Published online Jun 28, 2018. doi: 10.3748/wjg.v24.i24.2596
Peer-review started: March 23, 2018
First decision: April 21, 2018
Revised: April 26, 2018
Accepted: May 6, 2018
Article in press: May 6, 2018
Published online: June 28, 2018
To determine a panel of serum microRNAs (miRNAs) that could be used as novel biomarkers for diagnosis of hepatocellular carcinoma (HCC).
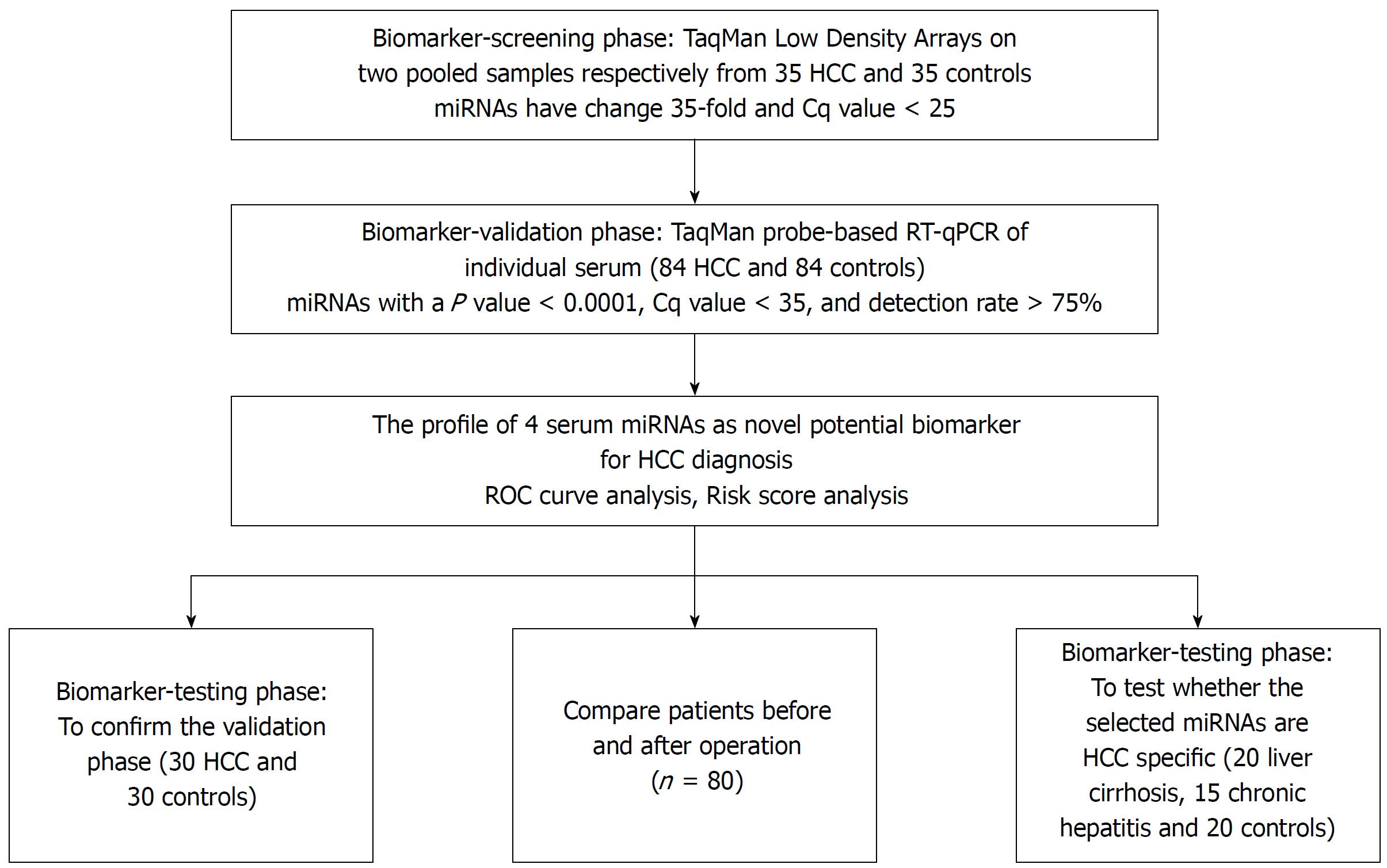
We initially screened 9 out of 754 serum miRNAs by TaqMan Low Density Array in two pooled samples respectively from 35 HCC and 35 normal controls, and then validated individually by RT-qPCR in another 114 patients and 114 controls arranged in two phases. The changes of the selected miRNAs after operation and their prognostic value were examined.
