Published online Oct 26, 2021. doi: 10.4252/wjsc.v13.i10.1595
Peer-review started: April 26, 2021
First decision: May 12, 2021
Revised: June 14, 2021
Accepted: August 23, 2021
Article in press: August 23, 2021
Published online: October 26, 2021
Senescence is characterized by a decline in hepatocyte function, with impairment of metabolism and regenerative capacity. Several models that duplicate liver functions in vitro are essential tools for studying drug metabolism, liver diseases, and organ regeneration. The human HepaRG cell line represents an effective model for the study of liver metabolism and hepatic progenitors. However, the impact of senescence on HepaRG cells is not yet known.
To characterize the effects of senescence on the transdifferentiation capacity and mitochondrial metabolism of human HepaRG cells.
We compared the transdifferentiation capacity of cells over 10 (passage 10 [P10]) vs P20. Aging was evaluated by senescence-associated (SA) beta-galactosidase activity and the comet assay. HepaRG transdifferentiation was analyzed by confocal microscopy and flow cytometry (expression of cluster of differentiation 49a [CD49a], CD49f, CD184, epithelial cell adhesion molecule [EpCAM], and cytokeratin 19 [CK19]), quantitative PCR analysis (expression of albumin, cytochrome P450 3A4 [CYP3A4], γ-glutamyl transpeptidase [γ-GT], and carcinoembryonic antigen [CEA]), and functional analyses (albumin secretion, CYP3A4, and γ-GT). Mitochondrial respiration and the ATP and nicotinamide adenine dinucleotide (NAD+)/NAD with hydrogen (NADH) content were also measured.
SA β-galactosidase staining was higher in P20 than P10 HepaRG cells; in parallel, the comet assay showed consistent DNA damage in P20 HepaRG cells. With respect to P10, P20 HepaRG cells exhibited a reduction of CD49a, CD49f, CD184, EpCAM, and CK19 after the induction of transdifferentiation. Furthermore, lower gene expression of albumin, CYP3A4, and γ-GT, as well as reduced albumin secretion capacity, CYP3A4, and γ-GT activity were reported in transdifferentiated P20 compared to P10 cells. By contrast, the gene expression level of CEA was not reduced by transdifferentiation in P20 cells. Of note, both cellular and mitochondrial oxygen consumption was lower in P20 than in P10 transdifferentiated cells. Finally, both ATP and NAD+/NADH were depleted in P20 cells with respect to P10 cells.
SA mitochondrial dysfunction may limit the transdifferentiation potential of HepaRG cells, with consequent impairment of metabolic and regenerative properties, which may alter applications in basic studies.
Core Tip: The human HepaRG cell line represents an effective model for the study of liver metabolism and hepatic progenitors. However, the impact of senescence on HepaRG cells is not known. We characterized the effects of senescence on the transdifferentiation capacity and mitochondrial metabolism of HepaRG cells. By using a replication protocol, we described higher senescence-associated markers and lower transdifferentiation markers in passage 20 (P20) than in P10 cells. Cellular and mitochondrial oxygen consumption, and ATP and nicotinamide adenine dinucleotide (NAD+)/NAD with hydrogen (NADH) content were lower in P20 than in P10 transdifferentiated cells. To conclude, senescence-associated mitochondrial dysfunction may limit the transdifferentiation potential of HepaRG cells.
- Citation: Bellanti F, di Bello G, Tamborra R, Amatruda M, Lo Buglio A, Dobrakowski M, Kasperczyk A, Kasperczyk S, Serviddio G, Vendemiale G. Impact of senescence on the transdifferentiation process of human hepatic progenitor-like cells. World J Stem Cells 2021; 13(10): 1595-1609
- URL: https://www.wjgnet.com/1948-0210/full/v13/i10/1595.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i10.1595
Primary human hepatocytes are the gold standard to study the biology, pharmacology, and toxicology of parenchymal liver cells[1]. Nevertheless, primary hepatocytes present with several limitations such as difficult isolation procedures, high variability between different donors, quick failure in biological function, and proliferation capacity[2]. To overcome these limitations, several human hepatoma cell lines were developed and characterized to provide a steady and unrestricted supply of hepatocyte-like cells. Of these, HepG2 and Huh-7 cells are the most widely used. HepG2 cells originated from an American patient[3], while Huh-7 were isolated from a Japanese patient[4], both affected by well-differentiated hepatocellular carcinoma. Even though these cell lines exhibit several hepatic functions, low expression of enzymes and cytochromes limit their use for metabolism and toxicity studies[5].
To study the metabolism, toxicology, and regeneration/differentiation processes, the HepaRG cell line was used as a replacement for primary hepatocytes, HepG2, and Huh-7 cells[6-8]. Isolated from an Edmonson grade I differentiated liver tumor, HepaRG cells exhibit a hepatocyte-like morphology and express hepatocyte-specific functions in defined culture conditions[9]. Nevertheless, HepaRG cells display features of human oval ductular bipotent hepatic progenitors at confluence[10,11]. Supplementation of dimethyl sulfoxide (DMSO) to confluent HepaRG cells triggers differentiation toward hepatocytes[10,12]. Thus, the behavior of HepaRG cells is exclusive; these cells can be cultured for several passages to proliferate, or stimulated to differentiate towards fully functional hepatocyte-like cells[13].
Cellular senescence consists of a steady cell cycle block occurring because of different harmful events, which include DNA damage, oxidative stress, or even replication[14]. Stem/progenitor cells undergoing senescence cause impairment of tissue homeostasis and regeneration, caused by defective stemness and differentiation processes[15]. The induction of senescence in both HepG2 and Huh-7 cells is associated with altered gene signature, which includes changes in cell cycle regulation, signal transduction, and metabolism[16,17]. Nevertheless, to the best of our knowledge, the impact of senescence on HepaRG cells has not yet been investigated.
Thus, this study investigated whether a replication protocol would induce senescence in HepaRG cells. In addition, we characterized the effects of senescence on the transdifferentiation capacity and mitochondrial metabolism.
The human cell line HepaRG was purchased by Merck Millipore (MMHPR116; Merck KGaA, Darmstadt, Germany). Undifferentiated HepaRG cells exhibit a fibroblast-like morphology, and the differentiation process induces both hepatocyte- and biliary-like epithelial phenotypes at confluence, indicating bipotent progenitor features[11,18]. HepaRG cells were seeded at 27000 cell/cm2 confluence in a base medium composed by William’s E Medium + GlutaMAX (3255-020; Thermo Fisher Scientific, Waltham, MA, United States) supplemented with 10% fetal bovine serum (F7524; Merck KGaA), 100 U/mL penicillin (13752; Merck KGaA), and 100 μg/mL streptomycin (P4333; Merck KGaA). Medium was changed twice a week and cells were passaged once every 7 d. Cells in passage 10 (P10, young cells) and P20 (senescent cells) were used for assays and compared. To obtain HepaRG differentiation, a two-step procedure was used as previously described. Cells were cultured in the medium for 2 wk and then in the presence of 2% DMSO for an additional 2 wk[11].
