Published online May 26, 2021. doi: 10.4252/wjsc.v13.i5.416
Peer-review started: March 10, 2021
First decision: April 19, 2021
Revised: April 20, 2021
Accepted: April 28, 2021
Article in press: April 28, 2021
Published online: May 26, 2021
Biological reactions require self-assembly of factors in the complex cellular milieu. Recent evidence indicates that intrinsically disordered, low-complexity sequence domains (LCDs) found in regulatory factors mediate diverse cellular processes from gene expression to DNA repair to signal transduction, by enriching specific biomolecules in membraneless compartments or hubs that may undergo liquid-liquid phase separation (LLPS). In this review, we discuss how embryonic stem cells take advantage of LCD-driven interactions to promote cell-specific transcription, DNA damage response, and DNA repair. We propose that LCD-mediated interactions play key roles in stem cell maintenance and safeguarding genome integrity.
Core Tip: This review article discusses recent findings regarding the role of low-complexity domain proteins and phase separation in regulating key cellular processes in embryonic stem cells, including transcriptional activation and repression, cellular signaling integration, DNA damage response, and DNA repair.
- Citation: Vodnala M, Choi EB, Fong YW. Low complexity domains, condensates, and stem cell pluripotency. World J Stem Cells 2021; 13(5): 416-438
- URL: https://www.wjgnet.com/1948-0210/full/v13/i5/416.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i5.416
Embryonic stem cells (ESCs) are derived from pluripotent cells in the inner cell mass of the blastocyst[1,2]. ESCs are highly proliferative cells that can self-renew indefinitely in vitro. In addition to replication stress due to an abbreviated cell cycle[3], it has been shown that ESCs are transcriptionally hyperactive[4]. The increased replication and transcriptional burdens in ESCs promote genome instability[5-8]. Therefore, ESCs are under increased pressure to conduct transcription and DNA repair efficiently to maintain stem cell identity and genome integrity. Deciphering the mechanisms by which ESCs safeguard transcriptional and genomic fidelity is important for understanding pluripotency, and for translating stem cell-based therapies.
In response to developmental signals, ESCs exit from self-renewal and undergo differentiation to generate every cell type in the body. This highly dynamic process requires coordinated changes in gene expression patterns. Genes required for stem cell self-renewal are silenced, while genes encoding developmental regulators that are normally repressed are reactivated to direct the differentiation of ESCs into cell types representing the three embryonic germ layers[9]. Wholesale changes in gene expression are accompanied by reconfiguration of chromatin structure in differentiating ESCs, whereby previously euchromatic regions associated with pluripotency genes are packaged into repressive heterochromatin[10,11]. Conversely, genomic loci associated with lineage-specific genes become euchromatic, thus permissive to transcriptional activation[12].
A fundamental problem in stem cell biology (and cell biology in general) is how complex biochemical reactions (e.g., transcription, DNA replication, repair, chromatin remodeling, and signal transduction) are organized and regulated inside a densely packed cellular space. While specific cellular reactions can be compartmentalized within classic membrane-enclosed organelles such as endoplasmic reticulum and Golgi apparatus, those that occur inside the nucleus present a unique challenge because the nucleus lacks such organelles to spatially and temporally control biological reactions, where inadvertent “mixing” of these reactions could prove fatal to a cell. Indeed, it has been shown that proteins in the nucleus are often enriched in discrete membraneless compartments. For example, factors involved in mRNA splicing are concentrated in the Cajal bodies to facilitate assembly of spliceosomal machinery[13]. Nucleoli are sites of ribosome biogenesis enriched in factors required for ribosomal RNA transcription and processing[14], and were recently identified as a protein quality control compartment[15]. Under specific conditions such as biomolecular concentration, temperature, pH, and salt concentration, biomolecules can coalesce and separate from bulk solution in cells, as condensates reminiscent of oil droplets in water[16-18]. This process, termed liquid-liquid phase separation (LLPS), underlies the formation of membraneless compartments such as the nucleolus and Cajal body[19,20]. Recent work also implicates biomolecular condensates in a wide range of cellular processes, enriching specific macromolecules within distinct compartments and increasing local concentration to overcome activation barriers[13,14,21-23].
In this review, we examine the emerging roles of protein hub formation and condensation in compartmentalizing and coordinating biochemical reactions in the complex nuclear environment. We discuss how protein condensates enhance cellular reactions critical for stem cell function, facilitate crosstalk between cellular processes to generate complex responses to changing cellular environment, and how these responses collectively safeguard stem cell fidelity.
Intrinsically disordered regions (IDRs) are prevalent in eukaryotic proteome, particularly among regulatory proteins such as transcription factors[24]. These unstructured regions are often composed of low-complexity sequences limited in amino acid diversity. Low complexity domains (LCDs) are enriched in glycine, and polar residues such as serine, asparagine, glutamine, and tyrosine. Other IDRs are characterized by clusters of positively and negative charged amino acid (e.g. lysine, glutamic acid) interspersed with hydrophobic residues such as phenylalanine[25]. These unique amino acid compositions found in LCDs have been shown to promote LLPS by polar or charge-charge intermolecular interactions in a concentration dependent manner[25,26]. In addition, the flexible nature of LCDs is thought to facilitate their interaction with multiple protein partners, by rapidly adopting an ensemble of conformations[27,28] Indeed, LCD’s ability to bind multiple proteins, also known as multivalency, is a major driving force of LLPS by lowering threshold concentration[29]. It is worth emphasizing that while LCDs are unstructured sequences, they do not always bind promiscuously to any proteins; instead, they can be selective for binding partners[30-32]. More importantly, because these selective multivalent interactions are usually weak and transient, as opposed to the high affinity (but low valency) “lock-and-key” interactions found in ligand-receptor complexes, they allow dynamic regulation of LLPS properties, condensate composition, and biochemical reactions that take place inside these bodies. In the following sections, we discuss examples wherein LCD-driven interactions play a critical role in regulating cellular processes relevant to ESC biology.
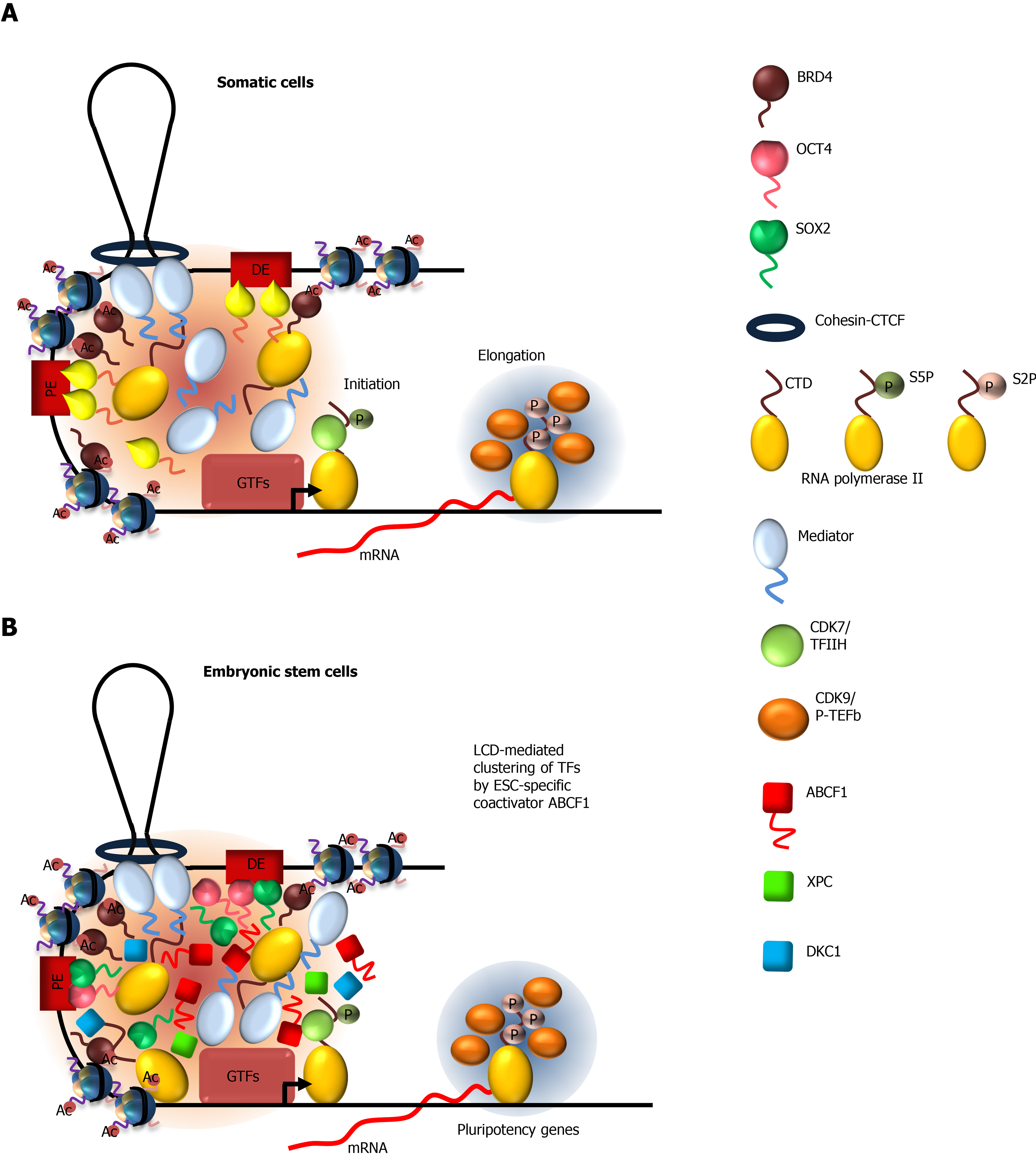
During early embryonic development, pluripotent cells in the inner cell mass of the blastocyst rapidly expand through self-renewal[33]. Buttressing this critical developmental period is a robust gene regulatory network that functions to maintain pluripotency in these cells[34-36]. High transcriptional activity in ESCs has been shown to skew towards genes that encode transcription factors and chromatin remodeling machinery[4], likely as an adaptive measure to meet the increased transcriptional demand. How expression of these factors stabilizes the pluripotent state in ESCs has become apparent through a number of seminal studies. Transcription factors octamer-binding transcription factor 4 (OCT4) and sex-determining region Y-box 2 (SOX2) play a pivotal role in activating stem cell pluripotency[37-42]. Cooperative binding of OCT4 and SOX2 along with a wide array of transcription factors and transcriptional coactivators at gene enhancers lead to the formation of “super enhancers.” Super enhancers differ from typical enhancers by their unusually high density of transcription factors spread over a relatively large genomic region measured in kilobases[43-45]. These transcription factor-rich domains are thought to fuel higher transcriptional output by the RNA polymerase II (Pol II) machinery. The cooperative nature of transcription factor assembly at super enhancers is thought to allow the formation or collapse of super enhancers over a relatively small concentration range of transcription factors[44], and is therefore proposed to play an important role in dynamic gene expression during ESC self-renewal and differentiation. Recent studies on LCDs, which are highly enriched in transcription factors, provide important insights into how these high-density transcription factor hubs are formed to drive cell-specific transcription in ESCs (Table 1) [46-54].