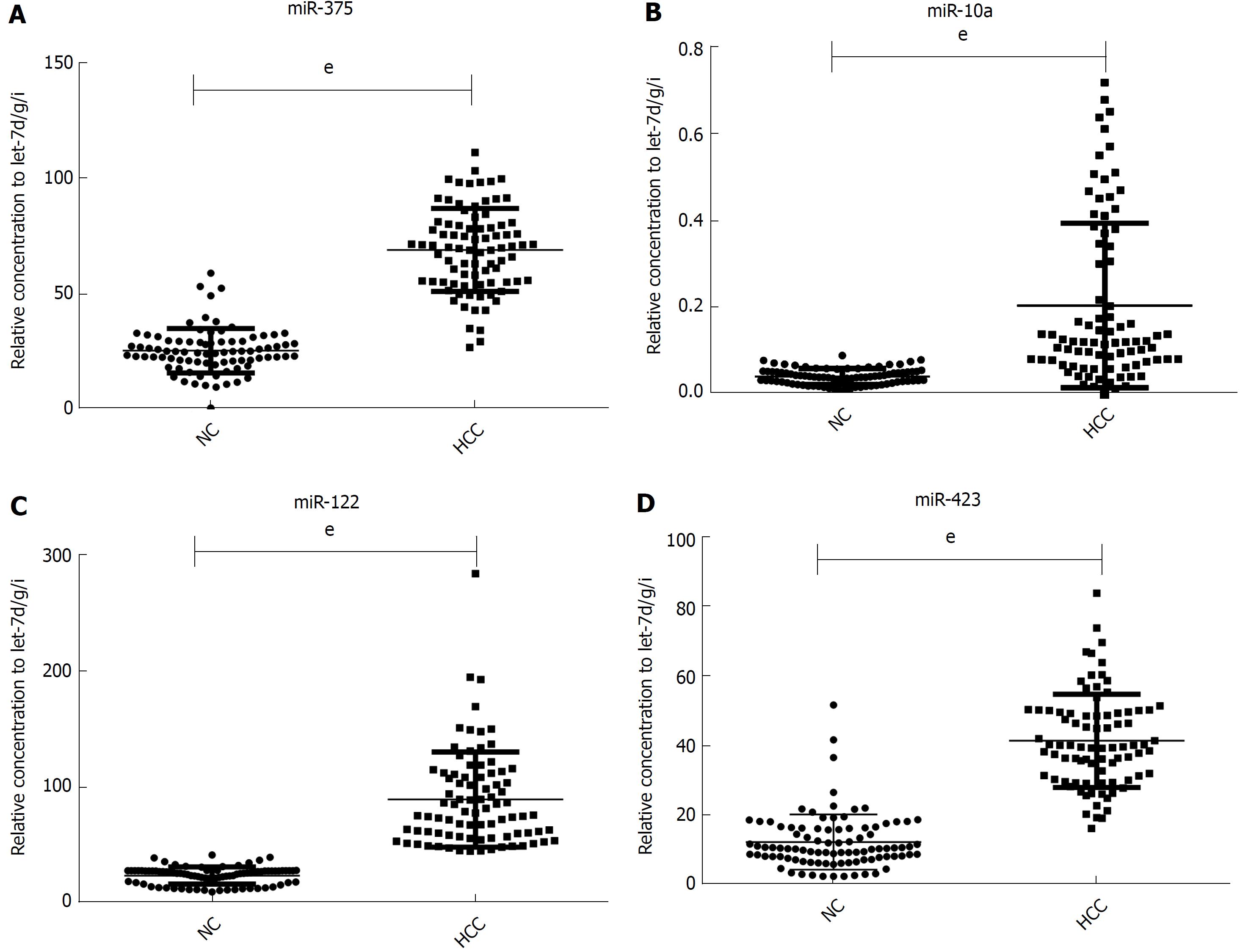
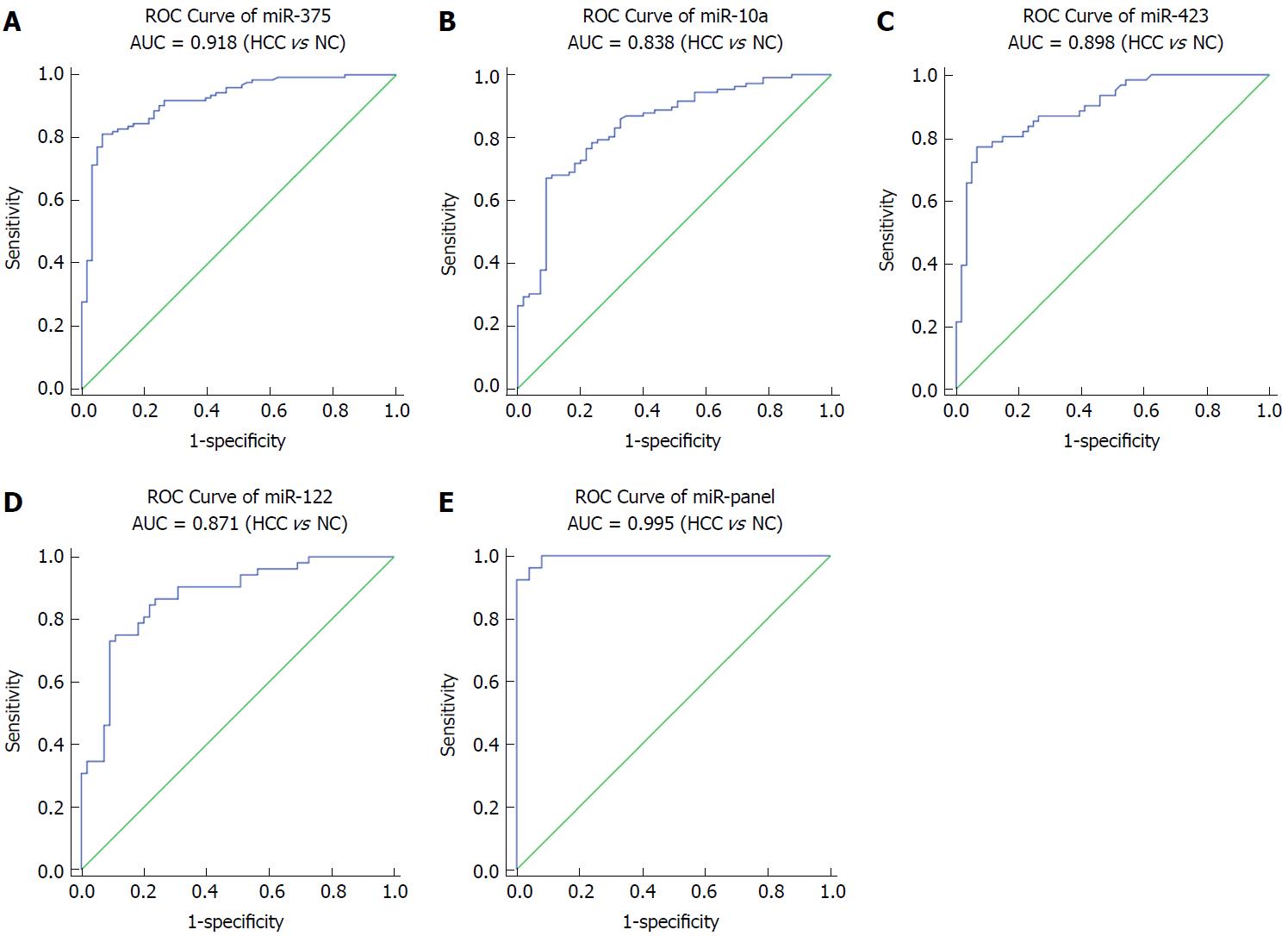
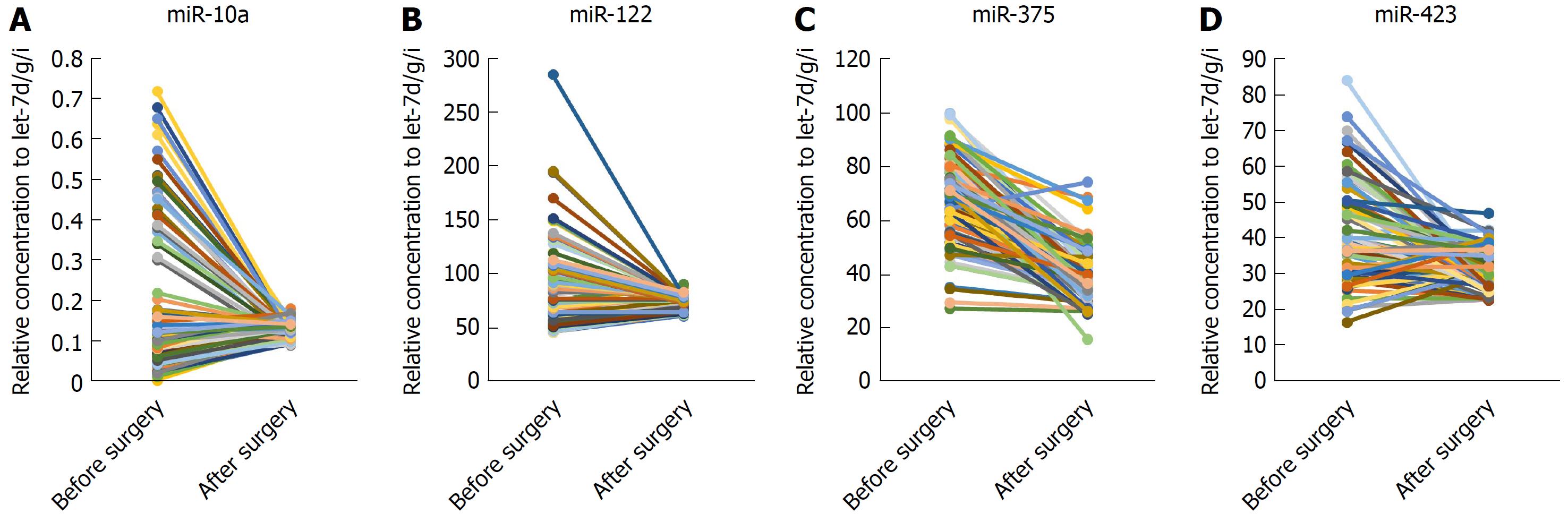
miR-375, miR-10a, miR-122 and miR-423 were found to be significantly higher in HCC than in controls (P < 0.0001), and the area under the receiver-operating-characteristic curve for the 4-miRNA panel was 0.995 (95%CI: 0.985-1). All the four miRNAs were significantly reduced after surgical removal of the tumors (P < 0.0001), while still higher than normal controls (at least P < 0.05)
The four serum miRNAs (miR-375, miR-10a, miR-122 and miR-423) could potentially serve as novel biomarkers for the diagnostic and prognostic of HCC.