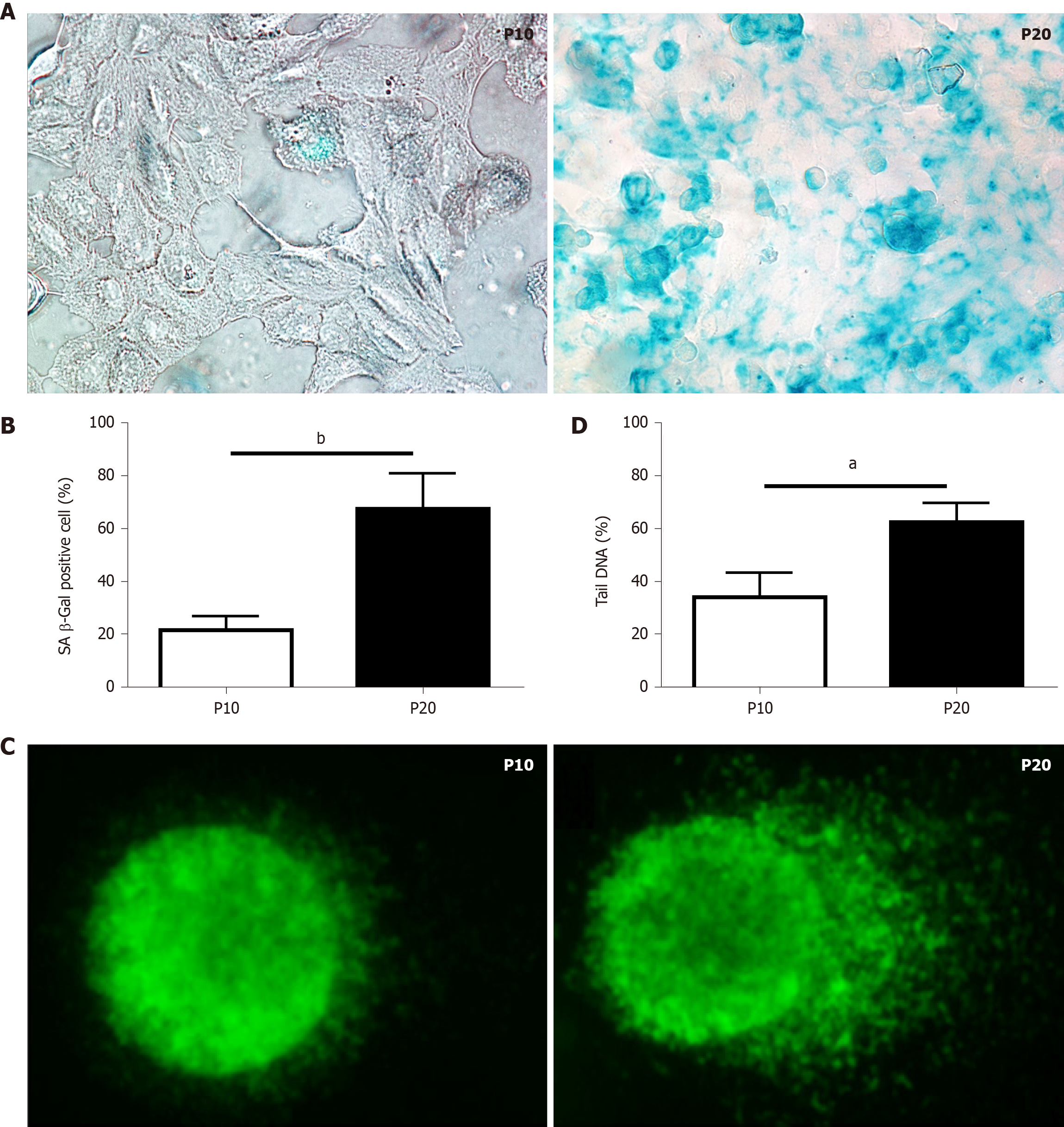
The senescence-associated (SA) β-galactosidase (SA-β-gal) activity assay was performed according to the manufacturer’s protocol (#9860; Cell Signaling Technology, Inc. Danvers, MA, United States). Briefly, P10 and P20 HepaRG cells grown on 6-well plates were fixed in 1X fixative solution containing 2% formaldehyde and 2% glutaraldehyde for 10 min, and then stained overnight at 37°C with the β-galactosidase staining solution at pH 6.0 for 15 h. Images were acquired using the Nikon Eclipse Ni-U microscope (Nikon, Tokyo, Japan).
The comet assay was performed as previously described[19]. DNA was stained with SYBR green (172-5271; Bio-Rad Laboratories, Hercules, CA, United States) just before blind slide scoring with the Nikon Eclipse Ni-U fluorescence microscope equipped with the CCD-200E video camera. At least 100 cells per sample were analyzed using the Comet Assay IV analysis software (Perceptive Instruments, Haverhill, Suffolk, United Kingdom). The extent of DNA damage in single cells was evaluated by the percentage of tail DNA.
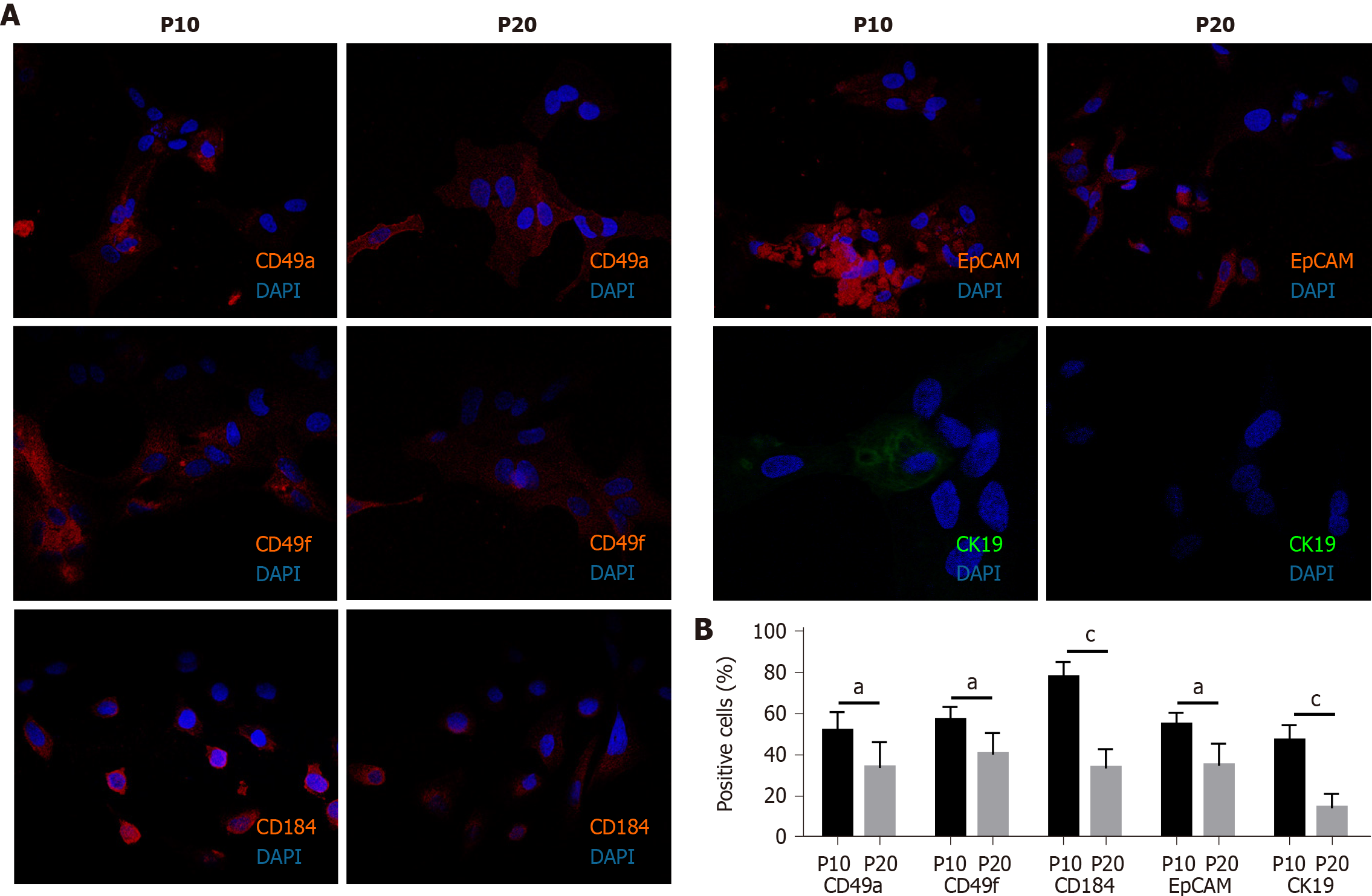
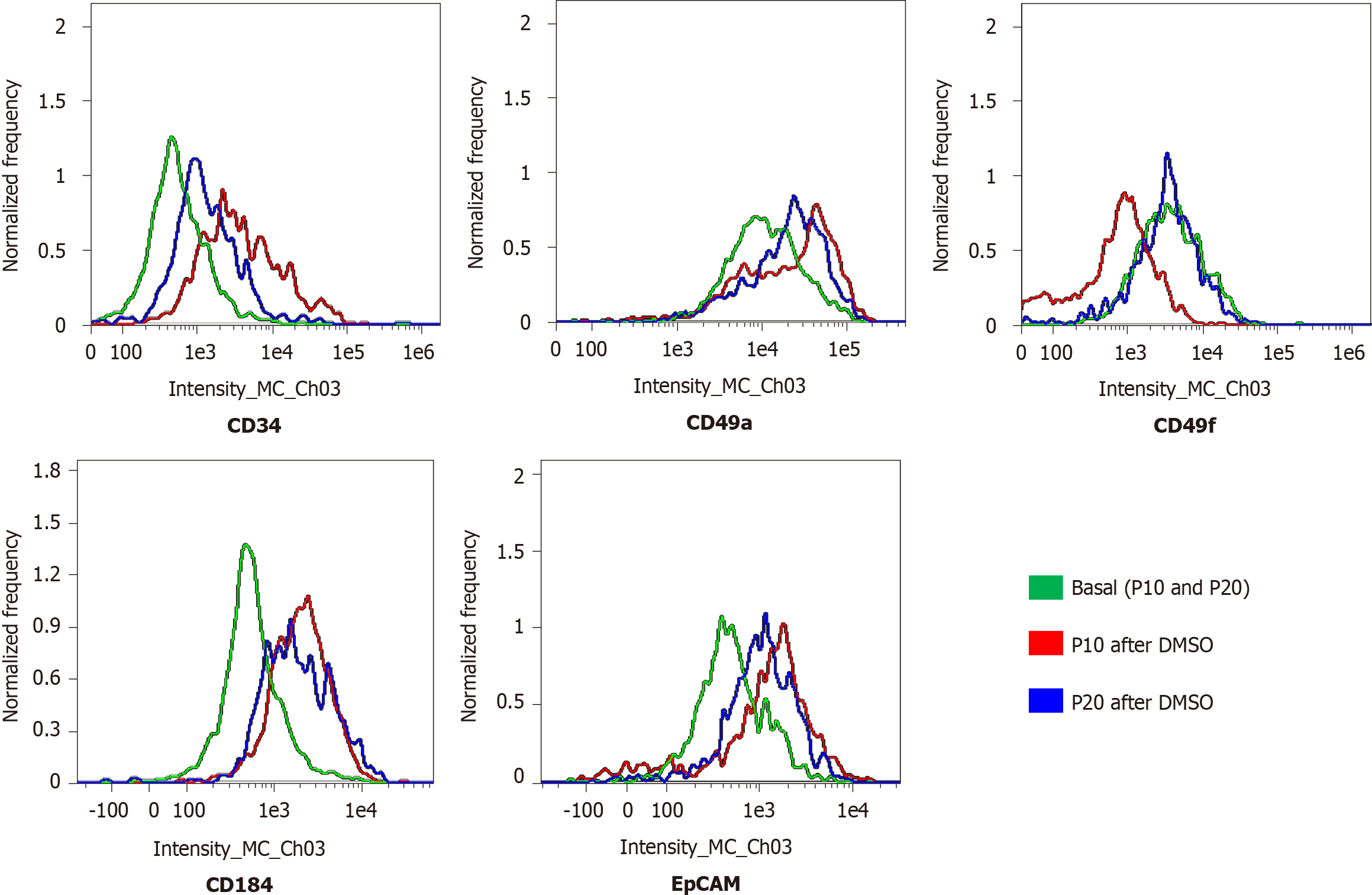
Cells (1.5 × 105 cells/well) were seeded on a glass coverslip in a 24-multiwell plate. The next day, cells were washed three times with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 10 min at room temperature (RT), and washed twice with PBS. Cells were first permeabilized with PBS + 0.1% X-100 Triton (93418; Merck KGaA) for 10 min, and then incubated in blocking buffer (3% bovine serum albumin, A7906; Merck KGaA) + 0.3 M glycine (G-7126-500-50; Merck KGaA) for 30 min at RT. Subsequently, cells were treated for 1.5 h in the dark at RT with the following labeled antibodies: anti-cluster of differentiation 49a (CD49a) (130-101-397), anti-CD49f (130-097-246), anti-CD184 (130-098-354), anti-epithelial cell adhesion molecule (EpCAM) (130-091-253), and anti-cytokeratin 19 (CK19; 130-080-101). All antibodies were purchased by Miltenyi Biotec B.V. & Co. KG (Bergisch Gladbach, North Rhine-Westphalia, Germany), and labeled with phycoerythrin, except anti-CK19, which was labeled with fluorescein isothiocyanate. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole included in mounting medium (ab104139; Abcam plc, Cambridge, United Kingdom). Cells were analyzed using the Nikon Eclipse Ti-E confocal microscope and by flow cytometry analysis using the FlowSight Cytometer (Amnis; Merck Millipore) and the IDEAS software.
To study the levels of genes expressed in P10 and P20 HepaRG cells after differentiation, RNA was extracted from 1.0 × 106 cells/sample and converted into cDNA, which was used as a template in the following RT-PCR. RNA extraction was performed by the “Pure Link RNA Mini Kit” (12183025; Thermo Fisher Scientific), according to the manufacturer’s protocol. RNA concentration was determined by spectrophotometer method at Nanodrop, measuring absorbance at λ = 260 nm. A260/A280 > 2 was evaluated to guarantee protein-free samples. Reverse transcription was performed using the High-Capacity cDNA Reverse Transcription Kit (4368814; Thermo Fisher Scientific), and SYBR Green (172-5271; Bio-Rad Laboratories, Hercules, CA, United States) was used as a fluorescent probe. The sequences of the forward and reverse primers for all of the genes studied are provided in Table 1.