| Ref. | LCD-Factors | LCD-domain | Nature of LCD | Experimentally confirmed | Modifications | |
| In vitro | In vivo | |||||
| Boija et al[46] | MED1 | C-terminus | Polar | Yes | Yes1 | NA |
| Boija et al[46] Brehm et al[47] | OCT4 | N- and C-terminal domains | Acidic amino acids (electrostatic) | Yes | Yes1 | Phosphorylation |
| Boija et al[46] Xue et al[48] | SOX2 | N-terminus | NA | Yes | NA | NA |
| Boija et al[46] Metallo et al[49] | c-MYC | Entire polypeptide | NA | Yes | NA | NA |
| Boija et al[46] Jenkins et al[50]; Oldfield et al[51] | p53 | N- and C-terminal domains | Acidic amino acids (N-terminus) | Yes | NA | Phosphorylation |
| Boija et al[46] Xue et al[48] | NANOG | N- and C-terminal domains | NA | Yes | NA | NA |
| Boija et al[46] | ER | NA | NA | Yes | NA | NA |
| Choi et al[30] | ABCF1 | N-terminus | NA | Yes | Yes2 | NA |
| Boehning et al[52] | RNA Pol II | C-terminus | Hydrophobic and electrostatic | Yes | Yes1 | Phosphorylation |
| Lau et al[53] | HP1α | N-terminus | Acidic amino acids | Yes | Yes1 | Phosphorylation |
| Plys et al[54] | CBX2 | Entire polypeptide | Positively charged | Yes | Yes2 | Phosphorylation |
The ubiquitous transcriptional coactivator Mediator is a large, multisubunit complex that is required for transcription of most Pol II genes, by virtue of its ability to interact with a wide array of transcription factors and Pol II[55,56]. Mediator stimulates transcription by functionally and physically connecting transcription factors at enhancers to the Pol II machinery at promoters[57], where distal enhancers are brought to proximity to their target promoters through DNA looping by cohesion-CTCF (CCCTC-binding factor)[58,59] (Figure 1A). Small hairpin RNA-mediated screens indicated that downregulation of subunits of the Mediator complex compromises expression of OCT4/SOX2-dependent genes in mouse ESCs[60]. Consistent with its role as a coactivator for OCT4/SOX2, Mediator colocalizes extensively with OCT4 and SOX2 across the ESC genome[61].
The mediator complex subunit 1 (MED1) of the Mediator complex contains an LCD at the C-terminus that is rich in serine residues[62]. Studies have shown that MED1 LCD and Mediator holocomplex undergo LLPS in vitro. Substitution of serine residues in MED1 with alanine abolishes phase separation, indicating the importance of serine-mediated polar intermolecular interactions in LLPS. To examine the mechanism by which Mediator interacts with OCT4 and SOX2, in vitro droplet assays showed that MED1 LLPS droplets readily incorporate OCT4 and SOX2[46]. Furthermore, mutations of acidic amino acids in the activation domain of OCT4, which abrogate transactivation activity, also compromise its ability to phase separate with MED1. These observations indicate a functional correlation between MED1-OCT4 LLPS and transcriptional activation, and suggest LCD-dependent phase separation as a potential mechanism by which activator-coactivator complexes are assembled at gene enhancers (Figure 1B). It is worth noting that diverse transcription factors (e.g. p53, myelocytomatosis viral oncogene homolog, NANOG, estrogen receptor) can also phase separate with MED1 in vitro[46]. These results demonstrate that the LCD of MED1 is rather promiscuous in binding, consistent with Mediator acting as a ubiquitous coactivator.
Biochemical studies demonstrated that Mediator interacts with RNA Pol II through the carboxy-terminal domain (CTD) of the largest subunit of Pol II complex[63,64]. Mammalian CTD contains 52 heptad repeats of the consensus sequence Y1S2P3T4S5P6S7. This LCD plays important roles at all steps of transcription from initiation to elongation to termination[65]. Initiation requires the assembly of the preinitiation complex (PIC), composed of general transcription factors (GTFs), Mediator, and Pol II with unphosphorylated CTD, at gene promoters[66,67]. As Pol II leaves the promoter and initiates transcription, the CTD becomes phosphorylated on serine 5 (Ser5) by GTF TFIIH-associated kinase, cyclin dependent kinase 7 (CDK7)[68-71]. It is known that Mediator interacts preferentially with the unphosphorylated CTD[72,73] (Figure 1A). This suggests that phosphorylation of Pol II CTD may disrupt its interaction with Mediator, thus providing a mechanism by which Pol II can dissociate from PIC to initiate transcription. Two recent studies support this notion and implicated LLPS in regulating Mediator-Pol II interaction[32,52]. They demonstrated that the ability of the CTD to undergo LLPS by itself, or with MED1, is disrupted by phosphorylation of Ser5 by CDK7. Therefore, Mediator-Pol II interaction and promoter-enhancer communication can be modulated by phosphorylation status of the CTD during the transcription cycle.
After Pol II escapes the promoter, the CTD becomes hyperphosphorylated at Ser2 by CDK9 of the positive transcription elongation factor b (P-TEFb), while Ser5 previously phosphorylated by TFIIH is gradually removed by phosphatases[74-76]. This switch in Ser phosphorylation pattern is thought to promote elongation by aiding the recruitment of elongation and chromatin-modifying factors to the transcribing Pol II[75] (Figure 1A). A recent study indicated that the histidine-rich LCD of the cyclin T1 subunit of P-TEFb (a heterodimer of CDK9 and cyclin T1) stabilizes the binding of P-TEFb to active genes and to the Pol II CTD to catalyze Ser2 hyperphosphorylation[77]. They showed that cyclin T1 forms liquid-like puncta in the nucleus in an LCD-dependent manner. Formation of these nuclear condensates and the ability of P-TEFb to hyperphosphorylate the CTD are sensitive to 1,6-hexanediol that disrupts LLPS. Consistent with these observations, the LCD is also required for phase separation of cyclin T1 with CTD in vitro. Interestingly, pre-phosphorylation of the CTD by CDK7/TFIIH significantly enhances cyclin T1-CTD LLPS, suggesting that a potential function of Ser5 phosphorylation by TFIIH after promoter escape is to prime LLPS of P-TEFb with the CTD, thereby increasing the efficiency of Ser2 phosphorylation. Taken together, these observations underscore the role of LCD-mediated interactions in regulating transition from transcriptional initiation to elongation.
Bromodomain-containing protein 4 (BRD4) is a critical transcriptional and epigenetic regulator in ESCs[78-80]. It contains two bromodomains that recognize acetylated lysines on histone H3 and H4 that are associated with active gene promoters[81]. BRD4 also acts as a scaffold for recruiting P-TEFb and chromatin remodeling proteins to facilitate transcription by Pol II[78,82,83]. BRD4 has been shown to colocalize with Mediator at super enhancers that control genes important for stem cell identity[62] (Figure 1B). BRD4 contains an LCD at its C-terminus with high proline and glutamine content. Studies showed that BRD4 LCD by itself can form LLPS droplets in vitro and can be incorporated into MED1 condensates. These results suggest that LLPS between Mediator and BRD4 represents a mechanism by which they are concentrated at super enhancers[62,84]. This is supported by the observation that treatment of cells with 1,6-hexanediol reduced their occupancy at enhancers. It would be interesting to examine whether binding of BRD4 to acetylated nucleosomal DNA promotes its LLPS with Mediator, due to increased valency (i.e., cooperativity) in interactions by BRD4[85]. While the mechanism by which BRD4 recruits P-TEFb to gene promoters is unknown, it is tempting to speculate that their interaction could be promoted by their respective LCD.
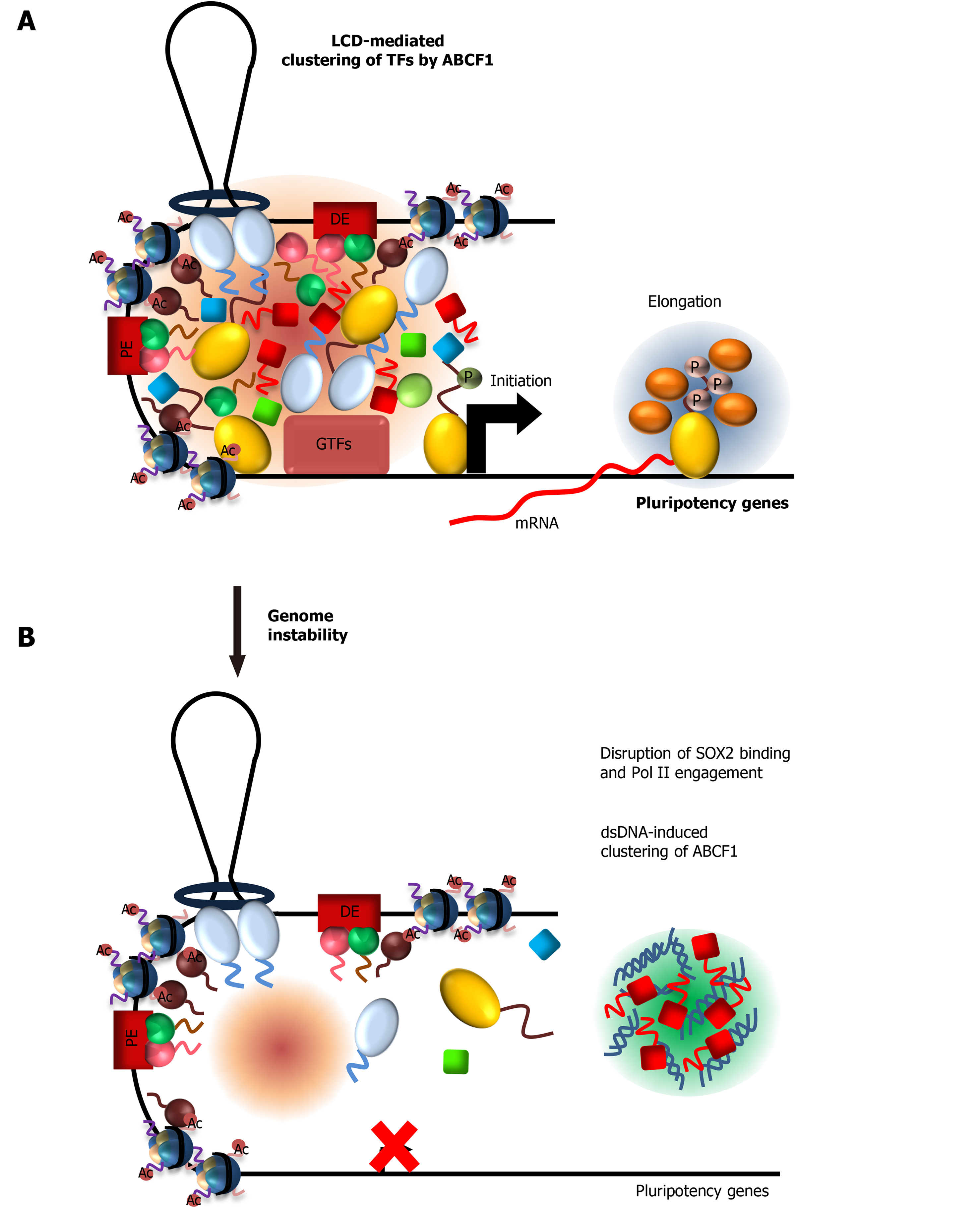
Most if not all of the regulatory factors described thus far are utilized by many transcription factors to activate their target genes in both ESCs and somatic cells. Our work and others indicated that robust transcriptional activation by OCT4 and SOX2 in ESCs requires additional coactivators that are distinct from Mediator[30,86-88]. Using a fully reconstituted in vitro transcription assay, we detected multiple novel coactivators that work in concert with OCT4 and SOX2 to activate pluripotency gene transcription. Biochemical purification of these coactivators led to the discovery of three stem cell-specific coactivators - the nucleotide excision repair protein xeroderma pigmentosum, complementation group C (XPC)[87-90], dyskerin (DKC1) ribonucleoprotein complex[86], and the ATP-binding cassette subfamily F member 1 (ABCF1)[30] (Figure 1B). We found that the ability of XPC and DKC1 to stimulate OCT4/SOX2-activated transcription is strongly dependent on ABCF1, indicating a pivotal role of ABCF1 in mediating stem cell-specific transcription.