Core tip: We aimed to determine a panel of serum microRNAs (miRNAs) that could be used as novel biomarkers for diagnosis of hepatocellular carcinoma (HCC). After TaqMan Low Density Array screening, and validation by RT-qPCR in two phases, we demonstrated that the four serum miRNAs (miR-375, miR-10a, miR-122 and miR-423) could potentially serve as novel biomarkers for the diagnostic and prognostic of HCC.
- Citation: An Y, Gao S, Zhao WC, Qiu BA, Xia NX, Zhang PJ, Fan ZP. Novel serum microRNAs panel on the diagnostic and prognostic implications of hepatocellular carcinoma. World J Gastroenterol 2018; 24(24): 2596-2604
- URL: https://www.wjgnet.com/1007-9327/full/v24/i24/2596.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i24.2596
According to the World Health Organization (WHO)[1,2], hepatocellular carcinoma (HCC) is the fifth most common malignancy in the world and the second leading cause of cancer-related deaths. HCC is one of the most common malignancies in China. Because early diagnosis is difficult, it often goes undetected, and once diagnosed, more than half of patients find themselves with HCC in the middle and late stages of the disease. If patients with liver cancer are not treated, the average survival time is approximately 3 mo. Thus, liver cancer remains one of the worst malignant tumors[3]. Despite this, early detection, early diagnosis, and early surgical treatment are still the keys to improved clinical outcomes in liver cancer treatment[4]. In recent years, with the advancement of diagnosis and treatment techniques for liver cancer, many patients have been diagnosed with liver cancer at early stages, and the detected tumors have been surgically removed, yielding a timely and effective treatment. However, statistics indicate that although the one-year survival rate of radical resection of hepatocellular carcinoma has increased from 39.3% to 87.0%, the 5-year survival rate after surgery is still only 15%-40%, and 62%-82% of patients relapse within two years[5].
MicroRNAs (miRNAs), are small, single-stranded RNAs of approximately 19-23 nucleotides in length. They are often located in noncoding regions of the genome and are highly conserved across evolution, allowing the regulation of gene expression at the post-transcriptional level. MiRNAs are extensive and diverse in eukaryotes, and they represent a group of noncoding RNAs. Several studies have found that miRNAs can resist the effects of RNase and are stably present in serum; additionally, the expression profiles of miRNAs in the serum of normal and cancer patients are significantly different[6-10]. MiRNAs appear to be useful biomarkers for cancer. As the 5-year survival rate of HCC patients is very poor, it is equally important to evaluate the prognostic or therapeutic efficacy such biomarkers. However, few studies of HCC to date analyze the dynamic changes of miRNA in patient serum.
In this study, Illumina next-generation sequencing (NGS) and reverse transcriptional quantitative PCR (RT-qPCR) were used to characterize the genome-wide miRNA expression profiles in sera from HCC patients and control groups, identifying a panel of serum miRNAs that could be used as novel biomarkers for diagnosis of HCC.
A total of 149 patients with HCC, 20 patients with liver cirrhosis and 15 patients with chronic hepatitis were treated from 2013 to 2016. All patients were newly diagnosed. Eighty of these patients had tumor resection before any adjuvant treatment. None of the patients in this study had acute infections or other types of cancer. In addition, 146 people who did not display any signs of disease at the time of participation in the physical examination were collected as noncancer controls. All participants have signed informed consent documents, and this study was approved by the ethics committee of hospital.
The preoperative serum of HCC patients was collected upon admission to the hospital, and the postoperative serum was collected 7 days after operation. Each patient had 5 mL of venous blood drawn, which was then centrifuged at 3000 × g for 5 min at 20 °C. The samples were then stored at -80 °C until analysis.
For TaqMan Low-Density Arrays (TLDA), equal volumes of sera from 35 HCC patients and 35 controls (400 μL each) with a similar age and sex distribution were pooled separately to form the case and control sample pools (each pool contained 14 mL total sera). Total RNA from each serum sample pool was extracted using TRIzol (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. The aqueous phase was extracted and then subjected to a 3-step acid phenol/chloroform purification to remove proteins, followed by isopropanol precipitation. The RNA pellet was solubilized in 30 μL of ribonuclease-free (RNase-free) water. RNA was then quantified using a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Fisher Scientific Inc). The amounts of the two RNAs extracted were 8.5 mg and 7.8 mg, respectively.
For RT-qPCR analysis, total RNA was extracted from 100 μL of serum. Briefly, 300 μL of RNase-free water was mixed into 100 μL of each serum and then mixed with 200 μL of acidic phenol and 200 μL of chloroform. The mixture was vortex-mixed vigorously and then incubated at room temperature for 15 min. After centrifugation at 12000 × g for 20 min at 4 °C, the phases were separated, and the aqueous layer was mixed with 2 volumes of isopropanol and 1/10 volume of 3 mol/L sodium acetate (pH 5.3) and then stored at 20 °C for 1 hour. After precipitation, samples were centrifuged at 16000 × g for 20 min at 4 °C to collect the RNA pellet, followed by washing once with 75% (V/V) ethanol and drying at room temperature for 10 min. Finally, 20 μL of RNase-free water was added to solubilize the RNA pellet, and then, the samples were stored at -80 °C until further analysis.
