anemia, Hanna and colleagues have successfully used genetically engineered skin-derived iPSCs to correct a genetic defect caused by the FANCA gene[73]. It is also possible to introduce the iPSC-differentiated endothelial progenitor cells into the livers of genetically defective hemophilic mice in order to cure bleeding disorder[81]. These studies offer exciting potential and optimism for advancing iPS technology for possible future clinical use in humans.
Dr. Yuying Liang is acknowledged for her critical reading of the manuscript.
Peer reviewer: Kumiko Saeki, MD, PhD, Department of Hematology, Research Institute, International Medical Center of Japan, 1-21-1, Toyama, Shinjuku-ku, Tokyo 162-8655, Japan
S- Editor Wang JL L- Editor Hughes D E- Editor Zheng XM
| 1. | Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41:181-190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1157] [Cited by in F6Publishing: 1151] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 2. | Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197-201. [PubMed] [Cited in This Article: ] |
| 3. | Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4007] [Cited by in F6Publishing: 3948] [Article Influence: 116.1] [Reference Citation Analysis (0)] |
| 4. | Lindsey J, McGill NI, Lindsey LA, Green DK, Cooke HJ. In vivo loss of telomeric repeats with age in humans. Mutat Res. 1991;256:45-48. [PubMed] [Cited in This Article: ] |
| 5. | Harley CB, Vaziri H, Counter CM, Allsopp RC. The telomere hypothesis of cellular aging. Exp Gerontol. 1992;27:375-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 362] [Cited by in F6Publishing: 345] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 6. | HAYFLICK L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37:614-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4095] [Cited by in F6Publishing: 3941] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 7. | Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493-517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 340] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 8. | Chen JL, Greider CW. Telomerase RNA structure and function: implications for dyskeratosis congenita. Trends Biochem Sci. 2004;29:183-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 9. | Dokal I. Dyskeratosis congenita in all its forms. Br J Haematol. 2000;110:768-779. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 412] [Cited by in F6Publishing: 358] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 10. | Alter BP, Giri N, Savage SA, Rosenberg PS. Cancer in dyskeratosis congenita. Blood. 2009;113:6549-6557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 332] [Cited by in F6Publishing: 336] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 11. | Alter BP, Baerlocher GM, Savage SA, Chanock SJ, Weksler BB, Willner JP, Peters JA, Giri N, Lansdorp PM. Very short telomere length by flow fluorescence in situ hybridization identifies patients with dyskeratosis congenita. Blood. 2007;110:1439-1447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 251] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 12. | Du HY, Pumbo E, Ivanovich J, An P, Maziarz RT, Reiss UM, Chirnomas D, Shimamura A, Vlachos A, Lipton JM. TERC and TERT gene mutations in patients with bone marrow failure and the significance of telomere length measurements. Blood. 2009;113:309-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Marrone A, Walne A, Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr Opin Genet Dev. 2005;15:249-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 101] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 14. | Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature. 1999;402:551-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 841] [Cited by in F6Publishing: 800] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 15. | Wong JM, Collins K. Telomerase RNA level limits telomere maintenance in X-linked dyskeratosis congenita. Genes Dev. 2006;20:2848-2858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 151] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 16. | Ding YG, Zhu TS, Jiang W, Yang Y, Bu DF, Tu P, Zhu XJ, Wang BX. Identification of a novel mutation and a de novo mutation in DKC1 in two Chinese pedigrees with Dyskeratosis congenita. J Invest Dermatol. 2004;123:470-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Heiss NS, Mégarbané A, Klauck SM, Kreuz FR, Makhoul E, Majewski F, Poustka A. One novel and two recurrent missense DKC1 mutations in patients with dyskeratosis congenita (DKC). Genet Couns. 2001;12:129-136. [PubMed] [Cited in This Article: ] |
| 18. | Hiramatsu H, Fujii T, Kitoh T, Sawada M, Osaka M, Koami K, Irino T, Miyajima T, Ito M, Sugiyama T. A novel missense mutation in the DKC1 gene in a Japanese family with X-linked dyskeratosis congenita. Pediatr Hematol Oncol. 2002;19:413-419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Kanegane H, Kasahara Y, Okamura J, Hongo T, Tanaka R, Nomura K, Kojima S, Miyawaki T. Identification of DKC1 gene mutations in Japanese patients with X-linked dyskeratosis congenita. Br J Haematol. 2005;129:432-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Knight SW, Heiss NS, Vulliamy TJ, Greschner S, Stavrides G, Pai GS, Lestringant G, Varma N, Mason PJ, Dokal I. X-linked dyskeratosis congenita is predominantly caused by missense mutations in the DKC1 gene. Am J Hum Genet. 1999;65:50-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 167] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Knight SW, Vulliamy TJ, Morgan B, Devriendt K, Mason PJ, Dokal I. Identification of novel DKC1 mutations in patients with dyskeratosis congenita: implications for pathophysiology and diagnosis. Hum Genet. 2001;108:299-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 22. | Kraemer DM, Goebeler M. Missense mutation in a patient with X-linked dyskeratosis congenita. Haematologica. 2003;88:ECR11. [PubMed] [Cited in This Article: ] |
| 23. | Lin JH, Lee JY, Tsao CJ, Chao SC. DKC1 gene mutation in a Taiwanese kindred with X-linked dyskeratosis congenita. Kaohsiung J Med Sci. 2002;18:573-577. [PubMed] [Cited in This Article: ] |