| Actin | Human | FOR | 5’-TGGACATCCGCAAAGACCTG-3’ |
| REV | 5’-GCCGATCCACACGGAGTACTT-3’ | ||
| Albumin | Human | FOR | 5’-CCTGTTGCCAAAGCTCGATG-3’ |
| REV | 5’-GAAATCTCTGGCTCAGGCGA-3’ | ||
| CYP3A4 | Human | FOR | 5’-CTTCATCCAATGGACTGCATAAAT-3’ |
| REV | 5’-TCCCAAGTATAACACTCTACACAG-3’ | ||
| CEA | Human | FOR | 5’-GGTCTTCAACCCAATCAGTAAGAAC-3’ |
| REV | 5’-ATGGCCCCAGGTGAGAGG-3’ | ||
| γ-GT | Human | FOR | 5’-TTTGGTGTGCTGCTGGATGAC-3’ |
| REV | 5’-ACCTGAGCTTCCCCACCTATG-3’ |
To assess the secretion of human albumin, supernatants of P10 and P20 HepaRG cells in basal conditions and after DMSO treatment were collected after 24 h of culture and assessed using a Human Albumin ELISA Kit, according to the manufacturer’s instructions (ab108788; Abcam, Cambridge, United Kingdom).
P10 and P20 HepaRG cells in basal conditions and after DMSO treatment were washed with 1× PBS, and 50 μL of 3 μM P450-Glo™ substrate (V8801; Promega, Waldorf, Germany) was added and incubated for 1 h at 37°C, 5% CO2. Then, 25 μL substrate medium was transferred to a 96-white plate, and CYP3A4 activity was measured according to the manufacturer’s protocol. HepaRG cells were homogenized in 200 μL ice-cold γ-glutamyl transpeptidase (γ-GT) assay buffer, and the γ-GT Activity Colorimetric Assay Kit (MAK089; Merck KGaA) was used according to the manufacturer’s protocol.
HepaRG cells (5.0 × 106 cells) were washed with PBS and resuspended in 10 mmol/L KH2PO4, 27 mmol/L KCl, 1 mmol/L MgCl2, 40 mmol/L HEPES, 0.5 mmol/L EGTA buffer (pH 7.1), and assayed for oxygen consumption by the Oxygraph Plus System (Hansatech Instruments, Norfolk, UK) at 37°C under continuous stirring. Oligomycin (8 μg/mL) was added followed the addition of valinomycin (2 μg/mL) after 5 min. The rates of oxygen consumption were corrected for 3 mmol/L potassium cyanide (KCN)-insensitive respiration and normalized to the cell number. Each experiment was repeated in triplicate.
Total intracellular ATP was quantified using the ENLITEN® ATP Assay System (FF2000; Promega Corporation, Madison, WI, United States), according to the manufacturer’s instructions. This assay is based on luciferase, and used as the catalyzing enzyme of the ATP reaction with d-Luciferin. ATP extraction is carried out with trichloroacetic acid reagent, which releases ATP from cells, preventing its enzymatic degradation. After addition of the enzyme reagent to the intracellular extract, light emission was detected by a luminometer at 560 nm (DTX 880 Microplate Reader; Beckman Coulter, Brea, CA, United States).
Total intracellular NAD+/NADH was quantified using the NAD/NADH-Glo Bioluminescent Assay Kit (G9072; Promega Corporation, United States), according to the manufacturer’s instructions. HepaRG cells were first lysed with dodecyl trimethyl ammonium bromide and treated to neutralize their counterparts. To measure NAD+, the extract was treated with 25 μL of 0.4 N HCl and heated at 60°C for 15 min, incubated at RT for 10 min, following the addition of 25 μL Trizma base. To quantify NADH, the extract was incubated at 60°C for 15 min followed by further incubation for 10 min at RT. Then 50 mL HCl/Trizma solution was added to the extract. In the presence of each species, a reductase reduced a pro-luciferin reductase to luciferin. The intensity of light (proportional to the amount of each metabolite) was detected by a luminometer (DTX 880 Microplate Reader; Beckman Coulter).