MicroRNA profiling was performed on 754 different human microRNAs using the TLDA (TaqMan Array Human MicroRNA A+B Cards Set v3.0) (Life Technologies). First, reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit and Megaplex RT Primers. Briefly, 3 μL of total RNA was added to 4.5 μL of the RT reaction mix (Megaplex RT Primers 10 ×, RNase Inhibitor 20 U/μL, MultiScribe Reverse Transcriptase 50 U/μL, 10 × RT Buffer, dNTPs with dTTP 100 mmol/L, nuclease-free water and MgCl2 25 mmol/L). Reverse transcription was performed in a thermal cycler (Applied Biosystems) after reactions were incubated on ice for 5 min. After reverse transcription, we performed a pre-amplification to increase the sensitivity of TLDA, using the 7900 HT Fast Real-Time PCR System (Applied Biosystems)[11]. The concentrations of miRNAs are presented as threshold cycle (Cq) values, and the calculated ΔCq of each miRNA is normalized to an internal reference suggested by the manufacturer. The equation 2-ΔΔCq was used to calculate the fold changes in miRNA expression.
qRT-PCR was performed according to the manufacturer’s instructions using the hydrolysis probe-based qRT-PCR using 7300 Sequence Detection System (Applied Biosystems). Reverse transcription was carried as previously described[8]. The reaction mixtures were incubated at 16 °C for 15 min, followed by 42 °C for 1 h and 85 °C for 5 min. Samples were then held at 4 °C for cDNA synthesis. The qRT-PCR was performed as previously described [8]. All experiments were carried out in triplicate. The qRT-PCR data was normalized to a combination of let-7d, let-7g and let-7i (let-7d/g/i) serving as an internal reference as previously described[12,13]. The 2-ΔΔCq method was performed to calculate the relative levels of miRNAs[12,14].
We performed statistical analyses using SPSS 16.0. The data were expressed as the mean ± standard deviation (SD). Differences in variables between two groups were compared using a student’s t-test or two-sided χ2 test, and P-value of < 0.05 was considered statistically significant. To evaluate the selected miRNAs’ predictive power, receiver-operating-characteristic (ROC) curves were also constructed and the area under the ROC curves (AUC) was calculated. Furthermore, to evaluate the associations between miRNAs and HCC, we performed a risk score analysis as previously described[9]. The risk score analysis was performed as previously described[9]. For each miRNA, according to a linear combination model of the expression level, a risk score function (RSF) to predict HCC was defined. There is an equation for the RSF for sample i: rsfi=Σ7j-1Wj.sij. In this equation, for a miRNA j, the weight of its risk score is represented as Wj, and its risk score in sample i is represented as sij. To determine the Ws, four univariate logistic regression models were fitted to each of the risk scores using the disease status[15,16]. To indicate a miRNAs’ contribution to the RSF, we used the regression coefficient of risk score as the weight. Samples were ranked according to their RSF and then divided into a high-risk group, representing the predicted HCC cases, and a low-risk group, representing the predicted control individuals. To evaluate the diagnostic effects of the miRNA panel, frequency tables and ROC curves were analyzed to find an appropriate cutoff point.
To determine the set of effective miRNAs that would be useful as biomarkers for monitoring tumor dynamics and diagnosing and forecasting the prognosis of HCC, we designed a case control study (Figure 1, Table 1). In total, 35 HCC and 35 normal control serum samples were pooled to analyze the expression of miRNA using TLDA. A miRNA was considered upregulated if its Cq value was < 25 in HCC and its concentration showed > 2-fold (overexpressed in HCC compared to the control). Comparative analysis of all 754 miRNAs revealed that 53 miRNAs (7.03%) were differentially expressed in the HCC set. The miRNAs that had Cq values that were < 25 and had expression levels in HCC more 35-fold higher than those of the normal controls were selected for additional RT-qPCR validation. Finally, 9 miRNAs including miR-375, miR-10a, miR-23a, miR-125b, miR-122, miR-320a, miR-320b, miR-99a and miR-423 were selected for further analyses.
| Variables | HCC (n = 149) | Normal controls (n = 149) | P value |
| Average age (yr) | 52.7 ± 10.84 | 51.5 ± 8.26 | 0.3101 |
| Age (yr) | 0.1582 | ||
| ≤ 59 | 56 (37.6) | 68 (45.6) | |
| > 59 | 93 (62.4) | 81 (54.4) | |
| Sex | 0.1482 | ||
| Male | 101 (67.8) | 89 (59.7) | |
| Female | 48 (32.2) | 60 (40.3) | |
| Alcohol consumption | 0.0692 | ||
| Ever or Current | 104 (69.8) | 89 (59.7) | |
| Never | 45 (30.2) | 60 (40.3) | |
| TNM Stage | |||
| I | 5 (3.4) | ||
| II | 12 (8.0) | ||
| IIIA | 53 (35.6) | ||
| IIIB | 63 (42.3) | ||
| IVA | 14 (9.4) | ||
| IVB | 2 (1.3) | ||
| Differentiation grade | |||
| High | 25 (16.8) | ||
| Middle | 81 (54.4) | ||
| Low | 43 (28.9) |
Although the TLDA is a RT-PCR-based assay, some inaccuracy in the measured Cq values is possible, due to a single sample in the pooled sample of the two groups measuring abnormally for a particular miRNA. Therefore, the array results needed to be validated using RT-qPCR analysis performed at the individual serum sample level. The 9 most dramatically differentially expressed miRNAs in TLDA were further examined in an independent cohort of 84 HCC patients using an individual RT-qPCR assay. As a result, 4 serum miRNAs (miR-375, miR-10a, miR-122 and miR-423) were found to be statistically significantly over expressed in HCC patients compared with those of the normal controls (at most P < 0.0001) (Figure 2A-D), and 3 miRNAs (miR-23a, miR-125b and miR-99a) were also increased in HCC patients but were not statistically significant. While the other 2 serum miRNAs (miR-320a and miR-320b) tended to be decreased in HCC patients, none of them showed a statistically significantly decrease in expression.