| 24. | Salowsky R, Heiss NS, Benner A, Wittig R, Poustka A. Basal transcription activity of the dyskeratosis congenita gene is mediated by Sp1 and Sp3 and a patient mutation in a Sp1 binding site is associated with decreased promoter activity. Gene. 2002;293:9-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 25. | Vulliamy TJ, Knight SW, Heiss NS, Smith OP, Poustka A, Dokal I, Mason PJ. Dyskeratosis congenita caused by a 3' deletion: germline and somatic mosaicism in a female carrier. Blood. 1999;94:1254-1260. [PubMed] [Cited in This Article: ] |
| 26. | Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 673] [Cited by in F6Publishing: 656] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 27. | He J, Navarrete S, Jasinski M, Vulliamy T, Dokal I, Bessler M, Mason PJ. Targeted disruption of Dkc1, the gene mutated in X-linked dyskeratosis congenita, causes embryonic lethality in mice. Oncogene. 2002;21:7740-7744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Carroll KA, Ly H. Telomere dysfunction in human diseases: the long and short of it! Int J Clin Exp Pathol. 2009;2:528-543. [PubMed] [Cited in This Article: ] |
| 29. | Vulliamy T, Marrone A, Szydlo R, Walne A, Mason PJ, Dokal I. Disease anticipation is associated with progressive telomere shortening in families with dyskeratosis congenita due to mutations in TERC. Nat Genet. 2004;36:447-449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 339] [Cited by in F6Publishing: 360] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 30. | Walne AJ, Vulliamy T, Marrone A, Beswick R, Kirwan M, Masunari Y, Al-Qurashi FH, Aljurf M, Dokal I. Genetic heterogeneity in autosomal recessive dyskeratosis congenita with one subtype due to mutations in the telomerase-associated protein NOP10. Hum Mol Genet. 2007;16:1619-1629. [PubMed] [Cited in This Article: ] |
| 31. | Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev. 2011;25:11-16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 189] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 32. | Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323:644-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 376] [Cited by in F6Publishing: 393] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 33. | Venteicher AS, Artandi SE. TCAB1: driving telomerase to Cajal bodies. Cell Cycle. 2009;8:1329-1331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 34. | Gurdon JB, Melton DA. Nuclear reprogramming in cells. Science. 2008;322:1811-1815. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 223] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 35. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17394] [Article Influence: 966.3] [Reference Citation Analysis (0)] |
| 36. | Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177-1181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 590] [Cited by in F6Publishing: 526] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 37. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2048] [Cited by in F6Publishing: 1877] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 38. | Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2182] [Cited by in F6Publishing: 2075] [Article Influence: 122.1] [Reference Citation Analysis (0)] |
| 39. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14327] [Cited by in F6Publishing: 13800] [Article Influence: 862.5] [Reference Citation Analysis (0)] |
| 40. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7589] [Cited by in F6Publishing: 7080] [Article Influence: 416.5] [Reference Citation Analysis (0)] |
| 41. | Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134-2139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 473] [Cited by in F6Publishing: 480] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 42. | Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136-1139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 739] [Cited by in F6Publishing: 749] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 43. | Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1004] [Cited by in F6Publishing: 1006] [Article Influence: 67.1] [Reference Citation Analysis (0)] |
| 44. | Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 841] [Cited by in F6Publishing: 861] [Article Influence: 57.4] [Reference Citation Analysis (0)] |
| 45. | Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149-1153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 785] [Cited by in F6Publishing: 791] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 46. | Shiels PG, Kind AJ, Campbell KH, Wilmut I, Waddington D, Colman A, Schnieke AE. Analysis of telomere length in Dolly, a sheep derived by nuclear transfer. Cloning. 1999;1:119-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 47. | Shiels PG, Kind AJ, Campbell KH, Waddington D, Wilmut I, Colman A, Schnieke AE. Analysis of telomere lengths in cloned sheep. Nature. 1999;399:316-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Lanza RP, Cibelli JB, Blackwell C, Cristofalo VJ, Francis MK, Baerlocher GM, Mak J, Schertzer M, Chavez EA, Sawyer N. Extension of cell life-span and telomere length in animals cloned from senescent somatic cells. Science. 2000;288:665-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 361] [Cited by in F6Publishing: 373] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 49. | Schaetzlein S, Rudolph KL. Telomere length regulation during cloning, embryogenesis and ageing. Reprod Fertil Dev. 2005;17:85-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 50. | Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 368] [Cited by in F6Publishing: 367] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 51. | Yehezkel S, Rebibo-Sabbah A, Segev Y, Tzukerman M, Shaked R, Huber I, Gepstein L, Skorecki K, Selig S. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics. 2011;6:63-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 52. | Mathew R, Jia W, Sharma A, Zhao Y, Clarke LE, Cheng X, Wang H, Salli U, Vrana KE, Robertson GP. Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. FASEB J. 2010;24:2702-2715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 53. | Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, Murthy S, Cibelli JB. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS One. 2009;4:e8124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 54. | Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature. 2010;464:292-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 236] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 55. | Feuerhahn S, Iglesias N, Panza A, Porro A, Lingner J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010;584:3812-3818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 56. | Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 378] [Cited by in F6Publishing: 408] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 57. | Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 436] [Cited by in F6Publishing: 451] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 58. | Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1768] [Cited by in F6Publishing: 1635] [Article Influence: 116.8] [Reference Citation Analysis (0)] |