Data are expressed as the mean ± standard deviation of three different experiments. Within-group variability was analyzed using Levene’s test for homogeneity of variances. Differences between two groups (P10 vs P20) were determined by the Student’s t-test, while two-way analysis of variance was used to test the main effects of senescence (S, P10 vs P20) or transdifferentiation (T, Basal vs DMSO) as between-subject factors; the interaction S × T was studied, and a Tukey’s test was used as a post hoc test for multiple comparisons. Statistical significance was accepted when P < 0.05. GraphPad Prism 6.0 software was used to perform the analyses.
Replication-induced senescence was first described in human fibroblasts because of serial culture passages[20]. Since increased SA-β-gal activity is a well-recognized marker of senescence[21], positive SA-β-gal-stained cells were counted to validate senescence in HepaRG passaged up to P10 compared to cells passaged up to P20. As shown in Figure 1A and B, replication-induced senescence in P20 HepaRG cells resulted in increased numbers of cells positive for SA-β-gal staining compared with P10 cells. A further marker of senescence is represented by the extent of DNA damage[22], determined by the comet assay, which measures the prevalence of single- and double-strand breaks. The level of DNA damage was calculated from the percentage of DNA in the tail of the comet formed after single-cell gel electrophoresis, as broken DNA moves faster in the current. The percentage of DNA in the tails vs the core was analyzed using Comet Assay IV software, and the results showed that P20 HepaRG cells presented with higher levels of damaged DNA than P10 cells (Figure 1C and D).
HepaRG cells are able to actively proliferate and commit toward hepatocyte and biliary differentiation pathway, reaching maximum cell differentiation after a 2-wk DMSO exposure[11]. Then we exposed P10 and P20 HepaRG cells to DMSO, and studied the expression of markers of both progenitor and differentiated cells.
The expression of adhesion molecules such as CD49a (limited to hepatocyte-like cells) and CD49f (associated with biliary-like cells) after the differentiation process induced by DMSO was lower in P20 than in P10 HepaRG cells. Even CD184/C-X-C motif chemokine receptor 4, a known definitive endoderm marker, was less expressed in P20 than in P10 HepaRG cells exposed to DMSO. Moreover, the differentiation protocol resulted in lower expression of EpCAM and CK19 (markers of hepatic progenitor cells[23]) in P20 than in P10 HepaRG cells (Figures 2 and 3).
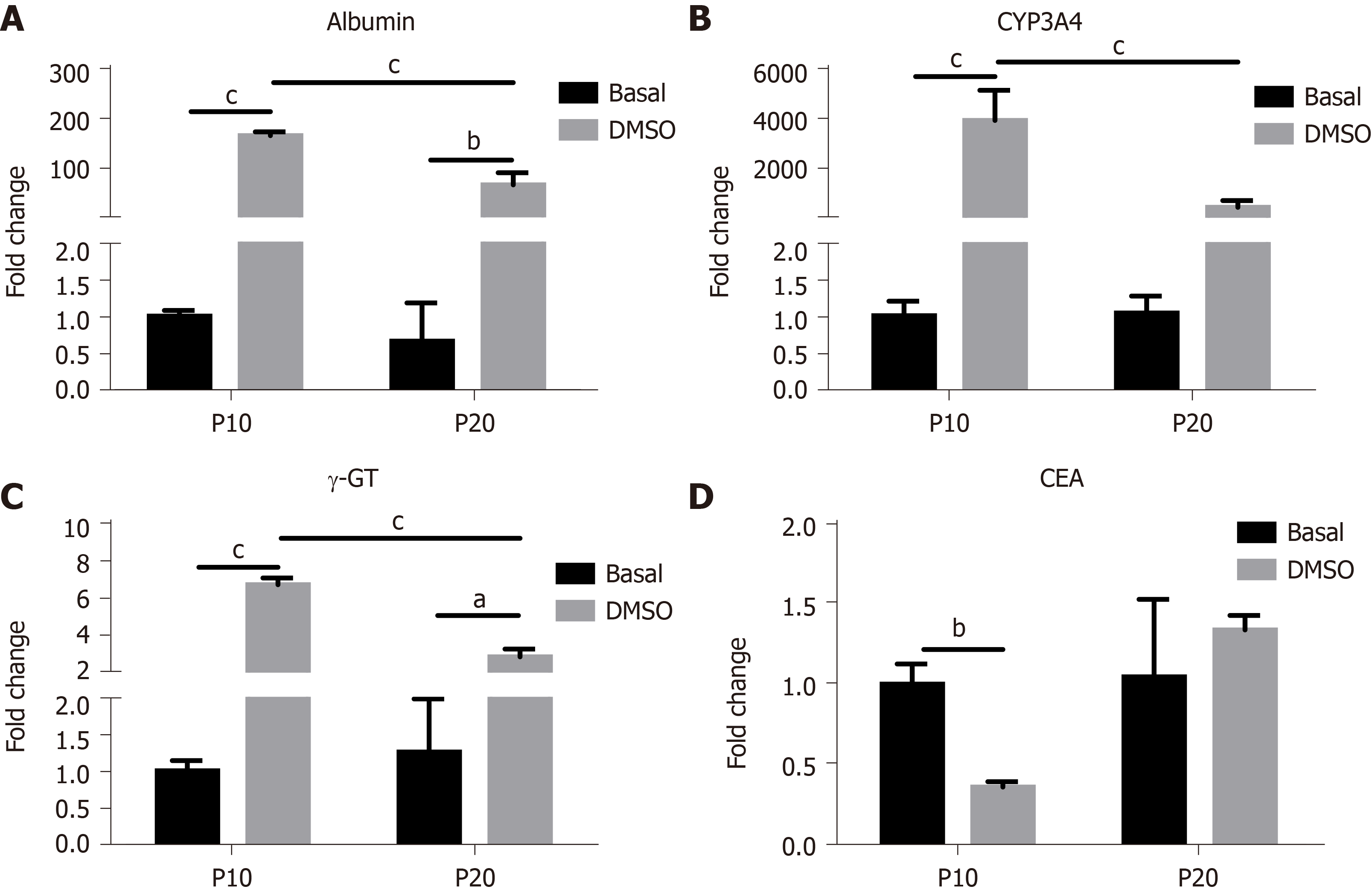
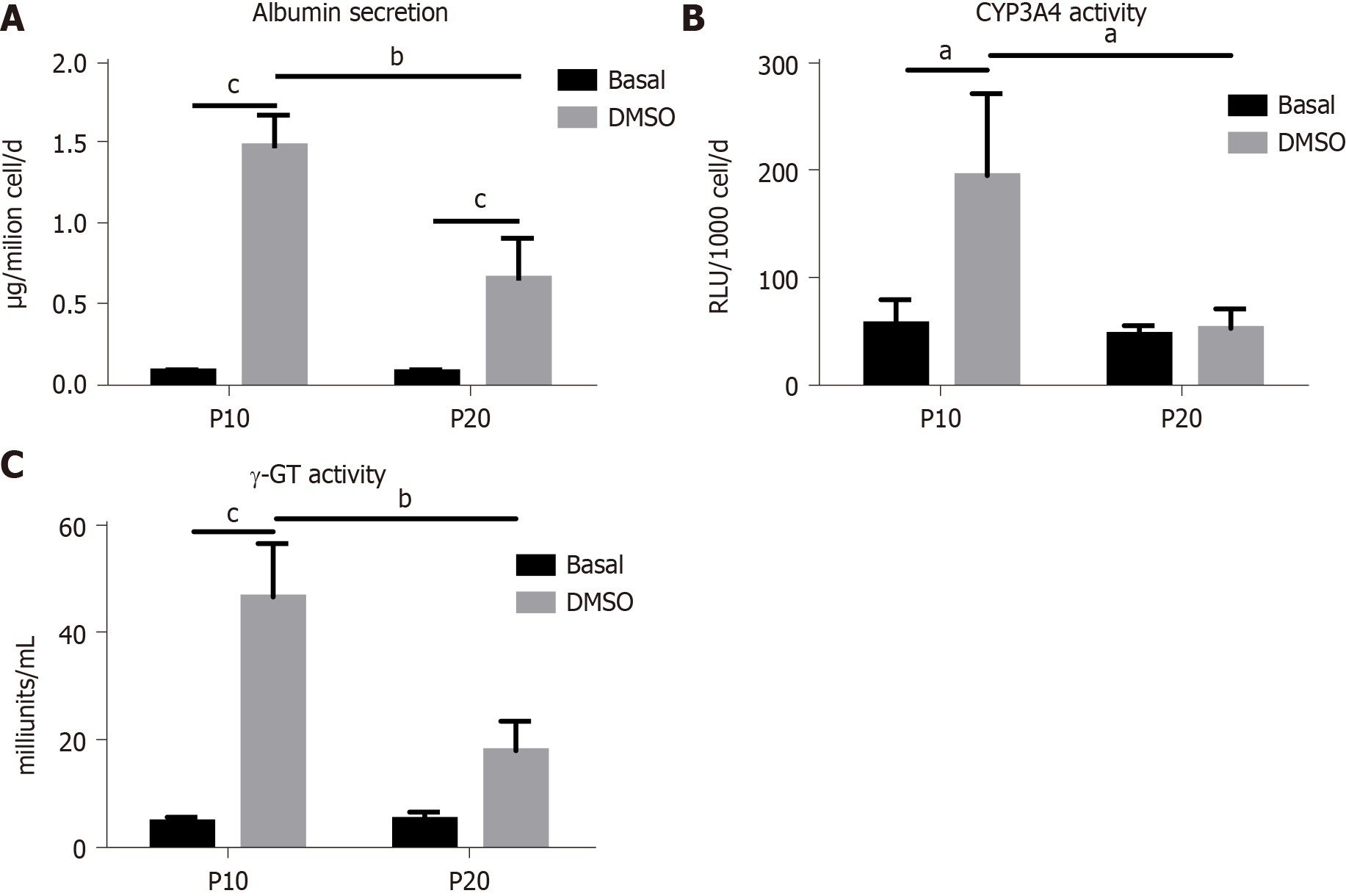
The expression of genes typical of a differentiated status, such as albumin, CYP3A4 and γ-GT, and of CEA as marker of undifferentiation was next determined in HepaRG cells before and after DMSO exposure. The results obtained by statistical analyses aimed to test the main effects of S, of T, and the interaction S × T. The main effect of S was significant for the expression of all the genes studied (albumin: F(1,8) = 44.54, P = 0.0002; CYP3A4: F(1,8) = 24.22, P = 0.0012; γ-GT: F(1,8) = 46.82, P = 0.0001; CEA: F(1,8) = 12.24, P = 0.0081). The main effect of T was significant for the expression of albumin (F(1,8) = 243.3, P < 0.0001), CYP3A4 (F(1,8) = 37.50, P = 0.0003), and γ-GT (F(1,8) = 190.2, P < 0.0001), but not for CEA. The interaction between S and T was significant for the expression of all the genes studied (albumin: F(1,8) = 43.95, P = 0.0002; CYP3A4: F(1,8) = 24.22, P = 0.0012; γ-GT: F(1,8) = 60.71, P < 0.0001; CEA: F(1,8) = 10.13, P = 0.0129). Post hoc analysis showed that the expression of albumin, CYP3A4 and γ-GT was induced by DMSO exposure both in P10 and in P20 HepaRG cells, supporting the transdifferentiation process; however, mRNA levels were lower in DMSO-treated P20 rather than P10 HepaRG cells (Figure 4A-C). Finally, the expression of CEA was reduced by DMSO exposure in P10 but not in P20 HepaRG cells (Figure 4D).
