Published online Jul 26, 2021. doi: 10.4252/wjsc.v13.i7.685
Peer-review started: January 27, 2021
First decision: February 28, 2021
Revised: March 9, 2021
Accepted: April 14, 2021
Article in press: April 14, 2021
Published online: July 26, 2021
Pediatric neuroblastomas (NBs) are heterogeneous, aggressive, therapy-resistant embryonal tumours that originate from cells of neural crest (NC) origin and in particular neuroblasts committed to the sympathoadrenal progenitor cell lineage. Therapeutic resistance, post-therapeutic relapse and subsequent metastatic NB progression are driven primarily by cancer stem cell (CSC)-like subpopulations, which through their self-renewing capacity, intermittent and slow cell cycles, drug-resistant and reversibly adaptive plastic phenotypes, represent the most important obstacle to improving therapeutic outcomes in unfavourable NBs. In this review, dedicated to NB CSCs and the prospects for their therapeutic eradication, we initiate with brief descriptions of the unique transient vertebrate embryonic NC structure and salient molecular protagonists involved NC induction, specification, epithelial to mesenchymal transition and migratory behaviour, in order to familiarise the reader with the embryonic cellular and molecular origins and background to NB. We follow this by introducing NB and the potential NC-derived stem/progenitor cell origins of NBs, before providing a comprehensive review of the salient molecules, signalling pathways, mechanisms, tumour microenvironmental and therapeutic conditions involved in promoting, selecting and maintaining NB CSC subpopulations, and that underpin their therapy-resistant, self-renewing metastatic behaviour. Finally, we review potential therapeutic strategies and future prospects for targeting and eradication of these bastions of NB therapeutic resistance, post-therapeutic relapse and metastatic progression.
Core Tip: Cancer stem cells promote tumour-heterogeneity, post-therapeutic relapse and metastatic progression in neuroblastomas (NBs), by way of their drug-resistant, self-renewing and reversibly plastic phenotypes, and are, therefore, critical targets to eliminate if improvements in therapeutic outcomes are to be realized. Despite progress in understanding the molecular mechanisms that underpin NB cancer stem cell (CSC) selection, promotion and maintenance, efficacious CSC targeting remains difficult. Here we review the molecular mechanisms considered to regulate NB CSCs, together with the therapeutic strategies and future prospects for their eradication, highlighting the need for a better understanding and targeting of polyploid giant cancer cell states, future prospects for chimeric antigen receptor (CAR) cell immunotherapies, the need to identify additional functional cell surface NB CSC-specific targets and to develop better models for NB CSC experimental therapeutics.
- Citation: Farina AR, Cappabianca LA, Zelli V, Sebastiano M, Mackay AR. Mechanisms involved in selecting and maintaining neuroblastoma cancer stem cell populations, and perspectives for therapeutic targeting. World J Stem Cells 2021; 13(7): 685-736
- URL: https://www.wjgnet.com/1948-0210/full/v13/i7/685.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i7.685
Cancer stem cells (CSCs) are subpopulations of aggressive, undifferentiated, multipotent, self-renewing tumour cells with similar behaviour and gene expression patterns to normal stem cells. In pediatric neuroblastomas (NBs), CSCs are considered to be responsible for tumour heterogeneity, genetic instability, therapeutic resistance, post-therapeutic relapse and metastatic progression, making them critical therapeutic targets, the eradication of which will improve therapeutic outcomes. In this review, dedicated to NB CSCs and the prospects for their therapeutic eradication, we provide initial descriptions of the unique transient vertebrate embryonic neural crest (NC) structure and salient molecular protagonists involved in NC formation and migratory behaviour, in order to familiarise the reader with the embryonic cellular and molecular origins and background to NB. We follow this with an introduction to NBs and their potential cellular origins, describe salient molecules, signalling pathways, mechanisms and conditions involved in promoting, selecting and maintaining NB CSC subpopulations, and that underpin their self-renewing, therapy-resistant and metastatic behaviour. We also review potential therapeutic strategies and future prospects for targeting and eradication of these bastions of therapeutic resistance, post-therapeutic relapse and metastatic progression.
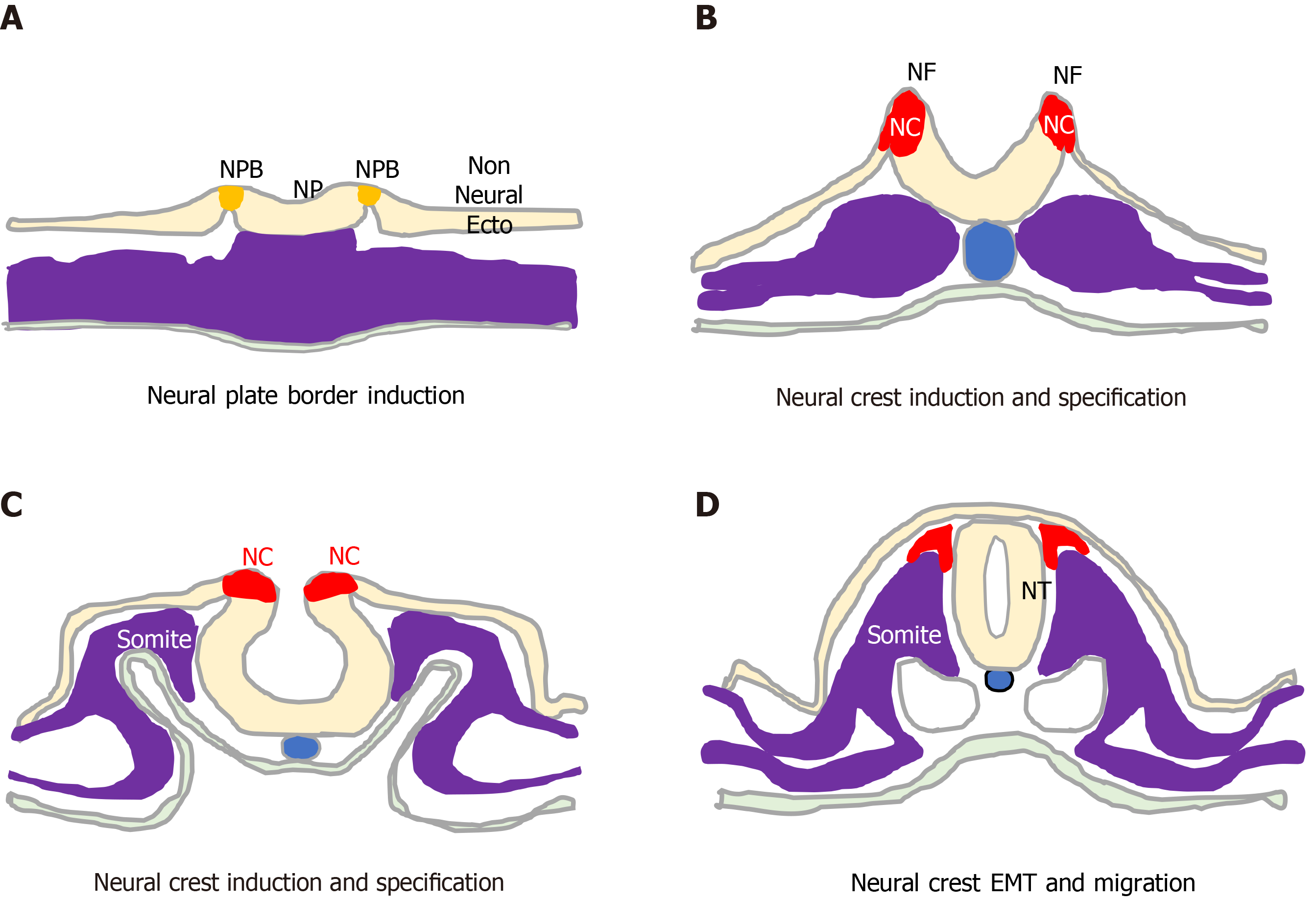
The unique vertebrate embryonic NC structure was first described by Wilhelm His in 1868 as a band of particular material lying between the presumptive epidermis “Hornblatt” and the neural tube, termed the “Zwischenstrang”, and the origin of spinal ganglia[1]. The NC, composed of multipotent NC cells, appears in humans towards the end of gastrulation at the dorsal edges of the neural folds and neural plate (NP) along the entire neuro-axis[2-5]. Once formed, NC cells undergo epithelial to mesen
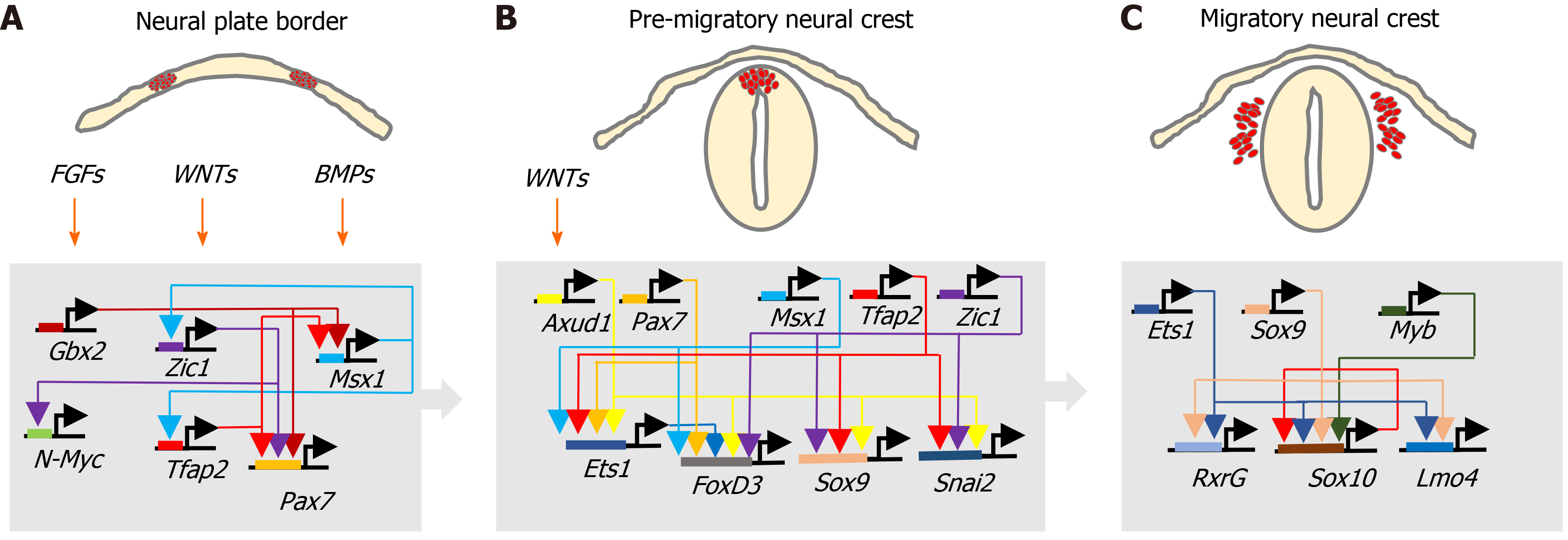
During development, ectoderm regional patterning forms the NP border and NP, from which the NC, neural tube (NT), nonneural ectoderm and placodes subsequently form[4,10]. This process is regulated by a morphogenic gradient of bone morphogenic protein (BMP) activity within the ectoderm, fibroblast growth factor and wingless/Int-1 (Wnt) signalling pathways[4,10,11]. Subsequent NC specification is an ordered multistep process[4,5], involving Snai2, FoxD3, Ets-1, Sox8, Sox9, Myc, Id2 and later Sox10 transcription factors[12-14], followed by EMT transition[6,10], delamination and NC cell migration throughout the developing embryo, for reviews see[4,6,11,15] (Figure 2).
NC cell EMT involves the loss of apical/basal polarity, altered expression of type 1 (E-cadherin and N-cadherin) and type 2 cadherins (cadherins 6, 7 and 8)[16-19], cytoskeletal reorganisation and proteolytic activity. It is initiated by Snai2, FoxD3, Sox9, Sox10 and Twist transcription factors, regulated by BMP, Wnt-planar cell polarity, Frizzled, RhoA-ROCK signalling in coordination with surrounding tissue morphogenesis[4,6,16], and involves: connexin-43[20], clatherin-mediated endocytosis[21]; cadherin 6B proteolysis[21-23] ; hypoxia-inducible factor (HIF)-a[24-26] ADAMs, γ-secretase and matrix metalloproteinases (MMPs) and matrix components, including collagen, fibronectin, laminin, tenascin, collagen IX, chondroitin-6-sulphate, aggrecan and versecan, which combine to lower intracellular adhesivity and increased motility[27-30]. With respect to the formation of NBs that originate from trunk NC cells, trunk NC cells that delaminate and migrate ventro-laterally via contact inhibited locomotion, co-attraction and chemotaxis, accumulate at the dorsal aorta, mix and then form bi-lateral sympathetic ganglia that go on to innervate various organs and skin[6,28-31].
The term NC stem cells (NCSCs) was introduced in 1992 by Stemple and Anderson[32], who demonstrated in vitro that rodent NC cells, in addition to differentiating into autonomic and sensory neurons, glia and smooth muscle cells, also exhibited self-renewing capacity[32]. At that time, NCSCs were considered a transient population in vivo and to be in vitro equivalents of embryonic stem cells from blastomeres. NCSCs were subsequently identified in post-natal sciatic nerve, dorsal root ganglion, the gut, bone marrow, cornea, heart, carotid body, dental pulp and periodontal ligament and skin tissues[15,33,34], as a multipotent self-renewing NCSC population resembling embryonic NCSCs in the adult organism[35]. This indicates that, despite the transient nature of the NC, the low self-renewal capacity of NC cells and rapid transition from multipotency to fate and differentiation restriction, undifferentiated NCSCs also populate migrating NC cell streams and post-embryonic tissues, providing an additional population of self-renewing NCSCs that, when necessary, can be called upon to differentiate into specific cell types in response to microenvironmental factors[36,37] and growth factor receptor activation[38], with self-renewal regulated by Wnt and BMP in early migratory NCSCs and later by responses to growth factors[39,40], representing a 4th germinal layer[15]. Multipotent NCSCs can be isolated from embryos and generated from human embryonic and pluripotent stem cells, with important implications for regenerative medicine and disease modelling[41-43]. Post-migratory NCSCs resemble embryonic counterparts in differentiation capacity, with stemness, migratory behaviour in migrating NC cell populations demonstrated at the single cell level by tracking, and purified cephalic NCSCs have been shown to differentiate into neurons, glia, melanocytes, chondrocytes, osteoblasts and smooth muscle cells[44,45]. A considerable fraction of the NC exhibits an SC phenotype, with fate decisions regulated later by environmental factors, including oxygenation status[46-49], exemplified by: Shh promotion of NC progenitors with mesenchymal skeletogenic, chondrogenic and neurogenic potential; stem cell factor promotion of NCSC survival and melanocyte lineage trophism, when combined with the neurotrophins nerve growth factor (NGF), brain-derived neurotrophic factor (BDNF) and NT3; endothelin-3 promotion of glial and melanocyte progenitor proliferation and survival, and basic fibroblast growth factor promotion of NCSC proliferation[46,47,50,51].
Although there are no specific individual markers for NC cells or NCSCs[4], gene expression patterns that identify potential NCSC populations, include VE-cadherin/ CD144, the epidermal growth factor (EGF) family member CFC1/Cripto, transcription factors Pax, Sox10, Hox, mash1, Phox2b; neurotrophic factor receptors p75NTR, RET and EDNRB, and the nerve-related proteins NF, NC-1, E/C8, HNK1, nestin and α4-integrin. RET expression identifies NCSCs within ganglia and is crucial for vagal NC development, P75NTR is used widely to purify NCSCs, Sox10 is considered to be a relatively specific and sensitive NCSC marker, and NSCSs in vitro express Sox10, P75NTR and RET[4,15,52,53].
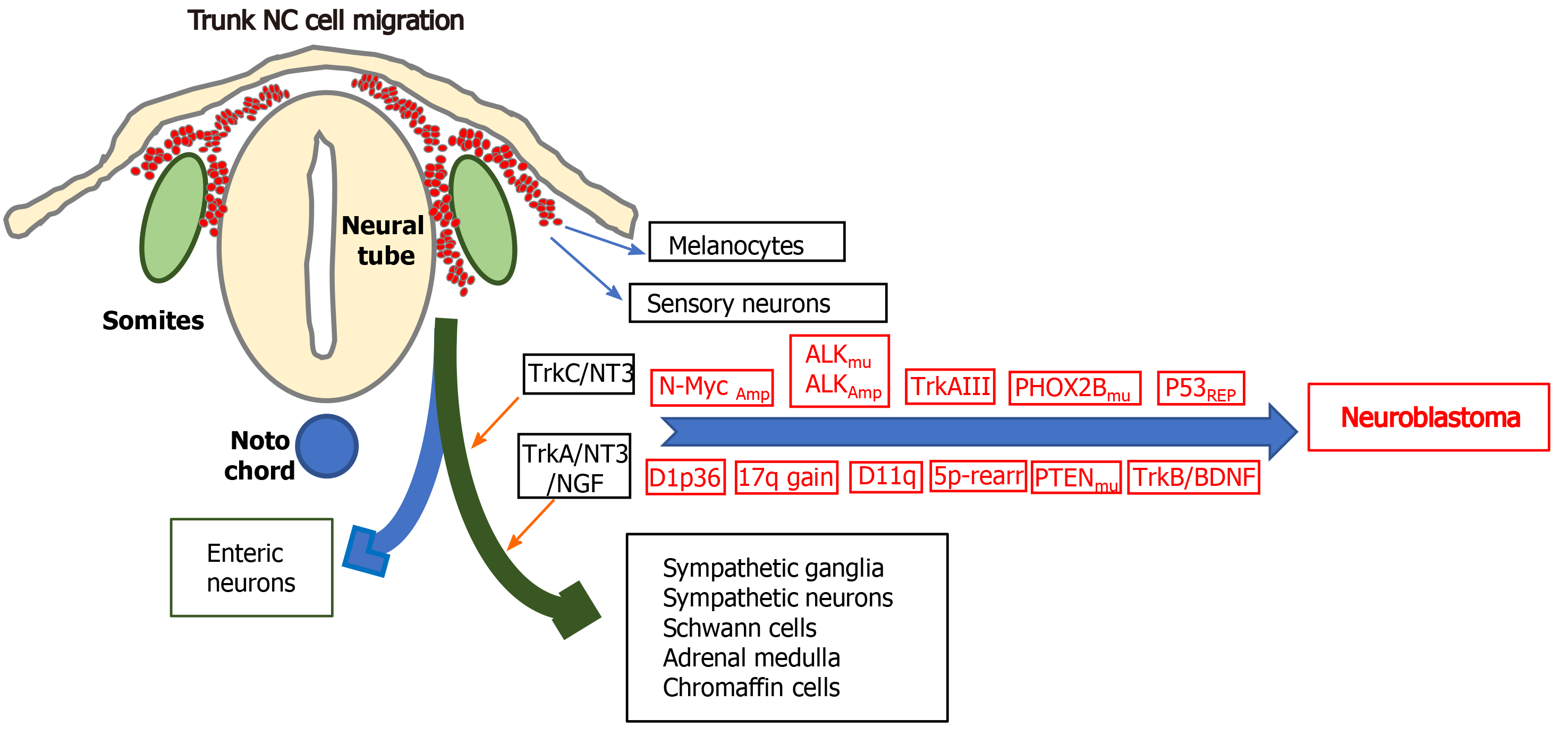
Neuroblastomas (NBs) are small round cell extracranial paediatric tumours that arise during embryonic development from trunk-derived NC cells of the sympathoadrenal lineage and account for approximately 15% of cancer-related childhood deaths. NBs develop anywhere along the sympathetic chain, are more frequent in the abdomen and adrenal medulla, exhibit broad clinical heterogeneity, ranging from spontaneous regression to aggressive metastatic disease and are highly refractory to therapy. Low and intermediate-risk NBs exhibit cure rates of 80%-90%, and < 50% for high-risk disease, with < 10% survival associated with relapsed recurrent disease, for recent reviews see[54,55].
Chromosome aberrations associated with high-risk NB, include hemizygous or homozygous 1p deletions, heterozygous 11q deletions, 17q gains, 5p15.33 rearrangements, and deoxyribonucleic acid (DNA) methylation[56,57]. Although NBs exhibit low somatic mutation rates and no single mutation can explain tumour initiation, activating mutations in the anaplastic lymphoma kinase (ALK) gene characterise familial NBs and 7%-10% of non-familial NBs, activating PTPN11 mutations are associated with approximately 3% of NBs, activating N-Myc mutations with approximately 2% of NBs and activating KRAS mutations with approximately 1% of NBs. Inactivating mutations that are associated with NB include, paired-like homeobox-2 mutations in familial NBs and approximately 4% high-risk NBs, ATRX transcriptional regulator mutations in approximately 2.5% of NBs, p53 mutations in approximately 2% of primary and approximately 10% of relapsed NBs, BRCA2 mutations in approximately 1% of NBs and ARIDIA1A/1B mutations in approximately 3% of NBs[58-63]. Despite low level p53 mutation, p53 inactivation is common in NB[64], and several long non-coding ribonucleic acids (lncRNAs) have been implicated in NB progression[65].
N-Myc overexpression combined with activating ALK mutations have been implicated in NB initiation. N-Myc regulates trunk NC cell migration, balances sympathoadrenal progenitor expansion with apoptosis and N-Myc gene amplification characterises approximately 50% of advanced stage high-risk NBs. Mice transgenic for tyrosine hydroxylase-promoted N-Myc expression form NBs in sympathetic ganglia[66], N-Myc overexpression induces NBs in zebra fish[67], and mice transgenic F1174F mutation-activated ALK develop NBs in the presence of high-level N-Myc expression[68]. Tyrosine kinase receptor A (TrkA) and tyrosine kinase B (TrkB) neurotrophin receptors have also been implicated in NB pathogenesis[16,69]. TrkA is required for sympathetic nervous system development and is expressed by NC cells in sympathetic ganglia where it regulates proliferation, survival, differentiation and culling under neurotrophin limiting conditions[16]. TrkA expression in NB associates with favourable prognosis, spontaneous regression and Schwann cell stroma-rich ganglioneuromas[16,69]. However, NBs exhibiting Ip36.2 deletions lose cell surface TrkA expression, in a potential tumour initiating mechanism based upon NC cell escape from neurotrophin-dependent differentiation and apoptosis[70]. Furthermore, oncogenic alternative TrkAIII splicing also represents a potential stress-regulated tumour promoting switch in NB, is exhibited by advanced stage metastatic and relapsed NBs, transforms NIH 3T3 fibroblasts and exhibits oncogenic activity in NB models. Both of which help to explain why TrkA expression is not always associated with a favourable prognosis in NB[16,69,71,72]. Other potential NB susceptibility genes include LIN28B, CASC15, NBAT-1, BARD1, DDX4, IL31RA, HSD17B12, HACE1, LIN28B and NEFL. Of these, LIN28B induces N-Myc expression and alters the expression of micro ribonucleic acids (miRNAs), RAN and Aurora A kinase, in a potential NB-inducing Lin28B-Ran-Aurora A kinase axis, BARD1 polymorphisms promote NB cell proliferation and invasiveness, identifying BARD1 as a potential NB tumour suppressor, and the lncRNAs CASC15 and NBAT1 interact with 17q gain-associated Sox9 and USP36 to inhibit NB differentiation[65].
The cellular origins of neuroblastoma: With respect to the cellular origins of NBs, NC progenitors and NCSCs have both been identified as potential origins by their behaviour in vitro and in vivo, by Myc or tamoxifen-inducible Myc-ERT gene expression patterns in immortalized murine NC progenitor cells engineered to express mutated ALK oncogenes[73,74] and by enforced N-Myc transformation of wild-type NC cells grown from murine NT explants, into NB forming cells[75]. A central nervous system (CNS) neural stem cell-like origin has also been proposed as an origin for some N-Myc amplified NBs, as ectopic N-Myc expression in avian NC cells promotes NC cell conversion to neural stem cells (SCs) that exhibit trunk migration and inadvertent colonization of sympathetic chain ganglia, as a potential primed NC origin for NB, helping to explain gene expression patterns in some N-Myc amplified NBs[76]. A sympathoadrenal progenitor origin for NB is supported by transgenic mouse models of NC targeted N-Myc and ALKF1174L expression, which results in perinatal lethality, indicating the NC cannot support oncogenic transformation in vivo, and the absence of the NCSC marker Sox10 and NSCS gene regulatory network expression in human NBs and NB cell lines, contrasting with Sox9 and Sox9-regulated gene expression, consistent with a sympathoadrenal (SA) rather than NC progenitor origin[77]. Furthermore, the migratory behaviour of human NB grafts in avian embryonic NC recapitulates the collective migratory behaviour of SA progenitors, avoiding endogenous no go areas and coalescing in sympathetic ganglia and adrenal medulla, which are SA progenitor target sites[78,79]. A potential Schwann cell progenitor origin for some NBs has also been proposed, considering that > 50% of NBs arise in the adrenal medulla and in sympathetic ganglia Schwann cell progenitor target sites, and exhibit distinct features[80]. This suggests different origins for NBs that develop within the sympathetic chain and adrenal medulla, further supported by differences in dissemination and migratory behaviour, illustrated by the migratory behaviour of NB cells transplanted onto the avian embryo NC, which migrate to form primary tumours in sympathetic ganglia, then exhibit secondary migration along nerves and blood vessels, reminiscent of Schwann progenitor migratory behaviour[9,78,79,81]. Malig
Neuroblastoma cancer stem cells: Consistent with potential NCSC, CNS SCs, sympatho
CSC markers differ between tumour types, many are also expressed by normal cells and cell surface molecular markers can be used for targeted therapy. It is important, therefore, to understand markers and patterns of marker expression that specifically define NB CSC populations, since private highly specific functional CSCs cell surface markers represent the best targets for monoclonal antibody and chimeric antigen receptor T-cell immunotherapy (CAR-T) therapy, and CSC-specific intracellular markers and signalling pathways may provide targets for small molecular inhibitors.
CD133, CD44 and CD24: The 5 transmembrane domain cell surface 120 kDa glycoprotein CD133 (AC133/Prominim-1) was originally identified as a haemato
The transmembrane glycoprotein CD44 mediates cell-cell interactions and cellular attachment to extracellular matrices by binging hyaluronic acid. CD44 is expressed as a variety of isoforms and in NB is a strong prognostic marker, together with N-Myc. Both CD44 expression and lack of expression have been associated with tumour initiating NB CSC-like properties[111,112]. CD44 negative NB populations express higher levels of the tumour initiation marker CD24, and exhibit a gene expression pattern characterised by reduced p21Cip1, p16INK4a and p15INK4b expression, consistent with uncontrolled cell cycle progression, reduced expression of apoptosis factor NAIP, Bcl-xl, Bax and IRF1, consistent with apoptosis evasion and are tumou
The GPI-anchored CD24 cell surface sialo-glycoprotein is expressed by differentiating neuroblasts, is crucial for neural development and is involved in neurite outgrowth and neurogenesis. CD24 interacts with β1-integrins, activates mitogen-activated protein kinase (MAPK) through focal adhesion kinase and Src kinases, is implicated in NF-κB, Notch and Shh signalling[113] and promotes metastasis by interacting with P-selectin on activated platelets and endothelial cells[114,115]. CD24+/CD34+ enriched NB populations are more tumourigenic in mice, CD44- CSC-like tumour initiating NB cells express high levels of CD24[111] and CD24+ CSC-like tumour initiating cells are present in bone marrow NB metastases, suggesting that CD24 enriches the tumourigenic potential of CSC-like tumour initiating NB cells within the bone marrow[99].
Delta-like 1 homolog and prohibitins: The development-regulated transmembrane protein delta-like 1 homolog (DLK1), which regulates stem/progenitor cells, is overexpressed in NBs and is robustly expressed in undifferentiated NB cells[116,117]. In NB cells small interfering ribonucleic acid (siRNA) inhibition of DLK1 expression promotes spontaneous neuronal differentiation, decreases clonogenic and colony forming capacity and suppresses tumourigenicity, and vice versa for DLK1 overexpression[116,117]. HIFs bind and activate the DLK1 promoter[117], linking DLK1 expression to tumour hypoxia and oncogenic HIF-1 and HIF-2 activation, explaining the localisation of DLK1+ NB cells to hypoxic niches[116]. DLK1 also interacts specifically with prohibitins (PHB) 1 and 2, regulating mitochondrial function and reactive oxygen species (ROS) production via a DLK1/PHB axis involved in NB CSC self-renewal and clonogenic capacity[118]. Conversely, high DLK1 expression is associated with NB cells arrested in relatively late-stage chromaffin lineage differentiation[119].
L1-CAM: The neural cell adhesion molecule L1-CAM regulates neuronal growth, survival, migration, axonal outgrowth and neurite extension[120], and has been implicated in CSC marker upregulation and aggressiveness in gliomas. Although L1-CAM expression has been reported to associate with better prognosis in NB[121], L1-CAM knockdown in NB cells down-regulates N-Myc expression, up-regulates PTEN expression, and inhibits self-renewal, tumour sphere formation and migration, suggesting that L1-CAM may indeed play an important role in NB CSC-like behaviour[122].
ABC transporters: Members of the membrane-associated adenosine triphosphate-binding cassette ABC transporter super-family promote molecular transport across extracellular and intracellular membranes, and are important drug-efflux pumps that increase multidrug-resistance[123,124]. ABCG2 and ABCA3 are co-expressed in Hoechst dye excluding CD133+ SP CSC-like NB cells that exhibit enhanced chemo-resistance and tumourigenic activity in vivo[97,124-126].
TRPM7: The transient receptor potential cation channel family member and kinase TRPM7 regulates mechanical cellular interactions within microenvironments by interacting with the cellular cytoskeleton, integrating adhesion and differentiation responses with microenvironmental stimuli. TRPM7 expression is essential for maintaining NC cell multipotency, contributes to NB by disrupting NB cell maturation and preserving progenitor CSC-like features, and is promoted by N-Myc in NB cells[127,128]. TRPM7 expression is closely associated with NB CSC-like cell migratory and metastatic behaviour[129], controls developmental transcription through SNAI2, helping to maintain progenitor-like features[130,131], enhances therapeutic resistance[132] and may also promote metastasis via Hsp90a/uPA/MMP-2 signalling[133].
Lamin A/C: The nuclear membrane-associated protein lamin A/C regulates nuclear stability. Knockdown of lamin A/C expression in NB cells promotes aggressive, self-renewing tumour-initiating CSC-like populations that exhibit a more drug-resistant phenotype[134], increases CSC marker Oct4, Nanog, Sox2 and N-Myc, CD133, Sox2 and SSEA4 expression, and enhances neuro sphere forming capacity[135].
Cryptic family member CFC1: The epidermal growth factor family member CFC1 is also considered a potential NB CSC marker, involved in NB cell growth, colony and tumour sphere formation in vitro and NB xenograft tumour formation in vivo. CFC1 inhibits NB differentiation induced by activin A and smad-2 phosphorylation, implicating collaborative signalling between EGF and CFC1 in NB[136].
Frizzled/Wnt signalling and LGR5/GRP49: Wnt signalling is critical for NC induction, specification, EMT and migration, occurs via canonical and non-canonical Wnt pathways, and has been closely implicated in NB progression[137,138]. In the canonical pathway, Wnt ligands bind Frizzled receptors inducing nuclear translocation of β-catenin, which binds and activates Tcf/Lef transcription factors, whilst blocking Tcf/Lef target gene repression, whereas non-canonical pathways include planar cell polarity and calcium mobilisation via G-protein activation. In NBs, Frizzled Wnt receptor Fzd6 expression predicts poor survival and marks rare HIF1/2a-positive CSC-like tumour initiating NB cell populations located in hypoxic tumour areas[139]. Fzd6 positive NB cells share biological features with CSCs including neurosphere formation, increased invasive capacity and resistance to chemotherapeutic agents, Fzd6 is also required for non-canonical Wnt pathway gene expression and is implicated in aggressive NB behaviour, consistent with a role in promoting and maintaining NB CSC-like subpopulations[140]. Wnt pathway promotion of NB CSCs also involves SLC34A2 transcription factor, which negatively correlates with overall and relapse-free survival in NB patients, promotes NB CSCs by binding the miR25 promoter and activating miR25 expression, which binds the Gsk3β messenger RNA (mRNA) 3’-UTR, reducing Gsk3β protein expression, resulting in Wnt signalling[141]. The genotoxic agent doxorubicin also enhances Wnt signalling in NB cells in association with up-regulated Frzd4 and 6 expression, implicating the DNA damage-response in NB CSC-like states[140] and subsequent chemoresistance[142,143]. Hyperactivation of the Wnt/β-catenin pathway in NB cells is also promoted by NEU4 Long sialidase, which enhances proliferation, G1 to S phase transition and promotes a more undifferentiated CSC-like phenotype, potentially via Myc, Nanog, Oct4, CD133 and Nestin pluripotency gene expression, identifying NEU4L as a novel potential therapeutic NB CSC target[144]. The SC marker LGR5/GPR49 also promotes canonical Wnt signalling and is overexpressed in advanced stage N-Myc amplified and ALK-mutated NBs. LGR5/GPR49 knockdown induces NB cell apoptosis in association with diminished MAPK signalling, identifying LGR5/GPR49 as a novel potential NB CSCs therapeutic target[145,146].