In another validation cohort including 30 HCC patients and 30 matched controls, we examined the expression of these 4 miRNAs by RT-qPCR. The results demonstrated that the 4 serum miRNAs were significantly increased in HCC cases over that seen in the controls (at least P < 0.05), similar to the former cohort.
Next, we conducted ROC curve analyses on each of the individual five serum miRNAs to assess the diagnostic value of each for discriminating between HCC and normal subjects in a validation cohort. The AUCs for these miRNAs were 0.918, 0.838, 0.898 and 0.871, respectively (Figure 3A-D). The ROC curve for the panel demonstrated a remarkable accuracy for the diagnosis of HCC, demonstrated by an AUC of 0.995 (95%CI: 0.985-1), which was much larger than the AUC values for each of the 4 individual miRNAs (Figure 3E). We then performed a risk score analysis to further evaluate the potential of these 4 miRNAs for distinguishing HCC patients from normal controls. In the validation phase, the cutoff value was 2.258, and 8 healthy controls had a risk score < 2.258, while 73 of the 84 HCC patients exhibited a risk score > 2.2584 (Table 2).
| Score | 0-2.258 | > 2.258 | PPV | NPV |
| Normal subject (n = 84) | 76 | 8 | 0.87 | |
| HCC (n = 84) | 11 | 73 | 0.90 | |
| Total | 87 | 81 |
To identify the specificity of the miRNAs selected in an additional validation cohort for HCC, we also tested the expression of these miRNAs in the serum of patients with liver cirrhosis and chronic hepatitis. We found that miR-10a and miR-122 were significantly increased in the serum of patients with chronic hepatitis compared with those of the normal controls (P < 0.001), while there was no significant difference between patients and normal controls for the expression of the two other miRNAs.
Next, we examined whether the four miRNAs could be used to assess tumor dynamics in patients with HCC. The expression of these miRNAs in serum samples that were divided into two paired groups was compared before and after surgery in 80 HCC patients. We found that the four miRNAs were all reduced after operation significantly (P < 0.0001) but still were higher than in the normal controls (at least P < 0.05) (Figure 4A-D).
In this study, through the use of a TaqMan Low Density Array (TLDA) and individual RT-qPCR validations, we examined the expression of serum miRNA in HCC systematically and found a new miRNA panel (miR-10a, miR-122, miR-375 and miR-423) that can distinguish HCC patients from normal controls effectively and that is significantly downregulated after surgery. It has been reported previously that miR-21, miR-122, mi-125a/b, miR-199a/b, miR-221, miR-222, miR-223, miR-224, miR-16-2-3p, miR-92a-3p, miR-107, and 3126-5p were all significantly altered in HCC patients compared with those of the controls[17-20]. In contrast, we found that the 4-miRNA panel in our study had a high AUC of ROC curves at 0.995. We also measured the expression of these miRNAs in the serum of patients with liver cirrhosis and chronic hepatitis and found that while miR-10a and miR-122 were significantly increased in the serum of patients with chronic hepatitis compared with those of normal controls (P < 0.001), there was no significant difference between the patients and normal controls in the expression of the two other miRNAs. Therefore, for diagnosis of HCC, the 4-miRNA panel discovered by our work is more specific than any single miRNA.
To date, there have been few reports about the differential expression of miRNAs in paired postoperative and preoperative plasma/serum samples from HCC patients. We compared the level of expression of the 4 miRNAs in paired serum samples before and after surgery in a large cohort (n = 80) of HCC patients and found that expression of all 4 was reduced significantly after surgery but was still higher than in the normal controls. Therefore, the 4 miRNAs were considered effective to monitor the tumor dynamics of HCC patients. Studies show that there are some tumor-derived miRNAs[10,21-23], and serum miRNAs secreted by tissues and circulating cells affected by this disease[3]. In cancer patients, the major reason for differential miRNA expression profiles may be the passive or active release of miRNAs by tumor cells. Although the levels of these 4 miRNAs in the serum after surgery did not return to basal levels, their expression levels were significantly reduced after surgery, demonstrating to some extent that tumors can secrete or release some miRNAs. However, further research is needed to confirm this.
In primary colorectal cancer (CRC), matrix metalloproteinase 14 (MMP14) and actin gamma 1 (ACTG1) were validated as target genes of miR-10a in CRC cells, and miR-10a can promote migration and invasion in vitro. miR-10a inhibited metastasis in vivo by regulating the epithelial-to-mesenchymal transition and anoikis[24]. Anti-dsDNA IgG Ab downregulated miR-10a expression in human mesangial cells (HMCs), resulting in the induction of various target genes involved in HMC proliferation and chemokine expression[25]. MiR-10a contributes to NSCLC by targeting PTEN[26]. There are also many studies on the role of miR-122 in hepatic diseases. A novel hepatitis B virus (HBV) mRNA-miR-122-PBF regulatory pathway that facilitates malignant hepatocyte growth and invasion in chronic hepatitis B (CHB) that may contribute to CHB-induced HCC development and progression has been reported recently[27]. Inflammation-induced miR-122 downregulation in hepatitis contributes to carcinogenesis, and it has been suggested that increasing miR-122 may be an effective strategy for preventing HCC development in CHB patients[28]. MiR-375 was involved in the Hippo pathway by targeting YAP1-TEAD4-CTGF axis to regulate the progress of gastric tumorigenesis[29] and may be negatively regulated by Snail and involved in gastric cancer cell migration and invasion by potentially targeting JAK2[30]. MiR-423 functions as an oncogene in glioma tissues by suppressing ING-4, and it has been implicated that it has therapeutic potential for the treatment of glioma[31]. Mir423 can also induce endoplasmic reticulum (ER) stress and oxidative stress by inhibiting GSTM1 and can suppresses repair after acute kidney injury[32]. Further investigation of these 4 miRNAs (miR-10a, miR-122, miR-375 and miR-423), including identifying the target genes and mechanisms involved in their altered expression is necessary for understanding the significance of their differential expression in HCC patients. However, there are still some limitations in our study. First, the specificity of AFP which were used often to evaluate the diagnostic value of HCC, but our diagnostic model were not compared with AFP. Second, the sample size was relatively small in our study, more samples are needed to evaluate the diagnostic value. Third, the target gene of the selected miRNAs were not detected.
When an RT-qPCR assay is performed, proper normalization by using a stably expressed internal reference gene is critical for accurately quantifying RNA levels. To date, no consensus on endogenous gene controls has been established in the study of circulating miRNAs. Different studies have used different genes as their endogenous controls, such as RNU6B, RNU44, RNU48, and miR-16[33,34], with different results in each case. In this study, to normalize serum miRNAs, we employed the combination of let-7d, let-7g and let-7i as reference genes, the approach of which is statistically superior to a single reference gene and can better correct for experimental changes[9]. Results demonstrated that between the HCC and control groups, let-7d/g/i remained relatively steady.
In summary, we found a novel serum miRNA panel (miR-10a, miR-122, miR-375 and miR-423) diagnostic for HCC, and all 4 of these serum miRNAs were increased in HCC patients compared to the levels in normal controls. Specifically, we have also demonstrated that these 4 miRNAs may serve as biomarkers for predicting the prognosis of HCC.
Detection, diagnosis, and surgical treatment are the key to the improvement of liver cancer treatment. However, there is little extensive analysis of the dynamic changes and prognostic value of serum microRNAs (miRNAs) in hepatocellular carcinoma (HCC) patients.
miRNAs can be used as useful biomarkers for cancer. With a very poor 5-year survival rate in HCC, markers to assess prognosis or treatment effect are equally important.
TaqMan Low Density Array was used to screen in two pooled serum samples respectively from 35 HCC and 35 normal controls, and then the screened miRNAs were validated individually by RT-qPCR in two phases. The selected miRNAs after operation and their prognostic value were also evaluated.
miR-375, miR-10a, miR-122 and miR-423 were found to be significantly higher in HCC than in controls, and tAUC for the 4-miRNA joint diagnostic panel was 0.995 (95%CI: 0.778-0.934). The four miRNAs were significantly decreased after surgical removal, but still was higher than normal controls.
The four serum miRNAs (miR-375, miR-10a, miR-122 and miR-423) could potentially serve as novel biomarkers for the diagnostic and prognostic of HCC.
This study provides an effective and non-invasive detection method for the detection of HCC, however, more samples and the early detection value should be investigated in the future study.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Hayes MJ, Kato S, Yao DF S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Yin SY
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18694] [Cited by in F6Publishing: 20838] [Article Influence: 2315.3] [Reference Citation Analysis (2)] |
| 2. | Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245-1255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3249] [Cited by in F6Publishing: 3477] [Article Influence: 289.8] [Reference Citation Analysis (3)] |
| 3. | Park HJ, Choi BI, Lee ES, Park SB, Lee JB. How to Differentiate Borderline Hepatic Nodules in Hepatocarcinogenesis: Emphasis on Imaging Diagnosis. Liver Cancer. 2017;6:189-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 51] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 4. | Girotra M, Soota K, Dhaliwal AS, Abraham RR, Garcia-Saenz-de-Sicilia M, Tharian B. Utility of endoscopic ultrasound and endoscopy in diagnosis and management of hepatocellular carcinoma and its complications: What does endoscopic ultrasonography offer above and beyond conventional cross-sectional imaging? World J Gastrointest Endosc. 2018;10:56-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 6. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3218] [Cited by in F6Publishing: 3383] [Article Influence: 211.4] [Reference Citation Analysis (0)] |
| 7. | Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, Dai J, Hu Z, Zhou X, Chen L. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871-1879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 251] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 8. | Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, Huang D, Chen X, Zhang H, Zhuang R. A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer. 2011;47:784-791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 308] [Cited by in F6Publishing: 342] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 9. | Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N. Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer. 2013;132:116-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 142] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 10. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5636] [Cited by in F6Publishing: 6046] [Article Influence: 377.9] [Reference Citation Analysis (0)] |
| 11. | Luo Y, Wang C, Chen X, Zhong T, Cai X, Chen S, Shi Y, Hu J, Guan X, Xia Z. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin Chem. 2013;59:658-666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Chen X, Liang H, Guan D, Wang C, Hu X, Cui L, Chen S, Zhang C, Zhang J, Zen K. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One. 2013;8:e79652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 13. | Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, Zhang Y, Chen X, Wang J, Zen K. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. 2014;9:e92292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 14. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117419] [Cited by in F6Publishing: 121468] [Article Influence: 5281.2] [Reference Citation Analysis (0)] |
| 15. | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [PubMed] [Cited in This Article: ] |
| 16. | Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245-5250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5406] [Cited by in F6Publishing: 4894] [Article Influence: 244.7] [Reference Citation Analysis (0)] |
| 17. | Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma X, Wei Q, Han L. Serum microRNA panel for early diagnosis of the onset of hepatocellular carcinoma. Medicine (Baltimore). 2017;96:e5642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 18. | Fiorino S, Bacchi-Reggiani ML, Visani M, Acquaviva G, Fornelli A, Masetti M, Tura A, Grizzi F, Zanello M, Mastrangelo L. MicroRNAs as possible biomarkers for diagnosis and prognosis of hepatitis B- and C-related-hepatocellular-carcinoma. World J Gastroenterol. 2016;22:3907-3936. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 46] [Cited by in F6Publishing: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 19. | Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21:9853-9862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 58] [Cited by in F6Publishing: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Greene CM, Varley RB, Lawless MW. MicroRNAs and liver cancer associated with iron overload: therapeutic targets unravelled. World J Gastroenterol. 2013;19:5212-5226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 40] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Uotani K, Fujiwara T, Yoshida A, Iwata S, Morita T, Kiyono M, Yokoo S, Kunisada T, Takeda K, Hasei J. Circulating MicroRNA-92b-3p as a Novel Biomarker for Monitoring of Synovial Sarcoma. Sci Rep. 2017;7:14634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 22. | Kikete S, Chu X, Wang L, Bian Y. Endogenous and tumour-derived microRNAs regulate cross-presentation in dendritic cells and consequently cytotoxic T cell function. Cytotechnology. 2016;68:2223-2233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Croset M, Kan C, Clézardin P. Tumour-derived miRNAs and bone metastasis. Bonekey Rep. 2015;4:688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 24. | Liu Y, Zhang Y, Wu H, Li Y, Zhang Y, Liu M, Li X, Tang H. miR-10a suppresses colorectal cancer metastasis by modulating the epithelial-to-mesenchymal transition and anoikis. Cell Death Dis. 2017;8:e2739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 25. | Tangtanatakul P, Thammasate B, Jacquet A, Reantragoon R, Pisitkun T, Avihingsanon Y, Leelahavanichkul A, Hirankarn N. Transcriptomic profiling in human mesangial cells using patient-derived lupus autoantibodies identified miR-10a as a potential regulator of IL8. Sci Rep. 2017;7:14517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Yu T, Liu L, Li J, Yan M, Lin H, Liu Y, Chu D, Tu H, Gu A, Yao M. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6:30239-30250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 100] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 27. | Li C, Wang Y, Wang S, Wu B, Hao J, Fan H, Ju Y, Ding Y, Chen L, Chu X. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J Virol. 2013;87:2193-2205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 117] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 28. | Li C, Deng M, Hu J, Li X, Chen L, Ju Y, Hao J, Meng S. Chronic inflammation contributes to the development of hepatocellular carcinoma by decreasing miR-122 levels. Oncotarget. 2016;7:17021-17034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 29. | Kang W, Huang T, Zhou Y, Zhang J, Lung RWM, Tong JHM, Chan AWH, Zhang B, Wong CC, Wu F. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018;9:92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 30. | Xu Y, Jin J, Liu Y, Huang Z, Deng Y, You T, Zhou T, Si J, Zhuo W. Snail-regulated MiR-375 inhibits migration and invasion of gastric cancer cells by targeting JAK2. PLoS One. 2014;9:e99516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | Li S, Zeng A, Hu Q, Yan W, Liu Y, You Y. miR-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017;19:55-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 32. | Yuan XP, Liu LS, Chen CB, Zhou J, Zheng YT, Wang XP, Han M, Wang CX. MicroRNA-423-5p facilitates hypoxia/reoxygenation-induced apoptosis in renal proximal tubular epithelial cells by targeting GSTM1 via endoplasmic reticulum stress. Oncotarget. 2017;8:82064-82077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 33. | Chang KH, Mestdagh P, Vandesompele J, Kerin MJ, Miller N. MicroRNA expression profiling to identify and validate reference genes for relative quantification in colorectal cancer. BMC Cancer. 2010;10:173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 184] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 34. | Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50:298-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 867] [Cited by in F6Publishing: 901] [Article Influence: 64.4] [Reference Citation Analysis (0)] |