The same analysis was performed on specific functional activities of HepaRG cells, studying albumin secretion, CYP3A4 and γ-GT activity, and the results are represented in Figure 5. In particular, we found a main effect of S (albumin secretion: F(1,8) = 20.95, P = 0.0018; CYP3A4 activity: F(1,8) = 10.24, P = 0.0126; γ-GT activity: F(1,8) = 18.13, P = 0.0028), T (albumin secretion: F(1,8) = 120.2, P < 0.0001, CYP3A4 activity: F(1,8) = 9.15, P = 0.0164; γ-GT activity: F(1,8) = 68.25, P < 0.0001), and interaction between S and T (albumin secretion: F(1,8) = 20.53, P = 0.0019; CYP3A4 activity: F(1,8) = 7.181, P = 0.0233; γ-GT activity: F(1,8) = 19.36, P = 0.0023). Post hoc analysis showed that albumin secretion, CYP3A4 and γ-GT activities were reduced in DMSO-treated P20 rather than P10 HepaRG cells.
Taken together, these results suggest that the transdifferentiation process triggered by DMSO in HepaRG cells is hampered by the replication-induced senescence.
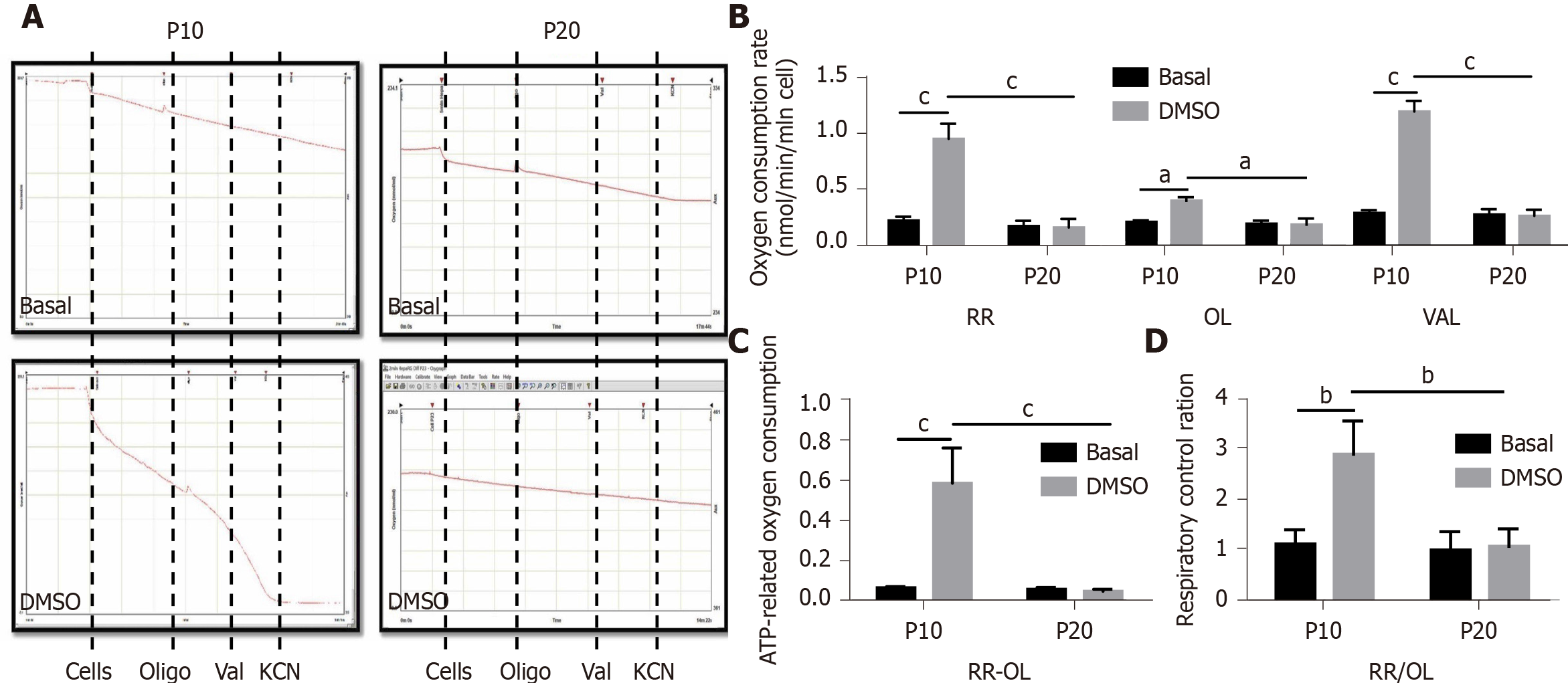
Mitochondrial dysfunction is recognized as one of the hallmarks of senescence[24]. Thus, we performed respirometry analyses in HepaRG by high-resolution oximetry. Figure 6A details the protocol used and is representative of the oxygraphic profiles in young (P10) and senescent (P20) HepaRG cells in basal conditions and after the transdifferentiation protocol (DMSO). The resting respiration (RR), which depends on endogenous substrates, was impacted by both the S and the T factor (S: F(1,8) = 78.38, P < 0.0001; T: F(1,8) = 57.12, P < 0.0001), and by the interaction S×T (F(1,8) = 60.86, P < 0.0001). Addition of oligomycin (a FoF1-ATP synthase inhibitor) reduced the oxygen uptake, suggesting that most of the mitochondrial respiration was coupled to the synthesis of ATP. Nevertheless, both the S and the T factor impacted this parameter (S: F(1,8) = 10.83, P = 0.0018; T: F(1,8) = 6.847, P = 0.0308), as well as the interaction S × T (F(1,8) = 8.104, P = 0.0216). Restoration of the oxygen uptake by the addition of valinomycin (a K+ ionophore which uncouples oxygen consumption from ATP synthesis) was also impacted by both the S and the T factor (S: F(1,8) = 59.58, P < 0.0001; T: F(1,8) = 53.09, P < 0.0001), and the interaction S × T (F(1,8) = 55.97, P < 0.0001). The post hoc analysis showed that the oxygen uptake in all the examined conditions was higher in P10 HepaRG cells after the transdifferentiation protocol with respect to the other samples (Figure 6B). The effects of both S and T, and their interaction, was observed on the ATP-dependent oxygen uptake, calculated as the difference between RR and oligomycin-induced respiration (S: F(1,8) = 28.94, P = 0.0007; T: F(1,8) = 25.35, P = 0.001; S × T: F(1,8) = 27.36, P = 0.0008). The post hoc analysis showed that the P10 HepaRG cells after the transdifferentiation protocol exhibited a higher ATP-dependent oxygen uptake than the other samples (Figure 6C). The respiratory control ratio (RCR), calculated as the ratio between RR and oligomycin-induced respiration, was also influenced by both S and T (S: F(1,8) = 14.76, P = 0.0049; T: F(1,8) = 13.43, P = 0.0064), and their interaction (S × T: F(1,8) = 11.38, P = 0.0097). The post hoc analysis resulted in a higher RCR for the P10 HepaRG cells after the transdifferentiation protocol compared to the other samples (Figure 6D). Taken together, these data suggest that the transdifferentiation induced by DMSO increases mitochondrial respiration in HepaRG cells; however, this does not occur in senescent cells.
