Copyright
©The Author(s) 2025.
World J Stem Cells. Jul 26, 2025; 17(7): 104607
Published online Jul 26, 2025. doi: 10.4252/wjsc.v17.i7.104607
Published online Jul 26, 2025. doi: 10.4252/wjsc.v17.i7.104607
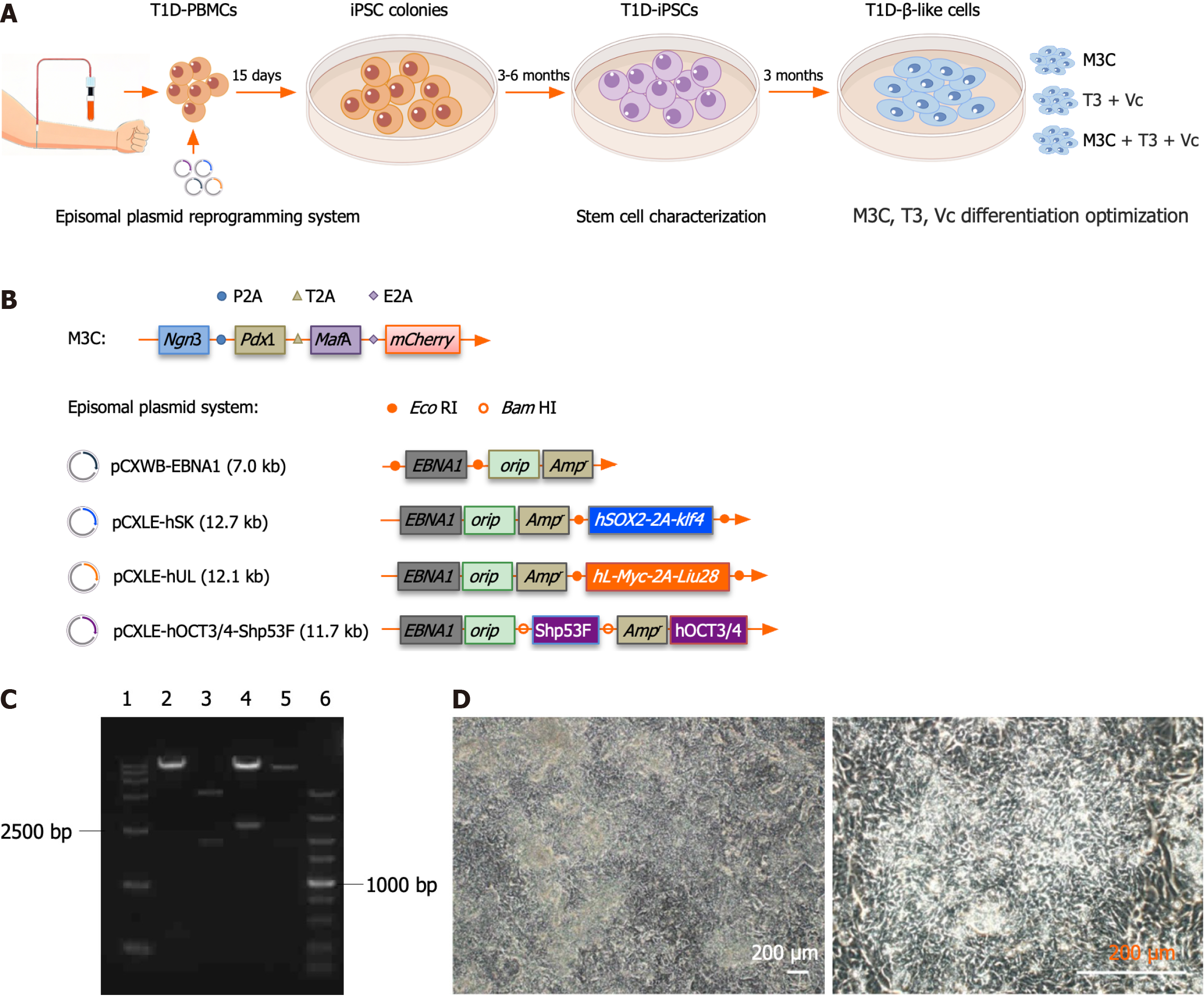
Figure 1 Reprogramming of type 1 diabetes peripheral blood mononuclear cells into type 1 diabetes-induced pluripotent stem cells.
A: Schematic of the type 1 diabetes (T1D)-β-like cell generation workflow. The diagram outlines the complete differentiation pipeline: (1) T1D patient-derived peripheral blood mononuclear cells were reprogrammed into induced pluripotent stem cells (iPSCs) using non-integrating episomal vectors, followed by 3-6 months of clone selection, expansion, and pluripotency verification, including appropriate morphology, differentiation potential, genomic integrity, and expression of pluripotency-associated genes; (2) iPSCs underwent 3-month directed differentiation using three optimized protocols [M3C, triiodothyronine (T3) + vitamin C (Vc), and M3C + T3 + Vc], with progression monitored through stage-specific markers (insulin 1, insulin 2, pancreatic duodenal homeobox-1, neurogenin 3, MAF bZIP transcription factor A, NeuroD, glucagon-like peptide-1 receptor, Nkx6.1, glucose transporter 2, and Kir62); and (3) Functional validation included glucose-stimulated insulin secretion assays and transplantation into streptozotocin-induced diabetic mice; B: Schematic representation of the M3C and episomal plasmid systems. The pAd-M3C plasmid was 40.2 kbp long and comprised a 36.6 kbp pAd-CMV/V5-DEST vector backbone with a 3.5 kbp M3C insert. The M3C insert (PubMed: 25402613) included pancreatic duodenal homeobox-1 (852 bp), neurogenin 3 (642 bp), MAF bZIP transcription factor A (1077 bp), and mCherry (711 bp). The junctions use restriction enzymes SalI (5’-GTCGAC-3’), SpeI (5’-ACTAGT-3’), BamHI (5’-GGATCC-3’), ClaI (5’-ATCGAT-3’), and NotI (5’-GCGGCCGC-3’). Synthetic sequences such as P2A (from porcine tetanus virus), T2A (porcine thymovirus type II), and E2A (coronavirus type II) are used for ligated inserts. The episomal plasmid system consisted of four plasmids: PCXWB-EBNA1 (7.0 kb) and its recombined counterparts, including pCXLE-hSK (12.7 kb), pCXLE-hUL (12.1 kb), and pCXLE-hOCT3/4-Shp53F (11.7 kb); C: Electrophoretic analysis of the episomal plasmid system post-restriction digestion. Agarose gel electrophoresis results: (1) DNA ladder DL15000; (2) pCXLE-hOCT3/4-Shp53F digested with BamHI, yielding fragments of 11.3 kb and 0.4 kb; (3) pCXWB-EBNA1 using EcoRI, with fragment sizes of 5 and 2 kb; (4) pCXLE-hSK digested with EcoRI, resulting in 10.2 kb and 2.5 kb fragments; (5) pCXLE-hUL with EcoRI giving 102 and 1.9 kb fragments; and (6) DNA ladder DL5000; D: Morphological characterization of T1D-iPSCs. T1D-iPSCs exhibit typical pluripotent stem cell morphology, including small cell size and compact colony formation (scale bar = 200 μm). T1D: Type 1 diabetes; PBMC: Peripheral blood mononuclear cell; iPSC: Induced pluripotent stem cells; T3: Triiodothyronine; Vc: Vitamin C; Ngn3: Neurogenin 3; Pdx1: Pancreatic duodenal homeobox-1; MafA: MAF bZIP transcription factor A.
- Citation: Wang K, Lin W, Han JY, Chen JY, Liu RH, Yu Z, Jin JJ. Differentiation of patient-specific induced pluripotent stem cells derived from type 1 diabetes peripheral blood mononuclear cells into pancreatic β-like cells. World J Stem Cells 2025; 17(7): 104607
- URL: https://www.wjgnet.com/1948-0210/full/v17/i7/104607.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v17.i7.104607