| 59. | Hewitt KJ, Shamis Y, Hayman RB, Margvelashvili M, Dong S, Carlson MW, Garlick JA. Epigenetic and phenotypic profile of fibroblasts derived from induced pluripotent stem cells. PLoS One. 2011;6:e17128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 60. | Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 711] [Cited by in F6Publishing: 662] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 61. | Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877-886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1655] [Cited by in F6Publishing: 1592] [Article Influence: 99.5] [Reference Citation Analysis (0)] |
| 62. | Batista LF, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature. 2011;474:399-402. [PubMed] [Cited in This Article: ] |
| 63. | Agarwal S, Daley GQ. Telomere dynamics in dyskeratosis congenita: the long and the short of iPS. Cell Res. 2011;21:1157-1160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 64. | Kiskinis E, Eggan K. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120:51-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 280] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 65. | Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1202] [Cited by in F6Publishing: 1124] [Article Influence: 86.5] [Reference Citation Analysis (0)] |
| 66. | Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 956] [Cited by in F6Publishing: 930] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 67. | Pasi CE, Dereli-Öz A, Negrini S, Friedli M, Fragola G, Lombardo A, Van Houwe G, Naldini L, Casola S, Testa G. Genomic instability in induced stem cells. Cell Death Differ. 2011;18:745-753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 113] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 68. | Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 722] [Cited by in F6Publishing: 681] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 69. | Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 684] [Cited by in F6Publishing: 657] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 70. | Robbins RD, Prasain N, Maier BF, Yoder MC, Mirmira RG. Inducible pluripotent stem cells: not quite ready for prime time? Curr Opin Organ Transplant. 2010;15:61-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 71. | Pfannkuche K, Liang H, Hannes T, Xi J, Fatima A, Nguemo F, Matzkies M, Wernig M, Jaenisch R, Pillekamp F. Cardiac myocytes derived from murine reprogrammed fibroblasts: intact hormonal regulation, cardiac ion channel expression and development of contractility. Cell Physiol Biochem. 2009;24:73-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 72. | Kuzmenkin A, Liang H, Xu G, Pfannkuche K, Eichhorn H, Fatima A, Luo H, Saric T, Wernig M, Jaenisch R. Functional characterization of cardiomyocytes derived from murine induced pluripotent stem cells in vitro. FASEB J. 2009;23:4168-4180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 101] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 73. | Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920-1923. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1104] [Cited by in F6Publishing: 1167] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 74. | Raya A, Rodríguez-Pizà I, Guenechea G, Vassena R, Navarro S, Barrero MJ, Consiglio A, Castellà M, Río P, Sleep E. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature. 2009;460:53-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 536] [Cited by in F6Publishing: 564] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 75. | Taura D, Sone M, Homma K, Oyamada N, Takahashi K, Tamura N, Yamanaka S, Nakao K. Induction and isolation of vascular cells from human induced pluripotent stem cells--brief report. Arterioscler Thromb Vasc Biol. 2009;29:1100-1103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 76. | Tateishi K, He J, Taranova O, Liang G, D'Alessio AC, Zhang Y. Generation of insulin-secreting islet-like clusters from human skin fibroblasts. J Biol Chem. 2008;283:31601-31607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 236] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 77. | Osakada F, Jin ZB, Hirami Y, Ikeda H, Danjyo T, Watanabe K, Sasai Y, Takahashi M. In vitro differentiation of retinal cells from human pluripotent stem cells by small-molecule induction. J Cell Sci. 2009;122:3169-3179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 297] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 78. | Buchholz DE, Hikita ST, Rowland TJ, Friedrich AM, Hinman CR, Johnson LV, Clegg DO. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427-2434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 332] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 79. | Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218-1221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1445] [Cited by in F6Publishing: 1386] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 80. | Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, Broccoli V, Constantine-Paton M, Isacson O, Jaenisch R. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc Natl Acad Sci U S A. 2008;105:5856-5861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 882] [Cited by in F6Publishing: 826] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 81. | Xu D, Alipio Z, Fink LM, Adcock DM, Yang J, Ward DC, Ma Y. Phenotypic correction of murine hemophilia A using an iPS cell-based therapy. Proc Natl Acad Sci U S A. 2009;106:808-813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 202] [Article Influence: 13.5] [Reference Citation Analysis (0)] |