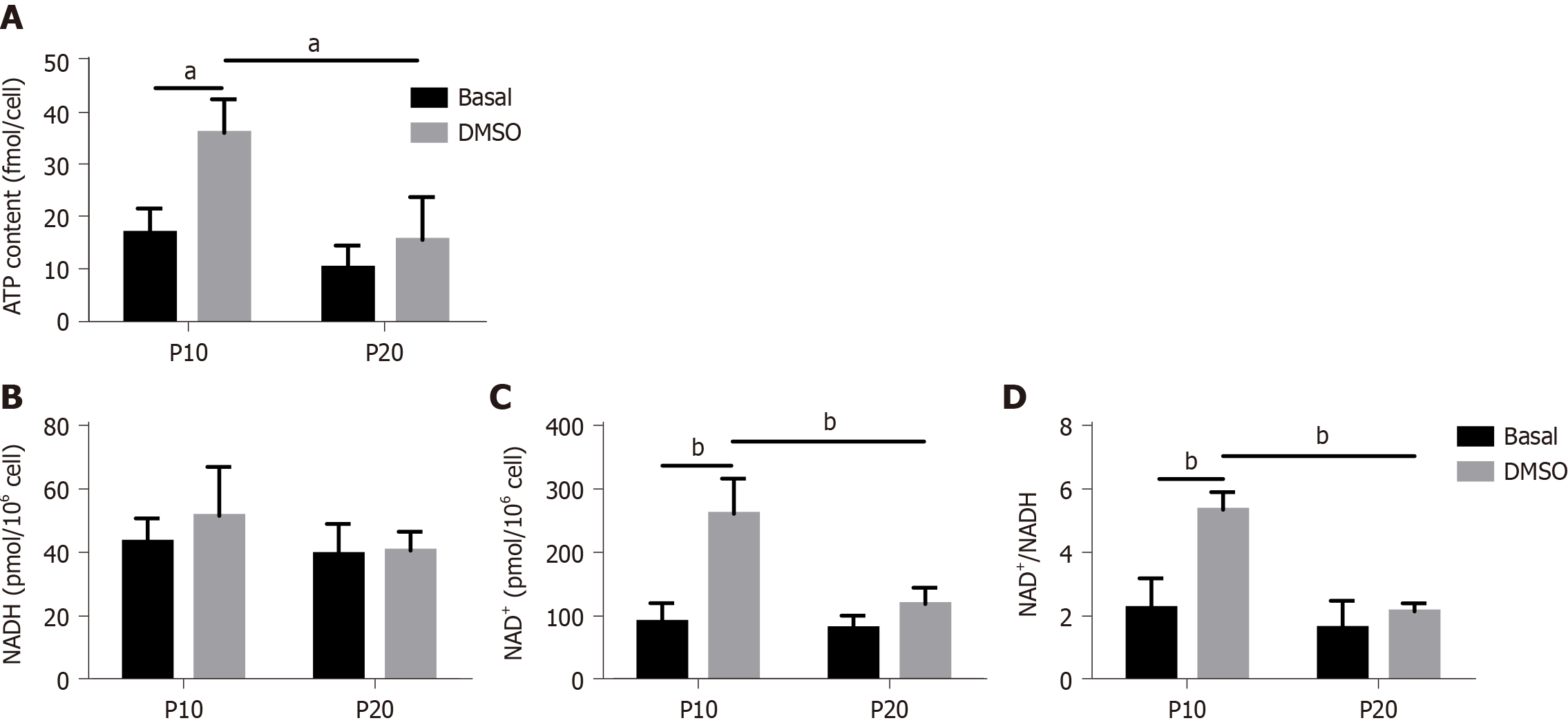
To confirm a defect in mitochondrial dysfunction of senescent HepaRG, the cellular ATP concentration was further measured. We observed a significant effect of both S and T (S:F(1,8) = 14.89, P = 0.0048; T:F(1,8) = 12.15, P = 0.0083), and the post hoc analysis showed that the ATP content of transdifferentiated P10 HepaRG cells was higher than the other samples (Figure 7A). SA mitochondrial dysfunction can be consequent to an exhaustion of the oxidized form of NAD+[25]. The analysis of NAD+/NADH content in HepaRG cells revealed no changes in NADH; nevertheless, the impact of S, T, and their interaction was observed on both NAD+ (S:F(1,8) = 13.84, P = 0.0059; T:F(1,8) = 26.00, P = 0.0009; S × T: F(1,8) = 10.47, P = 0.0120) and NAD+/NADH (S:F(1,8) = 22.23, P = 0.0015; T:F(1,8) = 19.75, P = 0.0022; S × T: F(1,8) = 10.02, P = 0.0133). The post hoc analysis resulted in higher NAD+ and NAD+/NADH in transdifferentiated P10 HepaRG cells with respect to the other samples (Figure 7B).
This study demonstrates that cellular replication in HepaRG cells is associated with both the expression of senescence markers and the reduction in transdifferentiation potential. Indeed, data related to phenotype, gene expression, functional analysis, and mitochondrial function suggest that the transdifferentiation process of HepaRG is preserved after a few passages, but it is altered by continuous replication.
HepaRG represents a unique cell line characterized by high expression of detoxifying and metabolizing enzymes, transport proteins, and nuclear receptors, as well as the ability to transdifferentiate toward hepatocyte-like and cholangiocyte-like cells[9-11]. Other than representing a viable tool for cell biology, drug metabolism, and virology studies, differentiated HepaRG cells are suitable to generate humanized liver in rodent models, allowing in vivo studies of liver development and physiology[26,27].
Cultured cells stop proliferating after a finite number of divisions, a phenomenon defined to as replicative senescence[28,29]. However, cells can escape replicative senescence by several mechanisms, including overexpression of viral genes such as simian virus 40 large T antigen (which inactivates p53), or telomerase reverse transcriptase protein (TERT, which elongates telomeres)[30,31]. Proliferating HepaRG cells show over-expression and hyper-activation of human TERT, as well as inhibition of p21 and p27, which in turn inhibit the catalytic activity of cyclin-dependent kinases and stop the cell cycle[32]. Nevertheless, our data demonstrate that a replication protocol induces the expression of senescence markers in HepaRG cells, such as positive SA-β-gal staining and DNA alterations. Thus, we hypothesize that the mechanisms that induce senescence in HepaRG cells are different from telomere shortening and p21/p27 activation.