ABCF1 contains an LCD at the N-terminus that is unusually rich in charged amino acids, of which about 40% are divided between lysine and glutamic acid residues. These clusters of positively and negatively charged amino acid, interspersed with hydrophobic residues such as phenylalanine, are known to promote LLPS[91,92]. Indeed, we showed that ABCF1 undergoes LLPS in an LCD-dependent manner. More importantly, the LCD is also required for transcriptional activity in vitro and in ESCs, due to its ability to selectively interact with SOX2 (but not OCT4), its co-dependent coactivators XPC and DKC1 as well as Pol II. These LCD-driven interactions are also detected at OCT4/SOX2-target gene enhancers and are sensitive to disruption by 1,6-hexanediol treatment. It is worth noting that the conformationally flexible XPC protein also contains several highly disordered regions that we found, however, to be dispensable for transcriptional activation[87-90]. These observations revealed the unique ability of ABCF1 LCD to integrate multiple lines of information encoded by SOX2, XPC, DKC1, and the Pol II machinery, likely by forming a hub of these factors at target gene promoters through selective multivalent interactions (Figure 1B). In summary, cell type-specific transcriptional activation in ESCs requires an interconnected network of LCD-driven interactions by both general and cell-specific coactivators for optimal and gene-specific transcriptional activation.
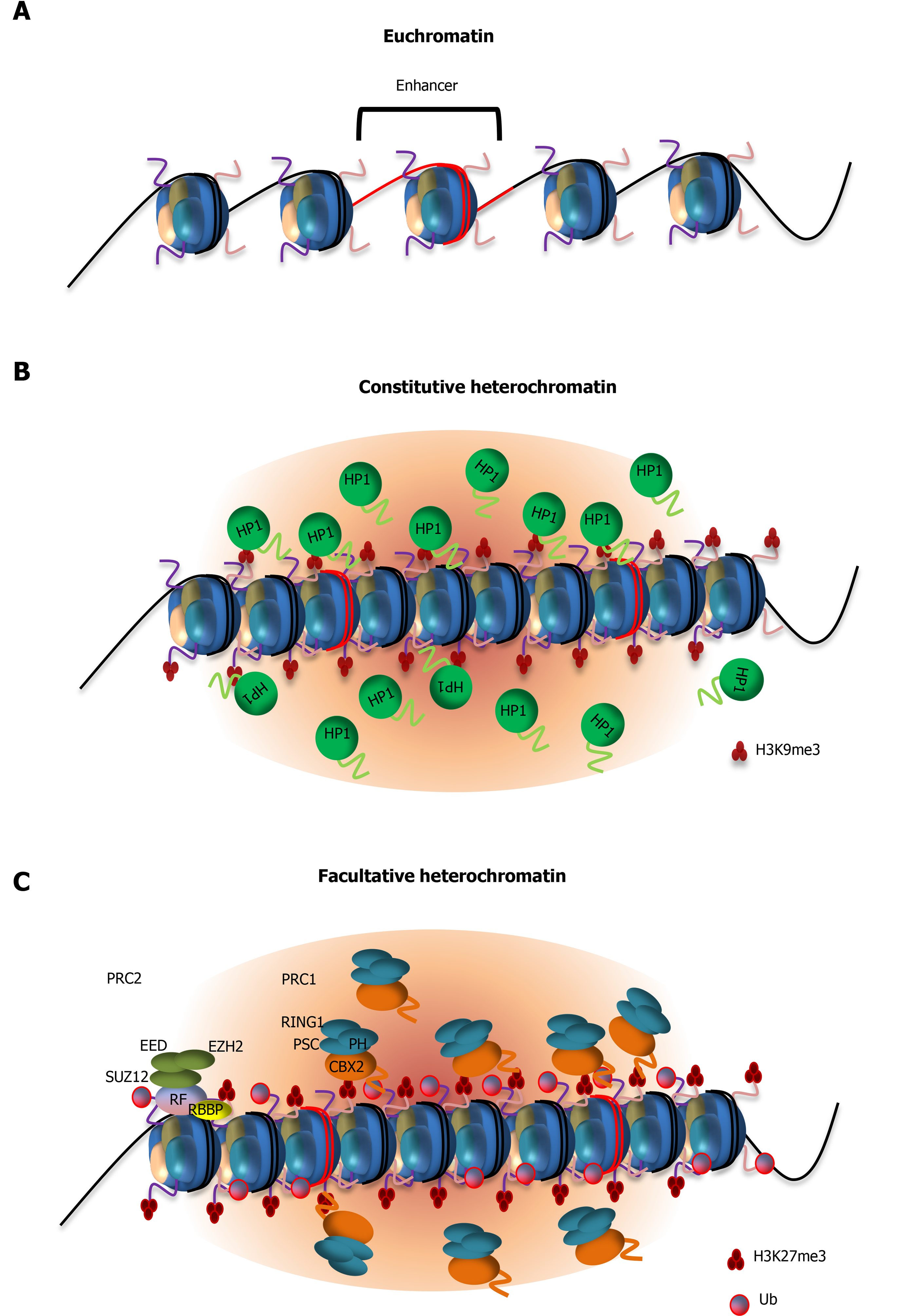
During stem cell self-renewal, developmental genes must be properly silenced. Failure to repress these genomic regions compromises stem cell identity and pluripotency of ESCs[93-96]. Studies have shown that heterochromatin is essential for silencing the autosomal imprinted genomic loci, HOX gene clusters and other differentiation-associated genes[97,98]. Heterochromatic regions are characterized by hypoacetylated histones and repressive modifications such as trimethylated histone H3 Lysine 9 (H3K9me3), trimethylated histone H3 Lysine 27 (H3K27me3), and mono-ubiquitination of histone H2A lysine 119 (H2AK119ub)[99-103]. These modifications not only control nucleosomal interactions but also regulate the association of non-histone chromosomal proteins that together influence nucleosomal packaging and gene repression. For example, heterochromatic regions are established and protected by chromatin components and trans-acting factors such as heterochromatin protein 1 (HP1) and Polycomb repressive complexes 1 and 2 (PRC1, PRC2)[104] (Table 1). Understanding how histone binding proteins and histone modifying enzymes are assembled at heterochromatin will elucidate the mechanisms by which a repressed chromatin state is initiated and maintained to silence developmental genes during stem cell self-renewal, and how these heterochromatic regions are decondensed to facilitate their reactivation when ESCs undergo differentiation (Figure 2A). The highly compact heterochromatin structure has led to a number of studies that evoke LLPS for heterochromatin domain formation.
Compaction of chromatin is a key process in maintaining the repressed state of heterochromatin. HP1 recognizes H3K9me3 modifications through its chromo shadow domain and nucleates chromatin condensation[105,106]. Underscoring a direct role of HP1 in chromatin condensation, artificial targeting of HP1 to a genomic locus is sufficient to cause local condensation and formation of high-order chromatin structure[107]. In mammals, HP1 exists in three isoforms: HP1α, β and γ. HP1α is commonly associated with silenced heterochromatic regions, while the other two isoforms appear to have both gene silencing and activating functions[108-111]. These HP1 proteins possess three LCDs (LCD1, 2, and 3). Interaction between LCD1 and LCD2 has been shown to contribute to multivalent interactions with nucleo
The recruitment of PRC1 complexes to chromatin drives nucleosome compaction and transcriptional silencing[10,103,120,121]. This is mediated by the chromobox 2 (CBX2) subunit of PRC1, which recognizes H3K27me3 that is deposited by histone methyltransferase Enhancer of zeste homolog 2 subunit of the PRC2 complex[122,123]. Once PRC1 is recruited to H3K27me3, it monoubiquitinates H2A at lysine 119 (H2AK119ub), which is essential for maintaining gene repression in ESCs[124]. It has long been observed that PRC1 complexes form concentrated nuclear compartments known as Polycomb bodies[54,125]. Recent studies indicated that CBX2 is responsible for PRC1 LLPS and chromatin compaction[54] (Figure 2C). CBX2 is a low-complexity disordered protein containing a serine-rich patch and positively charged amino acid rich region. It has been shown that phosphorylation of serine residues by casein kinase 2 enhances CBX2 LLPS in vitro, likely by facilitating electrostatic intermolecular interactions between phosphorylated serines and positively charged lysines. Consistent with this hypothesis, mutation of 23 Lysine and arginine residues to alanine abolishes CBX2 LLPS in vitro. Importantly, lysine to alanine substitutions in CBX2 result in axial patterning defects in mice, indicating altered Hox gene expression patterns during development[53]. Thus, these results support a functional link between CBX2 LLPS and gene silencing. Taken together, these studies suggest a role of LLPS in gene repression through LCD-driven chromatin condensation, and in the proper reactivation of developmental genes in a spatially and temporally regulated manner. It appears that LLPS may play a role in concentrating factors that are critical for chromatin compaction and maintenance of the repressed chromatin state, and in excluding factors that would otherwise gain access to these repressed domains and interfere with gene silencing[115].