Neurotrophin receptors P75NTR, TRKA, TRKAIII, TRKB, TRKC and sortilin: Neurotrophins NGF, BDNF, NT3 and NT4/5 and their receptors are critical for the development and maintenance of vertebrate central and peripheral nervous systems, and have been implicated in NB regression and metastatic progression. NT receptors include: the tropomyosin-related tyrosine kinases TrkA that binds NGF and NT3; TrkB that binds BDNF and NT4/5; TrkC that exclusively binds NT3; CD271/p75NTR that binds mature NGF with low affinity and all pro-form NTs with high affinity, and sortilin that binds mature NGF and pro-form NGF, BDNF and NT-3. NT/TrkA activation inhibits default apoptotic programmes to promote NT-dependent survival through PI3K/Akt/NF-κB signalling and Bcl-2 family induction and in the absence of NTs, TrkA and TrkC, but not TrkB, induce apoptosis through a CD271/p75NTR-dependent mechanism, identifying TrkA and TrkC as dependence receptors. TrkC is the only NT receptor expressed during early embryogenesis and during neurulation is detected in the NT and NP anlage, and in both pre-migratory and migrating NC cell subsets. NT3/TrkC activation is required for NC cell survival at target sites and, during NC migration, somites express NT-3 and sympathetic neuroblasts and neurons express NT3 and TrkC, promoting neuroblast survival, proliferation and subsequent differentiation. This temporary effect declines and switches to NGF-dependency, associated with reduced TrkC expression and increased TrkA and CD271/p75NTR expression, regulated in part by autocrine NT3 activity. NT3/TrkA interactions continue to promote sympathetic neuroblasts, neuron survival and target organ innervation, then changes gradually to dependence upon NGF/TrkA interaction[16,69].
CD271/p75NTR is a NC SC marker, and self-renewing CD271/p75NTR+ neural SCs co-express TrkA, TrkAIII, TrkB and TrkC. In general, NB expression of CD271/p75NTR is associated with favourable outcome and CD271/p75NTR exhibits tumour suppressor function in NB models, inducing differentiation, promoting apoptosis and reducing tumourigenic activity. NB CSC-like cells, however, express CD271/p75NTR, CD133+/CD271/p75NTR+ self-renewing, clonogenic NB cells produce more malignant tumours, and CD271/p75NTR has been closely linked to migratory and invasive behaviour of CSCs, implicating CD271/p75NTR in promoting malignant NB CSC-like behaviour[147].
Studies on NT receptors in NB initiated with the detection of an inverse relationship between high N-Myc and low TrkA expression is associated with poor prognosis and mid to high TrkA expression in non-N-Myc amplified disease[148]. Low TrkA expression combined with N-Myc amplification and expression, in general, characterises unfavourable NB but N-Myc amplified NBs also exhibit heterogeneity in TrkA expression, with a small number also exhibiting high TrkA expression, indicating that a more complicated relationship between N-Myc and TrkA in NB[16,69]. In general, moderate to high TrkA expression distinguishes non N-Myc and N-Myc amplified NBs but does not distinguish prognostic groups in non-N-Myc amplified NBs. Low TrkA expression in NBs may relate to a non-TrkA expressing sympathoadrenal cell origin in coalescing sympathetic ganglia, paraganglia or adrenal medulla anlage during development or N-Myc repression of TrkA expression post-transformation through promoter methylation. Conversely, high TrkA expression in NB may relate to an undifferentiated TrkA expressing NC progenitor origin within the sympathetic chain or adrenal primordia or to induction post-transformation by either NTs, growth factors and/or cytokines. In NB models, fully spliced TrkA receptors exhibit tumour suppressor activity and restore NGF-induced neuronal NB cell differentiation, via IGF2, c-Src, PKCe and Ras/MAPK/Erk signalling, reduce angiogenic factor expression, angiogenesis and tumourigenicity and promote genetic stability. TrkA also induces p53-dependent NB cell apoptosis through CCM2, Erk and caspase-7. NB cell responses to NT/TrkA activation are optimized by CD271/p75NTR, and TrkA/CD271/p75NTR co-expression in NB carries a good prognosis[16,69,148]. The discovery of the oncogenic alternative TrkAIII splice variant in advanced stage NBs and NB cell lines, however, challenges the hypothesis of an exclusively tumour suppressing role for TrkA in NB. TrkAIII alternative splicing is development and stress-regulated and generates an oncogenic TrkAIII isoform that transforms NIH3T3 cells and exhibits oncogenic activity in NB models[69,71,72]. In NB cells, TrkAIII expression induces centrosome amplification and increases genetic instability[149], promotes stress-induced aerobic glycolysis[150], enhances angiogenic factor expression, impedes NGF/TrkA signalling through Ras/MAPK, induces a survival adapted endoplasmic reticulum (ER)-stress response via retrograde ERGIC to ER recycling in vitro[72,151], and enhances tumour-associated angiogenesis, tumourigenesis and metastatic capacity in vivo but does not restore NGF responsiveness nor induce NB cell differentiation or apoptosis[69,71,72]. A potential role for TrkAIII in NB CSCs comes from observations that TrkAIII increases mitochondrial Superoxide dismutase 2 (SOD2) expression and activity in NB cells, enhancing resistance to ROS-induced death within the context of CSC-like phenotype[152]. Furthermore, TrkAIII promotes centrosome amplification, and microtubule polymerization resulting in polyploid and aneuploid de-differentiated anaplastic phenotypes[153], implicating TrkAIII in selecting and maintaining CSC-like NB subpopulations within stressful pathological and therapeutic tumour microenvironments[72].
TrkB has also been implicated in NB pathogenesis, progression and therapeutic resistance[16,154-156], exhibits a positive correlation with N-Myc amplification and expression, and is stimulated by activated Erb-B in NB cells. Unfavourable NBs express BDNF providing a potential autocrine/paracrine survival mechanism and TrkB is transiently expressed by sympathoblast subpopulations during sympathetic nervous system development, providing a potential origin for TrkB expressing NBs, with BDNF expression acquired at a later stage[157]. TrkB expression is increased by HIF-1 suggesting that tumour hypoxia may promote TrkB expression in NBs, and BDNF/TrkB interaction increases NB cell survival, chemoresistance, invasive capacity, angiogenesis factor expression, angiogenesis and metastatic behaviour[16,156,157]. Recently, an engineered TrkB receptor, devoid of Ig-like domains, analogous to TrkAIII, has been shown to be oncogenic[158], although a similar endogenous isoform(s) has not yet been described in NB.
TrkC expression in NB is associated with a favourable outcome. However, high level NT-3 and TrkC co-expression has been identified in a subset of advanced stage IV NBs, providing a potential paracrine/autocrine survival and proliferation mechanism for selection in tissues that do not express NT3, similar to migrating NC-derived sympathoblasts, as a potential cellular origin for this NB subset[16]. In further support of NT receptor involvement in NB stemness, I-Type CSC-like NB cells exhibit high-level TrkA, TrkB, TrkC and CD271/p75NTR expression[92].
ROR1: The receptor tyrosine kinase-like orphan receptor ROR-1 is related to CSC-like phenotypes[159], is expressed across all NB stages, correlates with survival and prognosis, and targeted anti-ROR-1 therapy results in additive cytotoxicity to NB cells, identifying ROR1 as a novel regulator of the NB CSC compartment and potential therapeutic target[160].
Apoptosis signal regulating kinase 1: The apoptosis signal-regulating kinase apoptosis signal regulating kinase 1 (ASK1) is a member of the MAPK family and activates JNK and p38MAPK in response to ER-stress, oxidative stress and calcium influx. ASK1 is co-expressed and associates with TLX/NR2E1 in CSC-like NB SP cells, particularly in hypoxic conditions within the CSC niche. ASK1 phosphorylation stabilises TLX/NR2E1, resulting in HIF-1α induction and VEGF-A expression, contributing to angiogenesis and NB CSC survival[161].
STAT3 granulocyte colony-stimulating factor receptor and CD114: STAT3 transcrip
FOXD3 and FOXM1: PDFG, Wnt and BMPs signalling regulate NC induction and specification through transcription factors, including FoxD3 (Teng et al[174], 2008). The forkhead family transcriptional repressor FoxD3, originally characterised in embryonic SCs and multipotent NC cells[175,176], is considered to be an early NC lineage marker that specifies fate by upregulating HNK1 and Slug expression, involved in maintaining the undifferentiated state of migrating NC cells and self-renewal potential[174]. In NB, FoxD3 acts as a tumour suppressor, regulating the expression of N-Myc downstream regulated 1 and downstream genes. FoxD3 repression promotes NB tumourigenic, invasive, metastatic and angiogenic activity in vitro and in vivo, and is associated with NB CSC phenotypes[177]. In contrast, FoxM1 is a proliferation-specific transcription factor, the knockdown of which induces NB cell differentiation, associated with Sox2 and Bmi1 repression. FoxM1 directly activates Sox2 expression in NB cells and regulates both Sox2 and Bmi1 expression, required for neural stem cell self-renewal and maintenance of an undifferentiated NB CSC-like state[176,178].
Bmi1 and Kif1Bβ: The polycomb group transcriptional repressor Bmi1, originally identified in murine lymphomas as an oncogenic partner of c-Myc, forms part of Polycomb repressor complex 1, involved in gene repression through chromatin modification[179,180]. Bmi1 is required for NCSC self-renewal, CSC self-renewal and tumourigenicity[181,182] and Bmil knockout reduces NCSC self-renewal and results in progressive loss of NCSCs in mice[183]. NB CSCs express Bmi1, and the Bmi1 promoter binds and is activated by E2F-1 transcription factor[184]. Knockdown of Bmi1 expression in NB cells induces differentiation and growth suppression, and the role of Bmi1 in NB tumourigenesis has been linked to FoxM1, which activates the expression of both Sox2 and Bmi1[178]. In I-type CSC-like NB cells, Bmi1 is required for self-renewal but also influences lineage commitment. Bmi1 repression promotes S-type differentiation, high Bmi1 expression induces N-type differentiation and Bmi1 over-expression induces neuronal differentiation, implicating fine concentration-dependent Bmi1 control of the delicate balance between NB CSC self-renewal and differentiation[182]. Bmi1 transiently inhibits the p53 response to N-Myc-induced oncogenic-stress in perinatal NB precursor cells by directly binding and promoting p53 ubiquitination and degradation, in a general mechanism for p53 inactivation in NBSC-like progenitors, susceptible to N-Myc oncogenesis during embryonal development[185]. Bmi1 is also a N-Myc target-gene involved in repressing expression of the NB tumour suppressors TSCL1 and Kif1Bβ, with implications for NB pathogenesis[186,187], and promotes IkBα degradation via ubiquitination within the context of the SCF complex, inducing CSC survival NF-κB signalling, tumour-associated inflammation, proliferation, MMP-9 expression and EMT[188-191].
The kinesin Kif1Bβ acts downstream of EglN3 to induce neuronal apoptosis and exhibits loss of function mutation or loss associated with Ip36.2 deletion in NBs, resulting in apoptosis evasion[192]. Kif1Bβ is also required for neuroblast differentiation and in mouse sympathetic neuroblasts deleted of Kif1Bβ and human NBs that lack Kif1Bβ, neural differentiation is ablated. Loss of Kif1Bβ expression contributes to undifferentiated aggressive NBs, characterised by high-level Sox10 expression, loss of cell surface TrkA expression, and up-regulated expression of ALK ligands and suspected MDK and NHLH2 NB oncogenes[70]. Furthermore, NB cells that lack cell surface TrkA, due to Kif1Bβ deletion or mutation, do not exhibit NGF-mediated differentiation and in the absence of neurotrophins, escape TrkA-induced apoptosis, compounding the Sox10 expressing de-differentiated NB CSC-like state[70]. Bmi1/Kif1Bβ involvement in NB CSC induction and maintenance has also been associated with a novel alternative extra-short form NF-YAx splice variant subunit of the ubiquitous NF-Y transcription factor. NF-YAx was originally identified in primary human NBs, is expressed during murine embryonic development at times corresponding to sympathetic neuroblast culling and is induced in NB cells by the genotoxic chemotherapeutic agent doxorubicin. NF-YAx expression in NB cells represses Bmi1 expression and enhances Kif1Bβ expression, resulting in necroptosis, suggesting a NB tumour suppressor function. However, chronic NF-YAx expression also selects tumourigenic NB cells that are resistant to NF-YAx-induced cytotoxicity, do not exhibit NF-YAx-induced Bmi1 repression and Kif1Bβ induction, and exhibit a CSC-like expression signature characterised by enhanced CD117, p75NTR, Nanog, Nestin, Sox2 and EglNL3 mRNA expression, indicating that NF-YAx selects NB CSC phenotypes by eliminating cells sensitive to NF-YAx-induced necroptosis, in a potential DNA-damage-induced CSC selection mechanism, with implications for NB genotoxic chemotherapy[193].
N-Myc, cMyc and ALK: Myc/Max complexes inhibit ESC and iPSC differentiation and have been implicated in the epigenetic reprogramming that enhances CSC phenotypes[194]. Both N-Myc and mutated ALK are important NB oncogenes and collaborate to promote NB progression. In embryonic sympathetic neuroblasts, N-Myc overexpression promotes proliferation but does not sustain survival, and long-term ALKF1174L activation promotes cell cycle arrest, differentiation and survival of post-mitotic neurons, in association with NEFM, RET and VACHT expression and decreased BMF, BIM, TH, CDC25A and CDK1 expression. N-Myc and ALKF1174L co-expression maintain neuroblast proliferation and promote a different differentiated phenotype, characterised by SKP2, CCNA2, E2F8 and DKC1 expression. This is consistent with initial steps towards NB development, and is supported by demonstration that N-Myc and ALKF1174L are sufficient to drive NB development from NC progenitor cells[73,195,196]. N-Myc also forms a positive feedback loop with its cis-antisense gene N-Cym, co-amplified with N-Myc, and Oct4 transcription factor[197]. This contributes to a more CSC-like aggressive phenotype in N-Myc amplified NBs, as N-Cym stabilises N-Myc, resulting in Oct4 and CSC-related Nanog, Sox2 and Lin28 expression[198]. N-Myc also promotes survival by up-regulating components of the error-prone non-canonical alternative nonhomologous end-joining DNA repair pathway, resulting in increased DNA repair activity to overcome DNA damage-induced death. This behaviour is shared with NCSCs, consistent with a NCSC status of N-Myc amplified NB cells[199]. N-Myc also promotes lysine 4 histone H3 trimethylation in stem cell-related genes, and in microarray studies, N-Myc amplified NBs exhibit Lif, Klf2, Klf4 and Lin28b ESC-related factor expression, with N-Myc observed to directly bind to the Lif promoter, implicating N-Myc in the regulation of overlapping SC-related gene expression, CSC-like pluripotency and self-renewal in NB[200]. Glutamine is also involved in N-Myc regulation of NB CSCs, as glutamine deprivation leads to N-Myc/c-Myc cooperation, resulting in CD133 expression in N-Myc amplified NB cells, associated with changes in acetaldehyde dehydrogenase (ALDH) activity[201].
NF-Y: The ubiquitous heterotrimeric transcription factor NF-Y binds inverted CCAAT-boxes in approximately 70% of gene promoters and is composed of NF-YA, NF-YB and NF-YC subunits[202-204]. Alternative splicing of the NF-YA subunit has been implicated in cellular staminality, differentiation, apoptosis and transformation, and the short alternatively spliced NF-YAs isoform forms part of the SC transcriptional circuitry, predominates in ESCs and is lost upon differentiation, whereas fully-spliced long-form NF-YAl promotes differentiation and NF-YA loss induces senescence or apoptosis[205-207]. Alternative NF-YAs splicing is promoted by oncogenes and converts tumour suppressing, differentiation promoting NF-Y complexes predominated by NF-YAl into tumour and CSC promoting complexes predominated by NF-YAs[208,209]. Recently, a novel developmental and doxorubicin-regulated extra short-form alternative NF-YAx splice variant has been identified in primary stage 2 and stage 3 NBs[210,211]. This novel isoform forms functional NF-Y complexes but has lost capacity to bind the ubiquitous cancer-associated transcription factor SP1[211], characterising NF-YAx as a functional NF-Y modifier. NF-YAx expression in NB cells promotes Kif1Bβ-dependent necroptosis and is expressed during murine embryo development at times corresponding to Kif1Bβ-dependent sympathetic neuroblast culling, consistent with a potential tumour suppressor function. However, NF-YAx also selects CSC-like subpopulations resistant to NF-YAx-induced death that exhibit enhanced resistance to the genotoxic agent doxorubicin, which also promotes alterna
Paired related homeobox 1 and notch-3: Cell types identified in NBs include interconverting de-differentiated, mesenchymal, multipotent NC-derived progenitor-like, adrenergic and intermediate cell types that exhibit distinct super enhancer and associated transcription factor networks and divergent gene expression profiles[212,213]. Expression of the embryonal-restricted transcription factor paired related homeobox 1 (PRRX1) in mesenchymal tissues, reprogrammes adrenergic NB cell super enhancer and mRNA landscapes towards the NC-derived progenitor-like mesen
Repressor element 1 silencing transcription factor, MUSASHI1 and C-KIT/CD117: Repressor element 1 silencing transcription factor (REST) zinc finger transcription factor is a master repressor of neuronal programmes, helps to maintain de-differentiated SC and CSC states and promotes genomic instability, contributing to cellular transformation[216,217]. In NBs, REST exhibits oncogenic activity and high REST expression is associated with poor prognosis, supporting a potential role in promoting NB CSC-like behaviour[218-220].
The mRNA-binding protein Musashi1 is a critical regulator of SC self-renewal and has been implicated in CSC-like self-renewal capacity in NB by promoting target mRNA translation and stem cell-fate transition[105,221]. Although the signalling mechanisms involved in Musashi1 function in stem/progenitor cells are unclear, Musashi1 is regulated by phosphorylation and by MAPK signalling in oocytes, and exhibits functional duality by promoting translation and repressing mRNA targets, in a similar way to Musashi2, which represses TGFβR1 mRNA, whilst activating translation of SMAD3 mRNA in proliferating K562 myelogenous leukemia cells[222].
The c-kit/CD117 stem cell factor receptor tyrosine kinase is a SC and CSC marker[91,223], and has been reported to be one of seven genes exhibiting elevated expression exclusively in NB CSCs[92].
JARID1B: The H3K4me3 histone demethylase JARID1B is involved in ESC fate and plays important roles in developmental gene expression and neural differentiation. In NB, JARID1B expression enhances CSC-like behaviour, therapeutic resistance and enables transit between distinct CSC-like cell populations via the PI3K pathway[224]. JARID1B knockdown increases NB cell sensitivity to cisplatin and reduces invasion and tumourigenic capacity, in association with down-regulation of Notch/Jagged signalling, consistent with a novel NB CSC modulator and therapeutic target[225].
TLX/NR2E1: The orphan nuclear receptor TLX/NRE2 promotes self-renewal in NB CSC-like cells and correlates with poor patient survival. In NB CSC-like cells, TLX/NRE2 is co-expressed with the neural SC/progenitor markers Nestin, CD133 and Oct 4, the neural SC/progenitor migration markers CD15 and MMP-2, and binds and activates Oct4 and MMP-2 promoters[226].
MMPs and TIMPS: MMP-9 is involved in almost all of the hallmarks of cancer, including the metastatic behaviour of CSC-like cells[227]. CSC-like I-type but not S-type NB cells exhibit constitutive MMP-9 expression and spontaneous S-type to I-type NB cell CSC-like EMT is associated with NF-κB-induced MMP-9 expression and enhanced MMP-9-dependent invasive capacity in vitro, implicating MMP-9 in NB CSC metastatic behaviour[93,227]. NB CSC-like cells also constitutively express the polycomb transcriptional repressor Bmi1[193], which promotes IkBα degradation, activates NF-κB[191] and increases tumour aggressiveness via the NF-κB/MMP-9 pathway[188,189]. Bcl2, which also exhibits enhanced expression in NB CSCs, also promotes MMP-9 expression by enhancing NF-κB activity[228], NB cells exhibit methylation-dependent repression of the metalloproteinase inhibitor TIMP-2 and secrete redox enzymes that inhibit TIMP activity, adding to an aberrant MMP/TIMP equilibrium in NB CSCs[210]. TLX/NR2E1-induced MMP-2 expression has also been implicated in the migratory and invasive behaviour of NB CSC-like cells[226].
Nestin: The neural stem/progenitor cell marker and intermediate filament protein nestin is considered to be a selective marker of bone marrow-derived mesenchymal SCs (MSC)[229] and CSCs[230]. Nestin is expressed in NBs, NB CSC-like subpopulations and is associated with aggressive NB behaviour[152,231]. Nestin expression is involved in SC, MSC and CSC self-renewal, angiogenesis and is promoted by Notch signalling, TrkAIII and N-Myc, identifying nestin as a NB-associated, oncogene-regulated, stemness gene[152,229,231].
SPDYA/SPY1: The cell cycle regulatory protein SPDYA/Spy1 controls CDK2 activity at the G1/S transition phase of the cell cycle and is a critical regulator of NB CSC-like behaviour. SPDYA/Spy1 regulates the self-renewal capacity of CD133+ NB CSC-like subpopulations[232], upregulates multipotency markers, drives NB tumourigenicity by activating cyclin dependent kinases, and its knockdown decreases c-Myc expression and impairs retinoic acid-induced differentiation of NB CSCs[233].
ALDH1 Isoenzymes: ALDHs are NAD(P)+-dependent enzymes that catalyze aldehyde oxidation to carboxylic acids, and are expressed by and active in haematopoietic SCs and NB CSCs[234]. NB cells express all 19 ALDH isoforms and ALDH1A2 and ALDG1A3 isoforms are potential markers of NB CSCs involved in promoting self-renewal, tumour sphere growth, colony formation, NB xenografts tumourigenicity and resistance to 13-cis-RA-induced differentiation[235,236].
LIN28B: The RNA binding protein LIN28B is also considered an NB oncogene. Lin28B represses the biogenesis of miRNAs let-7, miR-107, miR-143 and miR-200c by preventing miRNA maturation[237], and is regulated by N-Myc in NB cells via an N-Myc/miR-26a-5p/LIN28B regulatory axis, and direct binding and activation of the LIN28B promoter[238]. LIN28B binds and activates gene promoters by interacting with the zinc finger transcription factor ZNF143, activating downstream targets that regulate the core regulatory circuitry that controls the malignant state of NB cells, in addition to regulating Gsk3β and L1-CAM expression, involved in cell adhesion and migration[239].
miRNAs, long non-coding RNAs and endogenous retroviruses: Let-7 was the first suppressor miRNA reported to down-regulate tumour formation and metastasis[240], is involved in trunk NC cell migration, is regulated by LIN28B and is involved in LIN28B-mediated trunk sympathoadrenal precursor transformation, supporting roles for let-7 in NB initiation and metastatic potential[241]. Other miRNAs that are altered in NB, include the up-regulated miRNAs: mir-17-92 that targets DKK3, CDKN1A, BIM, MEF2D and ESR1; miR-21 that targets PTEN; miR128 that targets truncated TrkC and up-regulates Bcl-2 expression; miR-380-5p that targets p53; miR-558 that targets HPSE and miR-375 that targets ELAVL4. Down-regulated miRNAs, other than let-7, that are associated with NB include: miR-9 that targets MMP-14; miR-15 that targets Bcl2; miR-103 and miR-107 that target CDK5R1; miR-124 that targets Sox9 and PTPB1; miR-29a that targets cdk6 and CDC6; miR-152 that targets DNMT1; miR-125b that targets TrkC; miR-204 that targets Bcl2 and TrkB; miR-363 that targets ADAM15 and MYO1B and miR-335 that targets Sox4 and TNC. Embryonic SC-associated miR-380-5p cooperates with Ras in primary cell transformation and tumourigenesis, blocks cellular senescence in mice, correlates with N-Myc amplification and poor outcome in NBs and miR-380-5p inhibition promotes p53-dependent NB cell apoptosis and reduces tumourigenesis in an orthotopic mouse NB model[242-245]. A NB CSC suppressor role has been proposed for miR-128, not expressed in primary NBs, which represses the expression of Bmi1 and E2F3a pro-growth transcription factor, and reduces NB cell motility and invasiveness[246-249].
Long non-coding (lnc) RNAs have also been shown to promote and maintain CSCs, and regulate CSC behaviour[250]. LncRNAs implicated in NB CSCs[251-253], include the hypoxia-regulated lncRNA HOTAIR, up-regulated in NBs, which is associated with poor prognosis and CSC self-renewal, differentiation inhibition and high-level PRAF2, which promotes NB CSC proliferation and migration[254]. The hypoxia and N-Myc-regulated lncRNA MALAT-1 also contributes to CSC maintenance[255], promotes NB cell migration, invasive and angiogenic behaviour, and is induced by the N-Myc-regulated histone demethylase JMJD1A, which is also a hypoxia-inducible epigenetic regulator of SC behaviour, providing a lncRNA-mediated mechanism through which N-Myc can promote invasive, angiogenic and metastatic behaviour of NB CSCs[252,256,257]. Repression or polymorphisms in the epigenetic regulator H19 lncRNA have also been implicated in NB CSCs[251], and depletion of lncRNA NBAT1, implicated in retinoic acid-induced NB cell neuronal differentiation, prevents differentiation, and is repressed in undifferentiated CSCs in high-risk N-Myc amplified and non-amplified NB[252,258]. Human endogenous retroviruses (HERVs) have also been implicated in aggressive metastatic behaviour in a variety of tumours. HERV-K activation expands and maintains CD133+ CSC-like populations in NC-derived melanoma[259,260], HERV-H lncRNA is a specific marker of pluripotency in human SCs, that interacts with Oct4 to maintain embryonic SC gene expression[259,260] and HERV expression is up-regulated in NB cells by hypoxia and hypoxia mimics[261,262]. Although, no reports link HERV-K or HER-V to NB, it is likely that HERVs also play important roles in promoting and maintaining NB CSCs. Dysregulated HERV-W expression has, however, been reported in NB cells that express the HERV-W trace protein syncitin-1, which exhibits anti-apoptotic activity, promotes immune suppression, and induces EMT, invasion and metastasis, consistent with CSC promoting function. Furthermore, HERV-W induces c-AMP signalling in NB cells, resulting in activation of the syncitin-1 transcription factor CREB[263].
Metabolism, nuclear factor-erythroid 2-related factor 2 and redox regulators: CSCs exhibit unusual metabolism in order to maintain stemness and exhibit anabolic and catabolic metabolic flexibility, associated with alterations in signalling and redox activity, in response to changes within the tumour microenvironment. Normal SCs and CSCs exhibit enhanced glycolytic metabolism that promotes and maintains CSC traits through enhanced expression of glycolysis-related GLUTs, monocarboxylate transporters, hexokinases and pyruvate dehydrogenase kinase 1, the functional inhibition of which reduces tumourigenicity in vivo. Many of these genes are HIF-regulated, helping to explain CSC association with hypoxic tumour niches and pseudo-hypoxic states[264,265]. Glycolytic reprogramming in CSCs promotes metastatic progression, enhances therapeutic resistance and involves Snai1 suppression of phosphofructokinase, which switches metabolism from oxidative phosphorylation to glycolysis and promotes EMT. Oxidative phosphorylation is also activated in CSCs, in association with increased mitochondrial numbers and size. This metabolic flexibility is regulated by activated oncogenes, oncosuppressor inactivation, tumour cellular and chemical microenvironments, and result in co-existence of different CSC metabo-types, some fixed and others that exhibit reversible plasticity and can switch back and forth from glycolysis to oxidative phosphorylation, optimizing exploitation of changing tumour micro-environments[264,265]. TrkAIII expression in NB cells promotes metabolic flexibility within a CSC-like state, and in inactive-form localizes to outer mitochondrial membranes in un-stressed NB cells. Under conditions of ER-stress, inactive mitochondrial-associated TrkAIII exhibits Ca2+-dependent translocation to inner mitochondrial membranes, where it undergoes cleavage-dependent activation, resulting in tyrosine phosphorylation of mitochondrial pyruvate dehydrogenase kinase and a metabolic shift to aerobic glycolysis, implicating mitochondrial TrkAIII in an ER-stress-regulated mechanism for metabolic flexibility, within a CSC context and survival-adapted ER-stress response[72,150].
In order to survive the cytotoxic effects of increased ROS production from metabolic and therapeutic sources, CSCs also possess potent antioxidant systems that scavenge and regulate ROS production, ROS-dependent signalling pathways, ROS-regulated transcription factors and defend against ROS-induced cytotoxicity[266]. The nuclear factor-erythroid 2-related factor 2 (NRF2) transcription factor is a master regulator of cellular redox and metabolic homeostasis in normal SCs and CSCs, and is important for regulating CSC quiescent-pseudo-senescent-dormant states, survival, self-renewal, proliferation and differentiation[265,267]. ROS inactivate repressive Keap1/NRF2 interactions, activating the NRF2-dependent antioxidant system, which up-regulates the expression of the anti-oxidants superoxide dismutase, thioredoxin, thioredoxin reductase, glutathione peroxidase, glutathione reductase, ferritin, nicotinamide adenine dinucleotide phosphate, quinone oxi-reductase, glutamate-cysteine ligase catalytic subunit, glutamate-cysteine ligase modifier subunit and increases expression of the drug-metabolism and efflux genes ALDH and ABC transporters[265,267-269]. In tumours, NRF2 weakens oxidative phosphorylation and induces antioxidant gene expression, reducing ROS to levels optimal for CSCs stemness, self-renewal, proliferation, survival, metastatic progression and therapeutic resistance. NRF2 activity in CSCs is modulated by CD44 through p62[270], regulates Nestin, Bmi1, and Sox 2 expression and in NB cells promotes proliferation and inhibits neuronal differentiation[271,272], implicating NRF2 in fine tuning the metabolic/antioxidant system to ensure appropriate ROS levels for NB CSC maintenance. TrkAIII also regulates free-radical ROS levels in CSC-like NB cells by augmenting SOD2 expression, in a TrkAIII/SOD2 axis that increases resistance to mitochondrial free-radical ROS-induced cell death, within the context of a CSC-like phenotype. In this mechanism, increased mitochon
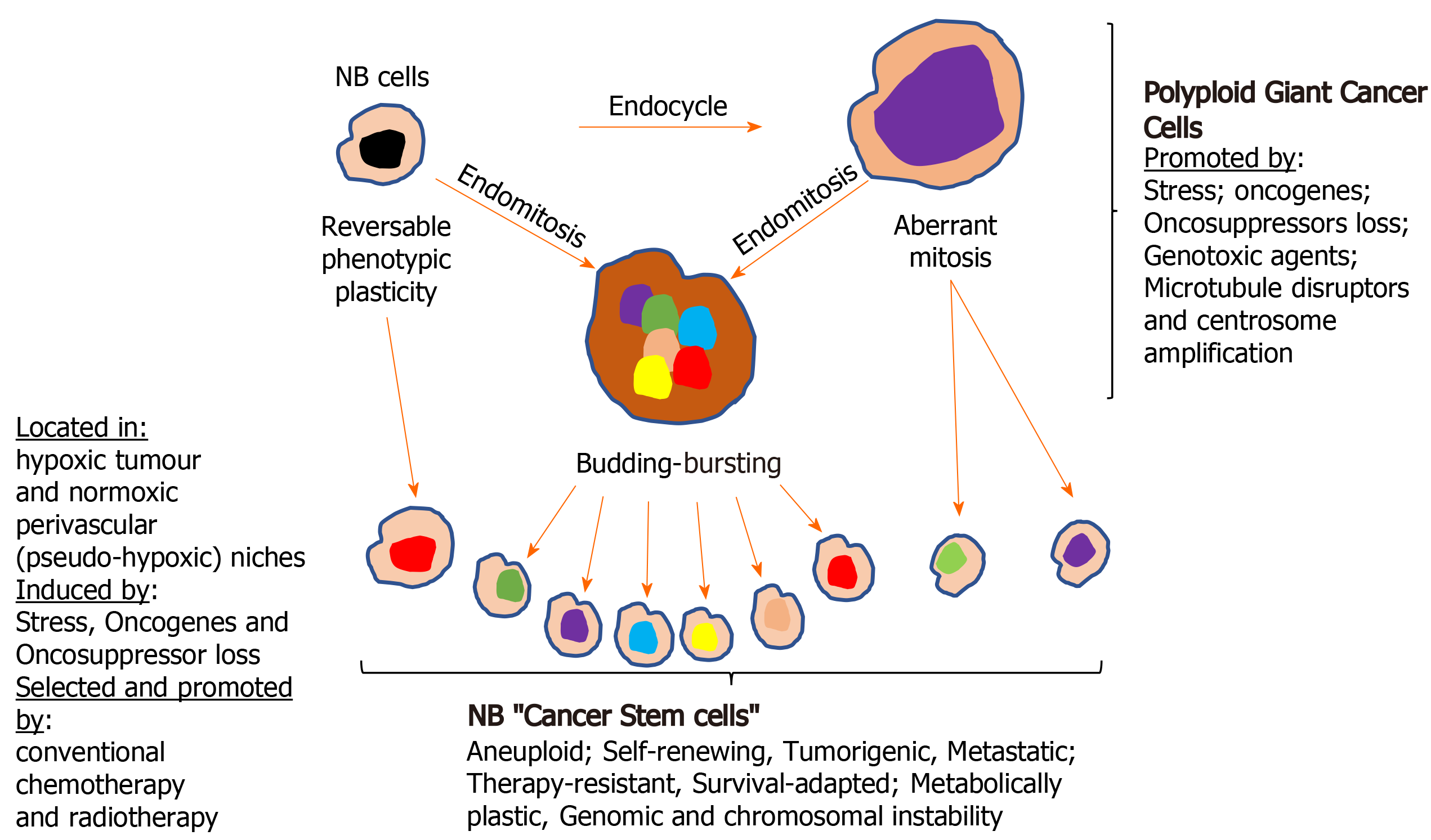
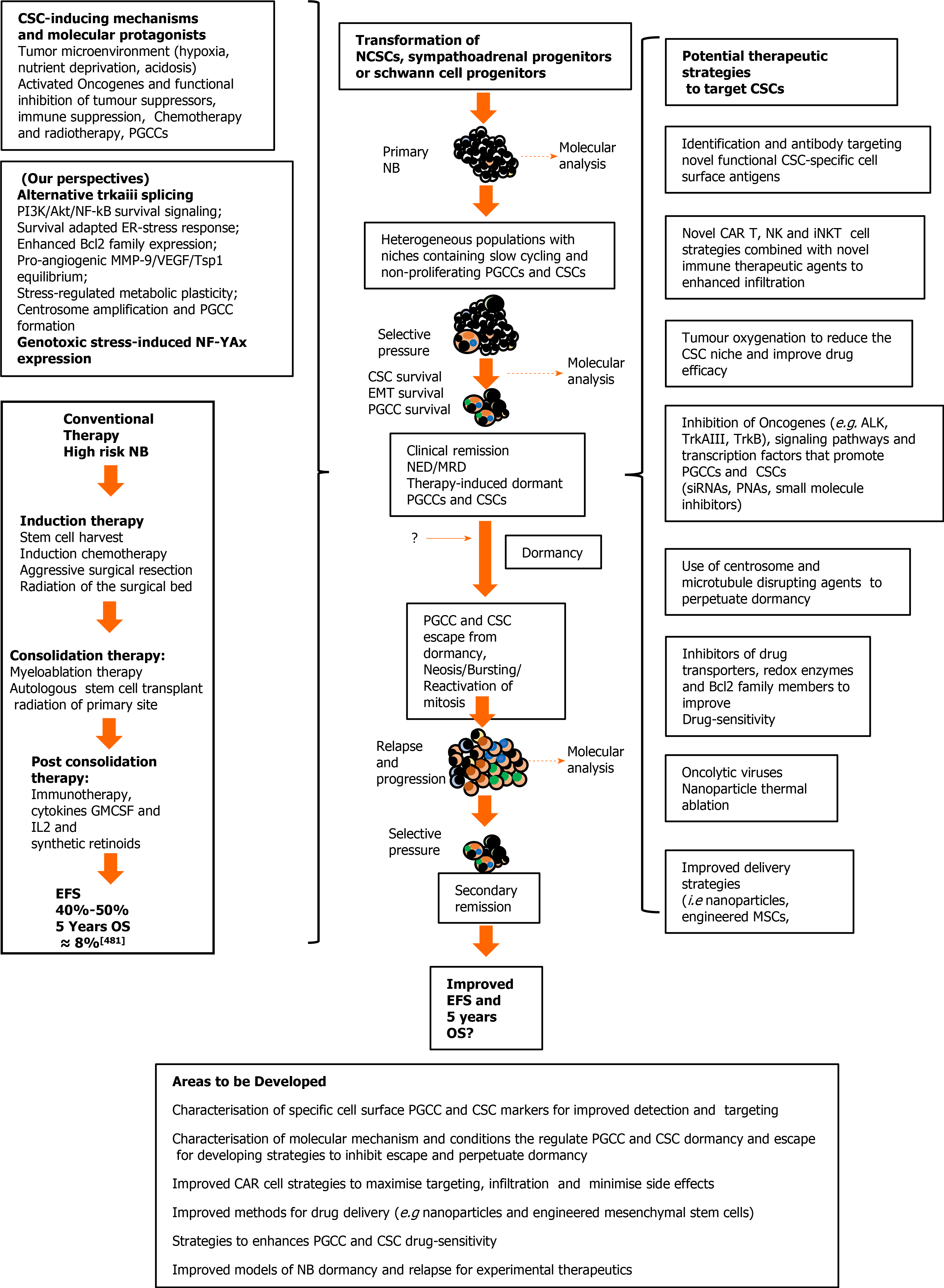
Cellular polyploidy: The tumour microenvironment, oncogene activation and oncosuppressor inactivation integrate to promote genomic instability and the formation of polyploid giant cancer cells[275], now considered to be an important source of aneuploid CSCs in tumours, including NBs. Theodor Boveri was the first to hypothesize chromosomal polyploidy and subsequent aneuploidy as a mechanism for malignant tumour formation, describing the origin of tumours from a single primordial cell with “abnormal chromosome constitution”, generated by abnormal stimuli that “might induce simultaneous multiple divisions of the centrosomes”, resulting in “tetraploid cells driven to divide by continuous proliferation“, which “opens the door for the production of sarcomatous or carcinomatous progenitor cells”[276]. Today, cellular polyploidy is no longer considered an exclusively pathological process but also a physiological mechanism for tissue homeostasis, that is conserved and subverted in tumour pathogenesis and progression[277]. Between 0.1%-20% of solid tumour volumes consist of polyploid cancer cells that increase in percentage with malignancy, contributing to tumour initiation, heterogeneity, EMT, invasion, metastasis, therapeutic resistance and the generation of CSC-like cells[278-281]. Previously, polyploid cancer cells were presumed to be non-dividing but recent observations confirm that these cells express genes involved in cell cycle regulation, in addition to expressing hypoxia-inducible, stem cell-regulating, chromatin remodelling, and invasion and metastasis promoting genes[278,280-282]. Cellular polyploidy can be achieved by cell fusion, an endocycle in which DNA synthesis and G-phases occur without M-phase or cytokinesis to generate single polyploid cells with giant nuclei that exhibit no mitotic features or by endomitosis caused by abortive mitosis that does not result in chromatid separation or cell division and is followed by reentry into S phase, generating multinucleated cells[282]. Most polyploid cells die but others survive initially in a quiescent G0 pseudo-senescent state but then escape this state to undergo asymmetrical division by blebbing and/or bursting to yield aneuploid progeny that go on to generate novel tumourigenic and therapy-resistant CSC-like cells by mitosis[283]. Polyploid tumour cells are generated by tumour microenvironmental stress, within a context of survival adapted cellular ER-stress responses and immunosuppression[279,280,284,285], and are induced by: gene overexpression and oncogene expression that increase centrosome numbers[149,279,280]; replication stress; genotoxic chemotherapeutic agents that induce abortive cell cycles[286-288], and by loss of tumour suppressor function[279,281,289,290]. Tumour cell polyploidy induced by hypoxia is associated with CSC and EMT gene expression patterns, resulting in a more adaptable, less disruptive phenotype within stressful and hypoxic microenvironments, with multiple gene copies lessening the need for chromosome segregation, promoting survival and facilitating tumour evolution and therapeutic resistance. Polyploid giant cancer cells evolve complex karyotypes and exhibit enhanced tumourigenic activity in vitro and in vivo implicating tumour cell polyploidy as a basis of aneuploidy[279,281]. Within the somatic context, long term passage of induced polyploid somatic cells eventually leads to transformation and tumourigenesis, in a cellular transformation process characterised by transition from diploid to aneuploid states via tetraploidy. Polyploid cancer cell survival depends upon repression of p53 function and surviving polyploid cells overcome oncogene-induced senescence by repressing p53 and overexpressing DNA repair genes, to form progeny with oncogenic karyotypes and phenotypes, and progressive chromosomal instability, explaining hyper-diploid karyotypes in many malignant tumours[279-282], including NBs[278,291]. Polyploid giant cancer cells exhibit chemo- and radiotherapeutic resistance that results from quiescent or pseudo-senescent states, infrequent slow cell cycles, increased expression of apoptosis inhibitors, p53 functional repression and enhanced DNA repair mechanisms. Resistant polyploid giant cancer cells follow a defined path of chromosome re-organisation and restructuring into polyploid nuclear states that can separate into secondary nuclei that form secondary progeny in a transformation process[279,281,282]. Secondary daughter cells exhibit anchorage-independent growth, undergo reductive mitotic division and produce smaller near-diploid aneuploid cells that proliferate and are tumourigenic, in a process likened to spores in lower organisms that permit survival in harsh conditions and rapid repopulation later. CSCs formed from polyploid cancer cells express CSC markers and exhibit embryonic-like stemness equivalent to a blastomere stage, form different tumour types, illustrating multipotent plasticity greater than marker-purified CSCs, and express fundamental genetic, cellular and developmental programmes for tumour initiation and progression that integrate with “normal” tumour cellular components[292]. CSC-like cells generated from hypoxia-induced CSC marker positive polyploid giant cancer cells, divide asymmetrically and cycle slowly to form aggressive mesenchymal treatment-resistant tumours[279,280,284].
Centrosome amplification: Theodore Boveri also considered that aberrant centrosome numbers were the driving force behind cellular tetraploidy, aneuploidy and malignant tumour formation[276]. Today, it is well recognized that aberrant centrosome numbers characterise polyploid and aneuploid CSCs in solid tumours, implicating mechanisms that promote centrosome amplification in CSCs formation by way of polypoid and aneuploid states. Centrosome amplification in tumour cells has been attributed to Aurora-A kinase overexpression[286]; drug-induced DNA damage-dependent activation of checkpoint kinases Chk1 and Chk-2[287,288]; enhanced polo kinase 4 expression and activity[289]; loss of BRCA1, APC and p53 function[290-292], and oncogenic alternative TrkAIII splicing[149]. In NB cells, spontaneously activated TrkAIII exhibits MT-dependent retrograde transport to the centrosome, where it phosphorylates Plk4, promoting centrosome amplification, augmenting microtubule polymerization, and increasing polyploid giant and aneuploid NB cell formation, chromosomal instability and sister chromatid exchanges[149,153], implicating hypoxia/stress-induced alternative TrkAIII splicing in promoting NB CSCs via polyploid giant cancer cell formation. This is supported by TrkAIII expression in neural SCs and CSC-like NB cells, associated with enhanced expression of Nanog, Nestin, Sox2, CD133 and CD117 mRNA, and by TrkAIII prevention of NB cell differentiation, promotion of NB xenograft tumourigenic and metastatic capacity, and therapeutic resistance[72], contrasting with fully spliced TrkA promotion of genomic stability in NB cells[293]. Therapy-resistant polyploid giant cancer cells express enhanced levels of Oct4, Nanog and Sox2, involved in self-renewal and apoptosis evasion, and re-proliferate upon pseudo bi-polar centrosome coalescence, resulting in CSC-like progeny that correlates with oncogene amplification, poor prognosis, post-therapeutic relapse, EMT, invasion, metastasis and therapeutic-resistance[280,281]. The formation of polyploid giant cancer cells is the result of centrosome amplification, therefore, represents a critical source of invasive, metastatic and therapy-resistant NB CSC-like phenotypes (Figure 4).
Nonsense mediated RNA decay: The multistep nonsense mediated RNA decay (NMD) pathway targets and rapidly destroys both mutated and non-mutated mRNAs, and has been implicated in preventing embryonic SC differentiation[294]. NMD is inhibited by hypoxia in an eIF2a-dependent manner. Stress-induced eIF2a phosphorylation activates stress-response gene expression, resulting in the stabilization of several NMD target mRNAs, stabilized by NMD repression, implicating hypoxia-induced NMD repression in promoting the cellular hypoxic stress-response. Hypoxic repression of NMD involves component re-localization to cytoplasmic stress granules and dynamically alters hypoxic gene expression, consistent with a potential role for NMD repression in hypoxic promotion of NB CSCs[295]. NMD is also repressed by c-Myc, stabilizing Myc target gene expression[296], implicating NMD repression in N-Myc/c-Myc cooperation, which regulates NB CSC phenotypes[201]. An NMD/miRNA-128 regulatory circuit has also been identified in which miR-128 represses NMD by targeting UPF1 and MLN51 NMD factors, and in NBs miR-128 exerts a tumour and CSC repressing role, inhibiting expression of the CSC self-renewal factor Bmi1 and the growth promoting transcription factor E2F3a, reducing NB cell motility and invasiveness[246-249].
The stressful hypoxic tumour microenvironment: Hypoxia is involved in NB initiation and progression and promotes NB CSC-like phenotypes[297]. The stressful tumour microenvironment continually selects survival-adapted tumour and tumour-associated “normal” cell populations, resulting in heterogeneous niches within highly aberrant vasculatures that promote episodes of hypoxic, nutrient, metabolic and redox stress. Oxygen pressure is lower in solid tumours than in normal tissues, hypoxic tumours are more aggressive than oxygenated tumours and tumour hypoxia is a common feature of solid tumours, including NB, and represents an independent prognostic marker for disease progression and poor clinical outcome[298-301]. Tumour hypoxia, oncogene activation and oncosuppressor inactivation integrate to promote tumour glycolytic metabolic adaptation, resulting in a reducing acidic tumour microenvironment that promotes further adaptation, EMT and progressive de-differentiation to CSC-like states with enhanced migratory, invasive, metastatic capacity, increased therapeutic resistance and augmented genomic instability[302-305]. In NB, hypoxia promotes more aggressive behaviour that is associated with CSC-like subpopulations that localize to hypoxic necrotic areas[306-308]. NB CSC-like cells exhibiting a pseudo-hypoxic phenotype also localize to oxygenated tumour perivascular niches[301,309]. Hypoxia inducible HIF-1α and HIF-2α transcription factors promote metastatic CSC-like phenotypes[310,311], share target genes but also exhibit target specificity and differences in regulation. HIF-1α proteins are stabilized by hypoxia but HIF-1α transcription is not induced by hypoxia, whereas hypoxia induces HIF-2α transcription. HIF-1α is involved in acute hypoxic responses and subsequently decreases, whereas HIF-2α function is maintained during prolonged hypoxia. Hypoxia-independent HIF expression is promoted by growth factor signalling, including IGF, VEGF and EGF signalling through the PI3K and Ras/MAPK pathways. In NBs, HIF-2α expression correlates with aggressive disease, HIF-2α+/HIF-1α- mesenchymal CSC-like NB cells populate perivascular NB regions and NB cells cultured at 5% O2 to mimic end capillary O2 diffusion levels, express HIF-2α but not HIF-1α, implicating HIF-2α in pseudo-hypoxic NB CSC-like phenotypes within peri-vascular NB areas, potentially driven by autocrine GF/GFR or oncogene-dependent signalling[301,309,312]. HIF-2α is also implicated in delta like noncanonical notch ligand 1 (DLK1) promotion of NB CSC-like phenotypes. DLK1 is regulated by HIF-1α and HIF-2α, induced by hypoxia, is a bona fide neural SC marker required for self-renewal[117,181] and is robustly expressed by undifferentiated tumour cells. In NB xenografts, DLK1+ cells preferentially localize to hypoxic areas[116,117], and together with HIF-2α promote aggressive tumour behaviour and undifferentiated CSC-like states[301,309,312]. HIF-1α also promotes NB CSC-like phenotypes within the context of an aberrant unfolded protein response, characterised by reduced ATF3 expression. In this mechanism, HIF-1α induces Id1 expression in NB cells that do not express ATF3, which down-regulates Id1 expression in most cell types[295]. Intermittent hypoxia also promotes NB CSC-like phenotypes involving HIF-1α and HIF-2α expression, resulting in enhanced survival and clonogenic activity, increased Oct4, CD133, Id2, Hes1, c-kit and Notch-1 expression, reduced expression of SNS differentiation markers NPY, Hash1 and dHand, and HIF-1α knockdown promotes neuronal differentiation-associated NeuN and NF-M expression[313]. Hypoxia-induced alternative splicing also promotes CSC-like phenotypes and EMT by promoting HIF-1α interaction with Zeb1 and a soluble sCD44 splice variant, induces DCLK1 alternative splicing to promote CSC self-renewal and chemoresistance, and promotes alternative TrkAIII splicing in NB cells, enhancing angiogenic, tumourigenic and metastatic behaviour associated with the context of a more CSC-like phenotype[314].
Mesenchymal stem cells: MSCS invade NB tumour tissues[315] and NB cells that express CXCR4 and CXCR7 chemokine receptors migrate towards MSCs that express SDF-1 in the bone marrow[316]. MSCs isolated from NBs promote tumourigenesis by interacting with NB and stromal cells, and are more frequently arrested in G0/G1 phases of the cell cycle, facilitating therapeutic-resistance[317]. Normal MSCs therefore are NB stromal components that regulate NB CSC tumourigenic and metastatic behaviour.
Considering the young age of NB patients, the aggressive nature of the disease and the concept that post-therapeutic disease-relapse, metastatic progression and eventual mortality result primarily from the selection of therapy-resistant CSC-like cells, it is of paramount importance to develop novel therapeutic strategies that target and eliminate NB CSC subpopulations in order to improve therapeutic response-duration and survival.
CSC-like NB subpopulations reflect not only the primitive embryonic origins of this tumour but also evolutionary processes promoted by accumulating genetic changes within a continuously evolving tumour microenvironment, also altered by conventional therapeutic strategies, which together select survival-adapted CSC-like phenotypes. CSC therapeutic resistance depends upon numerous mechanisms, including an intermittent slow cell cycle, enhanced expression of drug efflux ABC transporter and P-glycoprotein, enhanced expression of apoptosis inhibitors and repression of p53 function, a flexible pro-glycolytic metabo-type associated with high-level anti-oxidant expression, enhanced DNA repair mechanisms, survival-adapted ER-stress responses, formation of polyploid giant cancer cells, CSC localization to hypoxic niches, the present physical tumour barriers to therapy, and phenotypic plasticity, permitting CSC regeneration from non-CSC tumour cells[280,318-320]. These factors combined with CSC similarities to normal SCs, make specific therapeutic targeting and elimination of NB CSC populations a daunting problem.
In this next section, we review novel potential therapeutic strategies, currently aimed at targeting and eliminating NB CSC populations or that could be adapted for use in NB.
CSC cell surface marker targeting with antibodies and CAR cell strategies: CD44 was the first CSC marker to be targeted with monoclonal antibodies in acute myeloid leukemia, with successful disease eradication[321], suggesting that CD44 in an aggressive NB CSC-like subpopulation could also be targeted[112]. Targeting CD133, expressed by NB CSC-like cells[92], with antibodies conjugated to the cytotoxic agent monomethyl-auristatin has been reported to be internalized and to kill CD133+ liver cancer CSCs[322], and targeting CD144 with antibodies, or targeting STAT3, in CD114+ NB CSCs that exhibit self-renewal and high tumourigenic activity, depletes the NB CSC subpopulations, decreases tumourigenicity, reduces metastatic capacity and increases chemotherapeutic sensitivity[323].
CAR-T strategies eradicate tumour cells by recognizing cell surface antigens and, unlike natural T-cell receptor positive T cells, recognize antigens by an MHC-independent mechanism. CARs are comprised of an extracellular single chain variable tumour antigen-specific fragment linked to extracellular spacer, transmembrane and intracellular signalling domains. Latest generation CAR T-cells express inflammatory cytokines and can effectively and specifically eliminate CSCs. Pre-requisites for CSC CAR-T therapy include cell surface expression of CSC-specific, preferably functional, tumour antigens. CAR T-cells that specifically recognize CSC markers GD2, LGR5, IGFR-1, CD44, CD47, EpCAM, Dll4, FZD and CD123 have been generated, do not promote CSC enrichment and do not exhibit substantial tissue cytotoxicity, and CAR T-cells that recognize CD133, CD166, CD20, CD38, CLL-1, EpCAM, CD123, CD171, ROR1, CD44, CD47, CD117 and c-Met are presently being evaluated in clinical studies[159]. For NB therapy, targeting L1-CAM (CD171), which drives NB CSC invasive and migratory capacity, with L1-CAM CE7 epitope CAR T-cells developed for NB immunotherapy, is safe and efficacious in refractory recurrent NB, and CAR T-cells that target the CE7 epitope of CD171 (CE7-CAR-T cells), generated from NB patients with recurrent/refractory disease, has also been shown to be safe and efficacious[324].
Potential problems with CAR-T therapy include off-target toxicity due to low-level antigen expression in normal tissues. This can be overcome using CAR T-cells that recognize two or more antigens, enhancing tumour specificity, or CAR T-cells that express an inhibitory receptor against normal antigens in addition to recognizing a tumour specific antigen. Alternatively, considering that CAR antigen affinity is key to activating cytotoxicity, CAR T-cells with low affinity CARs that are only cytotoxic when they bind high density antigens may minimize off-target side effects, and CAR T-cells armed with caspase-9, sensitive to AP1903-induced activation, can ensure CAR T-cell elimination under conditions of aberrant cytotoxicity[325]. CAR T-cells engineered to contain a lenalidomide-inducible dimerization system that serves as an ON- and OFF-switch, characterised by lenalidomide-dependent anti-tumour activity in the presence of tumour antigen and lenalidomide-induced degradation in the absence, may further minimize potential off-target side effects[326]. Other problems associated with CSC-targeted therapy, include phenotypic plasticity, illustrated by reversible bidirectional conversion between CSC and non-CSC states induced by changes in the tumour microenvironment and therapeutic agents, which may render target-therapies, including CAR T-cells only transiently efficacious, until novel CSC-like subpopulations regenerate. In this case, efficacy could be maximized by combining CAR T-cell therapy with conventional chemo and/or radiotherapy therapy to kill both CSC and non-CSC tumour cells, supported by a report of synergistic efficacy in a rat glioma model, when NKG2D CAR-T therapy is combined with conventional radiotherapy[327], and when EpCAM CAR-NK92 CAR-T therapy is combined with regofinib plus salinomycin, metformin and S-phase kinase associated protein 2 inhibitors and low dose conventional chemotherapy[328,329]. Heterogeneity of CSC tumour antigen expression can also be mitigated using bi- or poly-specific CAR T-cells that target multiple tumour antigens, and CSC tumour antigen expression could be promoted to facilitate CAR T-cell recognition, exemplified by retinoic acid up-regulation of CD38 expression that sensitizes acute myeloid leukemia cells to CD38 CAR T-cells[330]. CAR T-cell exhaustion and senescence can be overcome using 4-IBB and ICOS co-stimulatory domains, CAR T-cell tumour infiltration and durability can be enhanced, and exhaustion and senescence reduced, combined with immune checkpoint PD-1, CTLA4, TIM-3, LAG-3 and adenosine 2A receptor inhibitors. CAR T-cells that express anti-apoptotic factors can reduce tumour-induced CAR T-cell apoptosis, CAR T-cells that express interleukin (IL)-7 and IL-15 can improve persistence, and CAR T-cells that express chemokine receptors improve tumour infiltration and homing to CSC niches, as does local or intra-tumour CAR T-cell delivery[159]. Physical tumour stromal barriers can be disrupted by CAR T-cell expressing extracellular matrix degrading enzymes targeted to tumour stromal FAP+ fibroblasts[331], and tumour oxygen supplementation can enhance infiltration and reduce immunosuppression[332]. Repression of CAR-T function by tumour-associated T-regulators, macrophages, neutrophils, myeloid-derived suppressor cells and inhibitory IL10, TGFβ, PGE2, arginase, cyclooxygenase-1, galectin-9 and idoleamine 2, 3 dioxygenase molecules, can be reduced using specific inhibitors[159]. Other modifications that may improve efficacy include adjuvant tumour de-bulking prior to CAR T-cell therapy to reduce toxicity due to cytokines and adjuvant chemotherapy with agents that induce T cell chemokine expression can pre-condition tumours for CAR T-cell therapy. Further characterisation of CSC-specific targets can reduce the risk of autoimmunity and improved understanding of how specific CSC antigens alter during disease progression will help fine-tune individualized CAR T-cell therapeutic approaches, which combined with targeting monoclonal antibodies, conventional chemotherapy and radiotherapy, unveil an important therapeutic future for CAR T-cell based immunotherapies in the eradication of NB CSCs.
Invariant natural killer T cells (INKTs) or natural killer cells (NKs) represent an alternative to CAR T-cell therapy, with advantages including potential clearing of tumour associated macrophages and better penetration. INKTs alter the behaviour of tumour-associated macrophages and myeloid derived suppressor cells, secrete cytokines that recruit immune effectors and kill neuroblasts and CSCs[333]. INKTs stimulated with a-galactosylceramide (GAg), adoptive iNKT transfer and CAR-iNKTs could all be utilized to target NB CSCs. GAg-stimulated iNKTs enhance NK-mediated NB cell killing, reinvigorate exhausted CD8+ T cells[334,335] and show good tumour infiltration when combined with immune checkpoint inhibitors. Adoptive iNKT transfer in humanized mice reprogrammes NB xenograft-associated macrophages from the M2 to the anti-tumour M1 phenotype[336], GD-2 specific CAR-iNKTs reduce NB xenograft growth in mice and prolong survival, and CAR-iNKTs are being evaluated in phase I clinical trials in relapsed or refractory NB[333]. With respect to NKs, Dinutuximab an anti-GD2 antibody was the first immunotherapy used for NB, and exerts its effects also by stimulating NK activity, implicating a potential use for NK activators, including novel bi- and tri-specific killer cell engagers that enhance NK cytotoxicity, in NB therapy. Alternatively, autologous adoptive transfer of activated NK cells combined with anti-GD2 antibodies could be used, which have been reported to increase survival in a mouse NB xenograft model[337]. Along the same theme, allogenic NK transfer combined with humanized anti-3F8 antibody and IL-2 is currently in clinical trial for NB[338] (NCT 02650648) and CAR-NKs evaluated in NB include GD2-CAR-NKs that exhibit specific robust GD2+ NB cell killing and NKG2D CAR-NKs that kill NB-associated myeloid derived suppressors[333].
Inhibition of immune checkpoint protagonists PD-1, CTLA4, TIM-3, LAG-3 and adenosine 2A receptor, and in particular PD-1/PD-L1 and CTLA4, also represents an important approach for restoring anti-tumour immunity and improving CAR T-cell, CAR-iNKT, CAR-NK, iNKT and NK immunotherapeutic approaches[339]. N-Myc amplified NBs exhibit high-level PD-L1 expression, which correlates with reduced tumour infiltrating T-cells and unfavourable prognosis, and can be reduced by shRNA-mediated functional N-Myc inhibition or the BET bromodomain inhibitor JQ1[340].
ABC and P-glycoprotein transporter inhibitors: ABC transporter inhibitors in general lack specificity and elicit important toxic side effects[123]. However, the ABC inhibitor probenecid sensitizes NB CSCs to cisplatin, increasing apoptosis and decreasing proliferation associated with caspase 3 expression, reduced ABC transporters MDR1, MDR2 and BCRP expression and reduced expression of CD133 and vimentin and snai1 EMT markers, forcing CSCs into a non-CSC state, associated with therapeutic re-sensitization[341]. The prophylactic antianginal agent Perhexiline induces non-coding Alu RNA neuroblastoma differentiation marker 29 expression in NB cells, represses ABC transporter expression, increases sensitivity to cisplatin, depletes CSCs through differentiation, reduces NB nodular formation and enhances survival in a mouse NB xenograft model[342]. The P-glycoprotein inhibitor vardenafil inhibits P-glycoprotein-mediated drug-efflux and increases the cytotoxicity of paclitaxel and vincristine in enriched CSCs[343].
NB CSC-associated oncogenes: Small RNA inhibitors that target E6 mRNA in CSC-enriched cervical cancers[344] and HMGA1 in glioblastoma CSCs[345] are proof of concept the NB CSC-associated oncogenes could be silenced. However, delivery of high enough concentrations of oncogene-specific small RNA inhibitors (antisense oligonucleotides, RNA interference, ribozymes) to tumours is a significant clinical problem, which may be overcome using nanoparticles, liposomes and PEGylated aptamers (see below) targeted to cell surface CSC markers (see above)[346]. NB CSC-associated Alk, TrkB and TrkAIII oncogenes could be inhibited by the Food and Drug Administration (FDA)-approved small molecule ALK inhibitors Crizotinib, Ceritinib, Alectinib, Brigatinib, Entrectinib and Lorlatinib[347] or by the FDA-approved small molecule Trk kinase inhibitors Entrectinib, Larotrectinib, Lestaurtinib, GNF-4256 or GNF-5837 either alone or in combination with conventional chemotherapy[72,348-350], with the drug-resistance evolution counteracted by parallel development of modified drugs specific for known and expected kinase-domain mutations, and efficacy enhanced when combined with conventional chemotherapy and/or radiotherapy. Alk, TrkB and TrkAIII oncoproteins could also be reduced using appropriately CSC-targeted delivery of gene-specific peptide nucleic acids or siRNAs[72]. Oncogenic alternative TrkAIII splicing could be reversed using new generation lentiviral vectors that express TrkAIII inhibitory siRNAs and full length fully-spliced TrkA complementary DNA, mutated to prevent inhibition of expression without altering the amino acid sequence, in order to block TrkAIII oncogenic activity and reinstate TrkA tumour-suppressing activity[72]. Ibrutinib and acalabrutinib inhibitors of Bruton’s Tyrosine kinase, which is highly expressed in N-Myc amplified NBs and is associated with poor prognosis, have been shown to inhibit the migratory, invasive and tumour sphere forming capacity of NB cell lines, and to synergize with cisplatin, identifying Bruton’s Tyrosine kinase as a potential druggable NB driver oncogene, of relevance to NB CSCs[351]. Survivin, an oncogenic inhibitor of apoptotic and autophagic cell-death, is also differentially expressed in CSCs, is associated with chromosome 17q gain and adverse outcome[352], and can be inhibited by a significant number of small molecule and mRNA inhibitors (antisense oligonucleotides, RNA interference, ribozymes)[353].
CSC-associated signalling pathways: CSC signalling pathway inhibitors could also be adapted for use in NB, including the Notch pathway inhibitor Psoralidin, which reduces tumour-sphere formation, induces apoptosis, inhibits proliferation and increases doxorubicin and docetaxel sensitivity in breast cancer CSCs[354]. Shh signalling is also essential for NB cell proliferation and tumourigenicity[355]. The Shh signalling antagonists cyclopamine, IPI-926 and GANT61 promote tumour regression and deplete CSC in cancers, including NB[356-359], decrease CD133 and CD15 double positive NB CSC-like cells and reduce NB tumourigenic activity in vitro[360]. Small molecule Wnt and CD44 signalling inhibitors that reduce breast cancer CSC self-renewal[361,362] and the cyclin dependent kinase inhibitor Roniciclib (BAY 1000394) that also targets CSCs, exhibit potent therapeutic effects in high-risk NB, by inhibiting Wnt/β-catenin signalling, inducing nucleolar-stress, down-regulating CSCs and inhibiting proliferation[363,364]. The Wnt/β-catenin signalling inhibitor Pimozide also blocks CSC EMT, inhibits CSC-associated STAT-3 and represses differentiation inhibiting gene expression[365]. Other inhibitors include, the irreversible Glycogen synthase kinase-3b serine/threonine kinase inhibitor Tideglusib, which reduces NB cell proliferation, migration and viability by eradicating the self-renewal capacity of highly resistant NB CSCs, reducing tumour sphere forming capacity and CD133 expression[366]. The AKT/mammalian target of rapamycin (mTOR) signalling inhibitors triciribine and rapamycin target NB CSCs, decreasing survival and tumour sphere-forming capacity[367], and the mTOR inhibitor temsirolimus, which combined with the P53 activating MDM inhibitors Nutlin 3a or RG7388, inhibits proliferation and induces apoptosis in in vivo NB models[368]. Dual inhibition of the AKT2/mTOR and MAPK pathways with specific CCT128930 AKT2 inhibitor and PD98059 MAPK inhibitor decreases NB CSC proliferation, migration, tumour sphere-forming and angiogenic capacity[369]. Retinoic acid combined with Dlk1 knockdown disrupts CSCs by reducing DLK1 interactions with PHB1 and PHB2 prohibitins, required for self-renewal and clonogenic capacity[118,370]. Targeting STAT3/JAK signalling with the natural polyphenol curcumin enhances NB chemosensitivity[371]. The highly oxidized plant steroid cucurbitacin-I either alone or in combination with the bi-phenol Honokiol inhibits NB tumourigenicity[372,373]. The selective Aurora B kinase inhibitor AZD1152 reduces NB cell proliferation and is selectively cytotoxic to tumour inducing NB CSC-like cells[374]. Inhibition of adenosine monophosphate activated protein kinase (AMPK) signalling also targets CSCs, the AMPK activator Metformin and AMPK pathway inhibitor vidarabine or 9-β-D-arabinofuranosyladenine reduce 2-D and 3-D NB CSC proliferation via Metformin inhibition of mTOR and Akt, and energy deprivation via Ara-a-mediated AMPK inhibition[375]. The PI3K/mTOR inhibitor NVP-BEZ235 decreases angiogenesis and increases survival in N-Myc-amplified orthotopic NB xenograft and transgenic N-Myc driven mouse NB models by reducing CSC-like activity[376].
N-Myc: NB CSC-associated N-Myc oncogene expression is stabilized and potentiated by interaction with ubiquitin-specific protease 7, and could be inhibited by the ubiquitin-specific protease 7 inhibitor P2207[377]. N-Myc expression in NB cells can also be reduced by small molecule BET bromodomain inhibitors JQ1 and I-BET151, which combined with HDAC inhibitor Panobinostat act synergistically to reduce N-Myc and LIN28B CSC-promoting oncogene expression in NB cells, resulting in apoptosis, reduced tumour growth and progression in NB-bearing mice[378]. Inhibitors of N-Myc expression would also reduce N-Myc regulated MALAT1 lncRNA which contributes to CSC induction and maintenance[255] and reduces the expression of N-Myc-regulated histone demethylase JMJD1A, a regulator of SC behaviour and CSC-associated MALAT1 expression in N-Myc amplified NB[251,256,257].
HIFs: Tumour hypoxia and pseudo-hypoxic phenotypes drive NB CSC phenotypes by activating HIF-1 and HIF-2 transcription factors, identifying HIFs are potential therapeutic targets in reducing NB CSC subpopulations. HIF inhibitors include inhibitors of HIF mRNA and protein expression, HIF dimerization, DNA-binding and transcriptional function and promoters of HIF-α degradation. HIF-1 mRNA and protein expression inhibitors include: lncRNA PIN1-v2[379]; S-TRPM2 calcium-permeable ion channel short variant[380] and EZN-2698 and EZN-2208 HIF-1α antisense oligonucleotides[381,382]. The topoisomerase inhibitor topecan inhibits HIF-1α protein translation and function[383], the natural flavonoid chrysin, soybean glyceollin phytoalexins and KC7F2 small molecule inhibit HIFα protein expression[384-386], and the estrogen metabolite 2-methoxy-estradiol inhibits HIF-1α and HIF-2α protein synthesis, nuclear translocation and transcriptional activity[387]. Hsp90 inhibitors GA, 17-AAG, 17DMAG and EC154, the HDAC inhibitor vorinostat and LW6t small molecule promote VHL-dependent HIF-α degradation, preventing accumulation and inhibiting transcriptional activity[388-391]. The small molecule PX12 inhibits HIF-1α accumulation by targeting thioredoxin-1[392,393], and BAY87–2243 suppresses HIF-1α and HIF-2α protein accumulation by inhibiting mitochondrial complex-1 (stopped in phase 1 trials)[394,395]. Inhibitors of HIF dimerization, include: cyclic CLLFVY that binds the HIF-1α PAS-B domain disrupting dimerization, transcriptional function and tumour cell hypoxic responses[396]; TC-S7009, a nanomo
HIF-1α silencing, when combined with retinoic acid formulations such as Al trans retinoic acid, used in the clinic to induce differentiation in high-risk NB following induction and consolidation chemotherapy[409,410], augment NB cell differentiation, senescence and chemosensitivity[411], and when combined with the proteasome inhibitor MG132 reduce NB CSC tumour sphere forming capacity, Nestin, Sox2 and Oct4 expression[412], highlighting the importance of combining HIF inhibitors with differentiation inducers to target malignant NB CSC subpopulations.
Targeting epigenetic transcription regulators: Targeting DNA methylation machine
Consistent with enhanced anti-oxidant activity in CSCs and potential extracellular roles for Trx and TrxR in altering the MMP/TIMP balance, disrupting extracellular matrices, promoting invasion and altering the angiogenic behaviour of endothelial cells[273,274], novel TrxR1 inhibitors have been reported to induce CSC death, suppress multidrug resistance by inhibiting the P-glycoprotein extrusion pump, to inhibit antioxidant defense, suppress invasion and migration and enhance sensitivity to chemotherapy[421]. Increased mitochondrial SOD2 expression and activity in NB CSCs, promoted by oncogenes, including TrkAIII, also enhances NB cell resistance to ROS-induced cytotoxicity[152], and could be targeted to enhance chemosensitivity. In this regard, the nonsteroidal anti-inflammatory drug Diclofenac has been shown to inhibit mitochondrial SOD2 expression and activity, and to promote NB cell apoptosis through the intrinsic mitochondrial pathway[422].
Other agents reported to target CSCs, include the plant alkaloid Berberine, which attenuates CD133, β-catenin, N-Myc, Sox2, Notch-2 and Nestin expression in Neuro2A NB cells impairing cancer stemness; down regulates PI3K/Akt and Ras/MAPK/Erk signalling reversing EMT in association with restored E-cadherin expression and reduced vimentin and fibronectin expression; represses cyclin and cyclin-dependent kinase expression potentiating G0/G1 cell cycle arrest and inhibiting proliferation, and modulates TGFβRI and RII receptors, promoting neuronal differentiation[423]. The polo-like kinase-1 inhibitor imidazotriazine GSK461346 (in clinical development), inhibits clonal expansion, promotes cell death and reduces tumourigenic activity in tumour initiating NB CSC models[424]. The small molecule tankyrase inhibitor XAV939 reverses NB CSC stemness and reduces migration capacity[425]. TRAIL induces apoptosis in TrkAIII expressing CSC-like NB cells through SHP/Src-mediated crosstalk with the TRAIL-receptor signalling pathway, suggesting that improved TRAIL formulation may also have a place in eradicating CSC populations, under certain circumstances[426]. The mitochondrial carnitine palmitoyl-transferase-1 inhibitor perhexiline promotes NB cell differentiation by inducing non-coding RNA NB differentiation marker 29 expression, resulting in reduced stemness, anchorage-dependent growth, reduced tumourigenic capacity and enhanced chemosensitivity via reduced ABC transporter expression and, combined with cisplatin, reduces nodule formation and increases survival in a mouse xenograft NB model, consistent with CSC depletion[342,427]. Stem cell-based small molecule screens have identified Dequalinium analogue C14 Linker, an analogue of the antibacterial mouthwash component Deca 10, as a nanomolar cytotoxic agent that kills tumour initiating CSC-like NB cells, reduces tumour sphere formation and targets NB CSCs in vivo by impairing mitochondrial function and inducing apoptosis[428]. Rapamycin has also been identified as an inhibitor of NB tumour initiating CSC-like survival and proliferation at nanomolar concentrations, and when combined with vinblastine inhibits NB xenograft tumour growth[428]. Additional compounds that exhibit selective activity against tumour initiating NB CSC-like cells include teniposide and vinblastine[428]. A drug library screen has also identified carbenoxolone as an inhibitor of FOXO-3-dependent therapeutic-resistance in high-stage NB cells, reducing survival and increasing sensitivity to chemotherapeutic agents in 2D and 3D models[429]. b-carotene also inhibits NB CSC self-renewal capacity and reduces CSC marker expression and resensitizes NB CSCs to cisplatin cytotoxicity, suggesting a potential use in NB chemotherapy[430]. Considering that neural-related tumour initiating CSC-like cells exhibit distinct telomere maintenance, NB CSC-like cells may also represent good targets for telomerase inhibitors[431].
Polyploid giant cells are therapy-induced, therapy-resistant tumour subpopulations capable of orchestrating post-therapeutic recurrence and disease progression through the generation of CSCs[280,281,283,432]. CSC Polyploidy is considered a druggable phenotype but mitosis targeting strategies may also promote polyploidy in surviving cells. Novel therapeutic strategies include targeting the higher energetic needs of polyploid giant cells[433]. In this regard, brain and acute myeloid leukemia polyploid giant cells are preferentially killed by the glycolysis inhibitor 2-deoxy-D-glucose[434,435]. Specific mTOR inhibitors induce NB cell osteogenic differentiation[436] and enhance the effect of Aurora kinase inhibitors in promoting apoptosis of polyploid giant cancer cells[433]. The inhibition of mTOR signalling prevents therapy-induced polyploid giant cell formation[437] and the mTOR inhibitor AZD8055 prevents therapy-induced formation of pancreatic cancer polyploid giant cancer cells[438], illustrating that metabolic inhibition in polyploid giant cells may not only reduce CSC generation but also enhance therapeutic sensitivity in tumours containing polyploid giant cells. The anti-fungal anti-oxidant resveratrol selectively reduces the fitness of polyploid giant cancer cells by slowing down the cell cycle and promoting apoptosis, an effect that is augmented when combined with AMPK stimulators. In this regard, resveratrol combined with aspirin reduces the formation of polyploid colon cancer cells that lack p53 function and in APCMin/+ mice, prevents polyploidy in intestinal cells and subsequent tumour formation[439,440]. Conversely, maintenance of polyploid cancer cells senescence may also be efficacious, illustrated by the small molecule inhibitor R1530, which promotes tubulin polymerization and mitotic checkpoint kinase BubR1 function, inducing senescent polyploid cancer cell formation and reducing tumour growth in a human H460 non-small-cell lung cancer xenograft model, suggesting that continual promotion of mitotic catastrophe in polyploid cancer cells may block NB CSC formation[441]. Along this theme, microtubule polymerizing taxanes and microtubule de-polymerizing vinca alkaloids promote mitotic catastrophe and death in cancer cells but lack specificity and induce severe side-effects; investigational Monastrol AZD4877, Ispinesib, and ARRY-520 (Phase 1 and II trials completed) Kinesin-5 motor protein inhibitors promote mitotic arrest, tumour cell death and are well tolerated; FDA-approved GSK923295 centrosome-associated protein-CENP-E inhibitor induces defective mitosis and inhibits proliferation; FDA-approved UCN-01/staurosporine and AZD7762 check-point kinase inhibitors induce death in p53-deficient tumours; WEE1 kinase and HDAC inhibitors combined with DNA damaging agents induce mitotic catastrophe; APC-Cdc20 targeting prevents cyclin B degradation and promotes mitotic exit; small molecule dynamin GTPase inhibitors induce cytokinesis failure and cell death, and c-myc repression promotes cancer cell mitotic catastrophe and death[442].
The central role of aberrant centrosome numbers and behaviour in polyploid giant cancer cell formation, continuous chromosomal instability, generation of aneuploid CSC-like cells, de-regulated microtubule organisation and irregular cell cycles, also makes the centrosome a promising therapeutic target for reducing tumour CSC populations[443,444]. Centrosomes and their pericentriolar matrices contain important proto-oncogenes and tumour suppressors that, when altered, drive centrosome amplification and subsequent chromosomal aberrations, the therapeutic inhibition of which can blunt centrosome involvement in tumour progression. Targeting centrosome Plk1 with BI2536 or BI6727 inhibitors has shown efficacy in acute myeloid leukemia[445]. Small molecular cyclin-dependent kinase inhibitors that de-regulate the centrosome cycle are in development[446], FDA-approved PD0332991 (Palbociclib), LEE011 (ribociclib) and LY2835219 (abemaciclib) cdk4/6 inhibitors are amongst the most recent[444] but may cause rare severe lung inflammation (https://www.fda.gov/drugs/drug-safety-and-availability/fda-warns-about-rare-severe-lung-inflammation-ibrance-kisqali-and-verzenio-breast-cancer). A number of Aurora kinase inhibitors are also in clinical trials aimed at reducing chromosomal instability caused by aberrant centrosome behaviour, and exhibit incremental efficacy when combined with conventional chemotherapeutic agents, restoring therapeutic sensitivity and reducing tumour progression[287,443]. The FDA-approved Plk4 inhibitor CFI-400945 promotes defective centrosome duplication, cell death and inhibits tumour xenograft growth[447] and the FDA-approved Mps1/TTK inhibitor BAY1161909 (Empesertib) blocks spindle assembly, chromosome attachment and exhibits marked tumour suppressing activity when combined with paclitaxel[448]. FDA-approved Nutlins restores P53 function by inhibiting interaction with MDM2 and PRIMA-1, which targets Y220C mutant p53, restores wild-type p53 function reducing centrosome amplification[449,450]. Centrosome clustering, required for pseudo-bipolar spindle formation, leads to mitosis and aneuploid CSCs formation in centrosome-amplified polyploid giant cancer cells. This can be prevented by the FDA-approved small molecule PARP-6 inhibitor AZ0108, which promotes multipolar spindle formation and apoptosis in centrosome-amplified cells and exhibits anti-tumour effects in cancer models[451,452]. Centrosome clustering can also be disrupted by the investigational small molecule CCB02, which disrupts the interaction between centrosomal-P4.1-associated protein and b-tubulin, resulting in prolonged multipolar mitosis and death of centrosome-amplified cancer cells[453].
Targeting the tumour microenvironment: Reprogramming of the hypoxic tumour niche improves the efficacy of chemotherapy, fractionated radiotherapy and CAR T-cell immunotherapy[159,454], and represses hypoxia-induced CSCs phenotypes. Tumour oxygenation can be improved by hyperbaric oxygenation, intra-tumoural lipid-stabilized oxygen microbubble injection[455,456], nanoparticle-mediated reoxygenation, oxygen-generating methods[457] or artificial red cells[458]. The “normalization” of aberrant tumour vasculatures may also improve tumour oxygenation, reduce tumour progression and enhance therapeutic efficacy, stemming from observations that vascular destruction promotes tumour hypoxia, reduces therapeutic efficacy and facilitates metastatic progression. Tumour vessels are immature, permeable, tortuous, have aberrant basement membranes and lack cellular and matrix components required for maturation and function. This results in increased interstitial hypertension that promotes rapid drug-efflux and generates hypoxic niches, which promote and select polyploid giant cancer cell and CSC-like phenotypes. Vascular “normalization” requires delicate rebalancing of angiogenic factor/inhibitor equilibria by careful selection and dosage of antiangiogenic agents. Inhibition of VEGFA, VEGFRs or PHD2 proline hydroxylase inhibitors can prune immature permeable vessels, reduce permeability, improve vessel maturity, blood flow and oxygenation, decrease interstitial pressure to improve drug penetration and chemosensitivity, reduce metastatic progression and potentially reduce hypoxic niche-associated CSC subpopulations[459,460]. Finally, tumour associated stromal and extracellular matrix barriers to CSC-targeted therapies delivered by nanoparticles or by engineered immune cells could be overcome using FAP+ tumour fibroblasts targeted CAR T-cells, or other CAR cell types, engineered to express matrix degrading enzymes to disrupt tumour stroma and improve therapeutic access[159].
Engineered MSCs: The close relationship between MSCs and NB CSCs in tumourigenesis and metastatic progression[319-321] has also led to novel strategies that utilize MSCs to deliver anti-NB therapeutic agents. In the mouse TH-N-Myc NB model, IFNβ-transduced mouse MSCs home to NBs, remain in tumours for up to 2 wk and augment tumour IFNβ levels[461]. Purified human adipose MSCs transfected with miR-124 and co-cultured with human M17 NB cells transfer miR-124 mimetic to NB cells by exosome delivery, resulting in miR-124-dependent reduced proliferation, increased apoptosis and neuronal differentiation, demonstrating that human adipose MSCs can be used effectively to deliver mimetic miRNAs to NB cells, supporting future therapeutic use of autonomous patient-derived MSCs as therapeutic vectors in NB[462]. Primary human MSCs engineered to express full length membrane-associated TRAIL[463], kill TRAIL receptor positive classic and primary NB cell lines, migrate to NB sites following intraperitoneal injection, and reduce NB xenograft growth in vivo, confirming drug delivery potential in NB[464]. However, the potential use of MSCs for drug delivery must take into account reports that MSCs also promote tumour growth and metastasis[465,466].
Oncolytic viruses: Genetically modified oncolytic virotherapy holds great promise as a novel anti-cancer therapy for eliminating CSCs[467]. The engineered oncolytic virus Nestin-targeted oHSV (rQNestin34.5) has been shown to kill doxorubicin-resistant CD133+ SP CSC-like NB cells and to eradicate both bulk and tumour initiating tumour spheres, significantly delaying tumour formation in in vivo models[468]. The first report of oncolytic virotherapy in NB was the direct intratumour injection of the Ad5/3-Cox2 oncolytic adenovirus, which induced a clinical response in a 6 year old stage IV NB patient[469]. The oncolytic adenovirus ICOVIR, that exhibits enhanced tumour selectivity by exploiting aberrant E2F expression, has been used to treat several children with stage IV refractory NB by intravenous injection of irradiation-inactivated ICOVIR infected autologous MSCs, with complete 5 year remission in the only patient to exhibit a response[470]. In a recent first in-human and in-child trial study of autologous Icovir-5 infected MSC treatment of NB, two patients exhibited disease stabilization associated with low toxicity[471]. The engineered measles virus Edmonston strain has also been shown to have significant cytopathic effects on human primary and classical NB cell lines and xenograft models, inducing apoptotic cell death[472].
Nanoparticles for improving delivery and efficacy: Nanoparticle drug delivery facilitates better control of drug-release kinetics, improves bio-distribution and prolongs systemic and localized drug longevity. Mesoporous silica nanoparticles loaded with Notch signalling inhibitors or carrying γ-secretase inhibitors efficiently target tumour CSCs[473,474]. Salinomycin conjugated to a hyaluronic acid-based nanogel targets drug-resistant CD44+ cells and enhances therapeutic efficacy[475]. Nanoparticles coated with antibodies to CSC markers improve targeting, polymeric (D, L lactide-co-glycolide) nanoparticles coated with an anti-CD133 antibody and loaded with paclitaxel reduce tumourigenicity in mouse xenograft models[476], and Fe2O3 nanoparticles coated with antibodies to ABCG2 transporter and loaded with paclitaxel target and kill myeloma cells[477]. With regard to NB, synthetic HDL nanoconjugates that target the SR-B1 receptor are taken up by validated NB CSCs, and reduce CSC viability, self-renewal, invasion and migration in vitro, and in vivo exhibit tumour-uptake and reduce tumour growth, supporting further evaluation as a therapeutic NB CSCs inhibitor[478]. Mitochondrial-targeted silver nanoparticle cytotoxicity could be enhanced in NB cells by inhibitors of humanin expression, reported to protect NB cells against silver nanoparticle-induced toxicity[479]. Low nanoparticle penetration, due to low-level extravasation, has also been intelligently addressed, based upon greater understanding of the permeable and immature nature of tumour micro-vasculatures, with the development of gold nanoshell nanoparticles that seed tumour vasculatures for photothermal tumour ablation. This approach has demonstrated remarkable efficacy in mouse xenograft tumour models and in a clinical pilot study of low and intermediate risk prostate cancer, extolling a 94% success rate in tumour ablation[480].
The embryonic origin of NBs from NC stem/progenitor cells and their stemness gene expression patterns, indicate that NBs contain significant CSC-like subpopulations from the outset, which combined with properties of self-renewal, tumourigenicity, multipotency, therapy-resistance and reversible plasticity play central roles in NB heterogeneity, therapeutic-resistance, post-therapeutic-relapse, metastatic progression and unfavourable outcome. Conventional induction, consolidation and post-consolidation therapeutic strategies for high risk unfavourable NBs, induce initial clinical remission to states of no evidence or minimal residual disease, but also select and promote the formation of therapy-resistant polyploid giant cancer cells (PGCCs) and CSC subpopulations, adding to the probability of post-therapeutic relapse and metastatic progression. It is, therefore, of paramount importance that novel therapeutic strategies are developed to specifically target and eliminate PGCCs and CSC subpopulations in order to improve the current ≈ 8% 5-year overall survival rate in high risk NB[481].
As illustrated in this review, major advances in understanding the molecular protagonists, signalling pathways and mechanisms responsible for the induction, selection, maintenance and behaviour of NB PGCCs and CSCs have been made, and novel therapeutic strategies are currently being developed to target these populations. From our own perspective, observations that NF-κB-induced MMP-9 expression and MMP-9-dependent invasion associate with spontaneous S to I (CSC) phenotype conversion in NB cells[93,175], and the NB-associated oncogenic alternative TrkAIII splice variant promotes NB cell MMP-9 expression within a CSC context, links MMP-9 and stress-regulated alternative TrkAIII splicing to more aggressive NB CSC behaviour and identifies MMP-9 and TrkAIII as potential therapeutic NB CSC targets[72,73]. TrkAIII promotion of PI3K/Akt/NF-κB survival signalling, Bcl2 family expression[480], a survival-adapted ER-stress response[219], mitochondrial SOD2 expression[198] and pro-angiogenic MMP-9/VEGF/TSP-1 equilibrium, within a NB CSC context, furthermore, provide important insights into oncogene-regulated apoptosis evasion, survival within stressful tumour microenvironments, protection against mitochondrial ROS-induced death and angiogenesis, likely to extend to other splice-activated, mutated and fusion Trk oncogenes. Stress-regulated TrkAIII-dependent metabolic plasticity, unveils a mechanism for NB CSC metabolic adaption to changing conditions within tumour microenvironments[218], and TrkAIII promotion of centrosome amplification, unveils a mechanism for enhancing therapeutic resistance and promoting dormancy through NB PGCC formation[217], which upon escape from dormancy can generate blastomere-like CSCs with highly aggressive metastatic karyotypes. Furthermore, our recent discovery of a novel NF-YAx splice variant, expressed by NBs and induced by doxorubicin, that selects NB CSC-like cells through differential cytotoxicity, unveils a potential mechanism for genotoxic drug-induced NB CSC selection and post-therapeutic relapse, which will be fully investigated in due course[136]. It is also our opinion that PGCCs formation and regulation of PGCC dormancy and escape from dormancy to generate potentially highly metastatic blastomere-stage CSCs, represent underestimated and understudied areas and should form the basis of novel pre-clinical models to study post-therapeutic NB relapse from PGCCs, with the aim of identifying single functional surface markers or marker-sets that uniformly and privately identify dormant NB PGCCs, counterparts that have escaped dormancy and their secondary CSC progeny, for better detection and therapeutic targeting and experimental therapeutics. Furthermore, current therapeutic strategies, which suffer from problems of delivery, uptake and efficacy due to aberrant tumour micro-vasculatures, physical barriers and acidic hypoxic microenvironmental conditions that promote drug-efflux, must also be improved to overcome these problems. In this review and within this context, we highlight the use of nanoparticles for improving drug kinetics or to facilitate thermal ablation; novel improved CSC-targeting CAR T-cells that modify inflammatory environments, modify tumour matrices and that can be switched off or eliminated to minimize side effects, with immune checkpoint inhibitors to improve infiltration; novel CAR NK, iNKT cells and engineered MSCs to target CSC niches; tumour oxygenation to reduce hypoxic CSC niches and enhance drug-sensitivity; centrosome dispersal and microtubule disrupting agents to perpetuate dormancy, and novel oncolytic viruses that exhibit improved CSC targeting and lytic activity (Figure 5). Continued research and development in these areas will hopefully lead to much needed improvements in event-free and overall survival in high risk unfavourable NBs.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Feng JX, Velasco-Velazquez M S-Editor: Zhang L L-Editor: Webster JR P-Editor: Li JH
| 1. | Horstadius S. The neural crest. Its properties and derivatives in light of experimental research. Geoffrey Cumberledge Oxford University press. 1950;. [DOI] [Cited in This Article: ] |
| 2. | O'Rahilly R, Müller F. The development of the neural crest in the human. J Anat. 2007;211:335-351. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Le Douarin NM, Dupin E. The "beginnings" of the neural crest. Dev Biol. 2018;444 Suppl 1:S3-S13. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Alkobtawi M, Monsoro-Burq AH. The neural crest, a vertebrate invention. In: Eames BF, Medeiros DM, Adamenko I. Evolving Neural Crest Cells. FL USA: CRC Press, 2020: 5-66. [Cited in This Article: ] |
| 5. | Yu JK, Su YH. The evolution of the neural border and peripheral nervous system-insights from invertebrate deuterostome animals. In: Eames BF, Medeiros DM, Adamenko I. Evolving Neural Crest Cells. FL USA: CRC Press, 2020: 103-136. [Cited in This Article: ] |
| 6. | York JR, Zehnder K, McCauley DW. The evolution of cellular EMT and migration. In: Eames BF, Medeiros DM, Adamenko I. Evolving Neural Crest Cells. FL USA: CRC Press, 2020: 67-102. [Cited in This Article: ] |
| 7. | Woodhoo A, Sommer L. Development of the Schwann cell lineage: from the neural crest to the myelinated nerve. Glia. 2008;56:1481-1490. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Monk KR, Feltri ML, Taveggia C. New insights on Schwann cell development. Glia. 2015;63:1376-1393. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Furlan A, Adameyko I. Schwann cell precursor: a neural crest cell in disguise? Dev Biol. 2018;444 Suppl 1:S25-S35. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Pla P, Monsoro-Burq AH. The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev Biol. 2018;444 Suppl 1:S36-S46. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Kobayashi GS, Musso CM, Moreira DP, Pontillo-Guimarães G, Hsia GSP, Caires-Júnior LC, Goulart E, Passos-Bueno MR. Recapitulation of Neural Crest Specification and EMT via Induction from Neural Plate Border-like Cells. Stem Cell Reports. 2020;15:776-788. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Sieber-Blum M, Zhang J-M. The neural crest and neural crest defects. Biomedical Reviews 2002; 13:29-37. [DOI] [Cited in This Article: ] |
| 13. | Huang X, Saint-Jeannet JP. Induction of the neural crest and the opportunities of life on the edge. Dev Biol. 2004;275:1-11. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Hackland JOS, Frith TJR, Thompson O, Marin Navarro A, Garcia-Castro MI, Unger C, Andrews PW. Top-Down Inhibition of BMP Signaling Enables Robust Induction of hPSCs Into Neural Crest in Fully Defined, Xeno-free Conditions. Stem Cell Reports. 2017;9:1043-1052. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Shakhova O, Sommer L. Neural crest-derived stem cells 2008. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Ruggeri P, Farina AR, Cappabianca L, Di Ianni N, Ragone M, Merolle S, Gulino A, Mackay AR. Neurotrophin and neurotrophin receptor involvement in human neuroblastoma. (May 29th 2013). [DOI] [Cited in This Article: ] |
| 17. | Vega-Lopez GA, Cerrizuela S, Aybar MJ. Trunk neural crest cells: formation, migration and beyond. Int J Dev Biol. 2017;61:5-15. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Scarpa E, Szabó A, Bibonne A, Theveneau E, Parsons M, Mayor R. Cadherin Switch during EMT in Neural Crest Cells Leads to Contact Inhibition of Locomotion via Repolarization of Forces. Dev Cell. 2015;34:421-434. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Taneyhill LA, Schiffmacher AT. Should I stay or should I go? Genesis. 2017;55. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Kotini M, Barriga EH, Leslie J, Gentzel M, Schambony A, Mayor R. Connexin43 controls N-cadherin transcription during collective cell migration. BioRxiv 2017. [DOI] [Cited in This Article: ] |
| 21. | Padmanabhan R, Taneyhill LA. Cadherin-6B undergoes macropinocytosis and clathrin-mediated endocytosis during cranial neural crest cell EMT. J Cell Sci. 2015;128:1773-1786. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Schiffmacher AT, Xie V, Taneyhill LA. Cadherin-6B proteolysis promotes the neural crest cell epithelial-to-mesenchymal transition through transcriptional regulation. J Cell Biol. 2016;215:735-747. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Schiffmacher AT, Adomako-Ankomah A, Xie V, Taneyhill LA. Cadherin-6B proteolytic N-terminal fragments promote chick cranial neural crest cell delamination by regulating extracellular matrix degradation. Dev Biol. 2018;444 Suppl 1:S237-S251. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Barriga EH, Maxwell PH, Reyes AE, Mayor R. The hypoxia factor Hif-1α controls neural crest chemotaxis and epithelial to mesenchymal transition. J Cell Biol. 2013;201:759-776. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Niklasson CU, Fredlund E, Monni E, Lindvall JM, Kokaia Z, Hammarlund EU, Bronner ME, Mohlin S. Hypoxia inducible factor-2α importance for migration, proliferation, and self-renewal of trunk neural crest cells. Dev Dyn. 2021;250:191-236. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Espina JA, Marchant CL, De Ferrari GV, Reyes AE. Pdgf1aa regulates zebrafish neural crest cells migration through Hif-1 in an oxygen-independent manner. Mech Dev. 2018;154:203-207. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Kulesa PM, Bailey CM, Kasemeier-Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Dev Biol. 2010;344:543-554. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Bronner ME, Simões-Costa M. The Neural Crest Migrating into the Twenty-First Century. Curr Top Dev Biol. 2016;116:115-134. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Li Y, Vieceli FM, Gonzalez WG, Li A, Tang W, Lois C, Bronner ME. In Vivo Quantitative Imaging Provides Insights into Trunk Neural Crest Migration. Cell Rep. 2019;26:1489-1500.e3. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Szabó A, Melchionda M, Nastasi G, Woods ML, Campo S, Perris R, Mayor R. In vivo confinement promotes collective migration of neural crest cells. J Cell Biol. 2016;213:543-555. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Lee VM, Hernandez S, Giang B, Chabot C, Hernandez J, de Bellard ME. Molecular Events Controlling Cessation of Trunk Neural Crest Migration and Onset of Differentiation. Front Cell Dev Biol. 2020;8:199. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Stemple DL, Anderson DJ. Isolation of a stem cell for neurons and glia from the mammalian neural crest. Cell. 1992;71:973-985. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Delfino-Machín M, Chipperfield TR, Rodrigues FS, Kelsh RN. The proliferating field of neural crest stem cells. Dev Dyn. 2007;236:3242-3254. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Vandamme N, Berx G. From neural crest cells to melanocytes: cellular plasticity during development and beyond. Cell Mol Life Sci. 2019;76:1919-1934. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Kruger GM, Mosher JT, Bixby S, Joseph N, Iwashita T, Morrison SJ. Neural crest stem cells persist in the adult gut but undergo changes in self-renewal, neuronal subtype potential, and factor responsiveness. Neuron. 2002;35:657-669. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Bixby S, Kruger GM, Mosher JT, Joseph NM, Morrison SJ. Cell-intrinsic differences between stem cells from different regions of the peripheral nervous system regulate the generation of neural diversity. Neuron. 2002;35:643-656. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Wong CE, Paratore C, Dours-Zimmermann MT, Rochat A, Pietri T, Suter U, Zimmermann DR, Dufour S, Thiery JP, Meijer D, Beermann F, Barrandon Y, Sommer L. Neural crest-derived cells with stem cell features can be traced back to multiple lineages in the adult skin. J Cell Biol. 2006;175:1005-1015. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | White PM, Morrison SJ, Orimoto K, Kubu CJ, Verdi JM, Anderson DJ. Neural crest stem cells undergo cell-intrinsic developmental changes in sensitivity to instructive differentiation signals. Neuron. 2001;29:57-71. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Kléber M, Lee HY, Wurdak H, Buchstaller J, Riccomagno MM, Ittner LM, Suter U, Epstein DJ, Sommer L. Neural crest stem cell maintenance by combinatorial Wnt and BMP signaling. J Cell Biol. 2005;169:309-320. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Fuchs S, Herzog D, Sumara G, Büchmann-Møller S, Civenni G, Wu X, Chrostek-Grashoff A, Suter U, Ricci R, Relvas JB, Brakebusch C, Sommer L. Stage-specific control of neural crest stem cell proliferation by the small rho GTPases Cdc42 and Rac1. Cell Stem Cell. 2009;4:236-247. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | Zhu Q, Lu Q, Gao R, Cao T. Prospect of Human Pluripotent Stem Cell-Derived Neural Crest Stem Cells in Clinical Application. Stem Cells Int. 2016;2016:7695836. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Zhang JT, Weng ZH, Tsang KS, Tsang LL, Chan HC, Jiang XH. MycN Is Critical for the Maintenance of Human Embryonic Stem Cell-Derived Neural Crest Stem Cells. PLoS One. 2016;11:e0148062. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Srinivasan A, Toh YC. Human Pluripotent Stem Cell-Derived Neural Crest Cells for Tissue Regeneration and Disease Modeling. Front Mol Neurosci. 2019;12:39. [PubMed] [DOI] [Cited in This Article: ] |
| 44. | Dupin E, Calloni G, Real C, Gonçalves-Trentin A, Le Douarin NM. Neural crest progenitors and stem cells. C R Biol. 2007;330:521-529. [PubMed] [DOI] [Cited in This Article: ] |
| 45. | Calloni GW, Le Douarin NM, Dupin E. High frequency of cephalic neural crest cells shows coexistence of neurogenic, melanogenic, and osteogenic differentiation capacities. Proc Natl Acad Sci USA. 2009;106:8947-8952. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Le Douarin NM, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development. 2004;131:4637-4650. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Sieber-Blum M. Growth factor synergism and antagonism in early neural crest development. Biochem Cell Biol. 1998;76:1039-1050. [PubMed] [Cited in This Article: ] |
| 48. | Sommer L. Growth factors regulating neural crest cell fate decisions. Adv Exp Med Biol. 2006;589:197-205. [PubMed] [DOI] [Cited in This Article: ] |
| 49. | Chen CC, Hsia CW, Ho CW, Liang CM, Chen CM, Huang KL, Kang BH, Chen YH. Hypoxia and hyperoxia differentially control proliferation of rat neural crest stem cells via distinct regulatory pathways of the HIF1α-CXCR4 and TP53-TPM1 proteins. Dev Dyn. 2017;246:162-185. [PubMed] [DOI] [Cited in This Article: ] |
| 50. | Langtimm-Sedlak CJ, Schroeder B, Saskowski JL, Carnahan JF, Sieber-Blum M. Multiple actions of stem cell factor in neural crest cell differentiation in vitro. Dev Biol. 1996;174:345-359. [PubMed] [Cited in This Article: ] |
| 51. | Lahav R, Dupin E, Lecoin L, Glavieux C, Champeval D, Ziller C, Le Douarin NM. Endothelin 3 selectively promotes survival and proliferation of neural crest-derived glial and melanocytic precursors in vitro. Proc Natl Acad Sci USA. 1998;95:14214-14219. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Zage PE, Whittle SB, Shohet JM. CD114: A New Member of the Neural Crest-Derived Cancer Stem Cell Marker Family. J Cell Biochem. 2017;118:221-231. [PubMed] [DOI] [Cited in This Article: ] |
| 53. | Tsubota S, Kadomatsu K. Neuroblastoma stem cells and CFC1. Oncotarget. 2017;8:45032-45033. [PubMed] [DOI] [Cited in This Article: ] |
| 54. | Tomolonis JA, Agarwal S, Shohet JM. Neuroblastoma pathogenesis: deregulation of embryonic neural crest development. Cell Tissue Res. 2018;372:245-262. [PubMed] [DOI] [Cited in This Article: ] |
| 55. | Johnsen JI, Dyberg C, Wickström M. Neuroblastoma-A Neural Crest Derived Embryonal Malignancy. Front Mol Neurosci. 2019;12:9. [PubMed] [DOI] [Cited in This Article: ] |
| 56. | Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann JM, Ikram F, Schmidt R, Ackermann S, Engesser A, Kahlert Y, Vogel W, Altmüller J, Nürnberg P, Thierry-Mieg J, Thierry-Mieg D, Mariappan A, Heynck S, Mariotti E, Henrich KO, Gloeckner C, Bosco G, Leuschner I, Schweiger MR, Savelyeva L, Watkins SC, Shao C, Bell E, Höfer T, Achter V, Lang U, Theissen J, Volland R, Saadati M, Eggert A, de Wilde B, Berthold F, Peng Z, Zhao C, Shi L, Ortmann M, Büttner R, Perner S, Hero B, Schramm A, Schulte JH, Herrmann C, O'Sullivan RJ, Westermann F, Thomas RK, Fischer M. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700-704. [PubMed] [DOI] [Cited in This Article: ] |
| 57. | Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, van Noesel MM, George RE, Tytgat GA, Molenaar JJ, Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411-1414. [PubMed] [DOI] [Cited in This Article: ] |
| 58. | Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, Laureys G, Speleman F, Kim C, Hou C, Hakonarson H, Torkamani A, Schork NJ, Brodeur GM, Tonini GP, Rappaport E, Devoto M, Maris JM. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930-935. [PubMed] [DOI] [Cited in This Article: ] |
| 59. | Garcia I, Mayol G, Ríos J, Domenech G, Cheung NK, Oberthuer A, Fischer M, Maris JM, Brodeur GM, Hero B, Rodríguez E, Suñol M, Galvan P, de Torres C, Mora J, Lavarino C. A three-gene expression signature model for risk stratification of patients with neuroblastoma. Clin Cancer Res. 2012;18:2012-2023. [PubMed] [DOI] [Cited in This Article: ] |
| 60. | Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK, Heguy A, Pappo AS, Federico S, Dalton J, Cheung IY, Ding L, Fulton R, Wang J, Chen X, Becksfort J, Wu J, Billups CA, Ellison D, Mardis ER, Wilson RK, Downing JR, Dyer MA; St Jude Children's Research Hospital–Washington University Pediatric Cancer Genome Project. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;307:1062-1071. [PubMed] [DOI] [Cited in This Article: ] |
| 61. | Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, Kim J, Lawrence MS, Lichenstein L, McKenna A, Pedamallu CS, Ramos AH, Shefler E, Sivachenko A, Sougnez C, Stewart C, Ally A, Birol I, Chiu R, Corbett RD, Hirst M, Jackman SD, Kamoh B, Khodabakshi AH, Krzywinski M, Lo A, Moore RA, Mungall KL, Qian J, Tam A, Thiessen N, Zhao Y, Cole KA, Diamond M, Diskin SJ, Mosse YP, Wood AC, Ji L, Sposto R, Badgett T, London WB, Moyer Y, Gastier-Foster JM, Smith MA, Guidry Auvil JM, Gerhard DS, Hogarty MD, Jones SJ, Lander ES, Gabriel SB, Getz G, Seeger RC, Khan J, Marra MA, Meyerson M, Maris JM. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279-284. [PubMed] [DOI] [Cited in This Article: ] |
| 62. | Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, Ebus ME, Haneveld F, Lakeman A, Schild L, Molenaar P, Stroeken P, van Noesel MM, Ora I, Santo EE, Caron HN, Westerhout EM, Versteeg R. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589-593. [PubMed] [DOI] [Cited in This Article: ] |
| 63. | Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nat Rev Clin Oncol. 2014;11:704-713. [PubMed] [DOI] [Cited in This Article: ] |
| 64. | Oh L, Hafsi H, Hainaut P, Ariffin H. p53, stem cell biology and childhood blastomas. Curr Opin Oncol. 2019;31:84-91. [PubMed] [DOI] [Cited in This Article: ] |
| 65. | Mondal T, Kanduri C. LncRNAs join hands together to regulate neuroblastoma progression. Mol Cell Oncol. 2019;6:1553697. [PubMed] [DOI] [Cited in This Article: ] |
| 66. | Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985-2995. [PubMed] [DOI] [Cited in This Article: ] |
| 67. | Zhu S, Thomas Look A. Neuroblastoma and Its Zebrafish Model. Adv Exp Med Biol. 2016;916:451-478. [PubMed] [DOI] [Cited in This Article: ] |
| 68. | Trigg RM, Turner SD. ALK in Neuroblastoma: Biological and Therapeutic Implications. Cancers (Basel). 2018;10. [PubMed] [DOI] [Cited in This Article: ] |
| 69. | Farina AR, Cappabianca L, Ruggeri P, Di Ianni N, Ragone M, Merolla S. Alternative TrkA splicing and neuroblastoma. In: Neuroblastoma - Present and Future. London: Prof Hiroyuki Shimada InTech, 2012: 111-136. [Cited in This Article: ] |
| 70. | Fell SM, Li S, Wallis K, Kock A, Surova O, Rraklli V, Höfig CS, Li W, Mittag J, Henriksson MA, Kenchappa RS, Holmberg J, Kogner P, Schlisio S. Neuroblast differentiation during development and in neuroblastoma requires KIF1Bβ-mediated transport of TRKA. Genes Dev. 2017;31:1036-1053. [PubMed] [DOI] [Cited in This Article: ] |
| 71. | Tacconelli A, Farina AR, Cappabianca L, Desantis G, Tessitore A, Vetuschi A, Sferra R, Rucci N, Argenti B, Screpanti I, Gulino A, Mackay AR. TrkA alternative splicing: a regulated tumor-promoting switch in human neuroblastoma. Cancer Cell. 2004;6:347-360. [PubMed] [DOI] [Cited in This Article: ] |
| 72. | Farina AR, Cappabianca L, Ruggeri P, Gneo L, Pellegrini C, Fargnoli MC, Mackay AR. The oncogenic neurotrophin receptor tropomyosin-related kinase variant, TrkAIII. J Exp Clin Cancer Res. 2018;37:119. [PubMed] [DOI] [Cited in This Article: ] |
| 73. | Schulte JH, Lindner S, Bohrer A, Maurer J, De Preter K, Lefever S, Heukamp L, Schulte S, Molenaar J, Versteeg R, Thor T, Künkele A, Vandesompele J, Speleman F, Schorle H, Eggert A, Schramm A. MYCN and ALKF1174L are sufficient to drive neuroblastoma development from neural crest progenitor cells. Oncogene. 2013;32:1059-1065. [PubMed] [DOI] [Cited in This Article: ] |
| 74. | Montavon G, Jauquier N, Coulon A, Peuchmaur M, Flahaut M, Bourloud KB, Yan P, Delattre O, Sommer L, Joseph JM, Janoueix-Lerosey I, Gross N, Mühlethaler-Mottet A. Wild-type ALK and activating ALK-R1275Q and ALK-F1174L mutations upregulate Myc and initiate tumor formation in murine neural crest progenitor cells. Oncotarget. 2014;5:4452-4466. [PubMed] [DOI] [Cited in This Article: ] |
| 75. | Olsen RR, Otero JH, García-López J, Wallace K, Finkelstein D, Rehg JE, Yin Z, Wang YD, Freeman KW. MYCN induces neuroblastoma in primary neural crest cells. Oncogene. 2017;36:5075-5082. [PubMed] [DOI] [Cited in This Article: ] |
| 76. | Kerosuo L, Neppala P, Hsin J, Mohlin S, Vieceli FM, Török Z, Laine A, Westermarck J, Bronner ME. Enhanced expression of MycN/CIP2A drives neural crest toward a neural stem cell-like fate: Implications for priming of neuroblastoma. Proc Natl Acad Sci USA. 2018;115:E7351-E7360. [PubMed] [DOI] [Cited in This Article: ] |
| 77. | Yang LN, Huang WK, Li XL, Bai YZ, Zhang SC. Sox10 Is a Specific Biomarker for Neural Crest Stem Cells in Immunohistochemical Staining in Wistar Rats. Dis Markers. 2020;2020:8893703. [PubMed] [DOI] [Cited in This Article: ] |
| 78. | Delloye-Bourgeois C, Bertin L, Thoinet K, Jarrosson L, Kindbeiter K, Buffet T, Tauszig-Delamasure S, Bozon M, Marabelle A, Combaret V, Bergeron C, Derrington E, Castellani V. Microenvironment-Driven Shift of Cohesion/Detachment Balance within Tumors Induces a Switch toward Metastasis in Neuroblastoma. Cancer Cell. 2017;32:427-443.e8. [PubMed] [DOI] [Cited in This Article: ] |
| 79. | Delloye-Bourgeois C, Castellani V. Hijacking of Embryonic Programs by Neural Crest-Derived Neuroblastoma: From Physiological Migration to Metastatic Dissemination. Front Mol Neurosci. 2019;12:52. [PubMed] [DOI] [Cited in This Article: ] |
| 80. | Vo KT, Matthay KK, Neuhaus J, London WB, Hero B, Ambros PF, Nakagawara A, Miniati D, Wheeler K, Pearson AD, Cohn SL, DuBois SG. Clinical, biologic, and prognostic differences on the basis of primary tumor site in neuroblastoma: a report from the international neuroblastoma risk group project. J Clin Oncol. 2014;32:3169-3176. [PubMed] [DOI] [Cited in This Article: ] |
| 81. | Aquino JB, Sierra R. Schwann cell precursors in health and disease. Glia. 2018;66:465-476. [PubMed] [DOI] [Cited in This Article: ] |
| 82. | Olsen TK, Otte J, Mei S, Kameneva P, Bjorklund A, Kryukov E, Johansson A, Sundstrom E, Martinsson T, Fransson S, Johnsen JI, Koger P, Adameyko I, Kharchenko PV, Baryawno N. Malignant Schwann cell precursors mediate intratumoral plasticity in human neuroblastoma. bioRevix 2020. [DOI] [Cited in This Article: ] |
| 83. | Bourdeaut F, Ribeiro A, Paris R, Pierron G, Couturier J, Peuchmaur M, Delattre O. In neuroblastic tumours, Schwann cells do not harbour the genetic alterations of neuroblasts but may nevertheless share the same clonal origin. Oncogene. 2008;27:3066-3071. [PubMed] [DOI] [Cited in This Article: ] |
| 84. | Weiss T, Tascher-Mandl S, Bileck A, Rifatbegovic F, Sorger H, Kauer M, Frech C, Windhager R, Gerner C, Ambros PF, Ambros IM. Schwann plasticity regulates neuroblastic differentiation. bioRevix 2020. [DOI] [Cited in This Article: ] |
| 85. | Pandian V, Ramraj S, Khan FH, Azim T, Aravindan N. Metastatic neuroblastoma cancer stem cells exhibit flexible plasticity and adaptive stemness signaling. Stem Cell Res Ther. 2015;6:2. [PubMed] [DOI] [Cited in This Article: ] |
| 86. | Cohen PS, Chan JP, Lipkunskaya M, Biedler JL, Seeger RC. Expression of stem cell factor and c-kit in human neuroblastoma. The Children's Cancer Group. Blood. 1994;84:3465-3472. [PubMed] [Cited in This Article: ] |
| 87. | Nervi B, Link DC, DiPersio JF. Cytokines and hematopoietic stem cell mobilization. J Cell Biochem. 2006;99:690-705. [PubMed] [DOI] [Cited in This Article: ] |
| 88. | Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res. 1989;49:219-225. [PubMed] [Cited in This Article: ] |
| 89. | Ross RA, Spengler BA, Domènech C, Porubcin M, Rettig WJ, Biedler JL. Human neuroblastoma I-type cells are malignant neural crest stem cells. Cell Growth Differ. 1995;6:449-456. [PubMed] [Cited in This Article: ] |
| 90. | Ross RA, Biedler JL, Spengler BA. A role for distinct cell types in determining malignancy in human neuroblastoma cell lines and tumors. Cancer Lett. 2003;197:35-39. [PubMed] [DOI] [Cited in This Article: ] |
| 91. | Walton JD, Kattan DR, Thomas SK, Spengler BA, Guo HF, Biedler JL, Cheung NK, Ross RA. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838-845. [PubMed] [DOI] [Cited in This Article: ] |
| 92. | Ross RA, Walton JD, Han D, Guo HF, Cheung NK. A distinct gene expression signature characterizes human neuroblastoma cancer stem cells. Stem Cell Res. 2015;15:419-426. [PubMed] [DOI] [Cited in This Article: ] |
| 93. | Farina AR, Tacconelli A, Vacca A, Maroder M, Gulino A, Mackay AR. Transcriptional up-regulation of matrix metalloproteinase-9 expression during spontaneous epithelial to neuroblast phenotype conversion by SK-N-SH neuroblastoma cells, involved in enhanced invasivity, depends upon GT-box and nuclear factor kappaB elements. Cell Growth Differ. 1999;10:353-367. [PubMed] [Cited in This Article: ] |
| 94. | Yang S, Zheng J, Xiao X, Xu T, Tang W, Zhu H, Yang L, Zheng S, Dong K, Zhou G, Wang Y. SOX2 promotes tumorigenicity and inhibits the differentiation of I-type neuroblastoma cells. Int J Oncol. 2015;46:317-323. [PubMed] [DOI] [Cited in This Article: ] |
| 95. | Han D, Spengler BA, Ross RA. Increased wild-type N-ras activation by neurofibromin down-regulation increases human neuroblastoma stem cell malignancy. Genes Cancer. 2011;2:1034-1043. [PubMed] [DOI] [Cited in This Article: ] |
| 96. | Long W, Zhao W, Ning B, Huang J, Chu J, Li L, Ma Q, Xing C, Wang HY, Liu Q, Wang RF. PHF20 collaborates with PARP1 to promote stemness and aggressiveness of neuroblastoma cells through activation of SOX2 and OCT4. J Mol Cell Biol. 2018;10:147-160. [PubMed] [DOI] [Cited in This Article: ] |
| 97. | Stepanova L, Sorrentino B. A limited role for p16Ink4a and p19Arf in the loss of hematopoietic stem cells during proliferative stress. Blood. 2005;106:827. [DOI] [Cited in This Article: ] |
| 98. | Ross RA, Spengler BA. Human neuroblastoma stem cells. Semin Cancer Biol. 2007;17:241-247. [PubMed] [DOI] [Cited in This Article: ] |
| 99. | Hansford LM, McKee AE, Zhang L, George RE, Gerstle JT, Thorner PS, Smith KM, Look AT, Yeger H, Miller FD, Irwin MS, Thiele CJ, Kaplan DR. Neuroblastoma cells isolated from bone marrow metastases contain a naturally enriched tumor-initiating cell. Cancer Res. 2007;67:11234-11243. [PubMed] [DOI] [Cited in This Article: ] |
| 100. | Takenobu H, Shimozato O, Nakamura T, Ochiai H, Yamaguchi Y, Ohira M, Nakagawara A, Kamijo T. CD133 suppresses neuroblastoma cell differentiation via signal pathway modification. Oncogene. 2011;30:97-105. [PubMed] [DOI] [Cited in This Article: ] |
| 101. | Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489-3498. [PubMed] [DOI] [Cited in This Article: ] |
| 102. | Lambertini C, Pantano S, Dotto GP. Differential control of Notch1 gene transcription by Klf4 and Sp3 transcription factors in normal vs cancer-derived keratinocytes. PLoS One. 2010;5:e10369. [PubMed] [DOI] [Cited in This Article: ] |
| 103. | Shum CK, Lau ST, Tsoi LL, Chan LK, Yam JW, Ohira M, Nakagawara A, Tam PK, Ngan ES. Krüppel-like factor 4 (KLF4) suppresses neuroblastoma cell growth and determines non-tumorigenic lineage differentiation. Oncogene. 2013;32:4086-4099. [PubMed] [DOI] [Cited in This Article: ] |
| 104. | Hilfenhaus G, Göhrig A, Pape UF, Neumann T, Jann H, Zdunek D, Hess G, Stassen JM, Wiedenmann B, Detjen K, Pavel M, Fischer C. Placental growth factor supports neuroendocrine tumor growth and predicts disease prognosis in patients. Endocr Relat Cancer. 2013;20:305-319. [PubMed] [DOI] [Cited in This Article: ] |
| 105. | Schiapparelli P, Enguita-Germán M, Balbuena J, Rey JA, Lázcoz P, Castresana JS. Analysis of stemness gene expression and CD133 abnormal methylation in neuroblastoma cell lines. Oncol Rep. 2010;24:1355-1362. [PubMed] [DOI] [Cited in This Article: ] |
| 106. | Ordóñez R, Gallo-Oller G, Martínez-Soto S, Legarra S, Pata-Merci N, Guegan J, Danglot G, Bernheim A, Meléndez B, Rey JA, Castresana JS. Genome-wide microarray expression and genomic alterations by array-CGH analysis in neuroblastoma stem-like cells. PLoS One. 2014;9:e113105. [PubMed] [DOI] [Cited in This Article: ] |
| 107. | Yin AH, Miraglia S, Zanjani ED, Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J, Buck DW. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002-5012. [PubMed] [Cited in This Article: ] |
| 108. | Liou GY. CD133 as a regulator of cancer metastasis through the cancer stem cells. Int J Biochem Cell Biol. 2019;106:1-7. [PubMed] [DOI] [Cited in This Article: ] |
| 109. | Tong QS, Zheng LD, Tang ST, Ruan QL, Liu Y, Li SW, Jiang GS, Cai JB. Expression and clinical significance of stem cell marker CD133 in human neuroblastoma. World J Pediatr. 2008;4:58-62. [PubMed] [DOI] [Cited in This Article: ] |
| 110. | Zhong ZY, Shi BJ, Zhou H, Wang WB. CD133 expression and MYCN amplification induce chemoresistance and reduce average survival time in pediatric neuroblastoma. J Int Med Res. 2018;46:1209-1220. [PubMed] [DOI] [Cited in This Article: ] |
| 111. | Siapati EK, Rouka E, Kyriakou D, Vassilopoulos G. Neuroblastoma cells negative for CD44 possess tumor-initiating properties. Cell Oncol (Dordr). 2011;34:189-197. [PubMed] [DOI] [Cited in This Article: ] |
| 112. | Vega FM, Colmenero-Repiso A, Gómez-Muñoz MA, Rodríguez-Prieto I, Aguilar-Morante D, Ramírez G, Márquez C, Cabello R, Pardal R. CD44-high neural crest stem-like cells are associated with tumour aggressiveness and poor survival in neuroblastoma tumours. EBioMedicine. 2019;49:82-95. [PubMed] [DOI] [Cited in This Article: ] |
| 113. | Gilliam DT, Menon V, Bretz NP, Pruszak J. The CD24 surface antigen in neural development and disease. Neurobiol Dis. 2017;99:133-144. [PubMed] [DOI] [Cited in This Article: ] |
| 114. | Aigner S, Sthoeger ZM, Fogel M, Weber E, Zarn J, Ruppert M, Zeller Y, Vestweber D, Stahel R, Sammar M, Altevogt P. CD24, a mucin-type glycoprotein, is a ligand for P-selectin on human tumor cells. Blood. 1997;89:3385-3395. [PubMed] [Cited in This Article: ] |
| 115. | Aigner S, Ramos CL, Hafezi-Moghadam A, Lawrence MB, Friederichs J, Altevogt P, Ley K. CD24 mediates rolling of breast carcinoma cells on P-selectin. FASEB J. 1998;12:1241-1251. [PubMed] [DOI] [Cited in This Article: ] |
| 116. | Begum A, Kim Y, Lin Q, Yun Z. DLK1, delta-like 1 homolog (Drosophila), regulates tumor cell differentiation in vivo. Cancer Lett. 2012;318:26-33. [PubMed] [DOI] [Cited in This Article: ] |
| 117. | Kim Y, Lin Q, Zelterman D, Yun Z. Hypoxia-regulated delta-like 1 homologue enhances cancer cell stemness and tumorigenicity. Cancer Res. 2009;69:9271-9280. [PubMed] [DOI] [Cited in This Article: ] |
| 118. | Begum A, Lin Q, Yu C, Kim Y, Yun Z. Interaction of delta-like 1 homolog (Drosophila) with prohibitins and its impact on tumor cell clonogenicity. Mol Cancer Res. 2014;12:155-164. [PubMed] [DOI] [Cited in This Article: ] |
| 119. | Van Limpt VA, Chan AJ, Van Sluis PG, Caron HN, Van Noesel CJ, Versteeg R. High delta-like 1 expression in a subset of neuroblastoma cell lines corresponds to a differentiated chromaffin cell type. Int J Cancer. 2003;105:61-69. [PubMed] [DOI] [Cited in This Article: ] |
| 120. | Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19-26. [PubMed] [DOI] [Cited in This Article: ] |
| 121. | Wachowiak R, Fiegel HC, Kaifi JT, Quaas A, Krickhahn A, Schurr PG, Erttmann R, Schachner M, Kluth D, Sauter G, Izbicki JR. L1 is associated with favorable outcome in neuroblastomas in contrast to adult tumors. Ann Surg Oncol. 2007;14:3575-3580. [PubMed] [DOI] [Cited in This Article: ] |
| 122. | Rached J, Nasr Z, Abdallah J, Abou-Antoun T. L1-CAM knock-down radiosensitizes neuroblastoma IMR-32 cells by simultaneously decreasing MycN, but increasing PTEN protein expression. Int J Oncol. 2016;49:1722-1730. [PubMed] [DOI] [Cited in This Article: ] |
| 123. | Choi YH, Yu AM. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 2014;20:793-807. [PubMed] [DOI] [Cited in This Article: ] |
| 124. | Aravindan N, Jain D, Somasundaram DB, Herman TS, Aravindan S. Cancer stem cells in neuroblastoma therapy resistance. Cancer Drug Resist. 2019;2:948-967. [PubMed] [DOI] [Cited in This Article: ] |
| 125. | An Y, Ongkeko WM. ABCG2: the key to chemoresistance in cancer stem cells? Expert Opin Drug Metab Toxicol. 2009;5:1529-1542. [PubMed] [DOI] [Cited in This Article: ] |
| 126. | Xing LL, Sha YL, Wu YM, Hu JM, Zhang M, Lv F. Preliminary analysis of stem cell-like cells in human neuroblastoma. World J Pediatr. 2015;11:54-60. [PubMed] [DOI] [Cited in This Article: ] |
| 127. | Lange I, Koomoa DL. MycN promotes TRPM7 expression and cell migration in neuroblastoma through a process that involves polyamines. FEBS Open Bio. 2014;4:966-975. [PubMed] [DOI] [Cited in This Article: ] |
| 128. | Zhang Z, Faouzi M, Huang J, Geerts D, Yu H, Fleig A, Penner R. N-Myc-induced up-regulation of TRPM6/TRPM7 channels promotes neuroblastoma cell proliferation. Oncotarget. 2014;5:7625-7634. [PubMed] [DOI] [Cited in This Article: ] |
| 129. | Hiraiwa T, Yamada TG, Miki N, Funahashi A, Hiroi N. Activation of cell migration via morphological changes in focal adhesions depends on shear stress in MYCN-amplified neuroblastoma cells. J R Soc Interface. 2019;16:20180934. [PubMed] [DOI] [Cited in This Article: ] |
| 130. | Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, Figdor CG, Moolenaar WH, Jalink K, van Leeuwen FN. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. EMBO J. 2006;25:290-301. [PubMed] [DOI] [Cited in This Article: ] |
| 131. | Middelbeek J, Visser D, Henneman L, Kamermans A, Kuipers AJ, Hoogerbrugge PM, Jalink K, van Leeuwen FN. TRPM7 maintains progenitor-like features of neuroblastoma cells: implications for metastasis formation. Oncotarget. 2015;6:8760-8776. [PubMed] [DOI] [Cited in This Article: ] |
| 132. | Lange I, Espinoza-Fuenzalida I, Ali MW, Serrano LE, Koomoa DT. FTY-720 induces apoptosis in neuroblastoma via multiple signaling pathways. Oncotarget. 2017;8:109985-109999. [PubMed] [DOI] [Cited in This Article: ] |
| 133. | Liu K, Xu SH, Chen Z, Zeng QX, Li ZJ, Chen ZM. TRPM7 overexpression enhances the cancer stem cell-like and metastatic phenotypes of lung cancer through modulation of the Hsp90α/uPA/MMP2 signaling pathway. BMC Cancer. 2018;18:1167. [PubMed] [DOI] [Cited in This Article: ] |
| 134. | Maresca G, Natoli M, Nardella M, Arisi I, Trisciuoglio D, Desideri M, Brandi R, D'Aguanno S, Nicotra MR, D'Onofrio M, Urbani A, Natali PG, Del Bufalo D, Felsani A, D'Agnano I. LMNA knock-down affects differentiation and progression of human neuroblastoma cells. PLoS One. 2012;7:e45513. [PubMed] [DOI] [Cited in This Article: ] |
| 135. | Nardella M, Guglielmi L, Musa C, Iannetti I, Maresca G, Amendola D, Porru M, Carico E, Sessa G, Camerlingo R, Dominici C, Megiorni F, Milan M, Bearzi C, Rizzi R, Pirozzi G, Leonetti C, Bucci B, Mercanti D, Felsani A, D'Agnano I. Down-regulation of the Lamin A/C in neuroblastoma triggers the expansion of tumor initiating cells. Oncotarget. 2015;6:32821-32840. [PubMed] [DOI] [Cited in This Article: ] |
| 136. | Chikaraishi K, Takenobu H, Sugino RP, Mukae K, Akter J, Haruta M, Kurosumi M, Endo TA, Koseki H, Shimojo N, Ohira M, Kamijo T. CFC1 is a cancer stemness-regulating factor in neuroblastoma. Oncotarget. 2017;8:45046-45059. [PubMed] [DOI] [Cited in This Article: ] |
| 137. | Hockman D, Chong-Morrison V, Green SA, Gavriouchkina D, Candido-Ferreira I, Ling ITC, Williams RM, Amemiya CT, Smith JJ, Bronner ME, Sauka-Spengler T. A genome-wide assessment of the ancestral neural crest gene regulatory network. Nat Commun. 2019;10:4689. [PubMed] [DOI] [Cited in This Article: ] |
| 138. | Becker J, Wilting J. WNT Signaling in Neuroblastoma. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] |
| 139. | Pietras A, Hansford LM, Johnsson AS, Bridges E, Sjölund J, Gisselsson D, Rehn M, Beckman S, Noguera R, Navarro S, Cammenga J, Fredlund E, Kaplan DR, Påhlman S. HIF-2αlpha maintains an undifferentiated state in neural crest-like human neuroblastoma tumor-initiating cells. Proc Natl Acad Sci USA. 2009;106:16805-16810. [PubMed] [DOI] [Cited in This Article: ] |
| 140. | Cantilena S, Pastorino F, Pezzolo A, Chayka O, Pistoia V, Ponzoni M, Sala A. Frizzled receptor 6 marks rare, highly tumourigenic stem-like cells in mouse and human neuroblastomas. Oncotarget. 2011;2:976-983. [PubMed] [DOI] [Cited in This Article: ] |
| 141. | Chen J, Wang P, Cai R, Peng H, Zhang C, Zhang M. SLC34A2 promotes neuroblastoma cell stemness via enhancement of miR-25/Gsk3β-mediated activation of Wnt/β-catenin signaling. FEBS Open Bio. 2019;9:527-537. [PubMed] [DOI] [Cited in This Article: ] |
| 142. | Vangipuram SD, Wang ZJ, Lyman WD. Resistance of stem-like cells from neuroblastoma cell lines to commonly used chemotherapeutic agents. Pediatr Blood Cancer. 2010;54:361-368. [PubMed] [DOI] [Cited in This Article: ] |
| 143. | Vangipuram SD, Buck SA, Lyman WD. Wnt pathway activity confers chemoresistance to cancer stem-like cells in a neuroblastoma cell line. Tumour Biol. 2012;33:2173-2183. [PubMed] [DOI] [Cited in This Article: ] |
| 144. | Tringali C, Cirillo F, Lamorte G, Papini N, Anastasia L, Lupo B, Silvestri I, Tettamanti G, Venerando B. NEU4L sialidase overexpression promotes β-catenin signaling in neuroblastoma cells, enhancing stem-like malignant cell growth. Int J Cancer. 2012;131:1768-1778. [PubMed] [DOI] [Cited in This Article: ] |
| 145. | Forgham H, Johnson D, Carter N, Veuger S, Carr-Wilkinson J. Stem Cell Markers in Neuroblastoma-An Emerging Role for LGR5. Front Cell Dev Biol. 2015;3:77. [PubMed] [DOI] [Cited in This Article: ] |
| 146. | Vieira GC, Chockalingam S, Melegh Z, Greenhough A, Malik S, Szemes M, Park JH, Kaidi A, Zhou L, Catchpoole D, Morgan R, Bates DO, Gabb PD, Malik K. LGR5 regulates pro-survival MEK/ERK and proliferative Wnt/β-catenin signalling in neuroblastoma. Oncotarget. 2015;6:40053-40067. [PubMed] [DOI] [Cited in This Article: ] |
| 147. | Wislet S, Vandervelden G, Rogister B. From Neural Crest Development to Cancer and Vice Versa: How p75NTR and (Pro)neurotrophins Could Act on Cell Migration and Invasion? Front Mol Neurosci. 2018;11:244. [PubMed] [DOI] [Cited in This Article: ] |
| 148. | Nakagawara A. Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer Lett. 2001;169:107-114. [PubMed] [DOI] [Cited in This Article: ] |
| 149. | Farina AR, Tacconelli A, Cappabianca L, Cea G, Panella S, Chioda A, Romanelli A, Pedone C, Gulino A, Mackay AR. The alternative TrkAIII splice variant targets the centrosome and promotes genetic instability. Mol Cell Biol. 2009;29:4812-4830. [PubMed] [DOI] [Cited in This Article: ] |
| 150. | Farina AR, Cappabianca L, Gneo L, Ruggeri P, Mackay AR. TrkAIII signals endoplasmic reticulum stress to the mitochondria in neuroblastoma cells, resulting in glycolytic metabolic adaptation. Oncotarget. 2018;9:8368-8390. [PubMed] [DOI] [Cited in This Article: ] |
| 151. | Farina AR, Cappabianca L, Ruggeri P, Gneo L, Maccarone R, Mackay AR. Retrograde TrkAIII transport from ERGIC to ER: a re-localisation mechanism for oncogenic activity. Oncotarget. 2015;6:35636-35651. [PubMed] [DOI] [Cited in This Article: ] |
| 152. | Ruggeri P, Farina AR, Di Ianni N, Cappabianca L, Ragone M, Ianni G, Gulino A, Mackay AR. The TrkAIII oncoprotein inhibits mitochondrial free radical ROS-induced death of SH-SY5Y neuroblastoma cells by augmenting SOD2 expression and activity at the mitochondria, within the context of a tumour stem cell-like phenotype. PLoS One. 2014;9:e94568. [PubMed] [DOI] [Cited in This Article: ] |
| 153. | Farina AR, Di Ianni N, Cappabianca L, Ruggeri P, Ragone M, Ianni G, Gulino A, Mackay AR. TrkAIII promotes microtubule nucleation and assembly at the centrosome in SH-SY5Y neuroblastoma cells, contributing to an undifferentiated anaplastic phenotype. Biomed Res Int. 2013;2013:740187. [PubMed] [DOI] [Cited in This Article: ] |
| 154. | Ho R, Eggert A, Hishiki T, Minturn JE, Ikegaki N, Foster P, Camoratto AM, Evans AE, Brodeur GM. Resistance to chemotherapy mediated by TrkB in neuroblastomas. Cancer Res. 2002;62:6462-6466. [PubMed] [Cited in This Article: ] |
| 155. | Thiele CJ, Li Z, McKee AE. On Trk--the TrkB signal transduction pathway is an increasingly important target in cancer biology. Clin Cancer Res. 2009;15:5962-5967. [PubMed] [DOI] [Cited in This Article: ] |
| 156. | Hua Z, Gu X, Dong Y, Tan F, Liu Z, Thiele CJ, Li Z. PI3K and MAPK pathways mediate the BDNF/TrkB-increased metastasis in neuroblastoma. Tumour Biol. 2016;. [PubMed] [DOI] [Cited in This Article: ] |
| 157. | Straub JA, Sholler GL, Nishi R. Embryonic sympathoblasts transiently express TrkB in vivo and proliferate in response to brain-derived neurotrophic factor in vitro. BMC Dev Biol. 2007;7:10. [PubMed] [DOI] [Cited in This Article: ] |
| 158. | Dewitt J, Ochoa V, Urschitz J, Elston M, Moisyadi S, Nishi R. Constitutively active TrkB confers an aggressive transformed phenotype to a neural crest-derived cell line. Oncogene. 2014;33:977-985. [PubMed] [DOI] [Cited in This Article: ] |
| 159. | Masoumi J, Jafarzadeh A, Abdolalizadeh J, Khan H, Philippe J, Mirzaei H, Mirzaei HR. Cancer stem cell-targeted chimeric antigen receptor (CAR)-T cell therapy: challenges and prospects. APSB. 2020;20. [DOI] [Cited in This Article: ] |
| 160. | Dave H, Butcher D, Anver M, Bollard CM. ROR1 and ROR2-novel targets for neuroblastoma. Pediatr Hematol Oncol. 2019;36:352-364. [PubMed] [DOI] [Cited in This Article: ] |
| 161. | Sobhan PK, Zhai Q, Green LC, Hansford LM, Funa K. ASK1 regulates the survival of neuroblastoma cells by interacting with TLX and stabilizing HIF-1α. Cell Signal. 2017;30:104-117. [PubMed] [DOI] [Cited in This Article: ] |
| 162. | Nichane M, Ren X, Bellefroid EJ. Self-regulation of Stat3 activity coordinates cell-cycle progression and neural crest specification. EMBO J. 2010;29:55-67. [PubMed] [DOI] [Cited in This Article: ] |
| 163. | Ho PL, Kurtova A, Chan KS. Normal and neoplastic urothelial stem cells: getting to the root of the problem. Nat Rev Urol. 2012;9:583-594. [PubMed] [DOI] [Cited in This Article: ] |
| 164. | Lin L, Liu A, Peng Z, Lin HJ, Li PK, Li C, Lin J. STAT3 is necessary for proliferation and survival in colon cancer-initiating cells. Cancer Res. 2011;71:7226-7237. [PubMed] [DOI] [Cited in This Article: ] |
| 165. | Ji J, Wang XW. Clinical implications of cancer stem cell biology in hepatocellular carcinoma. Semin Oncol. 2012;39:461-472. [PubMed] [DOI] [Cited in This Article: ] |
| 166. | Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27:2383-2392. [PubMed] [DOI] [Cited in This Article: ] |
| 167. | Neveu P, Kye MJ, Qi S, Buchholz DE, Clegg DO, Sahin M, Park IH, Kim KS, Daley GQ, Kornblum HI, Shraiman BI, Kosik KS. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell Stem Cell. 2010;7:671-681. [PubMed] [DOI] [Cited in This Article: ] |
| 168. | Crobu F, Latini V, Marongiu MF, Sogos V, Scintu F, Porcu S, Casu C, Badiali M, Sanna A, Manchinu MF, Ristaldi MS. Differentiation of single cell derived human mesenchymal stem cells into cells with a neuronal phenotype: RNA and microRNA expression profile. Mol Biol Rep. 2012;39:3995-4007. [PubMed] [DOI] [Cited in This Article: ] |
| 169. | Seo HS, Choi HS, Kim SR, Choi YK, Woo SM, Shin I, Woo JK, Park SY, Shin YC, Ko SG. Apigenin induces apoptosis via extrinsic pathway, inducing p53 and inhibiting STAT3 and NFκB signaling in HER2-overexpressing breast cancer cells. Mol Cell Biochem. 2012;366:319-334. [PubMed] [DOI] [Cited in This Article: ] |
| 170. | Melton C, Blelloch R. MicroRNA Regulation of Embryonic Stem Cell Self-Renewal and Differentiation. Adv Exp Med Biol. 2010;695:105-117. [PubMed] [DOI] [Cited in This Article: ] |
| 171. | Lu Y, Zhang K, Li C, Yao Y, Tao D, Liu Y, Zhang S, Ma Y. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One. 2012;7:e30999. [PubMed] [DOI] [Cited in This Article: ] |
| 172. | Hsu DM, Agarwal S, Benham A, Coarfa C, Trahan DN, Chen Z, Stowers PN, Courtney AN, Lakoma A, Barbieri E, Metelitsa LS, Gunaratne P, Kim ES, Shohet JM. G-CSF receptor positive neuroblastoma subpopulations are enriched in chemotherapy-resistant or relapsed tumors and are highly tumorigenic. Cancer Res. 2013;73:4134-4146. [PubMed] [DOI] [Cited in This Article: ] |
| 173. | Maris JM, Healy J, Park J, Ladenstein R, Pötschger U. G-CSF Is a Cancer Stem Cell-Specific Growth Factor-Letter. Cancer Res. 2015;75:3991. [PubMed] [DOI] [Cited in This Article: ] |
| 174. | Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615-1624. [PubMed] [DOI] [Cited in This Article: ] |
| 175. | Labosky PA, Kaestner KH. The winged helix transcription factor Hfh2 is expressed in neural crest and spinal cord during mouse development. Mech Dev. 1998;76:185-190. [PubMed] [DOI] [Cited in This Article: ] |
| 176. | Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1-23. [PubMed] [DOI] [Cited in This Article: ] |
| 177. | Li D, Mei H, Qi M, Yang D, Zhao X, Xiang X, Pu J, Huang K, Zheng L, Tong Q. FOXD3 is a novel tumor suppressor that affects growth, invasion, metastasis and angiogenesis of neuroblastoma. Oncotarget. 2013;4:2021-2044. [PubMed] [DOI] [Cited in This Article: ] |
| 178. | Wang Z, Park HJ, Carr JR, Chen YJ, Zheng Y, Li J, Tyner AL, Costa RH, Bagchi S, Raychaudhuri P. FoxM1 in tumorigenicity of the neuroblastoma cells and renewal of the neural progenitors. Cancer Res. 2011;71:4292-4302. [PubMed] [DOI] [Cited in This Article: ] |
| 179. | Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753-763. [PubMed] [DOI] [Cited in This Article: ] |
| 180. | van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737-752. [PubMed] [DOI] [Cited in This Article: ] |
| 181. | Cui H, Ma J, Ding J, Li T, Alam G, Ding HF. Bmi-1 regulates the differentiation and clonogenic self-renewal of I-type neuroblastoma cells in a concentration-dependent manner. J Biol Chem. 2006;281:34696-34704. [PubMed] [DOI] [Cited in This Article: ] |
| 182. | Cui H, Hu B, Li T, Ma J, Alam G, Gunning WT, Ding HF. Bmi-1 is essential for the tumorigenicity of neuroblastoma cells. Am J Pathol. 2007;170:1370-1378. [PubMed] [DOI] [Cited in This Article: ] |
| 183. | Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962-967. [PubMed] [DOI] [Cited in This Article: ] |
| 184. | Nowak K, Kerl K, Fehr D, Kramps C, Gessner C, Killmer K, Samans B, Berwanger B, Christiansen H, Lutz W. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 2006;34:1745-1754. [PubMed] [DOI] [Cited in This Article: ] |
| 185. | Calao M, Sekyere EO, Cui HJ, Cheung BB, Thomas WD, Keating J, Chen JB, Raif A, Jankowski K, Davies NP, Bekkum MV, Chen B, Tan O, Ellis T, Norris MD, Haber M, Kim ES, Shohet JM, Trahair TN, Liu T, Wainwright BJ, Ding HF, Marshall GM. Direct effects of Bmi1 on p53 protein stability inactivates oncoprotein stress responses in embryonal cancer precursor cells at tumor initiation. Oncogene. 2013;32:3616-3626. [PubMed] [DOI] [Cited in This Article: ] |
| 186. | Ochiai H, Takenobu H, Nakagawa A, Yamaguchi Y, Kimura M, Ohira M, Okimoto Y, Fujimura Y, Koseki H, Kohno Y, Nakagawara A, Kamijo T. Bmi1 is a MYCN target gene that regulates tumorigenesis through repression of Kif1Bβeta and TSLC1 in neuroblastoma. Oncogene. 2010;29:2681-2690. [PubMed] [DOI] [Cited in This Article: ] |
| 187. | Kamijo T. Role of stemness-related molecules in neuroblastoma. Pediatr Res. 2012;71:511-515. [PubMed] [DOI] [Cited in This Article: ] |
| 188. | Okuyama Y, Tanaka Y, Jiang JJ, Kamimura D, Nakamura A, Ota M, Ohki T, Higo D, Ogura H, Ishii N, Atsumi T, Murakami M. Bmi1 Regulates IκBα Degradation via Association with the SCF Complex. J Immunol. 2018;201:2264-2272. [PubMed] [DOI] [Cited in This Article: ] |
| 189. | Jiang L, Wu J, Yang Y, Liu L, Song L, Li J, Li M. Bmi-1 promotes the aggressiveness of glioma via activating the NF-kappaB/MMP-9 signaling pathway. BMC Cancer. 2012;12:406. [PubMed] [DOI] [Cited in This Article: ] |
| 190. | Li X, Yang Z, Song W, Zhou L, Li Q, Tao K, Zhou J, Wang X, Zheng Z, You N, Dou K, Li H. Overexpression of Bmi-1 contributes to the invasion and metastasis of hepatocellular carcinoma by increasing the expression of matrix metalloproteinase (MMP)2, MMP-9 and vascular endothelial growth factor via the PTEN/PI3K/Akt pathway. Int J Oncol. 2013;43:793-802. [PubMed] [DOI] [Cited in This Article: ] |
| 191. | Liu Y, Chu Z, Li Q, Peng B, Xu S, Lian CG, Geng S. Downregulation of Bmi-1 suppresses epithelialmesenchymal transition in melanoma. Oncol Rep. 2017;37:139-146. [PubMed] [DOI] [Cited in This Article: ] |
| 192. | Schlisio S, Kenchappa RS, Vredeveld LC, George RE, Stewart R, Greulich H, Shahriari K, Nguyen NV, Pigny P, Dahia PL, Pomeroy SL, Maris JM, Look AT, Meyerson M, Peeper DS, Carter BD, Kaelin WG Jr. The kinesin Kif1Bβeta acts downstream from EglN3 to induce apoptosis and is a potential 1p36 tumor suppressor. Genes Dev. 2008;22:884-893. [PubMed] [DOI] [Cited in This Article: ] |
| 193. | Cappabianca L, Farina AR, Di Marcotullio L, Infante P, De Simone D, Sebastiano M, Mackay AR. Discovery, characterization and potential roles of a novel NF-YAx splice variant in human neuroblastoma. J Exp Clin Cancer Res. 2019;38:482. [PubMed] [DOI] [Cited in This Article: ] |
| 194. | Yoshida GJ. Emerging roles of Myc in stem cell biology and novel tumor therapies. J Exp Clin Cancer Res. 2018;37:173. [PubMed] [DOI] [Cited in This Article: ] |
| 195. | Kramer M, Ribeiro D, Arsenian-Henriksson M, Deller T, Rohrer H. Proliferation and Survival of Embryonic Sympathetic Neuroblasts by MYCN and Activated ALK Signaling. J Neurosci. 2016;36:10425-10439. [PubMed] [DOI] [Cited in This Article: ] |
| 196. | Zhu S, Lee JS, Guo F, Shin J, Perez-Atayde AR, Kutok JL, Rodig SJ, Neuberg DS, Helman D, Feng H, Stewart RA, Wang W, George RE, Kanki JP, Look AT. Activated ALK collaborates with MYCN in neuroblastoma pathogenesis. Cancer Cell. 2012;21:362-373. [PubMed] [DOI] [Cited in This Article: ] |
| 197. | Suenaga Y, Islam SM, Alagu J, Kaneko Y, Kato M, Tanaka Y, Kawana H, Hossain S, Matsumoto D, Yamamoto M, Shoji W, Itami M, Shibata T, Nakamura Y, Ohira M, Haraguchi S, Takatori A, Nakagawara A. NCYM, a Cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3β resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet. 2014;10:e1003996. [PubMed] [DOI] [Cited in This Article: ] |
| 198. | Kaneko Y, Suenaga Y, Islam SM, Matsumoto D, Nakamura Y, Ohira M, Yokoi S, Nakagawara A. Functional interplay between MYCN, NCYM, and OCT4 promotes aggressiveness of human neuroblastomas. Cancer Sci. 2015;106:840-847. [PubMed] [DOI] [Cited in This Article: ] |
| 199. | Newman EA, Chukkapalli S, Bashllari D, Thomas TT, Van Noord RA, Lawlor ER, Hoenerhoff MJ, Opipari AW, Opipari VP. Alternative NHEJ pathway proteins as components of MYCN oncogenic activity in human neural crest stem cell differentiation: implications for neuroblastoma initiation. Cell Death Dis. 2017;8:3208. [PubMed] [DOI] [Cited in This Article: ] |
| 200. | Cotterman R, Knoepfler PS. N-Myc regulates expression of pluripotency genes in neuroblastoma including lif, klf2, klf4, and lin28b. PLoS One. 2009;4:e5799. [PubMed] [DOI] [Cited in This Article: ] |
| 201. | Le Grand M, Mukha A, Püschel J, Valli E, Kamili A, Vittorio O, Dubrovska A, Kavallaris M. Interplay between MycN and c-Myc regulates radioresistance and cancer stem cell phenotype in neuroblastoma upon glutamine deprivation. Theranostics. 2020;10:6411-6429. [PubMed] [DOI] [Cited in This Article: ] |
| 202. | Zambelli F, Pavesi G. Genome wide features, distribution and correlations of NF-Y binding sites. Biochim Biophys Acta Gene Regul Mech. 2017;1860:581-589. [PubMed] [DOI] [Cited in This Article: ] |
| 203. | Li G, Zhao H, Wang L, Wang Y, Guo X, Xu B. The animal nuclear factor Y: an enigmatic and important heterotrimeric transcription factor. Am J Cancer Res. 2018;8:1106-1125. [PubMed] [Cited in This Article: ] |
| 204. | Maity SN. NF-Y (CBF) regulation in specific cell types and mouse models. Biochim Biophys Acta Gene Regul Mech. 2017;1860:598-603. [PubMed] [DOI] [Cited in This Article: ] |
| 205. | Zhu J, Zhang Y, Joe GJ, Pompetti R, Emerson SG. NF-Ya activates multiple hematopoietic stem cell (HSC) regulatory genes and promotes HSC self-renewal. Proc Natl Acad Sci USA. 2005;102:11728-11733. [PubMed] [DOI] [Cited in This Article: ] |
| 206. | Dolfini D, Gatta R, Mantovani R. NF-Y and the transcriptional activation of CCAAT promoters. Crit Rev Biochem Mol Biol. 2012;47:29-49. [PubMed] [DOI] [Cited in This Article: ] |
| 207. | Dolfini D, Minuzzo M, Pavesi G, Mantovani R. The short isoform of NF-YA belongs to the embryonic stem cell transcription factor circuitry. Stem Cells. 2012;30:2450-2459. [PubMed] [DOI] [Cited in This Article: ] |
| 208. | Gu Z, Kuntz-Simon G, Rommelaere J, Cornelis J. Oncogenic transformation-dependent expression of a transcription factor NF-Y subunit. Mol Carcinog. 1999;24:294-299. [PubMed] [DOI] [Cited in This Article: ] |
| 209. | Gurtner A, Manni I, Piaggio G. NF-Y in cancer: Impact on cell transformation of a gene essential for proliferation. Biochim Biophys Acta Gene Regul Mech. 2017;1860:604-616. [PubMed] [DOI] [Cited in This Article: ] |
| 210. | Cappabianca L, Farina AR, Tacconelli A, Mantovani R, Gulino A, Mackay AR. Reconstitution of TIMP-2 expression in SH-SY5Y neuroblastoma cells by 5-azacytidine is mediated transcriptionally by NF-Y through an inverted CCAAT site. Exp Cell Res. 2003;286:209-218. [PubMed] [DOI] [Cited in This Article: ] |
| 211. | Suske G. NF-Y and SP transcription factors - New insights in a long-standing liaison. Biochim Biophys Acta Gene Regul Mech. 2017;1860:590-597. [PubMed] [DOI] [Cited in This Article: ] |
| 212. | van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, Broekmans M, Haneveld F, Nowakowska NE, Bras J, van Noesel CJM, Jongejan A, van Kampen AH, Koster L, Baas F, van Dijk-Kerkhoven L, Huizer-Smit M, Lecca MC, Chan A, Lakeman A, Molenaar P, Volckmann R, Westerhout EM, Hamdi M, van Sluis PG, Ebus ME, Molenaar JJ, Tytgat GA, Westerman BA, van Nes J, Versteeg R. Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet. 2017;49:1261-1266. [PubMed] [DOI] [Cited in This Article: ] |
| 213. | Boeva V, Louis-Brennetot C, Peltier A, Durand S, Pierre-Eugène C, Raynal V, Etchevers HC, Thomas S, Lermine A, Daudigeos-Dubus E, Geoerger B, Orth MF, Grünewald TGP, Diaz E, Ducos B, Surdez D, Carcaboso AM, Medvedeva I, Deller T, Combaret V, Lapouble E, Pierron G, Grossetête-Lalami S, Baulande S, Schleiermacher G, Barillot E, Rohrer H, Delattre O, Janoueix-Lerosey I. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet. 2017;49:1408-1413. [PubMed] [DOI] [Cited in This Article: ] |
| 214. | van Groningen T, Akogul N, Westerhout EM, Chan A, Hasselt NE, Zwijnenburg DA, Broekmans M, Stroeken P, Haneveld F, Hooijer GKJ, Savci-Heijink CD, Lakeman A, Volckmann R, van Sluis P, Valentijn LJ, Koster J, Versteeg R, van Nes J. A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat Commun. 2019;10:1530. [PubMed] [DOI] [Cited in This Article: ] |
| 215. | Påhlman S, Stockhausen MT, Fredlund E, Axelson H. Notch signaling in neuroblastoma. Semin Cancer Biol. 2004;14:365-373. [PubMed] [DOI] [Cited in This Article: ] |
| 216. | Conti L, Crisafulli L, Caldera V, Tortoreto M, Brilli E, Conforti P, Zunino F, Magrassi L, Schiffer D, Cattaneo E. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS One. 2012;7:e38486. [PubMed] [DOI] [Cited in This Article: ] |
| 217. | Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, Lasorella A, Iavarone A, Chang S, Hernando E, Pagano M. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365-369. [PubMed] [DOI] [Cited in This Article: ] |
| 218. | Majumder S. REST in good times and bad: roles in tumor suppressor and oncogenic activities. Cell Cycle. 2006;5:1929-1935. [PubMed] [DOI] [Cited in This Article: ] |
| 219. | Singh A, Rokes C, Gireud M, Fletcher S, Baumgartner J, Fuller G, Stewart J, Zage P, Gopalakrishnan V. Retinoic acid induces REST degradation and neuronal differentiation by modulating the expression of SCF(β-TRCP) in neuroblastoma cells. Cancer. 2011;117:5189-5202. [PubMed] [DOI] [Cited in This Article: ] |
| 220. | Liang J, Tong P, Zhao W, Li Y, Zhang L, Xia Y, Yu Y. The REST gene signature predicts drug sensitivity in neuroblastoma cell lines and is significantly associated with neuroblastoma tumor stage. Int J Mol Sci. 2014;15:11220-11233. [PubMed] [DOI] [Cited in This Article: ] |
| 221. | MacNicol AM, Hardy LL, Spencer HJ, MacNicol MC. Neural stem and progenitor cell fate transition requires regulation of Musashi1 function. BMC Dev Biol. 2015;15:15. [PubMed] [DOI] [Cited in This Article: ] |
| 222. | Park SM, Deering RP, Lu Y, Tivnan P, Lianoglou S, Al-Shahrour F, Ebert BL, Hacohen N, Leslie C, Daley GQ, Lengner CJ, Kharas MG. Musashi-2 controls cell fate, lineage bias, and TGF-β signaling in HSCs. J Exp Med. 2014;211:71-87. [PubMed] [DOI] [Cited in This Article: ] |
| 223. | Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. A distinct "side population" of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci USA. 2004;101:14228-14233. [PubMed] [DOI] [Cited in This Article: ] |
| 224. | Facompre ND, Harmeyer KM, Sole X, Kabraji S, Belden Z, Sahu V, Whelan K, Tanaka K, Weinstein GS, Montone KT, Roesch A, Gimotty PA, Herlyn M, Rustgi AK, Nakagawa H, Ramaswamy S, Basu D. JARID1B Enables Transit between Distinct States of the Stem-like Cell Population in Oral Cancers. Cancer Res. 2016;76:5538-5549. [PubMed] [DOI] [Cited in This Article: ] |
| 225. | Kuo YT, Liu YL, Adebayo BO, Shih PH, Lee WH, Wang LS, Liao YF, Hsu WM, Yeh CT, Lin CM. JARID1B Expression Plays a Critical Role in Chemoresistance and Stem Cell-Like Phenotype of Neuroblastoma Cells. PLoS One. 2015;10:e0125343. [PubMed] [DOI] [Cited in This Article: ] |
| 226. | Chavali PL, Saini RK, Zhai Q, Vizlin-Hodzic D, Venkatabalasubramanian S, Hayashi A, Johansson E, Zeng ZJ, Mohlin S, Påhlman S, Hansford L, Kaplan DR, Funa K. TLX activates MMP-2, promotes self-renewal of tumor spheres in neuroblastoma and correlates with poor patient survival. Cell Death Dis. 2014;5:e1502. [PubMed] [DOI] [Cited in This Article: ] |
| 227. | Farina AR, Mackay AR. Gelatinase B/MMP-9 in Tumour Pathogenesis and Progression. Cancers (Basel). 2014;6:240-296. [PubMed] [DOI] [Cited in This Article: ] |
| 228. | Ricca A, Biroccio A, Del Bufalo D, Mackay AR, Santoni A, Cippitelli M. bcl-2 over-expression enhances NF-kappaB activity and induces mmp-9 transcription in human MCF7(ADR) breast-cancer cells. Int J Cancer. 2000;86:188-196. [PubMed] [DOI] [Cited in This Article: ] |
| 229. | Xie L, Zeng X, Hu J, Chen Q. Characterization of Nestin, a Selective Marker for Bone Marrow Derived Mesenchymal Stem Cells. Stem Cells Int. 2015;2015:762098. [PubMed] [DOI] [Cited in This Article: ] |
| 230. | Neradil J, Veselska R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015;106:803-811. [PubMed] [DOI] [Cited in This Article: ] |
| 231. | Thomas SK, Messam CA, Spengler BA, Biedler JL, Ross RA. Nestin is a potential mediator of malignancy in human neuroblastoma cells. J Biol Chem. 2004;279:27994-27999. [PubMed] [DOI] [Cited in This Article: ] |
| 232. | Vora P, Venugopal C, Singh SK. Revealed: The spy who regulates neuroblastoma stem cells. Oncotarget. 2014;5:11014-11016. [PubMed] [DOI] [Cited in This Article: ] |
| 233. | Lubanska D, Porter LA. The atypical cell cycle regulator Spy1 suppresses differentiation of the neuroblastoma stem cell population. Oncoscience. 2014;1:336-348. [PubMed] [DOI] [Cited in This Article: ] |
| 234. | Vassalli G. Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem Cells Int. 2019;2019:3904645. [PubMed] [DOI] [Cited in This Article: ] |
| 235. | Hartomo TB, Van Huyen Pham T, Yamamoto N, Hirase S, Hasegawa D, Kosaka Y, Matsuo M, Hayakawa A, Takeshima Y, Iijima K, Nishio H, Nishimura N. Involvement of aldehyde dehydrogenase 1A2 in the regulation of cancer stem cell properties in neuroblastoma. Int J Oncol. 2015;46:1089-1098. [PubMed] [DOI] [Cited in This Article: ] |
| 236. | Flahaut M, Jauquier N, Chevalier N, Nardou K, Balmas Bourloud K, Joseph JM, Barras D, Widmann C, Gross N, Renella R, Mühlethaler-Mottet A. Aldehyde dehydrogenase activity plays a Key role in the aggressive phenotype of neuroblastoma. BMC Cancer. 2016;16:781. [PubMed] [DOI] [Cited in This Article: ] |
| 237. | Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066-1079. [PubMed] [DOI] [Cited in This Article: ] |
| 238. | Beckers A, Van Peer G, Carter DR, Gartlgruber M, Herrmann C, Agarwal S, Helsmoortel HH, Althoff K, Molenaar JJ, Cheung BB, Schulte JH, Benoit Y, Shohet JM, Westermann F, Marshall GM, Vandesompele J, De Preter K, Speleman F. MYCN-driven regulatory mechanisms controlling LIN28B in neuroblastoma. Cancer Lett. 2015;366:123-132. [PubMed] [DOI] [Cited in This Article: ] |
| 239. | Tao T, Shi H, Mariani L, Abraham BJ, Durbin AD, Zimmerman MW, Powers JT, Missios P, Ross KN, Perez-Atayde AR, Bulyk ML, Young RA, Daley GQ, Look AT. LIN28B regulates transcription and potentiates MYCN-induced neuroblastoma through binding to ZNF143 at target gene promotors. Proc Natl Acad Sci USA. 2020;117:16516-16526. [PubMed] [DOI] [Cited in This Article: ] |
| 240. | Sun H, Ding C, Zhang H, Gao J. Let7 miRNAs sensitize breast cancer stem cells to radiationinduced repression through inhibition of the cyclin D1/Akt1/Wnt1 signaling pathway. Mol Med Rep. 2016;14:3285-3292. [PubMed] [DOI] [Cited in This Article: ] |
| 241. | Corallo D, Donadon M, Pantile M, Sidarovich V, Cocchi S, Ori M, De Sarlo M, Candiani S, Frasson C, Distel M, Quattrone A, Zanon C, Basso G, Tonini GP, Aveic S. LIN28B increases neural crest cell migration and leads to transformation of trunk sympathoadrenal precursors. Cell Death Differ. 2020;27:1225-1242. [PubMed] [DOI] [Cited in This Article: ] |
| 242. | Swarbrick A, Woods SL, Shaw A, Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ, Judson RL, Huskey N, Blelloch R, Haber M, Norris MD, Lengyel P, Hackett CS, Preiss T, Chetcuti A, Sullivan CS, Marcusson EG, Weiss W, L'Etoile N, Goga A. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16:1134-1140. [PubMed] [DOI] [Cited in This Article: ] |
| 243. | Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, Aberdam D, Knight RA, Genchi G, Nicotera P, Melino G, Vasa-Nicotera M. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394:921-927. [PubMed] [DOI] [Cited in This Article: ] |
| 244. | Buhagiar A, Ayers D. Chemoresistance, cancer stem cells, and miRNA influences: the case for neuroblastoma. Anal Cell Pathol (Amst). 2015;2015:150634. [PubMed] [DOI] [Cited in This Article: ] |
| 245. | Samaraweera L, Grandinetti KB, Huang R, Spengler BA, Ross RA. MicroRNAs define distinct human neuroblastoma cell phenotypes and regulate their differentiation and tumorigenicity. BMC Cancer. 2014;14:309. [PubMed] [DOI] [Cited in This Article: ] |
| 246. | Karam R, Wilkinson M. A conserved microRNA/NMD regulatory circuit controls gene expression. RNA Biol. 2012;9:22-26. [PubMed] [DOI] [Cited in This Article: ] |
| 247. | Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125-9130. [PubMed] [DOI] [Cited in This Article: ] |
| 248. | Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, Gong Y, Yin B, Qiang B, Zhao J, Yuan J, Peng X. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl). 2009;87:43-51. [PubMed] [DOI] [Cited in This Article: ] |
| 249. | Evangelisti C, Florian MC, Massimi I, Dominici C, Giannini G, Galardi S, Buè MC, Massalini S, McDowell HP, Messi E, Gulino A, Farace MG, Ciafrè SA. MiR-128 up-regulation inhibits Reelin and DCX expression and reduces neuroblastoma cell motility and invasiveness. FASEB J. 2009;23:4276-4287. [PubMed] [DOI] [Cited in This Article: ] |
| 250. | Yan H, Bu P. Non-coding RNAs in cancer stem cells. Cancer Lett. 2018;421:121-126. [PubMed] [DOI] [Cited in This Article: ] |
| 251. | Pandey GK, Kanduri C. Long noncoding RNAs and neuroblastoma. Oncotarget. 2015;6:18265-18275. [PubMed] [DOI] [Cited in This Article: ] |
| 252. | Smith CM, Catchpoole D, Hutvagner G. Non-Coding RNAs in Pediatric Solid Tumors. Front Genet. 2019;10:798. [PubMed] [DOI] [Cited in This Article: ] |
| 253. | Rombaut D, Chiu HS, Decaesteker B, Everaert C, Yigit N, Peltier A, Janoueix-Lerosey I, Bartenhagen C, Fischer M, Roberts S, D'Haene N, De Preter K, Speleman F, Denecker G, Sumazin P, Vandesompele J, Lefever S, Mestdagh P. Integrative analysis identifies lincRNAs up- and downstream of neuroblastoma driver genes. Sci Rep. 2019;9:5685. [PubMed] [DOI] [Cited in This Article: ] |
| 254. | Yu GJ, Sun Y, Zhang DW, Zhang P. Long non-coding RNA HOTAIR functions as a competitive endogenous RNA to regulate PRAF2 expression by sponging miR-326 in cutaneous squamous cell carcinoma. Cancer Cell Int. 2019;19:270. [PubMed] [DOI] [Cited in This Article: ] |
| 255. | Zeng L, Cen Y, Chen J. Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol Lett. 2018;15:2117-2122. [PubMed] [DOI] [Cited in This Article: ] |
| 256. | Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, Pasquier E, Haber M, Norris MD, Suzuki T, Marshall GM, Liu T. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5:1793-1804. [PubMed] [DOI] [Cited in This Article: ] |
| 257. | Ueda J, Ho JC, Lee KL, Kitajima S, Yang H, Sun W, Fukuhara N, Zaiden N, Chan SL, Tachibana M, Shinkai Y, Kato H, Poellinger L. The hypoxia-inducible epigenetic regulators Jmjd1a and G9a provide a mechanistic link between angiogenesis and tumor growth. Mol Cell Biol. 2014;34:3702-3720. [PubMed] [DOI] [Cited in This Article: ] |
| 258. | Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyürek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26:722-737. [PubMed] [DOI] [Cited in This Article: ] |
| 259. | Matteucci C, Balestrieri E, Argaw-Denboba A, Sinibaldi-Vallebona P. Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol. 2018;53:17-30. [PubMed] [DOI] [Cited in This Article: ] |
| 260. | Lu X, Sachs F, Ramsay L, Jacques PÉ, Göke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423-425. [PubMed] [DOI] [Cited in This Article: ] |
| 261. | Hu L, Uzhameckis D, Hedborg F, Blomberg J. Dynamic and selective HERV RNA expression in neuroblastoma cells subjected to variation in oxygen tension and demethylation. APMIS. 2016;124:140-149. [PubMed] [DOI] [Cited in This Article: ] |
| 262. | Brütting C, Narasimhan H, Hoffmann F, Kornhuber ME, Staege MS, Emmer A. Investigation of Endogenous Retrovirus Sequences in the Neighborhood of Genes Up-regulated in a Neuroblastoma Model after Treatment with Hypoxia-Mimetic Cobalt Chloride. Front Microbiol. 2018;9:287. [PubMed] [DOI] [Cited in This Article: ] |
| 263. | Li S, Liu ZC, Yin SJ, Chen YT, Yu HL, Zeng J, Zhang Q, Zhu F. Human endogenous retrovirus W family envelope gene activates the small conductance Ca2+-activated K+ channel in human neuroblastoma cells through CREB. Neuroscience. 2013;247:164-174. [PubMed] [DOI] [Cited in This Article: ] |
| 264. | Luo M, Wicha MS. Targeting Cancer Stem Cell Redox Metabolism to Enhance Therapy Responses. Semin Radiat Oncol. 2019;29:42-54. [PubMed] [DOI] [Cited in This Article: ] |
| 265. | Tanabe A, Sahara H. The Metabolic Heterogeneity and Flexibility of Cancer Stem Cells. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] |
| 266. | Ding S, Li C, Cheng N, Cui X, Xu X, Zhou G. Redox Regulation in Cancer Stem Cells. Oxid Med Cell Longev. 2015;2015:750798. [PubMed] [DOI] [Cited in This Article: ] |
| 267. | Dai X, Yan X, Wintergerst KA, Cai L, Keller BB, Tan Y. Nrf2: Redox and Metabolic Regulator of Stem Cell State and Function. Trends Mol Med. 2020;26:185-200. [PubMed] [DOI] [Cited in This Article: ] |
| 268. | Kahroba H, Shirmohamadi M, Hejazi MS, Samadi N. The Role of Nrf2 signaling in cancer stem cells: From stemness and self-renewal to tumorigenesis and chemoresistance. Life Sci. 2019;239:116986. [PubMed] [DOI] [Cited in This Article: ] |
| 269. | Payandeh Z, Pirpour Tazehkand A, Barati G, Pouremamali F, Kahroba H, Baradaran B, Samadi N. Role of Nrf2 and mitochondria in cancer stem cells; in carcinogenesis, tumor progression, and chemoresistance. Biochimie. 2020;179:32-45. [PubMed] [DOI] [Cited in This Article: ] |
| 270. | Ryoo IG, Choi BH, Ku SK, Kwak MK. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: Implications for cancer stem cell resistance. Redox Biol. 2018;17:246-258. [PubMed] [DOI] [Cited in This Article: ] |
| 271. | Zhu J, Wang H, Sun Q, Ji X, Zhu L, Cong Z, Zhou Y, Liu H, Zhou M. Nrf2 is required to maintain the self-renewal of glioma stem cells. BMC Cancer. 2013;13:380. [PubMed] [DOI] [Cited in This Article: ] |
| 272. | de Miranda Ramos V, Zanotto-Filho A, de Bittencourt Pasquali MA, Klafke K, Gasparotto J, Dunkley P, Gelain DP, Moreira JCF. NRF2 Mediates Neuroblastoma Proliferation and Resistance to Retinoic Acid Cytotoxicity in a Model of In Vitro Neuronal Differentiation. Mol Neurobiol. 2016;53:6124-6135. [PubMed] [DOI] [Cited in This Article: ] |
| 273. | Farina AR, Tacconelli A, Cappabianca L, Masciulli MP, Holmgren A, Beckett GJ, Gulino A, Mackay AR. Thioredoxin alters the matrix metalloproteinase/tissue inhibitors of metalloproteinase balance and stimulates human SK-N-SH neuroblastoma cell invasion. Eur J Biochem. 2001;268:405-413. [PubMed] [DOI] [Cited in This Article: ] |
| 274. | Farina AR, Tacconelli A, Cappabianca L, DeSantis G, Gulino A, Mackay AR. Thioredoxin inhibits microvascular endothelial capillary tubule formation. Exp Cell Res. 2003;291:474-483. [PubMed] [DOI] [Cited in This Article: ] |
| 275. | Liang Y, Zhong Z, Huang Y, Deng W, Cao J, Tsao G, Liu Q, Pei D, Kang T, Zeng YX. Stem-like cancer cells are inducible by increasing genomic instability in cancer cells. J Biol Chem. 2010;285:4931-4940. [PubMed] [DOI] [Cited in This Article: ] |
| 276. | Boveri T. Concerning the origin of malignant tumours by theodor boveri: Translated and annotated by Henry Harris. J Cell Sci. 2008;121:1-81. [DOI] [Cited in This Article: ] |
| 277. | Holubcová Z, Matula P, Sedláčková M, Vinarský V, Doležalová D, Bárta T, Dvořák P, Hampl A. Human embryonic stem cells suffer from centrosomal amplification. Stem Cells. 2011;29:46-56. [PubMed] [DOI] [Cited in This Article: ] |
| 278. | Lundberg G, Jin Y, Sehic D, Øra I, Versteeg R, Gisselsson D. Intratumour diversity of chromosome copy numbers in neuroblastoma mediated by on-going chromosome loss from a polyploid state. PLoS One. 2013;8:e59268. [PubMed] [DOI] [Cited in This Article: ] |
| 279. | Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33:116-128. [PubMed] [DOI] [Cited in This Article: ] |
| 280. | Chen J, Niu N, Zhang J, Qi L, Shen W, Donkena KV, Feng Z, Liu J. Polyploid Giant Cancer Cells (PGCCs): The Evil Roots of Cancer. Curr Cancer Drug Targets. 2019;19:360-367. [PubMed] [DOI] [Cited in This Article: ] |
| 281. | Moein S, Adibi R, da Silva Meirelles L, Nardi NB, Gheisari Y. Cancer regeneration: Polyploid cells are the key drivers of tumor progression. Biochim Biophys Acta Rev Cancer. 2020;1874:188408. [PubMed] [DOI] [Cited in This Article: ] |
| 282. | Zhang S, Mercado-Uribe I, Hanash S, Liu J. iTRAQ-based proteomic analysis of polyploid giant cancer cells and budding progeny cells reveals several distinct pathways for ovarian cancer development. PLoS One. 2013;8:e80120. [PubMed] [DOI] [Cited in This Article: ] |
| 283. | Saleh T, Tyutyunyk-Massey L, Gewirtz DA. Tumor Cell Escape from Therapy-Induced Senescence as a Model of Disease Recurrence after Dormancy. Cancer Res. 2019;79:1044-1046. [PubMed] [DOI] [Cited in This Article: ] |
| 284. | Lopez-Sánchez LM, Jimenez C, Valverde A, Hernandez V, Peñarando J, Martinez A, Lopez-Pedrera C, Muñoz-Castañeda JR, De la Haba-Rodríguez JR, Aranda E, Rodriguez-Ariza A. CoCl2, a mimic of hypoxia, induces formation of polyploid giant cells with stem characteristics in colon cancer. PLoS One. 2014;9:e99143. [PubMed] [DOI] [Cited in This Article: ] |
| 285. | Bloy N, Sauvat A, Chaba K, Buqué A, Humeau J, Bravo-San Pedro JM, Bui J, Kepp O, Kroemer G, Senovilla L. Morphometric analysis of immunoselection against hyperploid cancer cells. Oncotarget. 2015;6:41204-41215. [PubMed] [DOI] [Cited in This Article: ] |
| 286. | D'Assoro AB, Haddad T, Galanis E. Aurora-A Kinase as a Promising Therapeutic Target in Cancer. Front Oncol. 2015;5:295. [PubMed] [DOI] [Cited in This Article: ] |
| 287. | Bourke E, Dodson H, Merdes A, Cuffe L, Zachos G, Walker M, Gillespie D, Morrison CG. DNA damage induces Chk1-dependent centrosome amplification. EMBO Rep. 2007;8:603-609. [PubMed] [DOI] [Cited in This Article: ] |
| 288. | Wang CY, Huang EY, Huang SC, Chung BC. DNA-PK/Chk2 induces centrosome amplification during prolonged replication stress. Oncogene. 2015;34:1263-1269. [PubMed] [DOI] [Cited in This Article: ] |
| 289. | Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, Foijer F, Holland AJ. Centrosome Amplification Is Sufficient to Promote Spontaneous Tumorigenesis in Mammals. Dev Cell. 2017;40:313-322.e5. [PubMed] [DOI] [Cited in This Article: ] |
| 290. | Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, Harris CC, Ried T, Deng CX. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389-395. [PubMed] [DOI] [Cited in This Article: ] |
| 291. | Fusco P, Esposito MR, Tonini GP. Chromosome instability in neuroblastoma. Oncol Lett. 2018;16:6887-6894. [PubMed] [DOI] [Cited in This Article: ] |
| 292. | Niu N, Mercado-Uribe I, Liu J. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene. 2017;36:4887-4900. [PubMed] [DOI] [Cited in This Article: ] |
| 293. | Schulte JH, Kuhfittig-Kulle S, Klein-Hitpass L, Schramm A, Biard DS, Pfeiffer P, Eggert A. Expression of the TrkA or TrkB receptor tyrosine kinase alters the double-strand break (DSB) repair capacity of SY5Y neuroblastoma cells. DNA Repair (Amst). 2008;7:1757-1764. [PubMed] [DOI] [Cited in This Article: ] |
| 294. | Lou CH, Shao A, Shum EY, Espinoza JL, Huang L, Karam R, Wilkinson MF. Posttranscriptional control of the stem cell and neurogenic programs by the nonsense-mediated RNA decay pathway. Cell Rep. 2014;6:748-764. [PubMed] [DOI] [Cited in This Article: ] |
| 295. | Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729-3741. [PubMed] [DOI] [Cited in This Article: ] |
| 296. | Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038-40043. [PubMed] [DOI] [Cited in This Article: ] |
| 297. | Huertas-Castaño C, Gómez-Muñoz MA, Pardal R, Vega FM. Hypoxia in the Initiation and Progression of Neuroblastoma Tumours. Int J Mol Sci. 2019;21. [PubMed] [DOI] [Cited in This Article: ] |
| 298. | Brizel DM, Dodge RK, Clough RW, Dewhirst MW. Oxygenation of head and neck cancer: changes during radiotherapy and impact on treatment outcome. Radiother Oncol. 1999;53:113-117. [PubMed] [DOI] [Cited in This Article: ] |
| 299. | Nordsmark M, Overgaard J. Tumor hypoxia is independent of hemoglobin and prognostic for loco-regional tumor control after primary radiotherapy in advanced head and neck cancer. Acta Oncol. 2004;43:396-403. [PubMed] [DOI] [Cited in This Article: ] |
| 300. | Hockel M, Schlenger K, Aral B, Mitze M, Schaffer U, Vaupel P. Association between tumor hypoxia and malignant progression in advanced cancer of the uterine cervix. Cancer Res. 1996;56:4509-4515. [PubMed] [Cited in This Article: ] |
| 301. | Påhlman S, Mohlin S. Hypoxia and hypoxia-inducible factors in neuroblastoma. Cell Tissue Res. 2018;372:269-275. [PubMed] [DOI] [Cited in This Article: ] |
| 302. | Philip B, Ito K, Moreno-Sánchez R, Ralph SJ. HIF expression and the role of hypoxic microenvironments within primary tumours as protective sites driving cancer stem cell renewal and metastatic progression. Carcinogenesis. 2013;34:1699-1707. [PubMed] [DOI] [Cited in This Article: ] |
| 303. | Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430-439. [PubMed] [DOI] [Cited in This Article: ] |
| 304. | Spill F, Reynolds DS, Kamm RD, Zaman MH. Impact of the physical microenvironment on tumor progression and metastasis. Curr Opin Biotechnol. 2016;40:41-48. [PubMed] [DOI] [Cited in This Article: ] |
| 305. | Petrova V, Annicchiarico-Petruzzelli M, Melino G, Amelio I. The hypoxic tumour microenvironment. Oncogenesis. 2018;7:10. [PubMed] [DOI] [Cited in This Article: ] |
| 306. | Axelson H, Fredlund E, Ovenberger M, Landberg G, Påhlman S. Hypoxia-induced dedifferentiation of tumor cells--a mechanism behind heterogeneity and aggressiveness of solid tumors. Semin Cell Dev Biol. 2005;16:554-563. [PubMed] [DOI] [Cited in This Article: ] |
| 307. | Das B, Tsuchida R, Malkin D, Koren G, Baruchel S, Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells. 2008;26:1818-1830. [PubMed] [DOI] [Cited in This Article: ] |
| 308. | Jögi A, Vallon-Christersson J, Holmquist L, Axelson H, Borg A, Påhlman S. Human neuroblastoma cells exposed to hypoxia: induction of genes associated with growth, survival, and aggressive behavior. Exp Cell Res. 2004;295:469-487. [PubMed] [DOI] [Cited in This Article: ] |
| 309. | Mohlin S, Wigerup C, Jögi A, Påhlman S. Hypoxia, pseudohypoxia and cellular differentiation. Exp Cell Res. 2017;356:192-196. [PubMed] [DOI] [Cited in This Article: ] |
| 310. | Gort EH, Groot AJ, van der Wall E, van Diest PJ, Vooijs MA. Hypoxic regulation of metastasis via hypoxia-inducible factors. Curr Mol Med. 2008;8:60-67. [PubMed] [DOI] [Cited in This Article: ] |
| 311. | Zhang W, Shi X, Peng Y, Wu M, Zhang P, Xie R, Wu Y, Yan Q, Liu S, Wang J. HIF-1α Promotes Epithelial-Mesenchymal Transition and Metastasis through Direct Regulation of ZEB1 in Colorectal Cancer. PLoS One. 2015;10:e0129603. [PubMed] [DOI] [Cited in This Article: ] |
| 312. | Cimmino F, Pezone L, Avitabile M, Persano L, Vitale M, Sassi M, Bresolin S, Serafin V, Zambrano N, Scaloni A, Basso G, Iolascon A, Capasso M. Proteomic Alterations in Response to Hypoxia Inducible Factor 2α in Normoxic Neuroblastoma Cells. J Proteome Res. 2016;15:3643-3655. [PubMed] [DOI] [Cited in This Article: ] |
| 313. | Bhaskara VK, Mohanam I, Rao JS, Mohanam S. Intermittent hypoxia regulates stem-like characteristics and differentiation of neuroblastoma cells. PLoS One. 2012;7:e30905. [PubMed] [DOI] [Cited in This Article: ] |
| 314. | Farina AR, Cappabianca L, Sebastiano M, Zelli V, Guadagni S, Mackay AR. Hypoxia-induced alternative splicing: the 11th Hallmark of Cancer. J Exp Clin Cancer Res. 2020;39:110. [PubMed] [DOI] [Cited in This Article: ] |
| 315. | Kimura K, Kishida T, Wakao J, Tanaka T, Higashi M, Fumino S, Aoi S, Furukawa T, Mazda O, Tajiri T. Tumor-homing effect of human mesenchymal stem cells in a TH-MYCN mouse model of neuroblastoma. J Pediatr Surg. 2016;51:2068-2073. [PubMed] [DOI] [Cited in This Article: ] |
| 316. | Ma M, Ye JY, Deng R, Dee CM, Chan GC. Mesenchymal stromal cells may enhance metastasis of neuroblastoma via SDF-1/CXCR4 and SDF-1/CXCR7 signaling. Cancer Lett. 2011;312:1-10. [PubMed] [DOI] [Cited in This Article: ] |
| 317. | Veschi V, Verona F, Thiele CJ. Cancer Stem Cells and Neuroblastoma: Characteristics and Therapeutic Targeting Options. Front Endocrinol (Lausanne). 2019;10:782. [PubMed] [DOI] [Cited in This Article: ] |
| 318. | Chen W, Dong J, Haiech J, Kilhoffer MC, Zeniou M. Cancer Stem Cell Quiescence and Plasticity as Major Challenges in Cancer Therapy. Stem Cells Int. 2016;2016:1740936. [PubMed] [DOI] [Cited in This Article: ] |
| 319. | Cojoc M, Mäbert K, Muders MH, Dubrovska A. A role for cancer stem cells in therapy resistance: cellular and molecular mechanisms. Semin Cancer Biol. 2015;31:16-27. [PubMed] [DOI] [Cited in This Article: ] |
| 320. | Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2:3. [PubMed] [DOI] [Cited in This Article: ] |
| 321. | Deonarain MP, Kousparou CA, Epenetos AA. Antibodies targeting cancer stem cells: a new paradigm in immunotherapy? MAbs. 2009;1:12-25. [PubMed] [DOI] [Cited in This Article: ] |
| 322. | Lee HY, Hong IS. Targeting Liver Cancer Stem Cells: An Alternative Therapeutic Approach for Liver Cancer. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] |
| 323. | Agarwal S, Lakoma A, Chen Z, Hicks J, Metelitsa LS, Kim ES, Shohet JM. G-CSF Promotes Neuroblastoma Tumorigenicity and Metastasis via STAT3-Dependent Cancer Stem Cell Activation. Cancer Res. 2015;75:2566-2579. [PubMed] [DOI] [Cited in This Article: ] |
| 324. | Künkele A, Taraseviciute A, Finn LS, Johnson AJ, Berger C, Finney O, Chang CA, Rolczynski LS, Brown C, Mgebroff S, Berger M, Park JR, Jensen MC. Preclinical Assessment of CD171-Directed CAR T-cell Adoptive Therapy for Childhood Neuroblastoma: CE7 Epitope Target Safety and Product Manufacturing Feasibility. Clin Cancer Res. 2017;23:466-477. [PubMed] [DOI] [Cited in This Article: ] |
| 325. | Budde LE, Berger C, Lin Y, Wang J, Lin X, Frayo SE, Brouns SA, Spencer DM, Till BG, Jensen MC, Riddell SR, Press OW. Combining a CD20 chimeric antigen receptor and an inducible caspase 9 suicide switch to improve the efficacy and safety of T cell adoptive immunotherapy for lymphoma. PLoS One. 2013;8:e82742. [PubMed] [DOI] [Cited in This Article: ] |
| 326. | Jan M, Scarfò I, Larson RC, Walker A, Schmidts A, Guirguis AA, Gasser JA, Słabicki M, Bouffard AA, Castano AP, Kann MC, Cabral ML, Tepper A, Grinshpun DE, Sperling AS, Kyung T, Sievers QL, Birnbaum ME, Maus MV, Ebert BL. Reversible ON- and OFF-switch chimeric antigen receptors controlled by lenalidomide. Sci Transl Med. 2021;13. [PubMed] [DOI] [Cited in This Article: ] |
| 327. | Weiss T, Weller M, Guckenberger M, Sentman CL, Roth P. NKG2D-Based CAR T Cells and Radiotherapy Exert Synergistic Efficacy in Glioblastoma. Cancer Res. 2018;78:1031-1043. [PubMed] [DOI] [Cited in This Article: ] |
| 328. | Zhang GN, Liang Y, Zhou LJ, Chen SP, Chen G, Zhang TP, Kang T, Zhao YP. Combination of salinomycin and gemcitabine eliminates pancreatic cancer cells. Cancer Lett. 2011;313:137-144. [PubMed] [DOI] [Cited in This Article: ] |
| 329. | Zhang Y, Zhang H, Wang X, Wang J, Zhang X, Zhang Q. The eradication of breast cancer and cancer stem cells using octreotide modified paclitaxel active targeting micelles and salinomycin passive targeting micelles. Biomaterials. 2012;33:679-691. [PubMed] [DOI] [Cited in This Article: ] |
| 330. | Yoshida T, Mihara K, Takei Y, Yanagihara K, Kubo T, Bhattacharyya J, Imai C, Mino T, Takihara Y, Ichinohe T. All-trans retinoic acid enhances cytotoxic effect of T cells with an anti-CD38 chimeric antigen receptor in acute myeloid leukemia. Clin Transl Immunology. 2016;5:e116. [PubMed] [DOI] [Cited in This Article: ] |
| 331. | Caruana I, Savoldo B, Hoyos V, Weber G, Liu H, Kim ES, Ittmann MM, Marchetti D, Dotti G. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat Med. 2015;21:524-529. [PubMed] [DOI] [Cited in This Article: ] |
| 332. | Hatfield SM, Kjaergaard J, Lukashev D, Schreiber TH, Belikoff B, Abbott R, Sethumadhavan S, Philbrook P, Ko K, Cannici R, Thayer M, Rodig S, Kutok JL, Jackson EK, Karger B, Podack ER, Ohta A, Sitkovsky MV. Immunological mechanisms of the antitumor effects of supplemental oxygenation. Sci Transl Med. 2015;7:277ra30. [PubMed] [DOI] [Cited in This Article: ] |
| 333. | McNerney KO, Karageorgos SA, Hogarty MD, Bassiri H. Enhancing Neuroblastoma Immunotherapies by Engaging iNKT and NK Cells. Front Immunol. 2020;11:873. [PubMed] [DOI] [Cited in This Article: ] |
| 334. | Mise N, Takami M, Suzuki A, Kamata T, Harada K, Hishiki T, Saito T, Terui K, Mitsunaga T, Nakata M, Ikeuchi T, Nakayama T, Yoshida H, Motohashi S. Antibody-dependent cellular cytotoxicity toward neuroblastoma enhanced by activated invariant natural killer T cells. Cancer Sci. 2016;107:233-241. [PubMed] [DOI] [Cited in This Article: ] |
| 335. | Bae EA, Seo H, Kim BS, Choi J, Jeon I, Shin KS, Koh CH, Song B, Kim IK, Min BS, Han YD, Shin SJ, Kang CY. Activation of NKT Cells in an Anti-PD-1-Resistant Tumor Model Enhances Antitumor Immunity by Reinvigorating Exhausted CD8 T Cells. Cancer Res. 2018;78:5315-5326. [PubMed] [DOI] [Cited in This Article: ] |
| 336. | Courtney AN, Tian G, Liu D, Marinova E, Heczey A, Xu X, Guo L, Gao X, Metelitsa LS. Cross-talk between NKT cells and tumor associated macrophages in the tumor microenvironment. J Immunol. 2016;196:142. [Cited in This Article: ] |
| 337. | Barry WE, Jackson JR, Asuelime GE, Wu HW, Sun J, Wan Z, Malvar J, Sheard MA, Wang L, Seeger RC, Kim ES. Activated Natural Killer Cells in Combination with Anti-GD2 Antibody Dinutuximab Improve Survival of Mice after Surgical Resection of Primary Neuroblastoma. Clin Cancer Res. 2019;25:325-333. [PubMed] [DOI] [Cited in This Article: ] |
| 338. | Modak S, Le Luduec JB, Cheung IY, Goldman DA, Ostrovnaya I, Doubrovina E, Basu E, Kushner BH, Kramer K, Roberts SS, O'Reilly RJ, Cheung NV, Hsu KC. Adoptive immunotherapy with haploidentical natural killer cells and Anti-GD2 monoclonal antibody m3F8 for resistant neuroblastoma: Results of a phase I study. Oncoimmunology. 2018;7:e1461305. [PubMed] [DOI] [Cited in This Article: ] |
| 339. | Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med. 2018;50:1-11. [PubMed] [DOI] [Cited in This Article: ] |
| 340. | Melaiu O, Mina M, Chierici M, Boldrini R, Jurman G, Romania P, D'Alicandro V, Benedetti MC, Castellano A, Liu T, Furlanello C, Locatelli F, Fruci D. PD-L1 Is a Therapeutic Target of the Bromodomain Inhibitor JQ1 and, Combined with HLA Class I, a Promising Prognostic Biomarker in Neuroblastoma. Clin Cancer Res. 2017;23:4462-4472. [PubMed] [DOI] [Cited in This Article: ] |
| 341. | Campos-Arroyo D, Maldonado V, Bahena I, Quintanar V, Patiño N, Carlos Martinez-Lazcano J, Melendez-Zajgla J. Probenecid Sensitizes Neuroblastoma Cancer Stem Cells to Cisplatin. Cancer Invest. 2016;34:155-166. [PubMed] [DOI] [Cited in This Article: ] |
| 342. | Vella S, Penna I, Longo L, Pioggia G, Garbati P, Florio T, Rossi F, Pagano A. Perhexiline maleate enhances antitumor efficacy of cisplatin in neuroblastoma by inducing over-expression of NDM29 ncRNA. Sci Rep. 2015;5:18144. [PubMed] [DOI] [Cited in This Article: ] |
| 343. | Ding PR, Tiwari AK, Ohnuma S, Lee JW, An X, Dai CL, Lu QS, Singh S, Yang DH, Talele TT, Ambudkar SV, Chen ZS. The phosphodiesterase-5 inhibitor vardenafil is a potent inhibitor of ABCB1/P-glycoprotein transporter. PLoS One. 2011;6:e19329. [PubMed] [DOI] [Cited in This Article: ] |
| 344. | Tyagi A, Vishnoi K, Mahata S, Verma G, Srivastava Y, Masaldan S, Roy BG, Bharti AC, Das BC. Cervical Cancer Stem Cells Selectively Overexpress HPV Oncoprotein E6 that Controls Stemness and Self-Renewal through Upregulation of HES1. Clin Cancer Res. 2016;22:4170-4184. [PubMed] [DOI] [Cited in This Article: ] |
| 345. | Colamaio M, Tosti N, Puca F, Mari A, Gattordo R, Kuzay Y, Federico A, Pepe A, Sarnataro D, Ragozzino E, Raia M, Hirata H, Gemei M, Mimori K, Del Vecchio L, Battista S, Fusco A. HMGA1 silencing reduces stemness and temozolomide resistance in glioblastoma stem cells. Expert Opin Ther Targets. 2016;20:1169-1179. [PubMed] [DOI] [Cited in This Article: ] |
| 346. | Mu Q, Wang H, Zhang M. Nanoparticles for imaging and treatment of metastatic breast cancer. Expert Opin Drug Deliv. 2017;14:123-136. [PubMed] [DOI] [Cited in This Article: ] |
| 347. | Umapathy G, Mendoza-Garcia P, Hallberg B, Palmer RH. Targeting anaplastic lymphoma kinase in neuroblastoma. APMIS. 2019;127:288-302. [PubMed] [DOI] [Cited in This Article: ] |
| 348. | Chen Y, Wang H, Chen Y, Wang M, Ding G, Li T. Trk inhibitor GNF5837 suppresses the tumor growth, survival and migration of renal cell carcinoma. Oncol Rep. 2019;42:2039-2048. [PubMed] [DOI] [Cited in This Article: ] |
| 349. | Croucher JL, Iyer R, Li N, Molteni V, Loren J, Gordon WP, Tuntland T, Liu B, Brodeur GM. TrkB inhibition by GNF-4256 slows growth and enhances chemotherapeutic efficacy in neuroblastoma xenografts. Cancer Chemother Pharmacol. 2015;75:131-141. [PubMed] [DOI] [Cited in This Article: ] |
| 350. | Iyer R, Evans AE, Qi X, Ho R, Minturn JE, Zhao H, Balamuth N, Maris JM, Brodeur GM. Lestaurtinib enhances the antitumor efficacy of chemotherapy in murine xenograft models of neuroblastoma. Clin Cancer Res. 2010;16:1478-1485. [PubMed] [DOI] [Cited in This Article: ] |
| 351. | Pikatan NW, Liu YL, Bamodu OA, Hsiao M, Hsu WM, Haryana SM, Sutaryo, Chao TY, Yeh CT. Aberrantly expressed Bruton's tyrosine kinase preferentially drives metastatic and stem cell-like phenotypes in neuroblastoma cells. Cell Oncol (Dordr). 2020;43:1067-1084. [PubMed] [DOI] [Cited in This Article: ] |
| 352. | Islam A, Kageyama H, Takada N, Kawamoto T, Takayasu H, Isogai E, Ohira M, Hashizume K, Kobayashi H, Kaneko Y, Nakagawara A. High expression of Survivin, mapped to 17q25, is significantly associated with poor prognostic factors and promotes cell survival in human neuroblastoma. Oncogene. 2000;19:617-623. [PubMed] [DOI] [Cited in This Article: ] |
| 353. | Warrier NM, Agarwal P, Kumar P. Emerging Importance of Survivin in Stem Cells and Cancer: the Development of New Cancer Therapeutics. Stem Cell Rev Rep. 2020;16:828-852. [PubMed] [DOI] [Cited in This Article: ] |
| 354. | Suman S, Das TP, Damodaran C. Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br J Cancer. 2013;109:2587-2596. [PubMed] [DOI] [Cited in This Article: ] |
| 355. | Xu L, Wang X, Wan J, Li T, Gong X, Zhang K, Yi L, Xiang Z, Xu M, Cui H. Sonic Hedgehog pathway is essential for neuroblastoma cell proliferation and tumor growth. Mol Cell Biochem. 2012;364:235-241. [PubMed] [DOI] [Cited in This Article: ] |
| 356. | Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A, Eberhart CG. Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells. 2007;25:2524-2533. [PubMed] [DOI] [Cited in This Article: ] |
| 357. | Lin TL, Wang QH, Brown P, Peacock C, Merchant AA, Brennan S, Jones E, McGovern K, Watkins DN, Sakamoto KM, Matsui W. Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PLoS One. 2010;5:e15262. [PubMed] [DOI] [Cited in This Article: ] |
| 358. | Fan X, Eberhart CG. Medulloblastoma stem cells. J Clin Oncol. 2008;26:2821-2827. [PubMed] [DOI] [Cited in This Article: ] |
| 359. | Miyazaki Y, Matsubara S, Ding Q, Tsukasa K, Yoshimitsu M, Kosai K, Takao S. Efficient elimination of pancreatic cancer stem cells by hedgehog/GLI inhibitor GANT61 in combination with mTOR inhibition. Mol Cancer. 2016;15:49. [PubMed] [DOI] [Cited in This Article: ] |
| 360. | Schiapparelli P, Shahi MH, Enguita-Germán M, Johnsen JI, Kogner P, Lázcoz P, Castresana JS. Inhibition of the sonic hedgehog pathway by cyplopamine reduces the CD133+/CD15+ cell compartment and the in vitro tumorigenic capability of neuroblastoma cells. Cancer Lett. 2011;310:222-231. [PubMed] [DOI] [Cited in This Article: ] |
| 361. | Xu X, Zhang M, Xu F, Jiang S. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol Cancer. 2020;19:165. [PubMed] [DOI] [Cited in This Article: ] |
| 362. | Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5:8. [PubMed] [DOI] [Cited in This Article: ] |
| 363. | Yang XH, Tang F, Shin J, Cunningham JM. A c-Myc-regulated stem cell-like signature in high-risk neuroblastoma: A systematic discovery (Target neuroblastoma ESC-like signature). Sci Rep. 2017;7:41. [PubMed] [DOI] [Cited in This Article: ] |
| 364. | Ognibene M, Pezzolo A. Roniciclib down-regulates stemness and inhibits cell growth by inducing nucleolar stress in neuroblastoma. Sci Rep. 2020;10:12902. [PubMed] [DOI] [Cited in This Article: ] |
| 365. | Gonçalves JM, Silva CAB, Rivero ERC, Cordeiro MMR. Inhibition of cancer stem cells promoted by Pimozide. Clin Exp Pharmacol Physiol. 2019;46:116-125. [PubMed] [DOI] [Cited in This Article: ] |
| 366. | Bahmad HF, Chalhoub RM, Harati H, Bou-Gharios J, Assi S, Ballout F, Monzer A, Msheik H, Araji T, Elajami MK, Ghanem P, Chamaa F, Kadara H, Abou-Antoun T, Daoud G, Fares Y, Abou-Kheir W. Tideglusib attenuates growth of neuroblastoma cancer stem/progenitor cells in vitro and in vivo by specifically targeting GSK-3β. Pharmacol Rep. 2021;73:211-226. [PubMed] [DOI] [Cited in This Article: ] |
| 367. | Bahmad HF, Mouhieddine TH, Chalhoub RM, Assi S, Araji T, Chamaa F, Itani MM, Nokkari A, Kobeissy F, Daoud G, Abou-Kheir W. The Akt/mTOR pathway in cancer stem/progenitor cells is a potential therapeutic target for glioblastoma and neuroblastoma. Oncotarget. 2018;9:33549-33561. [PubMed] [DOI] [Cited in This Article: ] |
| 368. | Moreno-Smith M, Lakoma A, Chen Z, Tao L, Scorsone KA, Schild L, Aviles-Padilla K, Nikzad R, Zhang Y, Chakraborty R, Molenaar JJ, Vasudevan SA, Sheehan V, Kim ES, Paust S, Shohet JM, Barbieri E. p53 Nongenotoxic Activation and mTORC1 Inhibition Lead to Effective Combination for Neuroblastoma Therapy. Clin Cancer Res. 2017;23:6629-6639. [PubMed] [DOI] [Cited in This Article: ] |
| 369. | Kim KW, Kim JY, Qiao J, Clark RA, Powers CM, Correa H, Chung DH. Dual-Targeting AKT2 and ERK in cancer stem-like cells in neuroblastoma. Oncotarget. 2019;10:5645-5659. [PubMed] [DOI] [Cited in This Article: ] |
| 370. | Theiss AL, Sitaraman SV. The role and therapeutic potential of prohibitin in disease. Biochim Biophys Acta. 2011;1813:1137-1143. [PubMed] [DOI] [Cited in This Article: ] |
| 371. | Goel A, Aggarwal BB. Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutr Cancer. 2010;62:919-930. [PubMed] [DOI] [Cited in This Article: ] |
| 372. | Gheeya JS, Chen QR, Benjamin CD, Cheuk AT, Tsang P, Chung JY, Metaferia BB, Badgett TC, Johansson P, Wei JS, Hewitt SM, Khan J. Screening a panel of drugs with diverse mechanisms of action yields potential therapeutic agents against neuroblastoma. Cancer Biol Ther. 2009;8:2386-2395. [PubMed] [DOI] [Cited in This Article: ] |
| 373. | Prasad R, Katiyar SK. Honokiol, an Active Compound of Magnolia Plant, Inhibits Growth, and Progression of Cancers of Different Organs. Adv Exp Med Biol. 2016;928:245-265. [PubMed] [DOI] [Cited in This Article: ] |
| 374. | Morozova O, Vojvodic M, Grinshtein N, Hansford LM, Blakely KM, Maslova A, Hirst M, Cezard T, Morin RD, Moore R, Smith KM, Miller F, Taylor P, Thiessen N, Varhol R, Zhao Y, Jones S, Moffat J, Kislinger T, Moran MF, Kaplan DR, Marra MA. System-level analysis of neuroblastoma tumor-initiating cells implicates AURKB as a novel drug target for neuroblastoma. Clin Cancer Res. 2010;16:4572-4582. [PubMed] [DOI] [Cited in This Article: ] |
| 375. | Mouhieddine TH, Nokkari A, Itani MM, Chamaa F, Bahmad H, Monzer A, El-Merahbi R, Daoud G, Eid A, Kobeissy FH, Abou-Kheir W. Metformin and Ara-a Effectively Suppress Brain Cancer by Targeting Cancer Stem/Progenitor Cells. Front Neurosci. 2015;9:442. [PubMed] [DOI] [Cited in This Article: ] |
| 376. | Chanthery YH, Gustafson WC, Itsara M, Persson A, Hackett CS, Grimmer M, Charron E, Yakovenko S, Kim G, Matthay KK, Weiss WA. Paracrine signaling through MYCN enhances tumor-vascular interactions in neuroblastoma. Sci Transl Med. 2012;4:115ra3. [PubMed] [DOI] [Cited in This Article: ] |
| 377. | Hasan K. N-Myc inhibition: advances in neuroblastoma treatment. Cell Bio 2017; 6: 27-34 . [DOI] [Cited in This Article: ] |
| 378. | Shahbazi J, Liu PY, Atmadibrata B, Bradner JE, Marshall GM, Lock RB, Liu T. The Bromodomain Inhibitor JQ1 and the Histone Deacetylase Inhibitor Panobinostat Synergistically Reduce N-Myc Expression and Induce Anticancer Effects. Clin Cancer Res. 2016;22:2534-2544. [PubMed] [DOI] [Cited in This Article: ] |
| 379. | Choi YJ, Kim I, Lee JE, Park JW. PIN1 transcript variant 2 acts as a long non-coding RNA that controls the HIF-1-driven hypoxic response. Sci Rep. 2019;9:10599. [PubMed] [DOI] [Cited in This Article: ] |
| 380. | Chen SJ, Hoffman NE, Shanmughapriya S, Bao L, Keefer K, Conrad K, Merali S, Takahashi Y, Abraham T, Hirschler-Laszkiewicz I, Wang J, Zhang XQ, Song J, Barrero C, Shi Y, Kawasawa YI, Bayerl M, Sun T, Barbour M, Wang HG, Madesh M, Cheung JY, Miller BA. A splice variant of the human ion channel TRPM2 modulates neuroblastoma tumor growth through hypoxia-inducible factor (HIF)-1/2α. J Biol Chem. 2014;289:36284-36302. [PubMed] [DOI] [Cited in This Article: ] |
| 381. | Jeong W, Rapisarda A, Park SR, Kinders RJ, Chen A, Melillo G, Turkbey B, Steinberg SM, Choyke P, Doroshow JH, Kummar S. Pilot trial of EZN-2968, an antisense oligonucleotide inhibitor of hypoxia-inducible factor-1 alpha (HIF-1α), in patients with refractory solid tumors. Cancer Chemother Pharmacol. 2014;73:343-348. [PubMed] [DOI] [Cited in This Article: ] |
| 382. | Coltella N, Valsecchi R, Ponente M, Ponzoni M, Bernardi R. Synergistic Leukemia Eradication by Combined Treatment with Retinoic Acid and HIF Inhibition by EZN-2208 (PEG-SN38) in Preclinical Models of PML-RARα and PLZF-RARα-Driven Leukemia. Clin Cancer Res. 2015;21:3685-3694. [PubMed] [DOI] [Cited in This Article: ] |
| 383. | Rapisarda A, Uranchimeg B, Sordet O, Pommier Y, Shoemaker RH, Melillo G. Topoisomerase I-mediated inhibition of hypoxia-inducible factor 1: mechanism and therapeutic implications. Cancer Res. 2004;64:1475-1482. [PubMed] [DOI] [Cited in This Article: ] |
| 384. | Fu B, Xue J, Li Z, Shi X, Jiang BH, Fang J. Chrysin inhibits expression of hypoxia-inducible factor-1alpha through reducing hypoxia-inducible factor-1alpha stability and inhibiting its protein synthesis. Mol Cancer Ther. 2007;6:220-226. [PubMed] [DOI] [Cited in This Article: ] |
| 385. | Lee SH, Jee JG, Bae JS, Liu KH, Lee YM. A group of novel HIF-1α inhibitors, glyceollins, blocks HIF-1α synthesis and decreases its stability via inhibition of the PI3K/AKT/mTOR pathway and Hsp90 binding. J Cell Physiol. 2015;230:853-862. [PubMed] [DOI] [Cited in This Article: ] |
| 386. | Narita T, Yin S, Gelin CF, Moreno CS, Yepes M, Nicolaou KC, Van Meir EG. Identification of a novel small molecule HIF-1αlpha translation inhibitor. Clin Cancer Res. 2009;15:6128-6136. [PubMed] [DOI] [Cited in This Article: ] |
| 387. | Poch A, Villanelo F, Henriquez S, Kohen P, Muñoz A, Strauss JF 3rd, Devoto L. Molecular modelling predicts that 2-methoxyestradiol disrupts HIF function by binding to the PAS-B domain. Steroids. 2019;144:21-29. [PubMed] [DOI] [Cited in This Article: ] |
| 388. | Lang SA, Moser C, Gaumann A, Klein D, Glockzin G, Popp FC, Dahlke MH, Piso P, Schlitt HJ, Geissler EK, Stoeltzing O. Targeting heat shock protein 90 in pancreatic cancer impairs insulin-like growth factor-I receptor signaling, disrupts an interleukin-6/signal-transducer and activator of transcription 3/hypoxia-inducible factor-1alpha autocrine loop, and reduces orthotopic tumor growth. Clin Cancer Res. 2007;13:6459-6468. [PubMed] [DOI] [Cited in This Article: ] |
| 389. | Bohonowych JE, Peng S, Gopal U, Hance MW, Wing SB, Argraves KM, Lundgren K, Isaacs JS. Comparative analysis of novel and conventional Hsp90 inhibitors on HIF activity and angiogenic potential in clear cell renal cell carcinoma: implications for clinical evaluation. BMC Cancer. 2011;11:520. [PubMed] [DOI] [Cited in This Article: ] |
| 390. | Hutt DM, Roth DM, Vignaud H, Cullin C, Bouchecareilh M. The histone deacetylase inhibitor, Vorinostat, represses hypoxia inducible factor 1 alpha expression through translational inhibition. PLoS One. 2014;9:e106224. [PubMed] [DOI] [Cited in This Article: ] |
| 391. | Lee K, Kang JE, Park SK, Jin Y, Chung KS, Kim HM, Lee K, Kang MR, Lee MK, Song KB, Yang EG, Lee JJ, Won M. LW6, a novel HIF-1 inhibitor, promotes proteasomal degradation of HIF-1αlpha via upregulation of VHL in a colon cancer cell line. Biochem Pharmacol. 2010;80:982-989. [PubMed] [DOI] [Cited in This Article: ] |
| 392. | Mukherjee A, Martin SG. The thioredoxin system: a key target in tumour and endothelial cells. Br J Radiol. 2008;81 Spec No 1:S57-S68. [PubMed] [DOI] [Cited in This Article: ] |
| 393. | Yu X, Zhao R, Lin S, Bai X, Zhang L, Yuan S, Sun L. CXCL16 induces angiogenesis in autocrine signaling pathway involving hypoxia-inducible factor 1α in human umbilical vein endothelial cells. Oncol Rep. 2016;35:1557-1565. [PubMed] [DOI] [Cited in This Article: ] |
| 394. | Sica V, Bravo-San Pedro JM, Izzo V, Pol J, Pierredon S, Enot D, Durand S, Bossut N, Chery A, Souquere S, Pierron G, Vartholomaiou E, Zamzami N, Soussi T, Sauvat A, Mondragón L, Kepp O, Galluzzi L, Martinou JC, Hess-Stumpp H, Ziegelbauer K, Kroemer G, Maiuri MC. Lethal Poisoning of Cancer Cells by Respiratory Chain Inhibition plus Dimethyl α-Ketoglutarate. Cell Rep. 2019;27:820-834.e9. [PubMed] [DOI] [Cited in This Article: ] |
| 395. | Griggio V, Vitale C, Todaro M, Riganti C, Kopecka J, Salvetti C, Bomben R, Bo MD, Magliulo D, Rossi D, Pozzato G, Bonello L, Marchetti M, Omedè P, Kodipad AA, Laurenti L, Del Poeta G, Mauro FR, Bernardi R, Zenz T, Gattei V, Gaidano G, Foà R, Massaia M, Boccadoro M, Coscia M. HIF-1α is over-expressed in leukemic cells from TP53-disrupted patients and is a promising therapeutic target in chronic lymphocytic leukemia. Haematologica. 2020;105:1042-1054. [PubMed] [DOI] [Cited in This Article: ] |
| 396. | Miranda E, Nordgren IK, Male AL, Lawrence CE, Hoakwie F, Cuda F, Court W, Fox KR, Townsend PA, Packham GK, Eccles SA, Tavassoli A. A cyclic peptide inhibitor of HIF-1 heterodimerization that inhibits hypoxia signaling in cancer cells. J Am Chem Soc. 2013;135:10418-10425. [PubMed] [DOI] [Cited in This Article: ] |
| 397. | Scheuermann TH, Li Q, Ma HW, Key J, Zhang L, Chen R, Garcia JA, Naidoo J, Longgood J, Frantz DE, Tambar UK, Gardner KH, Bruick RK. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9:271-276. [PubMed] [DOI] [Cited in This Article: ] |
| 398. | Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci USA. 2009;106:17910-17915. [PubMed] [DOI] [Cited in This Article: ] |
| 399. | Pang Y, Yang C, Schovanek J, Wang H, Bullova P, Caisova V, Gupta G, Wolf KI, Semenza GL, Zhuang Z, Pacak K. Anthracyclines suppress pheochromocytoma cell characteristics, including metastasis, through inhibition of the hypoxia signaling pathway. Oncotarget. 2017;8:22313-22324. [PubMed] [DOI] [Cited in This Article: ] |
| 400. | Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A, Fisher RJ, Shoemaker RH, Melillo G. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res. 2005;65:9047-9055. [PubMed] [DOI] [Cited in This Article: ] |
| 401. | Staab A, Loeffler J, Said HM, Diehlmann D, Katzer A, Beyer M, Fleischer M, Schwab F, Baier K, Einsele H, Flentje M, Vordermark D. Effects of HIF-1 inhibition by chetomin on hypoxia-related transcription and radiosensitivity in HT 1080 human fibrosarcoma cells. BMC Cancer. 2007;7:213. [PubMed] [DOI] [Cited in This Article: ] |
| 402. | Minegishi H, Fukashiro S, Ban HS, Nakamura H. Discovery of Indenopyrazoles as a New Class of Hypoxia Inducible Factor (HIF)-1 Inhibitors. ACS Med Chem Lett. 2013;4:297-301. [PubMed] [DOI] [Cited in This Article: ] |
| 403. | Li SH, Shin DH, Chun YS, Lee MK, Kim MS, Park JW. A novel mode of action of YC-1 in HIF inhibition: stimulation of FIH-dependent p300 dissociation from HIF-1{alpha}. Mol Cancer Ther. 2008;7:3729-3738. [PubMed] [DOI] [Cited in This Article: ] |
| 404. | Moreno-Manzano V, Rodríguez-Jiménez FJ, Aceña-Bonilla JL, Fustero-Lardíes S, Erceg S, Dopazo J, Montaner D, Stojkovic M, Sánchez-Puelles JM. FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status. J Biol Chem. 2010;285:1333-1342. [PubMed] [DOI] [Cited in This Article: ] |
| 405. | Chau NM, Rogers P, Aherne W, Carroll V, Collins I, McDonald E, Workman P, Ashcroft M. Identification of novel small molecule inhibitors of hypoxia-inducible factor-1 that differentially block hypoxia-inducible factor-1 activity and hypoxia-inducible factor-1alpha induction in response to hypoxic stress and growth factors. Cancer Res. 2005;65:4918-4928. [PubMed] [DOI] [Cited in This Article: ] |
| 406. | Ban HS, Kim BK, Lee H, Kim HM, Harmalkar D, Nam M, Park SK, Lee K, Park JT, Kim I, Hwang GS, Won M. The novel hypoxia-inducible factor-1α inhibitor IDF-11774 regulates cancer metabolism, thereby suppressing tumor growth. Cell Death Dis. 2017;8:e2843. [PubMed] [DOI] [Cited in This Article: ] |
| 407. | Lee YM, Lim JH, Yoon H, Chun YS, Park JW. Antihepatoma activity of chaetocin due to deregulated splicing of hypoxia-inducible factor 1α pre-mRNA in mice and in vitro. Hepatology. 2011;53:171-180. [PubMed] [DOI] [Cited in This Article: ] |
| 408. | Isham CR, Tibodeau JD, Bossou AR, Merchan JR, Bible KC. The anticancer effects of chaetocin are independent of programmed cell death and hypoxia, and are associated with inhibition of endothelial cell proliferation. Br J Cancer. 2012;106:314-323. [PubMed] [DOI] [Cited in This Article: ] |
| 409. | Craig BT, Rellinger EJ, Alvarez AL, Dusek HL, Qiao J, Chung DH. Induced differentiation inhibits sphere formation in neuroblastoma. Biochem Biophys Res Commun. 2016;477:255-259. [PubMed] [DOI] [Cited in This Article: ] |
| 410. | Duffy DJ, Krstic A, Halasz M, Schwarzl T, Konietzny A, Iljin K, Higgins DG, Kolch W. Retinoic acid and TGF-β signalling cooperate to overcome MYCN-induced retinoid resistance. Genome Med. 2017;9:15. [PubMed] [DOI] [Cited in This Article: ] |
| 411. | Cimmino F, Pezone L, Avitabile M, Acierno G, Andolfo I, Capasso M, Iolascon A. Inhibition of hypoxia inducible factors combined with all-trans retinoic acid treatment enhances glial transdifferentiation of neuroblastoma cells. Sci Rep. 2015;5:11158. [PubMed] [DOI] [Cited in This Article: ] |
| 412. | Hämmerle B, Yañez Y, Palanca S, Cañete A, Burks DJ, Castel V, Font de Mora J. Targeting neuroblastoma stem cells with retinoic acid and proteasome inhibitor. PLoS One. 2013;8:e76761. [PubMed] [DOI] [Cited in This Article: ] |
| 413. | Wongtrakoongate P. Epigenetic therapy of cancer stem and progenitor cells by targeting DNA methylation machineries. World J Stem Cells. 2015;7:137-148. [PubMed] [DOI] [Cited in This Article: ] |
| 414. | Zheng X, Naiditch J, Czurylo M, Jie C, Lautz T, Clark S, Jafari N, Qiu Y, Chu F, Madonna MB. Differential effect of long-term drug selection with doxorubicin and vorinostat on neuroblastoma cells with cancer stem cell characteristics. Cell Death Dis. 2013;4:e740. [PubMed] [DOI] [Cited in This Article: ] |
| 415. | Coffey DC, Kutko MC, Glick RD, Butler LM, Heller G, Rifkind RA, Marks PA, Richon VM, La Quaglia MP. The histone deacetylase inhibitor, CBHA, inhibits growth of human neuroblastoma xenografts in vivo, alone and synergistically with all-trans retinoic acid. Cancer Res. 2001;61:3591-3594. [PubMed] [Cited in This Article: ] |
| 416. | Shah RD, Jagtap JC, Mruthyunjaya S, Shelke GV, Pujari R, Das G, Shastry P. Sodium valproate potentiates staurosporine-induced apoptosis in neuroblastoma cells via Akt/survivin independently of HDAC inhibition. J Cell Biochem. 2013;114:854-863. [PubMed] [DOI] [Cited in This Article: ] |
| 417. | Almeida VR, Vieira IA, Buendia M, Brunetto AT, Gregianin LJ, Brunetto AL, Klamt F, de Farias CB, Abujamra AL, Lopez PLDC, Roesler R. Combined Treatments with a Retinoid Receptor Agonist and Epigenetic Modulators in Human Neuroblastoma Cells. Mol Neurobiol. 2017;54:7610-7619. [PubMed] [DOI] [Cited in This Article: ] |
| 418. | Teitz T, Wei T, Valentine MB, Vanin EF, Grenet J, Valentine VA, Behm FG, Look AT, Lahti JM, Kidd VJ. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529-535. [PubMed] [DOI] [Cited in This Article: ] |
| 419. | Charlet J, Schnekenburger M, Brown KW, Diederich M. DNA demethylation increases sensitivity of neuroblastoma cells to chemotherapeutic drugs. Biochem Pharmacol. 2012;83:858-865. [PubMed] [DOI] [Cited in This Article: ] |
| 420. | Fulda S, Debatin KM. 5-Aza-2'-deoxycytidine and IFN-gamma cooperate to sensitize for TRAIL-induced apoptosis by upregulating caspase-8. Oncogene. 2006;25:5125-5133. [PubMed] [DOI] [Cited in This Article: ] |
| 421. | Ikegaki N, Shimada H, Fox AM, Regan PL, Jacobs JR, Hicks SL, Rappaport EF, Tang XX. Transient treatment with epigenetic modifiers yields stable neuroblastoma stem cells resembling aggressive large-cell neuroblastomas. Proc Natl Acad Sci USA. 2013;110:6097-6102. [PubMed] [DOI] [Cited in This Article: ] |
| 422. | Cecere F, Iuliano A, Albano F, Zappelli C, Castellano I, Grimaldi P, Masullo M, De Vendittis E, Ruocco MR. Diclofenac-induced apoptosis in the neuroblastoma cell line SH-SY5Y: possible involvement of the mitochondrial superoxide dismutase. J Biomed Biotechnol. 2010;2010:801726. [PubMed] [DOI] [Cited in This Article: ] |
| 423. | Naveen CR, Gaikwad S, Agrawal-Rajput R. Berberine induces neuronal differentiation through inhibition of cancer stemness and epithelial-mesenchymal transition in neuroblastoma cells. Phytomedicine. 2016;23:736-744. [PubMed] [DOI] [Cited in This Article: ] |
| 424. | Grinshtein N, Datti A, Fujitani M, Uehling D, Prakesch M, Isaac M, Irwin MS, Wrana JL, Al-Awar R, Kaplan DR. Small molecule kinase inhibitor screen identifies polo-like kinase 1 as a target for neuroblastoma tumor-initiating cells. Cancer Res. 2011;71:1385-1395. [PubMed] [DOI] [Cited in This Article: ] |
| 425. | Tian X, Hou W, Bai S, Fan J, Tong H, Xu H. XAV939 inhibits the stemness and migration of neuroblastoma cancer stem cells via repression of tankyrase 1. Int J Oncol. 2014;45:121-128. [PubMed] [DOI] [Cited in This Article: ] |
| 426. | Gneo L, Ruggeri P, Cappabianca L, Farina AR, Di Ianni N, Mackay AR. TRAIL induces pro-apoptotic crosstalk between the TRAIL-receptor signaling pathway and TrkAIII in SH-SY5Y cells, unveiling a potential therapeutic "Achilles heel" for the TrkAIII oncoprotein in neuroblastoma. Oncotarget. 2016;7:80820-80841. [PubMed] [DOI] [Cited in This Article: ] |
| 427. | Castelnuovo M, Massone S, Tasso R, Fiorino G, Gatti M, Robello M, Gatta E, Berger A, Strub K, Florio T, Dieci G, Cancedda R, Pagano A. An Alu-like RNA promotes cell differentiation and reduces malignancy of human neuroblastoma cells. FASEB J. 2010;24:4033-4046. [PubMed] [DOI] [Cited in This Article: ] |
| 428. | Smith KM, Datti A, Fujitani M, Grinshtein N, Zhang L, Morozova O, Blakely KM, Rotenberg SA, Hansford LM, Miller FD, Yeger H, Irwin MS, Moffat J, Marra MA, Baruchel S, Wrana JL, Kaplan DR. Selective targeting of neuroblastoma tumour-initiating cells by compounds identified in stem cell-based small molecule screens. EMBO Mol Med. 2010;2:371-384. [PubMed] [DOI] [Cited in This Article: ] |
| 429. | Salcher S, Spoden G, Hagenbuchner J, Führer S, Kaserer T, Tollinger M, Huber-Cantonati P, Gruber T, Schuster D, Gust R, Zwierzina H, Müller T, Kiechl-Kohlendorfer U, Ausserlechner MJ, Obexer P. A drug library screen identifies Carbenoxolone as novel FOXO inhibitor that overcomes FOXO3-mediated chemoprotection in high-stage neuroblastoma. Oncogene. 2020;39:1080-1097. [PubMed] [DOI] [Cited in This Article: ] |
| 430. | Lee HA, Park S, Kim Y. Effect of β-carotene on cancer cell stemness and differentiation in SK-N-BE(2)C neuroblastoma cells. Oncol Rep. 2013;30:1869-1877. [PubMed] [DOI] [Cited in This Article: ] |
| 431. | Castelo-Branco P, Zhang C, Lipman T, Fujitani M, Hansford L, Clarke I, Harley CB, Tressler R, Malkin D, Walker E, Kaplan DR, Dirks P, Tabori U. Neural tumor-initiating cells have distinct telomere maintenance and can be safely targeted for telomerase inhibition. Clin Cancer Res. 2011;17:111-121. [PubMed] [DOI] [Cited in This Article: ] |
| 432. | Mittal K, Donthamsetty S, Kaur R, Yang C, Gupta MV, Reid MD, Choi DH, Rida PCG, Aneja R. Multinucleated polyploidy drives resistance to Docetaxel chemotherapy in prostate cancer. Br J Cancer. 2017;116:1186-1194. [PubMed] [DOI] [Cited in This Article: ] |
| 433. | Coward J, Harding A. Size Does Matter: Why Polyploid Tumor Cells are Critical Drug Targets in the War on Cancer. Front Oncol. 2014;4:123. [PubMed] [DOI] [Cited in This Article: ] |
| 434. | Liu LL, Long ZJ, Wang LX, Zheng FM, Fang ZG, Yan M, Xu DF, Chen JJ, Wang SW, Lin DJ, Liu Q. Inhibition of mTOR pathway sensitizes acute myeloid leukemia cells to aurora inhibitors by suppression of glycolytic metabolism. Mol Cancer Res. 2013;11:1326-1336. [PubMed] [DOI] [Cited in This Article: ] |
| 435. | Donovan P, Cato K, Legaie R, Jayalath R, Olsson G, Hall B, Olson S, Boros S, Reynolds BA, Harding A. Hyperdiploid tumor cells increase phenotypic heterogeneity within Glioblastoma tumors. Mol Biosyst. 2014;10:741-758. [PubMed] [DOI] [Cited in This Article: ] |
| 436. | Carpentieri A, Cozzoli E, Scimeca M, Bonanno E, Sardanelli AM, Gambacurta A. Differentiation of human neuroblastoma cells toward the osteogenic lineage by mTOR inhibitor. Cell Death Dis. 2015;6:e1974. [PubMed] [DOI] [Cited in This Article: ] |
| 437. | Sharma S, Zeng JY, Zhuang CM, Zhou YQ, Yao HP, Hu X, Zhang R, Wang MH. Small-molecule inhibitor BMS-777607 induces breast cancer cell polyploidy with increased resistance to cytotoxic chemotherapy agents. Mol Cancer Ther. 2013;12:725-736. [PubMed] [DOI] [Cited in This Article: ] |
| 438. | Zeng JY, Sharma S, Zhou YQ, Yao HP, Hu X, Zhang R, Wang MH. Synergistic activities of MET/RON inhibitor BMS-777607 and mTOR inhibitor AZD8055 to polyploid cells derived from pancreatic cancer and cancer stem cells. Mol Cancer Ther. 2014;13:37-48. [PubMed] [DOI] [Cited in This Article: ] |
| 439. | Mahmoud NN, Dannenberg AJ, Mestre J, Bilinski RT, Churchill MR, Martucci C, Newmark H, Bertagnolli MM. Aspirin prevents tumors in a murine model of familial adenomatous polyposis. Surgery. 1998;124:225-231. [PubMed] [DOI] [Cited in This Article: ] |
| 440. | Lissa D, Senovilla L, Rello-Varona S, Vitale I, Michaud M, Pietrocola F, Boilève A, Obrist F, Bordenave C, Garcia P, Michels J, Jemaà M, Kepp O, Castedo M, Kroemer G. Resveratrol and aspirin eliminate tetraploid cells for anticancer chemoprevention. Proc Natl Acad Sci USA. 2014;111:3020-3025. [PubMed] [DOI] [Cited in This Article: ] |
| 441. | Tovar C, Higgins B, Deo D, Kolinsky K, Liu JJ, Heimbrook DC, Vassilev LT. Small-molecule inducer of cancer cell polyploidy promotes apoptosis or senescence: Implications for therapy. Cell Cycle. 2010;9:3364-3375. [PubMed] [DOI] [Cited in This Article: ] |
| 442. | Mc Gee MM. Targeting the Mitotic Catastrophe Signaling Pathway in Cancer. Mediators Inflamm. 2015;2015:146282. [PubMed] [DOI] [Cited in This Article: ] |
| 443. | Rivera-Rivera Y, Saavedra HI. Centrosome - a promising anti-cancer target. Biologics. 2016;10:167-176. [PubMed] [DOI] [Cited in This Article: ] |
| 444. | Sabat-Pośpiech D, Fabian-Kolpanowicz K, Prior IA, Coulson JM, Fielding AB. Targeting centrosome amplification, an Achilles' heel of cancer. Biochem Soc Trans. 2019;47:1209-1222. [PubMed] [DOI] [Cited in This Article: ] |
| 445. | Gjertsen BT, Schöffski P. Discovery and development of the Polo-like kinase inhibitor volasertib in cancer therapy. Leukemia. 2015;29:11-19. [PubMed] [DOI] [Cited in This Article: ] |
| 446. | Cicenas J, Kalyan K, Sorokinas A, Jatulyte A, Valiunas D, Kaupinis A, Valius M. Highlights of the Latest Advances in Research on CDK Inhibitors. Cancers (Basel). 2014;6:2224-2242. [PubMed] [DOI] [Cited in This Article: ] |
| 447. | Mason JM, Lin DC, Wei X, Che Y, Yao Y, Kiarash R, Cescon DW, Fletcher GC, Awrey DE, Bray MR, Pan G, Mak TW. Functional characterization of CFI-400945, a Polo-like kinase 4 inhibitor, as a potential anticancer agent. Cancer Cell. 2014;26:163-176. [PubMed] [DOI] [Cited in This Article: ] |
| 448. | Wengner AM, Siemeister G, Koppitz M, Schulze V, Kosemund D, Klar U, Stoeckigt D, Neuhaus R, Lienau P, Bader B, Prechtl S, Raschke M, Frisk AL, von Ahsen O, Michels M, Kreft B, von Nussbaum F, Brands M, Mumberg D, Ziegelbauer K. Novel Mps1 Kinase Inhibitors with Potent Antitumor Activity. Mol Cancer Ther. 2016;15:583-592. [PubMed] [DOI] [Cited in This Article: ] |
| 449. | Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282-288. [PubMed] [DOI] [Cited in This Article: ] |
| 450. | Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical Overview of MDM2/X-Targeted Therapies. Front Oncol. 2016;6:7. [PubMed] [DOI] [Cited in This Article: ] |
| 451. | Johannes JW, Almeida L, Daly K, Ferguson AD, Grosskurth SE, Guan H, Howard T, Ioannidis S, Kazmirski S, Lamb ML, Larsen NA, Lyne PD, Mikule K, Ogoe C, Peng B, Petteruti P, Read JA, Su N, Sylvester M, Throner S, Wang W, Wang X, Wu J, Ye Q, Yu Y, Zheng X, Scott DA. Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering. Bioorg Med Chem Lett. 2015;25:5743-5747. [PubMed] [DOI] [Cited in This Article: ] |
| 452. | Wang Z, Grosskurth SE, Cheung T, Petteruti P, Zhang J, Wang X, Wang W, Gharahdaghi F, Wu J, Su N, Howard RT, Mayo M, Widzowski D, Scott DA, Johannes JW, Lamb ML, Lawson D, Dry JR, Lyne PD, Tate EW, Zinda M, Mikule K, Fawell SE, Reimer C, Chen H. Pharmacological Inhibition of PARP6 Triggers Multipolar Spindle Formation and Elicits Therapeutic Effects in Breast Cancer. Cancer Res. 2018;78:6691-6702. [PubMed] [DOI] [Cited in This Article: ] |
| 453. | Mariappan A, Soni K, Schorpp K, Zhao F, Minakar A, Zheng X, Mandad S, Macheleidt I, Ramani A, Kubelka T, Dawidowski M, Golfmann K, Wason A, Yang C, Simons J, Schmalz HG, Hyman AA, Aneja R, Ullrich R, Urlaub H, Odenthal M, Büttner R, Li H, Sattler M, Hadian K, Gopalakrishnan J. Inhibition of CPAP-tubulin interaction prevents proliferation of centrosome-amplified cancer cells. EMBO J. 2019;38. [PubMed] [DOI] [Cited in This Article: ] |
| 454. | Qiu GZ, Jin MZ, Dai JX, Sun W, Feng JH, Jin WL. Reprogramming of the Tumor in the Hypoxic Niche: The Emerging Concept and Associated Therapeutic Strategies. Trends Pharmacol Sci. 2017;38:669-686. [PubMed] [DOI] [Cited in This Article: ] |
| 455. | Fix SM, Papadopoulou V, Velds H, Kasoji SK, Rivera JN, Borden MA, Chang S, Dayton PA. Oxygen microbubbles improve radiotherapy tumor control in a rat fibrosarcoma model - A preliminary study. PLoS One. 2018;13:e0195667. [PubMed] [DOI] [Cited in This Article: ] |
| 456. | Ho YJ, Chu SW, Liao EC, Fan CH, Chan HL, Wei KC, Yeh CK. Normalization of Tumor Vasculature by Oxygen Microbubbles with Ultrasound. Theranostics. 2019;9:7370-7383. [PubMed] [DOI] [Cited in This Article: ] |
| 457. | Wang H, Li J, Wang Y, Gong X, Xu X, Wang J, Li Y, Sha X, Zhang Z. Nanoparticles-mediated reoxygenation strategy relieves tumor hypoxia for enhanced cancer therapy. J Control Release. 2020;319:25-45. [PubMed] [DOI] [Cited in This Article: ] |
| 458. | Qu J, Guo X, Li W, Hou W, Zhang H, Luo L, Zhu C, Yang J, Yin X, Tao Y, Du Y, Lou Y, Chen D, You J. Preparation of artificial red cell and its application on alleviation of tumor hypoxia. Colloids Surf B Biointerfaces. 2017;160:446-454. [PubMed] [DOI] [Cited in This Article: ] |
| 459. | Mazzone M, Dettori D, de Oliveira RL, Loges S, Schmidt T, Jonckx B, Tian YM, Lanahan AA, Pollard P, de Almodovar CR, De Smet F, Vinckier S, Aragonés J, Debackere K, Luttun A, Wyns S, Jordan B, Pisacane A, Gallez B, Lampugnani MG, Dejana E, Simons M, Ratcliffe P, Maxwell P, Carmeliet P. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell. 2009;136:839-851. [PubMed] [DOI] [Cited in This Article: ] |
| 460. | Farnsworth RH, Lackmann M, Achen MG, Stacker SA. Vascular remodeling in cancer. Oncogene. 2014;33:3496-3505. [PubMed] [DOI] [Cited in This Article: ] |
| 461. | Maniwa J, Fumino S, Kimura K, Tanaka T, Higashi M, Kishida T, Mazda O, Tajiri T. Novel mesenchymal stem cell delivery system as targeted therapy against neuroblastoma using the TH-MYCN mouse model. J Pediatr Surg. 2019;54:2600-2605. [PubMed] [DOI] [Cited in This Article: ] |
| 462. | Sharif S, Ghahremani MH, Soleimani M. Differentiation Induction and Proliferation Inhibition by A Cell-Free Approach for Delivery of Exogenous miRNAs to Neuroblastoma Cells Using Mesenchymal Stem Cells. Cell J. 2021;22:556-564. [PubMed] [DOI] [Cited in This Article: ] |
| 463. | Yuan Z, Kolluri KK, Sage EK, Gowers KH, Janes SM. Mesenchymal stromal cell delivery of full-length tumor necrosis factor-related apoptosis-inducing ligand is superior to soluble type for cancer therapy. Cytotherapy. 2015;17:885-896. [PubMed] [DOI] [Cited in This Article: ] |
| 464. | Nieddu V, Piredda R, Bexell D, Barton J, Anderson J, Sebire N, Kolluri K, Janes SM, Karteris E, Sala A. Engineered human mesenchymal stem cells for neuroblastoma therapeutics. Oncol Rep. 2019;42:35-42. [PubMed] [DOI] [Cited in This Article: ] |
| 465. | Chen J, Ji T, Wu D, Jiang S, Zhao J, Lin H, Cai X. Human mesenchymal stem cells promote tumor growth via MAPK pathway and metastasis by epithelial mesenchymal transition and integrin α5 in hepatocellular carcinoma. Cell Death Dis. 2019;10:425. [PubMed] [DOI] [Cited in This Article: ] |
| 466. | Krueger TE, Thorek DLJ, Meeker AK, Isaacs JT, Brennen WN. Tumor-infiltrating mesenchymal stem cells: Drivers of the immunosuppressive tumor microenvironment in prostate cancer? Prostate. 2019;79:320-330. [PubMed] [DOI] [Cited in This Article: ] |
| 467. | Li L, Liu S, Han D, Tang B, Ma J. Delivery and Biosafety of Oncolytic Virotherapy. Front Oncol. 2020;10:475. [PubMed] [DOI] [Cited in This Article: ] |
| 468. | Mahller YY, Williams JP, Baird WH, Mitton B, Grossheim J, Saeki Y, Cancelas JA, Ratner N, Cripe TP. Neuroblastoma cell lines contain pluripotent tumor initiating cells that are susceptible to a targeted oncolytic virus. PLoS One. 2009;4:e4235. [PubMed] [DOI] [Cited in This Article: ] |
| 469. | Pesonen S, Helin H, Nokisalmi P, Escutenaire S, Ribacka C, Sarkioja M, Cerullo V, Guse K, Bauerschmitz G, Laasonen L, Kantola T, Ristimaki A, Rajecki M, Oksanen M, Haavisto E, Kanerva A, Joensuu T, Hemminki A. Oncolytic adenovirus treatment of a patient with refractory neuroblastoma. Acta Oncol. 2010;49:117-119. [PubMed] [DOI] [Cited in This Article: ] |
| 470. | Ramírez M, García-Castro J, Alemany R. Oncolytic virotherapy for neuroblastoma. Discov Med. 2010;10:387-393. [PubMed] [Cited in This Article: ] |
| 471. | Ruano D, López-Martín JA, Moreno L, Lassaletta Á, Bautista F, Andión M, Hernández C, González-Murillo Á, Melen G, Alemany R, Madero L, García-Castro J, Ramírez M. First-in-Human, First-in-Child Trial of Autologous MSCs Carrying the Oncolytic Virus Icovir-5 in Patients with Advanced Tumors. Mol Ther. 2020;28:1033-1042. [PubMed] [DOI] [Cited in This Article: ] |
| 472. | Zhang SC, Cai WS, Zhang Y, Jiang KL, Zhang KR, Wang WL. Engineered measles virus Edmonston strain used as a novel oncolytic viral system against human neuroblastoma through a CD46 and nectin 4-independent pathway. Cancer Lett. 2012;325:227-237. [PubMed] [DOI] [Cited in This Article: ] |
| 473. | Mamaeva V, Rosenholm JM, Bate-Eya LT, Bergman L, Peuhu E, Duchanoy A, Fortelius LE, Landor S, Toivola DM, Lindén M, Sahlgren C. Mesoporous silica nanoparticles as drug delivery systems for targeted inhibition of Notch signaling in cancer. Mol Ther. 2011;19:1538-1546. [PubMed] [DOI] [Cited in This Article: ] |
| 474. | Mamaeva V, Niemi R, Beck M, Özliseli E, Desai D, Landor S, Gronroos T, Kronqvist P, Pettersen IK, McCormack E, Rosenholm JM, Linden M, Sahlgren C. Inhibiting Notch Activity in Breast Cancer Stem Cells by Glucose Functionalized Nanoparticles Carrying γ-secretase Inhibitors. Mol Ther. 2016;24:926-936. [PubMed] [DOI] [Cited in This Article: ] |
| 475. | Wei X, Senanayake TH, Warren G, Vinogradov SV. Hyaluronic acid-based nanogel-drug conjugates with enhanced anticancer activity designed for the targeting of CD44-positive and drug-resistant tumors. Bioconjug Chem. 2013;24:658-668. [PubMed] [DOI] [Cited in This Article: ] |
| 476. | Swaminathan SK, Roger E, Toti U, Niu L, Ohlfest JR, Panyam J. CD133-targeted paclitaxel delivery inhibits local tumor recurrence in a mouse model of breast cancer. J Control Release. 2013;171:280-287. [PubMed] [DOI] [Cited in This Article: ] |
| 477. | Yang C, Xiong F, Dou J, Xue J, Zhan X, Shi F, Li M, Wu S, Luo S, Zhang T, Zhang Y, Ming J, Gu N. Target therapy of multiple myeloma by PTX-NPs and ABCG2 antibody in a mouse xenograft model. Oncotarget. 2015;6:27714-27724. [PubMed] [DOI] [Cited in This Article: ] |
| 478. | Subramanian C, White PT, Kuai R, Kalidindi A, Castle VP, Moon JJ, Timmermann BN, Schwendeman A, Cohen MS. Synthetic high-density lipoprotein nanoconjugate targets neuroblastoma stem cells, blocking migration and self-renewal. Surgery. 2018;. [PubMed] [DOI] [Cited in This Article: ] |
| 479. | Gurunathan S, Jeyaraj M, Kang MH, Kim JH. Mitochondrial Peptide Humanin Protects Silver Nanoparticles-Induced Neurotoxicity in Human Neuroblastoma Cancer Cells (SH-SY5Y). Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] |
| 480. | Rastinehad AR, Anastos H, Wajswol E, Winoker JS, Sfakianos JP, Doppalapudi SK, Carrick MR, Knauer CJ, Taouli B, Lewis SC, Tewari AK, Schwartz JA, Canfield SE, George AK, West JL, Halas NJ. Gold nanoshell-localized photothermal ablation of prostate tumors in a clinical pilot device study. Proc Natl Acad Sci USA. 2019;116:18590-18596. [PubMed] [DOI] [Cited in This Article: ] |