Senescence impairs the transdifferentiation of several cell lines[33-36]. Senescent cells secrete a large variety of molecules that change the surrounding microenvironment, with consequent alterations of differentiation and tissue regeneration[37,38]. These compounds include a wide range of cytokines, growth factors, and signaling molecules that are included in the SA secretory phenotype (SASP)[39]. Our data clearly demonstrate that replicative senescence alters the transdifferentiation process of HepaRG cells. Nevertheless, we did not analyze the SASP in our study, since this marker is strictly linked with telomere shortening[40]. By contrast, we focused on mitochondria, since the homeostasis of these organelles is crucial for several aspects of senescence including SASP[41]. Indeed, the impairment of mitochondrial oxidative phosphorylation (OxPhos) is mainly involved in the early steps of cell senescence[42]. Moreover, senescent cells exhibit severe metabolic alterations associated with mitochondrial metabolites, such as oxidized to reduced NAD ratios (NAD+/NADH)[43]. The novel and straight findings of this study demonstrate the higher mitochon
The concentration of the oxidized form of NAD+ is determinant for mitochondrial homeostasis[44,45]. Moreover, NAD+ is crucial in several metabolic pathways, including glycolysis, citric acid cycle, and OxPhos, with consequent implications for both stemness/differentiation and cell senescence[46]. Cells that maintain a physiological quiescent state to preserve long-term self-renewal capacity are characterized by low NAD+ levels to obtain energy from glycolysis; on the contrary, cell differentiation leads to reduced glycolysis and increased OhPhos, which require high NAD+ levels[47]. Age-related reductions of both NAD+ levels and NAD+/NADH ratio are evolutionarily preserved, and consistent evidence for low NAD+ has been provided for several old mammalian tissues[48]. Our data show that HepaRG transdifferentiation is associated with increased NAD+ and relatively stable NADH, with consequent high NAD+/NADH. However, the raise of both NAD+ and NAD+/NADH is not observed in HepaRG undergoing replicative senescence. Even though the effect of a treatment goes beyond the scope of our study, it is conceivable that a replenishment of NAD+ would protect mitochondria and improve the transdifferentiation process of senescent HepaRG cells, as already described for other cell types[49-51].
In conclusion, the present report demonstrates that HepaRG cells undergo replicative senescence, which is associated with impairment in transdifferentiation, mitochondrial dysfunction, and NAD+ depletion. Further investigations are required to refine the molecular mechanism underlying such observation. The limitations in the transdifferentiation potential of senescent HepaRG cells, with consequent alteration of metabolic and regenerative properties, may have serious implications when this cell line is applied for basic studies.
The HepaRG cell line is used to study metabolism, toxicology, and the regeneration /differentiation processes, as a replacement to primary hepatocytes, HepG2, and Huh-7 cells. These cells exhibit a hepatocyte-like morphology and express hepatocyte-specific functions in defined culture conditions; furthermore, HepaRG display features of human oval ductular bipotent hepatic progenitors.
Cellular senescence consists in a steady cell cycle block occurring because of different harmful events, leading to defective stemness and differentiation processes, as well as changes in cell cycle regulation, signal transduction, and metabolism. The impact of senescence on HepaRG cells has not yet been investigated.
This study investigated whether a replication protocol would induce senescence in HepaRG cells. In addition, we characterized the effects of senescence on transdifferentiation capacity and mitochondrial metabolism.
The transdifferentiation capacity of HepaRG cells over passage 10 (P10) vs passage 20 (P20) was compared. To stimulate transdifferentiation, HepaRG cells were treated with dimethyl sulfoxide (DMSO). Aging was evaluated by senescence-associated (SA) β-galactosidase activity and the comet assay. HepaRG transdifferentiation was analyzed by confocal microscopy and flow cytometry (expression of cluster of differentiation 49a [CD49a], CD49f, CD184, epithelial cell adhesion molecule [EpCAM], and cytokeratin 19 [CK19]), by quantitative PCR analysis (expression of albumin, cytochrome P450 3A4 [CYP3A4], γ-glutamyl transpeptidase [γ-GT] and carcinoembryonic antigen [CEA]) and functional analysis (albumin secretion, CYP3A4 and γ-GT). Mitochondrial respiration, the ATP and the NAD+/NADH content were also measured.
We first observed that replication induces the expression of senescence markers in HepaRG cells, since SA β-galactosidase staining was higher in P20 than in P10 HepaRG cells, and the comet assay showed a consistent DNA damage in P20 HepaRG cells. We further reported that transdifferentiation towards bipotent progenitors is altered in senescent HepaRG cells, as P20 HepaRG cells exhibited a reduction of CD49a, CD49f, CD184, EpCAM and CK19 – with respect to P10 – after DMSO treatment. Furthermore, the lower gene expression of albumin, CYP3A4, and γ-GT, as well as the reduced albumin secretion capacity, CYP3A4, and γ-GT activity were reported in transdifferentiated P20 compared to P10 cells. By contrast, the gene expression level of CEA was not reduced by transdifferentiation in P20 cells. Finally, we show that senescence-induced impairment of HepaRG transdifferentiation is associated with mitochondrial dysfunction, since both cellular and mitochondrial oxygen consumption were lower in P20 than in P10 transdifferentiated cells, and both ATP and NAD+/NADH were depleted in P20 cells with respect to P10 cells.
The present study demonstrates that HepaRG cells undergo replicative senescence, with consequent impairment in transdifferentiation, functional activity, mitochondrial dysfunction, and NAD+ depletion.
Further studies will define the molecular mechanisms underlying our observations. The limitations in the transdifferentiation potential of senescent HepaRG cells, with consequent alteration of metabolic and regenerative properties, may have serious implications when this cell line is applied for basic studies.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chen S S-Editor: Wu XYJ L-Editor: Filipodia P-Editor: Wang LYT
| 1. | Fabre G, Combalbert J, Berger Y, Cano JP. Human hepatocytes as a key in vitro model to improve preclinical drug development. Eur J Drug Metab Pharmacokinet. 1990;15:165-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 2. | Madan A, Graham RA, Carroll KM, Mudra DR, Burton LA, Krueger LA, Downey AD, Czerwinski M, Forster J, Ribadeneira MD, Gan LS, LeCluyse EL, Zech K, Robertson P Jr, Koch P, Antonian L, Wagner G, Yu L, Parkinson A. Effects of prototypical microsomal enzyme inducers on cytochrome P450 expression in cultured human hepatocytes. Drug Metab Dispos. 2003;31:421-431. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 264] [Cited by in F6Publishing: 254] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 3. | Javitt NB. Hep G2 cells as a resource for metabolic studies: lipoprotein, cholesterol, and bile acids. FASEB J. 1990;4:161-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 277] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 4. | Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858-3863. [PubMed] [Cited in This Article: ] |
| 5. | Bulutoglu B, Rey-Bedón C, Mert S, Tian L, Jang YY, Yarmush ML, Usta OB. A comparison of hepato-cellular in vitro platforms to study CYP3A4 induction. PLoS One. 2020;15:e0229106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 6. | McGill MR, Yan HM, Ramachandran A, Murray GJ, Rollins DE, Jaeschke H. HepaRG cells: a human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology. 2011;53:974-982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 235] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 7. | Anthérieu S, Chesné C, Li R, Guguen-Guillouzo C, Guillouzo A. Optimization of the HepaRG cell model for drug metabolism and toxicity studies. Toxicol In Vitro. 2012;26:1278-1285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 121] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 8. | Dubuquoy L, Louvet A, Lassailly G, Truant S, Boleslawski E, Artru F, Maggiotto F, Gantier E, Buob D, Leteurtre E, Cannesson A, Dharancy S, Moreno C, Pruvot FR, Bataller R, Mathurin P. Progenitor cell expansion and impaired hepatocyte regeneration in explanted livers from alcoholic hepatitis. Gut. 2015;64:1949-1960. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 9. | Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, Guyomard C, Lucas J, Trepo C, Guguen-Guillouzo C. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99:15655-15660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 926] [Cited by in F6Publishing: 1065] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 10. | Parent R, Marion MJ, Furio L, Trépo C, Petit MA. Origin and characterization of a human bipotent liver progenitor cell line. Gastroenterology. 2004;126:1147-1156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 176] [Cited by in F6Publishing: 179] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 11. | Cerec V, Glaise D, Garnier D, Morosan S, Turlin B, Drenou B, Gripon P, Kremsdorf D, Guguen-Guillouzo C, Corlu A. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRG cell line through bipotent progenitor. Hepatology. 2007;45:957-967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 265] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 12. | Troadec MB, Glaise D, Lamirault G, Le Cunff M, Guérin E, Le Meur N, Détivaud L, Zindy P, Leroyer P, Guisle I, Duval H, Gripon P, Théret N, Boudjema K, Guguen-Guillouzo C, Brissot P, Léger JJ, Loréal O. Hepatocyte iron loading capacity is associated with differentiation and repression of motility in the HepaRG cell line. Genomics. 2006;87:93-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Guillouzo A, Corlu A, Aninat C, Glaise D, Morel F, Guguen-Guillouzo C. The human hepatoma HepaRG cells: a highly differentiated model for studies of liver metabolism and toxicity of xenobiotics. Chem Biol Interact. 2007;168:66-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 445] [Cited by in F6Publishing: 438] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 14. | Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018;28:436-453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 953] [Cited by in F6Publishing: 1241] [Article Influence: 206.8] [Reference Citation Analysis (0)] |