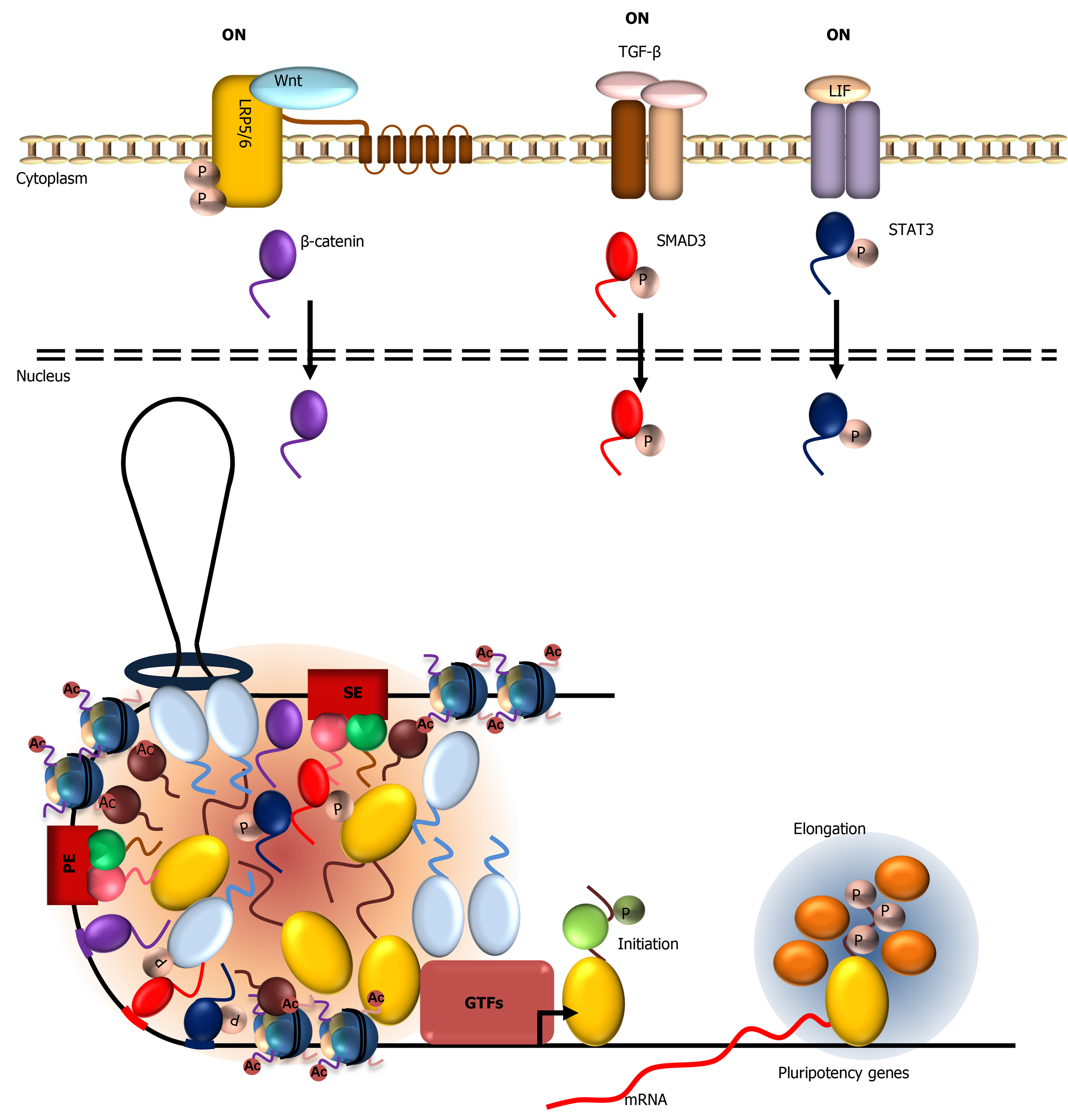
In mouse ESCs, Hippo/Yes-associated protein (YAP)/transcriptional coactivator with PDZ-binding motif (TAZ), Janus kinase (JAK)/signal transducer and activator of transcription (STAT), Wingless-related integration site (Wnt)/β-catenin, and transforming growth factor beta (TGF-β) pathways play important roles in supporting stem cell self-renewal and pluripotency[126-130]. How ESCs integrate and interpret these signals and generate an appropriate transcriptional response to these cues are key to understanding fundamental mechanism governing self-renewal vs differentiation cell fate decision.
The Hippo pathway controls cell proliferation and survival by regulating the activity of YAP, a transcriptional coactivator for transcriptional enhancer factors (TEFs)[131-133]. The Hippo pathway regulates YAP activity primarily by controlling its nucleocytoplasmic shuttling through phosphorylation. Activation of the Hippo pathway by signals derived from cell-cell contact, mechanosensing (i.e., substrate stiffness), and cellular stress inhibits YAP by phosphorylation at serine 127, leading to its sequestration in the cytoplasm. When Hippo signaling is inactivated, YAP translocates to the nucleus and stimulates TEF-activated transcription by forming complexes with Mediator and BRD4/P-TEFb[134,135]. Other studies added complexity to this model, by showing that hyperosmotic stress also activates nemo-like kinase, which leads to YAP phosphorylation at serine 128 and, unexpectedly, translocation to the nucleus and activation of YAP-dependent genes, despite simultaneous phosphorylation at serine 127 by the Hippo pathway[136,137].
YAP is enriched in pluripotent ESCs but its level significantly decreases upon differentiation and is further inactivated by phosphorylation at serine 127[127]. YAP supports stem cell maintenance by binding to key pluripotency-associated genes such as Nanog, Oct4, and Sox2 and regulate their expression. How YAP stimulates the transcription of these genes was unknown but recent studies implicated phase separation of YAP and its paralogue TAZ as a key mechanism. In one study, YAP was shown to form liquid-like condensates with TAZ and TEF in the nucleus upon hyperosmotic stress[138]. In another study, TAZ but not YAP was shown to undergo LLPS when the Hippo pathway is inhibited, even though YAP and TAZ show extensive sequence similarities[139]. Formation of TAZ condensates in cells is regulated by Hippo pathway, where signals that promote nuclear retention of TAZ induce the formation of nuclear puncta that colocalize with Pol II, BRD4, MED1 and CDK9/P-TEFb, indicating that these condensates likely represent transcriptionally active compartments. Protein domain swapping experiments demonstrated that the ww and coiled-coil (cc) domains of TAZ (but not YAP) contribute to LLPS. This result is in contrast to studies by Cai et al[138] showing that YAP can in fact phase separate in vitro. Differences in protein preparation, concentration, and in vitro droplet formation assay condition may explain the apparent discrepancy. Nevertheless, both studies demonstrated that the ability of YAP or TAZ to activate its target genes requires their LCDs, suggesting that transcriptional activation by TEF is facilitated by LCD-mediated interaction with YAP/TAZ.
Master transcription factors such as OCT4 and SOX2 define ESC identity in part by integrating extracellular signals at gene enhancers to drive cell-specific transcription. It has been shown that terminal effectors of the Wnt, TGF-β, and JAK/STAT signaling pathways, β-catenin, small mothers against decapentaplegics (SMADs), and STAT3, respectively, converge onto cell-specific super enhancers[140]. How these enhancers “hijack” signal-regulated transcription factors are not well-understood. A recent study showed that these signaling effectors synergize with OCT4, SOX2, and Mediator by forming transcription condensates at super enhancers[141]. Upon activation of the signaling pathways, β-catenin, SMADs, and STAT3 translocate to the nucleus and form condensates at super enhancer at the Nanog locus in mouse ESCs (Figure 3). By contrast, activation of Wnt signaling was not sufficient to target β-catenin to the transcriptionally silenced Nanog locus in the muscle cell line C2C12. These results indicate that recruitment of β-catenin to Nanog enhancer likely requires open chromatin, active transcription, and presence of other transcription factors bound at active enhancers. Perhaps the high density of transcription factors and abundance of LCD-mediated multivalent interactions at super enhancers promote efficient concentration of signal-dependent transcription factors. Indeed, β-catenin, SMADs, and STAT3 were shown to form condensates with Mediator in vitro through their LCDs. Mutations that disrupt β-catenin LLPS also compromise recruitment to its target gene enhancers and transcriptional activation, supporting a functional correlation between LLPS propensity and transcription factor recruitment and gene activation. Compartmentalization of these signaling effectors not only concentrates these factors at the appropriate enhancers but may also insulate these factors from activating the wrong targets. These LCD-dependent multivalent interactions at enhancers likely permit dynamic regulation of transcription - a key feature of regulated gene expression in response to extracellular signaling.
Unlike terminally differentiated somatic cells, the fast replication rate of ESCs makes them prone to replication stress-induced DNA damage such as double strand breaks (DSBs)[142-144]. At the same time, high proliferation rate poses significant challenge to DNA repair because DNA lesions that are left unrepaired prior to cell division will be inherited by daughter stem cells and then propagated to their progenitors, likely leading to deleterious effect in development[145]. Therefore, ESCs are under increased pressure to efficiently and accurately repair DNA damages. Indeed, it has been shown that ESCs express higher levels of DNA repair factors and favor high fidelity DSB repair by homologous recombination (HR)[28]. It has also been shown that ESCs are hypersensitive to DNA damage and readily undergo spontaneous differentiation and apoptosis[145,146]. This is likely a fail-safe mechanism by eliminating compromised ESCs from the self-renewing population. In the following sections, we will examine the role of LCD and LLPS in DNA repair and DNA damage response (DDR) and discuss how they safeguard stem cell genome integrity.
The abundance of repetitive sequences in heterochromatin poses unique challenges to DNA repair due to increased risks of aberrant recombination induced by DSBs, which can lead to deletion, duplication, and translocation[147]. Cells have developed elaborate mechanisms to promote efficient and error-free DNA repair by taking advantage of LLPS. Upon DSB, it has been shown that phosphorylation of threonine 51 in HP1 Leads to dissociation of HP1 from heterochromatin, as evidenced by loss of binding to H3K9me3 and dispersal of HP1 nuclear puncta[115]. Dissociation of HP1 Likely alters LLPS status at DSBs, which in turn facilitates chromatin relaxation and engagement of downstream effectors to initiate DNA repair[148]. In another study using Drosophila as a model system, phase separated heterochromatic domain at DSBs appears to be able to exclude repair factors such as Ku80 that are involved in error-prone non-homologous end joining (NHEJ), and enrich factors required for the initial steps of HR repair[149]. It was proposed that such exclusion mechanism favors repair by error-free HR, assuming that HR repair factors can still efficiently assess the damaged site. It will be interesting to test whether this exclusion mechanism is mediated by selective interaction between HP1 condensates and repair factors in HR but not NHEJ pathway.
Expansion of the HP1-organized heterochromatin domain is also thought to facilitate the physical relocation of the DSB DNAs to the nuclear periphery in Drosophila or to the heterochromatin domain periphery in mouse cells, locations that are believed to be more conducive to repair by HR[150,151]. Studies demonstrated that physical movement of heterochromatic DSBs depends on polymerization of F-actin and mobilization of DSB DNAs by tethering the DNA and ‘walking’ along the F-actin filaments by myosins[150]. It has been shown that F-actin crosslinked by filamin spontaneously assembled into phase-separated F-actin filament bundles that can extend and contract[152]. We speculate that changes in actin filament dynamics driven by phase separation could facilitate the relocation of heterochromatic DSBs to appropriate subcellular compartments as DSB repair progresses.
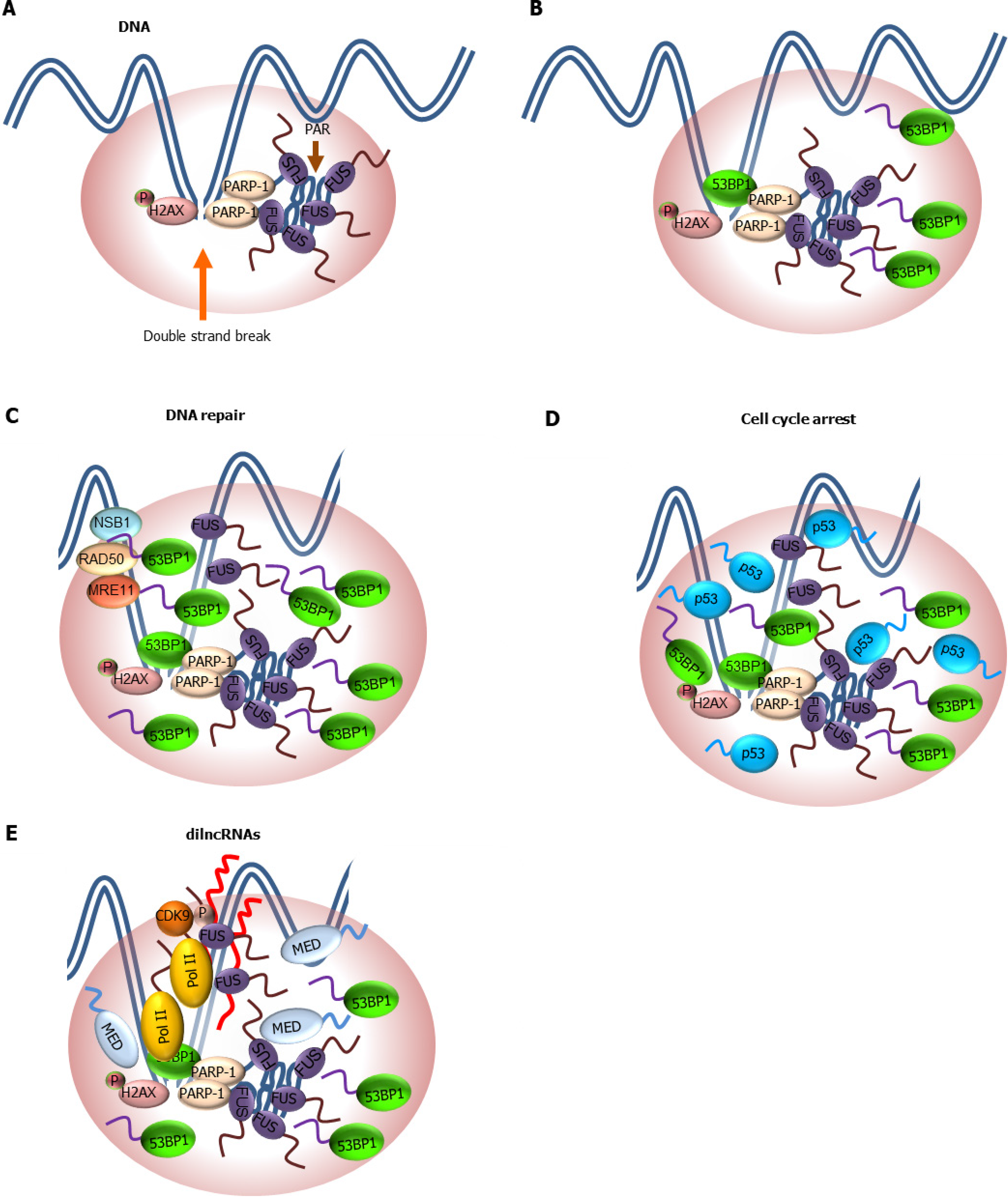
Fused in sarcoma, phosphorylation, and poly-adenosine diphosphate (ADP)-ribosylation: Fused in sarcoma (FUS, also known as translocated in liposarcoma, TLS) is one of the most studied proteins known to undergo phase separation. Its unstructured N-terminal prion-like domain is required for phase separation[153-155]. In addition to its role in RNA metabolism, recent studies highlighted a role of FUS in DDR. Upon DNA damage, FUS is rapidly recruited to DSB sites[156,157]. It has been shown that poly-ADP-ribosylation (PARylation) at DSBs by poly-ADP-ribose polymerase enzymes triggers the translocation of cytoplasmic FUS to the nucleus and formation of large phase separated FUS-containing compartments at DSB sites[158-160] (Figure 4A). These FUS compartments are thought to contribute to DNA repair by facilitating the recruitment of downstream effectors of DNA repair such as p53-binding protein 1 (53BP1)[161] (Figure 4B). The C-terminal arginine-glycine-glycine repeat (RGG) domain of FUS likely plays a role in LLPS by directly binding PAR[159]. Therefore, the high propensity of FUS to generate large phase separated domains in cells could be due to increased valency in interaction using both the N-terminal prion-like LCD as well as C-terminal RGG domain when PAR accumulates at DSBs. It is worth to stress that these phase separated compartments are not static structures. Indeed, it has been shown that multivalent interactions by FUS can be destabilized by phosphorylation of the prion-like LCD[162] and PAR hydrolysis by PARG[158]. The reversible nature of FUS LLPS compartments is likely a necessary feature of DNA repair where recruitment of repair factors to and exclusion from damaged sites must be dynamically regulated.
53BP1: In addition to PARylation at DSBs, phosphorylation of histone variant 2AX is another early event in DDR and is required for the recruitment of 53BP1 to DSB sites[163] (Figure 4B). 53BP1 has been shown to generate sizeable chromatin domains in the nucleus that persist throughout the repair process[164] and is thought to recruit downstream effectors to regulate DDR and repair (Figure 4C)[163]. Recent studies demonstrated that these 53BP1 domains display liquid-like properties[164,165]. In one study, it showed that 53BP1 can concentrate p53 into 53BP1 condensates and activates p53-target gene expression, thereby inducing a cell cycle checkpoint DDR[164] (Figure 4D). They showed that conditions that perturb 53BP1 condensate formation also compromise p53 signaling, indicating that the recruitment of p53 to 53BP1 condensates is likely important for proper activation of a p53 response in damaged cells. It will be interesting to examine whether 53BP1-dependent activation of p53 contributes to repression of pluripotency genes and activation of differentiation-associated genes observed in damaged ESCs[166]. Surprisingly, while 53BP1 contains a largely unstructured N-terminal domain, it is dispensable for LLPS in vitro[164]. Rather, the structured Tudor domain is required for phase separation by 53BP1. It is speculated that multivalent interactions between tyrosines (Y) and arginines (R) in Tudor domain promote LLPS, similar to what was observed regarding the role of Y/R in phase separation of the FET (FUS, Ewing sarcoma breakpoint region 1, TATA-box binding protein associated factor 15) protein family[167]. Future mutagenesis studies should help clarify the LLPS mechanism employed by 53BP1. The ability of 53BP1 Tudor domain to undergo LLPS demonstrated that structured domains can also contribute to phase separation. Another study highlighted the involvement of damaged-induced long non-coding RNA (dilncRNA) at DSBs in organizing 53BP1 condensates[165]. They showed that PIC assembly at DSBs containing Pol II, Mediator, and P-TEFb, and transcription of dilncRNAs facilitate molecular crowding and phase separation of DDR factors including 53BP1 (Figure 4E). Supporting this notion, inhibition of dilncRNA transcription reduces the size 53BP1 condensates and repair efficiency. Given that FUS binds RNA[168] and phase separates with Pol II CTD[155], it is tempting to speculate that transcription of dilncRNAs by Pol II at DSBs may also facilitate the incorporation of FUS into repair condensates.
ABCF1 and intracellular DNA sensing: ABCF1 was previously identified as a sensor for intracellular DNAs that arise from infection or DNA damage[169]. Binding of these DNAs by ABCF1 triggers an innate immune response in somatic cells. However, because ESCs lack a canonical innate immune response to DNAs[170-172], the functional consequence of DNA sensing by ABCF1 in ESCs is unknown. Our identification of ABCF1 as a critical stem cell coactivator prompted us to examine whether ABCF1 can couple DNA sensing with stem cell transcription in response to DNA damage[30] (Figure 5A). We found that ABCF1 specifically binds double-stranded (ds) but not single-stranded (ss) DNAs in an LCD-dependent manner. Remarkably, binding of ABCF1 to dsDNAs dramatically stimulates LLPS in vitro. These results suggest that upon DNA damage, ABCF1 may preferentially form condensates with dsDNAs in damaged ESCs instead of binding SOX2 and Pol II. Consistent with this model, we found that ABCF1’s interaction with SOX2 and assembly of Pol II transcription machinery at pluripotency gene promoters are disrupted upon DNA damage, resulting in downregulation of pluripotency genes critical for stem cell maintenance (Figure 5B). We propose that ESCs may leverage ABCF1’s ability to switch between transcription factor and dsDNA condensates to modulate pluripotency gene transcription. Direct coupling of DNA sensing and stem cell-specific transcription via ABCF1 may represent an effective strategy to safeguard genome integrity by eliminating compromised ESCs from the self-renewing population through enforced differentiation.
A growing number of factors have been shown to form condensates with the MED1 subunit of the Mediator complex. Less clear are the mechanisms by which MED1 forms these numerous, functionally distinct condensates. Changes in their composition upon signaling pathway activation, and at different stages of gene transcription where “cargoes” are handed off from one condensate (e.g. initiation) to another (e.g. elongation) must be tightly regulated. A key challenge is how to avoid accidental mixing of these MED1 condensates. Post-translational modifications of the CTD of Pol II provide one such strategy wherein different phosphorylated forms of the CTD (Ser5 vs Ser2) condense preferentially with regulatory factors in initiation or elongation. In addition, we propose that the requirement of coactivators such as ABCF1 to form stem cell-specific multivalent interactions adds another layer of specificity for gene regulation in ESCs.
Evidence of LLPS in cells, particularly with respect to transcription factors, relies in part on observations of their phase separation behaviors in vitro, that they are spherical in shape, can fuse and fission, and allow exchange of biomolecules. However, these properties are not unique to LLPS. Indeed, a recent study on Pol II compartment formation during herpes simplex virus type 1 infection highlighted that, despite sharing several properties that are consistent with phase separated condensates, these Pol II compartments are formed by non-specific interactions with viral genomic DNA, distinct from behaviors typically attributed to Pol II condensates[173]. In another study, it was shown that at physiological concentration, TFs activate Pol II transcription at endogenous genomic loci by forming dynamic LCD-driven hubs in the absence of LLPS[31]. Therefore, there are likely multiple pathways with which clustering of biomolecules in cells can be achieved without undergoing LLPS. In fact, a recent study provides evidence that formation of transcription factor droplets can actually be counterproductive to gene activation[174], suggesting that the topology and binding dynamics of multivalent interactions are critical for protein function in transcription and likely other cellular processes. For discussion on the role of phase separation in biological reactions, we recommend several excellent reviews on evidence for and against LLPS in cells[23,175-177].
Whether or not these LCD-driven domains in cells meet the criteria of LLPS, it is evident that an intricate network of multivalent interactions controls various steps in transcription, their integration with signaling pathways, and in DNA repair and DDR -processes essential for maintenance of stem cell pluripotency and genome integrity. Transient and weak protein-protein and protein-nucleic acid interactions mediated by LCDs in regulatory factors enhance efficiency of biological reactions by enriching relevant factors in distinct hubs or compartments, specificity by combinatorial assembly, and dynamic regulation in response to changing cellular environment by modulating LCD-LCD interaction affinity and specificity.
We apologize to colleagues whose work was not mentioned in this manuscript, due to the limited scope of this review. The authors thank Agarwal S, Chong S, and Zhang Z for valuable discussion.
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chen H, Jiang W S-Editor: Liu M L-Editor: A P-Editor: Xing YX
| 1. | Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5956] [Cited by in F6Publishing: 5283] [Article Influence: 122.9] [Reference Citation Analysis (0)] |
| 2. | Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634-7638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3879] [Cited by in F6Publishing: 3474] [Article Influence: 80.8] [Reference Citation Analysis (0)] |
| 3. | Kapinas K, Grandy R, Ghule P, Medina R, Becker K, Pardee A, Zaidi SK, Lian J, Stein J, van Wijnen A, Stein G. The abbreviated pluripotent cell cycle. J Cell Physiol. 2013;228:9-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 4. | Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, Gingeras TR, Misteli T, Meshorer E. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437-447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 506] [Cited by in F6Publishing: 496] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 5. | Fong YW, Cattoglio C, Tjian R. The intertwined roles of transcription and repair proteins. Mol Cell. 2013;52:291-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Burhans WC, Weinberger M. DNA replication stress, genome instability and aging. Nucleic Acids Res. 2007;35:7545-7556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 7. | Carr AM, Lambert S. Replication stress-induced genome instability: the dark side of replication maintenance by homologous recombination. J Mol Biol. 2013;425:4733-4744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 133] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 8. | Marnef A, Cohen S, Legube G. Transcription-Coupled DNA Double-Strand Break Repair: Active Genes Need Special Care. J Mol Biol. 2017;429:1277-1288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 9. | Pekkanen-Mattila M, Pelto-Huikko M, Kujala V, Suuronen R, Skottman H, Aalto-Setälä K, Kerkelä E. Spatial and temporal expression pattern of germ layer markers during human embryonic stem cell differentiation in embryoid bodies. Histochem Cell Biol. 2010;133:595-606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 10. | Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 188] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 11. | Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol. 2008;20:201-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 12. | Golob JL, Paige SL, Muskheli V, Pabon L, Murry CE. Chromatin remodeling during mouse and human embryonic stem cell differentiation. Dev Dyn. 2008;237:1389-1398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Morris GE. The Cajal body. Biochim Biophys Acta. 2008;1783:2108-2115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 14. | Yao RW, Xu G, Wang Y, Shan L, Luan PF, Wu M, Yang LZ, Xing YH, Yang L, Chen LL. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Mol Cell 2019; 76: 767-783. e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 15. | Frottin F, Schueder F, Tiwary S, Gupta R, Körner R, Schlichthaerle T, Cox J, Jungmann R, Hartl FU, Hipp MS. The nucleolus functions as a phase-separated protein quality control compartment. Science. 2019;365:342-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 286] [Article Influence: 57.2] [Reference Citation Analysis (0)] |
| 16. | André AAM, Spruijt E. Liquid-Liquid Phase Separation in Crowded Environments. Int J Mol Sci. 2020;21:5908. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 114] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 17. | Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L, Tompa P, Fuxreiter M. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018;28:420-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1138] [Cited by in F6Publishing: 1144] [Article Influence: 190.7] [Reference Citation Analysis (0)] |
| 18. | Posey AE, Holehouse AS, Pappu RV. Phase Separation of Intrinsically Disordered Proteins. Methods Enzymol. 2018;611:1-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 19. | Lafontaine DLJ. Birth of Nucleolar Compartments: Phase Separation-Driven Ribosomal RNA Sorting and Processing. Mol Cell. 2019;76:694-696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165:1686-1697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1059] [Cited by in F6Publishing: 1150] [Article Influence: 143.8] [Reference Citation Analysis (0)] |
| 21. | Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2487] [Cited by in F6Publishing: 2970] [Article Influence: 424.3] [Reference Citation Analysis (0)] |
| 22. | Tosolini D, Antoniali G, Dalla E, Tell G. Is there a role of phase partitioning in coordinating DNA damage response? Biomol Concepts. 2020;11:209-220. [DOI] [Cited in This Article: ] |
| 23. | Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A Phase Separation Model for Transcriptional Control. Cell. 2017;169:13-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1013] [Cited by in F6Publishing: 1031] [Article Influence: 147.3] [Reference Citation Analysis (0)] |
| 24. | Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359:1137-1149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 193] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 25. | Bianchi G, Longhi S, Grandori R, Brocca S. Relevance of Electrostatic Charges in Compactness, Aggregation, and Phase Separation of Intrinsically Disordered Proteins. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 26. | Gomes E, Shorter J. The molecular language of membraneless organelles. J Biol Chem. 2019;294:7115-7127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 405] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 27. | Zhang Z, Tjian R. Measuring dynamics of eukaryotic transcription initiation: Challenges, insights and opportunities. Transcription. 2018;9:159-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Choi UB, Sanabria H, Smirnova T, Bowen ME, Weninger KR. Spontaneous Switching among Conformational Ensembles in Intrinsically Disordered Proteins. Biomolecules. 2019;9:114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Feng Z, Chen X, Wu X, Zhang M. Formation of biological condensates via phase separation: Characteristics, analytical methods, and physiological implications. J Biol Chem. 2019;294:14823-14835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 121] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 30. | Choi EB, Vodnala M, Zerbato M, Wang J, Ho J, Inouye C, Fong Y. ATP-binding cassette protein ABCF1 couples gene transcription with maintenance of genome integrity in embryonic stem cells. bioRxiv. 2020;Preprint. [DOI] [Cited in This Article: ] |
| 31. | Chong S, Dugast-Darzacq C, Liu Z, Dong P, Dailey GM, Cattoglio C, Heckert A, Banala S, Lavis L, Darzacq X, Tjian R. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 732] [Cited by in F6Publishing: 588] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 32. | Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall'Agnese A, Hannett NM, Spille JH, Afeyan LK, Zamudio AV, Shrinivas K, Abraham BJ, Boija A, Decker TM, Rimel JK, Fant CB, Lee TI, Cisse II, Sharp PA, Taatjes DJ, Young RA. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543-548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 377] [Article Influence: 75.4] [Reference Citation Analysis (0)] |
| 33. | Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701-713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 461] [Cited by in F6Publishing: 428] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 34. | Li M, Izpisua Belmonte JC. Deconstructing the pluripotency gene regulatory network. Nat Cell Biol. 2018;20:382-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 35. | Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940-954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 872] [Cited by in F6Publishing: 857] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 36. | Fong YW, Cattoglio C, Yamaguchi T, Tjian R. Transcriptional regulation by coactivators in embryonic stem cells. Trends Cell Biol. 2012;22:292-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 37. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17152] [Article Influence: 952.9] [Reference Citation Analysis (0)] |
| 38. | Dunn SJ, Martello G, Yordanov B, Emmott S, Smith AG. Defining an essential transcription factor program for naïve pluripotency. Science. 2014;344:1156-1160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 265] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 39. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7589] [Cited by in F6Publishing: 7028] [Article Influence: 413.4] [Reference Citation Analysis (0)] |
| 40. | Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 594] [Cited by in F6Publishing: 664] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 41. | Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567-582. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1088] [Cited by in F6Publishing: 988] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 42. | Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, Bar-Nur O, Cheloufi S, Stadtfeld M, Figueroa ME, Robinton D, Natesan S, Melnick A, Zhu J, Ramaswamy S, Hochedlinger K. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617-1632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 651] [Cited by in F6Publishing: 642] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 43. | Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 451] [Cited by in F6Publishing: 462] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 44. | Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2538] [Cited by in F6Publishing: 2669] [Article Influence: 242.6] [Reference Citation Analysis (0)] |
| 45. | Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1873] [Cited by in F6Publishing: 2041] [Article Influence: 185.5] [Reference Citation Analysis (0)] |
| 46. | Boija A, Klein IA, Sabari BR, Dall'Agnese A, Coffey EL, Zamudio AV, Li CH, Shrinivas K, Manteiga JC, Hannett NM, Abraham BJ, Afeyan LK, Guo YE, Rimel JK, Fant CB, Schuijers J, Lee TI, Taatjes DJ, Young RA. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018; 175: 1842-1855. e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1066] [Cited by in F6Publishing: 952] [Article Influence: 158.7] [Reference Citation Analysis (0)] |
| 47. | Brehm A, Ohbo K, Schöler H. The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol Cell Biol. 1997;17:154-162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 80] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 48. | Xue B, Oldfield CJ, Van YY, Dunker AK, Uversky VN. Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst. 2012;8:134-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 49. | Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr Opin Chem Biol. 2010;14:481-488. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 220] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 50. | Jenkins LM, Durell SR, Mazur SJ, Appella E. p53 N-terminal phosphorylation: a defining layer of complex regulation. Carcinogenesis. 2012;33:1441-1449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 83] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 51. | Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9 Suppl 1:S1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 406] [Cited by in F6Publishing: 438] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 52. | Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, McSwiggen DT, Kokic G, Dailey GM, Cramer P, Darzacq X, Zweckstetter M. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol. 2018;25:833-840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 345] [Cited by in F6Publishing: 359] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 53. | Lau MS, Schwartz MG, Kundu S, Savol AJ, Wang PI, Marr SK, Grau DJ, Schorderet P, Sadreyev RI, Tabin CJ, Kingston RE. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science. 2017;355:1081-1084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 54. | Plys AJ, Davis CP, Kim J, Rizki G, Keenen MM, Marr SK, Kingston RE. Phase separation of Polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019;33:799-813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 213] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 55. | Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010;35:315-322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 253] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 56. | Kim YJ, Björklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 815] [Cited by in F6Publishing: 836] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 57. | El Khattabi L, Zhao H, Kalchschmidt J, Young N, Jung S, Van Blerkom P, Kieffer-Kwon P, Kieffer-Kwon KR, Park S, Wang X, Krebs J, Tripathi S, Sakabe N, Sobreira DR, Huang SC, Rao SSP, Pruett N, Chauss D, Sadler E, Lopez A, Nóbrega MA, Aiden EL, Asturias FJ, Casellas R. A Pliable Mediator Acts as a Functional Rather Than an Architectural Bridge between Promoters and Enhancers. Cell 2019; 178: 1145-1158. e20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 58. | Hansen AS, Hsieh TS, Cattoglio C, Pustova I, Saldaña-Meyer R, Reinberg D, Darzacq X, Tjian R. Distinct Classes of Chromatin Loops Revealed by Deletion of an RNA-Binding Region in CTCF. Mol Cell 2019; 76: 395-411. e13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 142] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 59. | Thiecke MJ, Wutz G, Muhar M, Tang W, Bevan S, Malysheva V, Stocsits R, Neumann T, Zuber J, Fraser P, Schoenfelder S, Peters JM, Spivakov M. Cohesin-Dependent and -Independent Mechanisms Mediate Chromosomal Contacts between Promoters and Enhancers. Cell Rep. 2020;32:107929. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 83] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 60. | Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1582] [Cited by in F6Publishing: 1421] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 61. | Göke J, Jung M, Behrens S, Chavez L, O'Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS Comput Biol. 2011;7:e1002304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 62. | Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1495] [Cited by in F6Publishing: 1356] [Article Influence: 226.0] [Reference Citation Analysis (0)] |
| 63. | Näär AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 2002;16:1339-1344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 127] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 64. | Robinson PJ, Trnka MJ, Bushnell DA, Davis RE, Mattei PJ, Burlingame AL, Kornberg RD. Structure of a Complete Mediator-RNA Polymerase II Pre-Initiation Complex. Cell 2016; 166: 1411-1422. e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 164] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 65. | Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119-2137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 452] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 66. | Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551-2569. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 544] [Cited by in F6Publishing: 532] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 67. | Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922-2936. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 556] [Cited by in F6Publishing: 552] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 68. | Dahmus ME. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171-182. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 101] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 69. | Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol Cell Biol. 2009;29:5455-5464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 243] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 70. | Larochelle S, Amat R, Glover-Cutter K, Sansó M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol. 2012;19:1108-1115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 287] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 71. | Rimel JK, Taatjes DJ. The essential and multifunctional TFIIH complex. Protein Sci. 2018;27:1018-1037. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 72. | Søgaard TM, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113-14120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 93] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 73. | Svejstrup JQ, Li Y, Fellows J, Gnatt A, Bjorklund S, Kornberg RD. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075-6078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 100] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 74. | Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 451] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 75. | Harlen KM, Churchman LS. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol. 2017;18:263-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 280] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 76. | Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483-508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 301] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 77. | Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, Zhou Q. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature. 2018;558:318-323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 404] [Cited by in F6Publishing: 347] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 78. | Liu W, Stein P, Cheng X, Yang W, Shao NY, Morrisey EE, Schultz RM, You J. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 2014;21:1950-1960. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 79. | Wu T, Pinto HB, Kamikawa YF, Donohoe ME. The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Reports. 2015;4:390-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 58] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 80. | Finley LWS, Vardhana SA, Carey BW, Alonso-Curbelo D, Koche R, Chen Y, Wen D, King B, Radler MR, Rafii S, Lowe SW, Allis CD, Thompson CB. Pluripotency transcription factors and Tet1/2 maintain Brd4-independent stem cell identity. Nat Cell Biol. 2018;20:565-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 81. | Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA. 2003;100:8758-8763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 502] [Cited by in F6Publishing: 508] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 82. | Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 801] [Cited by in F6Publishing: 838] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 83. | Wu T, Kamikawa YF, Donohoe ME. Brd4's Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1. Cell Rep. 2018;25:1756-1771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 84. | Han X, Yu D, Gu R, Jia Y, Wang Q, Jaganathan A, Yang X, Yu M, Babault N, Zhao C, Yi H, Zhang Q, Zhou MM, Zeng L. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat Struct Mol Biol. 2020;27:333-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 85. | Miller TC, Simon B, Rybin V, Grötsch H, Curtet S, Khochbin S, Carlomagno T, Müller CW. A bromodomain-DNA interaction facilitates acetylation-dependent bivalent nucleosome recognition by the BET protein BRDT. Nat Commun. 2016;7:13855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 78] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 86. | Fong YW, Ho JJ, Inouye C, Tjian R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. Elife. 2014;3:e03573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 87. | Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147:120-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 133] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 88. | Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731-24737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 777] [Cited by in F6Publishing: 810] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 89. | Cattoglio C, Zhang ET, Grubisic I, Chiba K, Fong YW, Tjian R. Functional and mechanistic studies of XPC DNA-repair complex as transcriptional coactivator in embryonic stem cells. Proc Natl Acad Sci USA. 2015;112:E2317-E2326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 90. | Zhang ET, He Y, Grob P, Fong YW, Nogales E, Tjian R. Architecture of the human XPC DNA repair and stem cell coactivator complex. Proc Natl Acad Sci USA. 2015;112:14817-14822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 91. | Pak CW, Kosno M, Holehouse AS, Padrick SB, Mittal A, Ali R, Yunus AA, Liu DR, Pappu RV, Rosen MK. Sequence Determinants of Intracellular Phase Separation by Complex Coacervation of a Disordered Protein. Mol Cell. 2016;63:72-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 625] [Cited by in F6Publishing: 494] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 92. | Homma K, Fukuchi S, Nishikawa K, Sakamoto S, Sugawara H. Intrinsically disordered regions have specific functions in mitochondrial and nuclear proteins. Mol Biosyst. 2012;8:247-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 93. | Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525-2534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 228] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 94. | Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061-4071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 664] [Cited by in F6Publishing: 675] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 95. | Schumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;384:648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 62] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 96. | Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci USA. 2003;100:2468-2473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 270] [Cited by in F6Publishing: 264] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 97. | Gentile C, Kmita M. Polycomb Repressive Complexes in Hox Gene Regulation: Silencing and Beyond: The Functional Dynamics of Polycomb Repressive Complexes in Hox Gene Regulation. Bioessays. 2020;42:e1900249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 98. | Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 981] [Cited by in F6Publishing: 1007] [Article Influence: 59.2] [Reference Citation Analysis (0)] |
| 99. | Tamburri S, Lavarone E, Fernández-Pérez D, Conway E, Zanotti M, Manganaro D, Pasini D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol Cell 2020; 77: 840-856. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 100. | Chaturvedi CP, Somasundaram B, Singh K, Carpenedo RL, Stanford WL, Dilworth FJ, Brand M. Maintenance of gene silencing by the coordinate action of the H3K9 methyltransferase G9a/KMT1C and the H3K4 demethylase Jarid1a/KDM5A. Proc Natl Acad Sci USA. 2012;109:18845-18850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 101. | Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664-678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 472] [Cited by in F6Publishing: 426] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 102. | Peixoto P, Castronovo V, Matheus N, Polese C, Peulen O, Gonzalez A, Boxus M, Verdin E, Thiry M, Dequiedt F, Mottet D. HDAC5 is required for maintenance of pericentric heterochromatin, and controls cell-cycle progression and survival of human cancer cells. Cell Death Differ. 2012;19:1239-1252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 103. | Schoenfelder S, Sugar R, Dimond A, Javierre BM, Armstrong H, Mifsud B, Dimitrova E, Matheson L, Tavares-Cadete F, Furlan-Magaril M, Segonds-Pichon A, Jurkowski W, Wingett SW, Tabbada K, Andrews S, Herman B, LeProust E, Osborne CS, Koseki H, Fraser P, Luscombe NM, Elderkin S. Polycomb repressive complex PRC1 spatially constrains the mouse embryonic stem cell genome. Nat Genet. 2015;47:1179-1186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 320] [Cited by in F6Publishing: 255] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 104. | Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 342] [Cited by in F6Publishing: 331] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 105. | Canzio D, Chang EY, Shankar S, Kuchenbecker KM, Simon MD, Madhani HD, Narlikar GJ, Al-Sady B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 210] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 106. | Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080-2083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 640] [Cited by in F6Publishing: 635] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 107. | Verschure PJ, van der Kraan I, de Leeuw W, van der Vlag J, Carpenter AE, Belmont AS, van Driel R. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol Cell Biol. 2005;25:4552-4564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 143] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 108. | Smallwood A, Hon GC, Jin F, Henry RE, Espinosa JM, Ren B. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22:1426-1436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 109. | Billur M, Bartunik HD, Singh PB. The essential function of HP1 beta: a case of the tail wagging the dog? Trends Biochem Sci. 2010;35:115-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 110. | Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014;24:377-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 111. | Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1γamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381-391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 534] [Cited by in F6Publishing: 533] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 112. | Zhao T, Heyduk T, Allis CD, Eissenberg JC. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J Biol Chem. 2000;275:28332-28338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 86] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 113. | Meehan RR, Kao CF, Pennings S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 2003;22:3164-3174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 110] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 114. | Munari F, Soeroes S, Zenn HM, Schomburg A, Kost N, Schröder S, Klingberg R, Rezaei-Ghaleh N, Stützer A, Gelato KA, Walla PJ, Becker S, Schwarzer D, Zimmermann B, Fischle W, Zweckstetter M. Methylation of lysine 9 in histone H3 directs alternative modes of highly dynamic interaction of heterochromatin protein hHP1β with the nucleosome. J Biol Chem. 2012;287:33756-33765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 115. | Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, Narlikar GJ. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1023] [Cited by in F6Publishing: 1066] [Article Influence: 152.3] [Reference Citation Analysis (0)] |
| 116. | Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X, Karpen GH. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1421] [Cited by in F6Publishing: 1128] [Article Influence: 161.1] [Reference Citation Analysis (0)] |
| 117. | Bosch-Presegué L, Raurell-Vila H, Thackray JK, González J, Casal C, Kane-Goldsmith N, Vizoso M, Brown JP, Gómez A, Ausió J, Zimmermann T, Esteller M, Schotta G, Singh PB, Serrano L, Vaquero A. Mammalian HP1 Isoforms Have Specific Roles in Heterochromatin Structure and Organization. Cell Rep. 2017;21:2048-2057. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 118. | Musselman CA, Kutateladze TG. Characterization of functional disordered regions within chromatin-associated proteins. iScience. 2021;24:102070. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 119. | Erdel F, Rademacher A, Vlijm R, Tünnermann J, Frank L, Weinmann R, Schweigert E, Yserentant K, Hummert J, Bauer C, Schumacher S, Al Alwash A, Normand C, Herten DP, Engelhardt J, Rippe K. Mouse Heterochromatin Adopts Digital Compaction States without Showing Hallmarks of HP1-Driven Liquid-Liquid Phase Separation. Mol Cell 2020; 78: 236-249. e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 170] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 120. | Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 240] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 121. | Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697-708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 902] [Cited by in F6Publishing: 1000] [Article Influence: 66.7] [Reference Citation Analysis (0)] |
| 122. | Laugesen A, Højfeldt JW, Helin K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol Cell. 2019;74:8-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 342] [Cited by in F6Publishing: 327] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 123. | Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718-2731. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 214] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 124. | Moussa HF, Bsteh D, Yelagandula R, Pribitzer C, Stecher K, Bartalska K, Michetti L, Wang J, Zepeda-Martinez JA, Elling U, Stuckey JI, James LI, Frye SV, Bell O. Canonical PRC1 controls sequence-independent propagation of Polycomb-mediated gene silencing. Nat Commun. 2019;10:1931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 43] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 125. | Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887-898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 228] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 126. | Tamm C, Böwer N, Annerén C. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD2 signaling pathway downstream of LIF. J Cell Sci. 2011;124:1136-1144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 165] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 127. | Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LS, Abujarour R, Ding S, Guan KL. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106-1118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 569] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 128. | Merrill BJ. Wnt pathway regulation of embryonic stem cell self-renewal. Cold Spring Harb Perspect Biol. 2012;4:a007971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 129. | Theka I, Sottile F, Cammisa M, Bonnin S, Sanchez-Delgado M, Di Vicino U, Neguembor MV, Arumugam K, Aulicino F, Monk D, Riccio A, Cosma MP. Wnt/β-catenin signaling pathway safeguards epigenetic stability and homeostasis of mouse embryonic stem cells. Sci Rep. 2019;9:948. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 130. | Mullen AC, Wrana JL. TGF-β Family Signaling in Embryonic and Somatic Stem-Cell Renewal and Differentiation. Cold Spring Harb Perspect Biol. 2017;9:a022186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 131. | Hong W, Guan KL. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785-793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 349] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 132. | Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schübeler D, Bauer A. YAP1 Exerts Its Transcriptional Control via TEAD-Mediated Activation of Enhancers. PLoS Genet. 2015;11:e1005465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 265] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 133. | Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229-1241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 520] [Cited by in F6Publishing: 540] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 134. | Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, Calogero RA, Camargo FD. YAP Drives Growth by Controlling Transcriptional Pause Release from Dynamic Enhancers. Mol Cell. 2015;60:328-337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 231] [Cited by in F6Publishing: 205] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 135. | Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, Quaranta E, Di Biagio D, Sigismondo G, Guzzardo V, Lejeune P, Haendler B, Krijgsveld J, Fassan M, Bicciato S, Cordenonsi M, Piccolo S. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med. 2018;24:1599-1610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 205] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 136. | Hong AW, Meng Z, Yuan HX, Plouffe SW, Moon S, Kim W, Jho EH, Guan KL. Osmotic stress-induced phosphorylation by NLK at Ser128 activates YAP. EMBO Rep. 2017;18:72-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 102] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 137. | Moon S, Kim W, Kim S, Kim Y, Song Y, Bilousov O, Kim J, Lee T, Cha B, Kim M, Kim H, Katanaev VL, Jho EH. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18:61-71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 138. | Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N, Sukenik S, Liu Z, Lippincott-Schwartz J. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol. 2019;21:1578-1589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 206] [Article Influence: 41.2] [Reference Citation Analysis (0)] |
| 139. | Lu Y, Wu T, Gutman O, Lu H, Zhou Q, Henis YI, Luo K. Phase separation of TAZ compartmentalizes the transcription machinery to promote gene expression. Nat Cell Biol. 2020;22:453-464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 184] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 140. | Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2647] [Cited by in F6Publishing: 2392] [Article Influence: 217.5] [Reference Citation Analysis (0)] |
| 141. | Zamudio AV, Dall'Agnese A, Henninger JE, Manteiga JC, Afeyan LK, Hannett NM, Coffey EL, Li CH, Oksuz O, Sabari BR, Boija A, Klein IA, Hawken SW, Spille JH, Decker TM, Cisse II, Abraham BJ, Lee TI, Taatjes DJ, Schuijers J, Young RA. Mediator Condensates Localize Signaling Factors to Key Cell Identity Genes. Mol Cell 2019; 76: 753-766. e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 142. | Atashpaz S, Samadi Shams S, Gonzalez JM, Sebestyén E, Arghavanifard N, Gnocchi A, Albers E, Minardi S, Faga G, Soffientini P, Allievi E, Cancila V, Bachi A, Fernández-Capetillo Ó, Tripodo C, Ferrari F, López-Contreras AJ, Costanzo V. ATR expands embryonic stem cell fate potential in response to replication stress. Elife. 2020;9:e54756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 143. | Técher H, Koundrioukoff S, Nicolas A, Debatisse M. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat Rev Genet. 2017;18:535-550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 170] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 144. | P. Maintaining genomic stability in pluripotent stem cells. Genome Instab Dis. 2020;1:92-97. [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 145. | Heyer BS, MacAuley A, Behrendtsen O, Werb Z. Hypersensitivity to DNA damage leads to increased apoptosis during early mouse development. Genes Dev. 2000;14:2072-2084. [PubMed] [Cited in This Article: ] |
| 146. | Roos WP, Christmann M, Fraser ST, Kaina B. Mouse embryonic stem cells are hypersensitive to apoptosis triggered by the DNA damage O(6)-methylguanine due to high E2F1 regulated mismatch repair. Cell Death Differ. 2007;14:1422-1432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 147. | Rawal CC, Caridi CP, Chiolo I. Actin' between phase separated domains for heterochromatin repair. DNA Repair (Amst). 2019;81:102646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 148. | Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 308] [Cited by in F6Publishing: 320] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 149. | Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1α domain to complete recombinational repair. Cell. 2011;144:732-744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 396] [Article Influence: 30.5] [Reference Citation Analysis (0)] |
| 150. | Caridi CP, Plessner M, Grosse R, Chiolo I. Nuclear actin filaments in DNA repair dynamics. Nat Cell Biol. 2019;21:1068-1077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 151. | Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401-1411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 173] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 152. | Weirich KL, Banerjee S, Dasbiswas K, Witten TA, Vaikuntanathan S, Gardel ML. Liquid behavior of cross-linked actin bundles. Proc Natl Acad Sci USA. 2017;114:2131-2136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 153. | Sukhanova MV, Singatulina AS, Pastré D, Lavrik OI. Fused in Sarcoma (FUS) in DNA Repair: Tango with Poly(ADP-ribose) Polymerase 1 and Compartmentalisation of Damaged DNA. Int J Mol Sci. 2020;21:7020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 154. | Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell. 2013;155:1049-1060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 390] [Cited by in F6Publishing: 406] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 155. | Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1439] [Cited by in F6Publishing: 1441] [Article Influence: 120.1] [Reference Citation Analysis (0)] |
| 156. | Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731-24741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 177] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 157. | Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gómez-Herreros F, Hafezparast M, Caldecott KW. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307-314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 158. | Singatulina AS, Hamon L, Sukhanova MV, Desforges B, Joshi V, Bouhss A, Lavrik OI, Pastré D. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep 2019; 27: 1809-1821. e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 159. | Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grøfte M, Rask MD, Streicher W, Jungmichel S, Nielsen ML, Lukas J. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun. 2015;6:8088. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 373] [Cited by in F6Publishing: 390] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 160. | Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, Pozniakovski A, Poser I, Maghelli N, Royer LA, Weigert M, Myers EW, Grill S, Drechsel D, Hyman AA, Alberti S. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 2015;162:1066-1077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1585] [Cited by in F6Publishing: 1739] [Article Influence: 193.2] [Reference Citation Analysis (0)] |
| 161. | Levone BR, Lenzken SC, Antonaci M, Maiser A, Rapp A, Conte F, Reber S, Ronchi AE, Mühlemann O, Leonhardt H, Cardoso MC, Ruepp M, Barabino SML. FUS-dependent liquid-liquid phase separation is an early event in double-strand break repair. 2020 Preprint. Available from: preLights. [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 68] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 162. | Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH, O'Meally R, Dignon GL, Conicella AE, Zheng W, Best RB, Cole RN, Mittal J, Shewmaker F, Fawzi NL. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951-2967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 406] [Cited by in F6Publishing: 441] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 163. | Mirman Z, de Lange T. 53BP1: a DSB escort. Genes Dev. 2020;34:7-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 139] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 164. | Kilic S, Lezaja A, Gatti M, Bianco E, Michelena J, Imhof R, Altmeyer M. Phase separation of 53BP1 determines liquid-like behavior of DNA repair compartments. EMBO J. 2019;38:e101379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 253] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 165. | Pessina F, Giavazzi F, Yin Y, Gioia U, Vitelli V, Galbiati A, Barozzi S, Garre M, Oldani A, Flaus A, Cerbino R, Parazzoli D, Rothenberg E, d'Adda di Fagagna F. Functional transcription promoters at DNA double-strand breaks mediate RNA-driven phase separation of damage-response factors. Nat Cell Biol. 2019;21:1286-1299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 197] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 166. | Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol Cell. 2012;46:30-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 167. | Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D, Poser I, Pappu RV, Alberti S, Hyman AA. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018; 174: 688-699. e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 890] [Cited by in F6Publishing: 1057] [Article Influence: 176.2] [Reference Citation Analysis (0)] |
| 168. | Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 147] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 169. | Lee MN, Roy M, Ong SE, Mertins P, Villani AC, Li W, Dotiwala F, Sen J, Doench JG, Orzalli MH, Kramnik I, Knipe DM, Lieberman J, Carr SA, Hacohen N. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol. 2013;14:179-185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 170. | Wu X, Dao Thi VL, Huang Y, Billerbeck E, Saha D, Hoffmann HH, Wang Y, Silva LAV, Sarbanes S, Sun T, Andrus L, Yu Y, Quirk C, Li M, MacDonald MR, Schneider WM, An X, Rosenberg BR, Rice CM. Intrinsic Immunity Shapes Viral Resistance of Stem Cells. Cell 2018; 172: 423-438. e25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 226] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 171. | Wang R, Wang J, Paul AM, Acharya D, Bai F, Huang F, Guo YL. Mouse embryonic stem cells are deficient in type I interferon expression in response to viral infections and double-stranded RNA. J Biol Chem. 2013;288:15926-15936. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 172. | D'Angelo W, Gurung C, Acharya D, Chen B, Ortolano N, Gama V, Bai F, Guo YL. The Molecular Basis for the Lack of Inflammatory Responses in Mouse Embryonic Stem Cells and Their Differentiated Cells. J Immunol. 2017;198:2147-2155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 173. | McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, Heckert AB, Umemoto KK, Dugast-Darzacq C, Tjian R, Darzacq X. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 182] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 174. | Trojanowski J, Frank L, Rademacher A, Grigaitis P, Rippe K. Transcription activation is enhanced by multivalent interactions independent of liquid-liquid phase separation. bioRxiv. 2021;Preprint. [DOI] [Cited in This Article: ] |
| 175. | McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev. 2019;33:1619-1634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 422] [Cited by in F6Publishing: 361] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 176. | A P, Weber SC. Evidence for and against Liquid-Liquid Phase Separation in the Nucleus. Noncoding RNA. 2019;5:50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 177. | Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176:419-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1048] [Cited by in F6Publishing: 1336] [Article Influence: 267.2] [Reference Citation Analysis (0)] |
| 178. | van Mierlo G, Veenstra GJC, Vermeulen M, Marks H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019;29:660-671. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 142] [Article Influence: 28.4] [Reference Citation Analysis (0)] |