| 15. | van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509:439-446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1437] [Cited by in F6Publishing: 1701] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 16. | Yildiz G, Arslan-Ergul A, Bagislar S, Konu O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H, Erdal E, Karademir S, Sagol O, Mizrak D, Bozkaya H, Ilk HG, Ilk O, Bilen B, Cetin-Atalay R, Akar N, Ozturk M. Genome-wide transcriptional reorganization associated with senescence-to-immortality switch during human hepatocellular carcinogenesis. PLoS One. 2013;8:e64016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 17. | Aravinthan A, Challis B, Shannon N, Hoare M, Heaney J, Alexander GJM. Selective insulin resistance in hepatocyte senescence. Exp Cell Res. 2015;331:38-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 18. | Aninat C, Piton A, Glaise D, Le Charpentier T, Langouët S, Morel F, Guguen-Guillouzo C, Guillouzo A. Expression of cytochromes P450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab Dispos. 2006;34:75-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 499] [Cited by in F6Publishing: 488] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 19. | Jossé R, Aninat C, Glaise D, Dumont J, Fessard V, Morel F, Poul JM, Guguen-Guillouzo C, Guillouzo A. Long-term functional stability of human HepaRG hepatocytes and use for chronic toxicity and genotoxicity studies. Drug Metab Dispos. 2008;36:1111-1118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Hayflick L, MOORHEAD PS. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5393] [Cited by in F6Publishing: 5098] [Article Influence: 196.1] [Reference Citation Analysis (0)] |
| 21. | Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113:3613-3622. [PubMed] [Cited in This Article: ] |
| 22. | Kuilman T, Michaloglou C, Mooi WJ, Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463-2479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1416] [Cited by in F6Publishing: 1487] [Article Influence: 106.2] [Reference Citation Analysis (0)] |
| 23. | Yovchev MI, Grozdanov PN, Zhou H, Racherla H, Guha C, Dabeva MD. Identification of adult hepatic progenitor cells capable of repopulating injured rat liver. Hepatology. 2008;47:636-647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 24. | López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194-1217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7855] [Cited by in F6Publishing: 8861] [Article Influence: 805.5] [Reference Citation Analysis (0)] |
| 25. | Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L, White JP, Teodoro JS, Wrann CD, Hubbard BP, Mercken EM, Palmeira CM, de Cabo R, Rolo AP, Turner N, Bell EL, Sinclair DA. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624-1638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 899] [Cited by in F6Publishing: 989] [Article Influence: 98.9] [Reference Citation Analysis (0)] |
| 26. | Jiang L, Li JG, Lan L, Wang YM, Mao Q, You JP. Human hepatoma HepaRG cell line engraftment in severe combined immunodeficient × beige mice using mouse-specific anti-Fas antibody. Transplant Proc. 2010;42:3773-3778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Higuchi Y, Kawai K, Yamazaki H, Nakamura M, Bree F, Guguen-Guillouzo C, Suemizu H. The human hepatic cell line HepaRG as a possible cell source for the generation of humanized liver TK-NOG mice. Xenobiotica. 2014;44:146-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | HAYFLICK L. THE LIMITED IN VITRO LIFETIME OF HUMAN DIPLOID CELL STRAINS. Exp Cell Res. 1965;37:614-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4095] [Cited by in F6Publishing: 3887] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 29. | Coates PJ. Markers of senescence? J Pathol. 2002;196:371-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 30. | Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349-352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3573] [Cited by in F6Publishing: 3337] [Article Influence: 128.3] [Reference Citation Analysis (0)] |
| 31. | Ali SH, DeCaprio JA. Cellular transformation by SV40 Large T antigen: interaction with host proteins. Semin Cancer Biol. 2001;11:15-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 272] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 32. | Sirma H, Kumar M, Meena JK, Witt B, Weise JM, Lechel A, Ande S, Sakk V, Guguen-Guillouzo C, Zender L, Rudolph KL, Günes C. The promoter of human telomerase reverse transcriptase is activated during liver regeneration and hepatocyte proliferation. Gastroenterology. 2011;141:326-337, 337.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 33. | Sun CK, Zhou D, Zhang Z, He L, Zhang F, Wang X, Yuan J, Chen Q, Wu LG, Yang Q. Senescence impairs direct conversion of human somatic cells to neurons. Nat Commun. 2014;5:4112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 34. | García-Prat L, Muñoz-Cánoves P. Aging, metabolism and stem cells: Spotlight on muscle stem cells. Mol Cell Endocrinol. 2017;445:109-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 35. | Moimas S, Salton F, Kosmider B, Ring N, Volpe MC, Bahmed K, Braga L, Rehman M, Vodret S, Graziani ML, Wolfson MR, Marchetti N, Rogers TJ, Giacca M, Criner GJ, Zacchigna S, Confalonieri M. miR-200 family members reduce senescence and restore idiopathic pulmonary fibrosis type II alveolar epithelial cell transdifferentiation. ERJ Open Res. 2019;5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 36. | Han L, Zhang Y, Zhang M, Guo L, Wang J, Zeng F, Xu D, Yin Z, Xu Y, Wang D, Zhou H. Interleukin-1β-Induced Senescence Promotes Osteoblastic Transition of Vascular Smooth Muscle Cells. Kidney Blood Press Res. 2020;45:314-330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 37. | Baar MP, Perdiguero E, Muñoz-Cánoves P, de Keizer PL. Musculoskeletal senescence: a moving target ready to be eliminated. Curr Opin Pharmacol. 2018;40:147-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 38. | Nicaise AM, Wagstaff LJ, Willis CM, Paisie C, Chandok H, Robson P, Fossati V, Williams A, Crocker SJ. Cellular senescence in progenitor cells contributes to diminished remyelination potential in progressive multiple sclerosis. Proc Natl Acad Sci U S A. 2019;116:9030-9039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 130] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 39. | Coppé JP, Patil CK, Rodier F, Sun Y, Muñoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853-2868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2346] [Cited by in F6Publishing: 2638] [Article Influence: 175.9] [Reference Citation Analysis (0)] |
| 40. | Razdan N, Vasilopoulos T, Herbig U. Telomere dysfunction promotes transdifferentiation of human fibroblasts into myofibroblasts. Aging Cell. 2018;17:e12838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 41. | Birch J, Barnes PJ, Passos JF. Mitochondria, telomeres and cell senescence: Implications for lung ageing and disease. Pharmacol Ther. 2018;183:34-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 89] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 42. | Vasileiou PVS, Evangelou K, Vlasis K, Fildisis G, Panayiotidis MI, Chronopoulos E, Passias PG, Kouloukoussa M, Gorgoulis VG, Havaki S. Mitochondrial Homeostasis and Cellular Senescence. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 119] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 43. | Correia-Melo C, Passos JF. Mitochondria: Are they causal players in cellular senescence? Biochim Biophys Acta. 2015;1847:1373-1379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 106] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 44. | Houtkooper RH, Cantó C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 637] [Cited by in F6Publishing: 642] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 45. | Cantó C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 806] [Cited by in F6Publishing: 842] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 46. | Johnson S, Imai SI. NAD + biosynthesis, aging, and disease. F1000Res. 2018;7:132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 47. | Shyh-Chang N, Ng HH. The metabolic programming of stem cells. Genes Dev. 2017;31:336-346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 194] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 48. | Yoshino J, Baur JA, Imai SI. NAD+ Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018;27:513-528. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 591] [Cited by in F6Publishing: 547] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 49. | Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D'Amico D, Ropelle ER, Lutolf MP, Aebersold R, Schoonjans K, Menzies KJ, Auwerx J. NAD⁺ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436-1443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 708] [Cited by in F6Publishing: 776] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 50. | Igarashi M, Miura M, Williams E, Jaksch F, Kadowaki T, Yamauchi T, Guarente L. NAD+ supplementation rejuvenates aged gut adult stem cells. Aging Cell. 2019;18:e12935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 84] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 51. | Lees JG, Gardner DK, Harvey AJ. Nicotinamide adenine dinucleotide induces a bivalent metabolism and maintains pluripotency in human embryonic stem cells. Stem Cells. 2020;38:624-638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |