Published online Nov 28, 2015. doi: 10.3748/wjg.v21.i44.12605
Peer-review started: May 25, 2015
First decision: June 25, 2015
Revised: July 26, 2015
Accepted: October 12, 2015
Article in press: October 13, 2015
Published online: November 28, 2015
AIM: To achieve a better understanding of the molecular mechanisms of microRNA expression changes involved in hepatocellular carcinoma.
METHODS: In this research process, patients were not treated with antivirals, immunosuppressants or immunomodulators for at least 6 mo before collecting serum. The study population was composed of 35 outpatient hepatitis B virus (HBV) cases and 12 healthy control cases from the Affiliated Hospital of Inner Mongolia Medical University (Inner Mongolia, China) from July 2013 to April 2014. The 35 HBV cases were divided into two groups: a hepatocirrhosis group with 20 cases and a liver cancer group with 15 cases. All 35 cases carried HBsAg. The diagnostic criteria followed the European Association for the Study of the Liver 2012 (EASL2012) standards. MicroRNA (miRNA) was extracted from a control group of patients, a group with hepatocirrhosis and a group with liver cancer and its quality was analyzed using the human V2 microRNA expression beadchip. Cluster analysis and a radar chart were then applied to the miRNA changes.
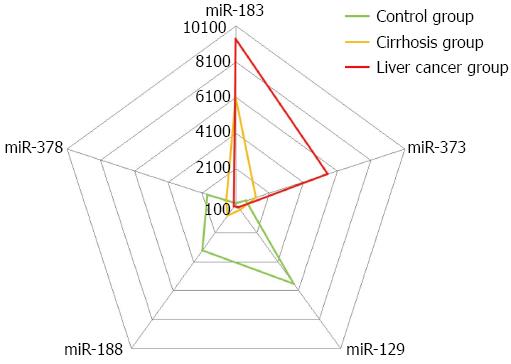
RESULTS: The miRNA-qualified rate of human serum samples was 93%. The concentration of a single sample was > 200 ng/μL and the volume was > 5 μL. All miRNA serum samples were uncontaminated by the genome. The Mann-Whitney test showed significant differences in miRNA between each group, with a detection P-value of < 0.05. Illumina software was set up with Diff Score set to ± 13, meaning that P = 0.001.There were significant changes in miRNA expression between the three groups. miRNA-183 was the most up-regulated, followed by miRNA-373. miRNA-129 and miRNA-188 were both strongly down-regulated and miRNA-378 was down-regulated a small amount. The liver cancer group had greater changes, which indicated that changes in miRNA expression levels were caused by hepatocirrhosis. The liver cancer disease course then further increased these changes. In the pentagon created by these five miRNAs, three groups showed significant deviation. The liver cancer group had a bigger deviation trend. The chart indicated that miRNA expression changes occurred in the hepatocirrhosis group, which increased in the liver cancer disease course and were irreversible.
CONCLUSION: There was a significant relationship between the irreversible up-regulation of miRNA-183/373 and down-regulation of miRNA-129/188/378 and incidences of hepatocirrhosis and liver cancer.
Core tip: There was a better understanding of the molecular mechanisms of microRNA (miRNA) expression changes involved in hepatocellular carcinoma, associated patients with aggressive malignancy and poor prognosis. The 35 hepatitis B virus cases were divided into two groups, a hepatocirrhosis group with 20 cases and a liver cancer group with 15 cases. 12 healthy people were used as a control. There were significant changes in miRNA expression between the three groups. miRNA-183 was the most up-regulated, followed by miRNA-373. miRNA-129 and miRNA-188 were both strongly down-regulated and miRNA-378 was down-regulated a small amount. The liver cancer group had greater changes, which indicated that changes in miRNA expression levels were caused by hepatocirrhosis. The liver cancer disease course then further increased these changes.
- Citation: Niu JX, Meng XK, Ren JJ. Studied microRNA gene expression in human hepatocellular carcinoma by microRNA microarray techniques. World J Gastroenterol 2015; 21(44): 12605-12611
- URL: https://www.wjgnet.com/1007-9327/full/v21/i44/12605.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i44.12605
Hepatocellular carcinoma (HCC) is one of the most common human malignancies worldwide, especially in East Asia and South Africa[1,2]. Due to its high malignant potential, HCC ranks as the third leading cause of cancer death in the world, resulting in almost 600000 deaths each year. Despite great improvements in treatment options, the long-term survival of patients with HCC remains unsatisfactory, with a 5-year survival rate of 20% to 30% reported in the literature. The treatment of advanced and metastatic HCC presents many challenging problems[3-5].
Carcinogenesis and the progression of HCC are multistage processes that involve many growth factors, oncogenes and tumor suppressor genes. Elucidating the molecular events underlying the tumorigenesis of HCC is important for its screening, prevention and treatment. In recent decades, many factors involved in the pathogenesis of HCC have been identified, including p53, Rb, PI3K, Akt, MAPK and many others[6-10]. However, the molecular mechanisms of HCC are still largely unknown.
Recent studies using DNA microarray techniques have identified unique gene expression profiles in hepatitis B and C virus-associated HCC[11,12]. Gene expression profiling also allows us to distinguish HCC from normal tissue or preneoplastic lesions and to evaluate metastatic or recurrent potential. These unique genes or gene products associated with malignant transformation and recurrent or metastatic potential may serve as molecular markers for early diagnosis and allow prediction of prognosis and responsiveness to therapy[13-15].
Although vaccination could reduce the incidence of hepatitis B, hepatocirrhosis and liver cancer remain major global health problems, the treatment of which poses many challenges. The World Health Organization reported 3.5 million new cases of chronic hepatitis in 2013 and 0.7 million deaths of chronic hepatitis patients from liver failure, cirrhosis and primary hepatocellular carcinoma[16-18]. The pathogenesis of HCC remains unclear. The inhibition microenvironment, comprising inhibitory receptors, inhibitory cells and immunosuppressant cytokines, can accelerate disease progression. In this study, the serum from hepatitis B virus (HBV) infected patients was used to analyze the different disease courses of HCC.
MicroRNAs (miRNAs) are short non-coding RNA molecules, similar to siRNAs. miRNAs consist of 20-25 nucleotides and their sequences are complementary to the 3’ untranslated regions of messenger RNAs. Binding results in inhibition of translation and gene silencing. Research has shown that thousands of human protein coding genes are regulated by miRNA[19-22].
It is suggested that miRNA plays an important role in the regulation processes of cell growth and development. miRNA is widely expressed in tissues and also in different types of cancer tissue. Research has shown that miRNAs can regulate the expression of target genes, thereby regulating liver cell proliferation and differentiation[20,21].
As such, miRNAs are significant in the ability of the liver to maintain normal physiological processes. To study the biological functions of miRNA, miRNA target genes must first be identified. Although miRNA has been studied intensely, the identification of miRNA target genes is still very challenging because each miRNA has hundreds of target genes and each miRNA can have a different regulatory process for each target gene. In recent years, studies have found that miRNA can bind to the 5’-UTR and the promoter region. This greatly increases the difficulty of studying miRNA but also provides ideas for understanding its complex biological functions[19-21].
The development of microarray technology, which allows us to undertake parallel analyses of many genes, has led to a new era in medical science. New genomic high-throughput technologies, such as RNA microarrays, considerably facilitate the molecular profiling of human tumors. Thousands of genes can now be analyzed in a simple hybridization microarray. The expression profile of a single tumor reflects the state of events of an individual malignancy at a certain time point. To generalize the findings and provide conclusive evidence for the involvement of a molecular alteration, it is often necessary to analyze several hundred tumors. Using traditional molecular pathology, such verification could take several months or even years to complete. To facilitate translational research in a large scale manner, technology was developed recently for making high density arrays of tumor tissue specimens. These arrays can be used for rapid miRNA evaluation of gene copy number and expression in thousands of tumors simultaneously. Current research is mostly concerned with the differentiation of one particular variable, such as HBV infection, environmental carcinogens, metastasis and recurrence, and sensitivity to chemo agents. It is rare for five kinds of gene expression profiling to be used. We have combined RNA microarray and TMA techniques to identify differentially expressed genes in the development and progression of human HCC[23-25]. In this study, we investigated the correlations between miRNA and clinical pathological characterization and our findings provide a comprehensive understanding of the molecular mechanisms of HCC and some new potential therapeutic targets.
The study population was composed of 35 outpatient HBV cases and 12 healthy control cases from the Affiliated Hospital of Inner Mongolia Medical University (Inner Mongolia, China) from July 2013 to April 2014.
The 35 HBV cases were divided into two groups, a hepatocirrhosis group with 20 cases and a liver cancer group with 15 cases. All 35 cases carried HBsAg. The diagnostic criteria followed the European Association for the Study of the Liver 2012 (EASL2012) standards.
Patients with other hepatitis or HIV infections, other causes of liver damage, autoimmune disorders or neoplasm were excluded from the study, as well as pregnant or lactating women. In this research process, patients were not treated with antivirals, immunosuppressants or immunomodulators for at least 6 mo before collecting serum.
Medical Ethics Committee of Inner Mongolia Medical College approval was obtained and all patients involved had previously provided their written, informed consent to have their clinical and pathogenic information used for research.
Blood samples were collected at the first hospital visit. Serum was separated and stored under -80 °C. MiRNApure Mini Kit (Cat. No. CW0627) was purchased from CWBiotech Ltd. Nucleic acids and miRNA were extracted separately from the serum in accordance with the instructions.
Electrophoresis apparatus and slot were DYY-6B and CQU-200, the gel imaging instrument was Gel Doc 2000, and the spectrophotometer was NanoDrop 2000.
Logarithms were used to convert data with positive skew into a normal distribution. For homogenous data, analysis of variance, Student-Newman-Keuls and Pearson’s correlation were used. For inhomogeneous data, Kruskal-Wallis, Games-Howell and Spearman’s correlation analysis were used. All analyses were carried out using SPSS 17.0 software (SPSS Inc, Chicago, IL, United States). Values less than 0.05 were considered to be statistically significant.
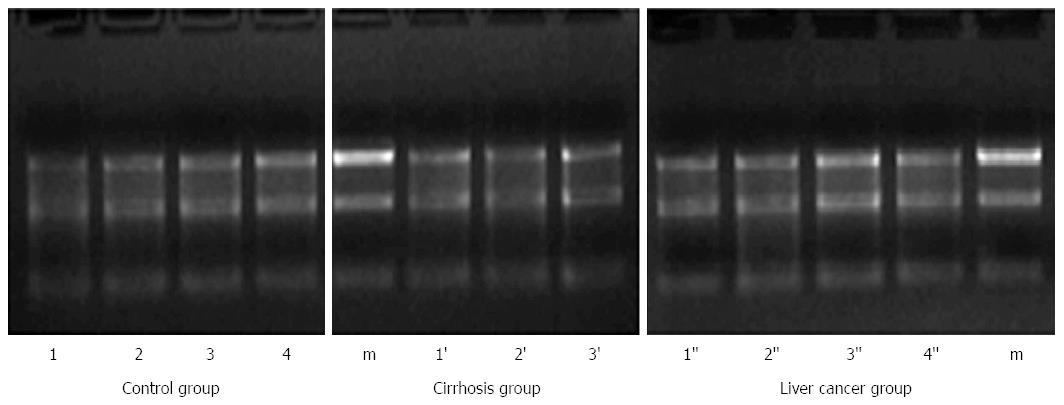
The miRNA-qualified rate of human serum samples was 93%. Three serum samples did not meet the requirements. Optical densities at 260-280 nm were between 1.7 and 2.1. The concentration of a single sample was > 200 ng/μL and the volume was > 5 μL. All miRNA serum samples were uncontaminated by the genome. The results of electrophoresis of miRNA in a 1.2% agarose gel are shown in Figure 1, displaying the correct molecular weight and a high concentration of miRNA.
Human miRNA from the Illumine Corporation were used as the gene chip, tested on the Illumine BeadArray. The human V2 miRNA expression beadchip was tested for quality control, including a negative control, a PAP control, query oligo annealing controls, mismatch controls, array hybridization controls and contamination controls.
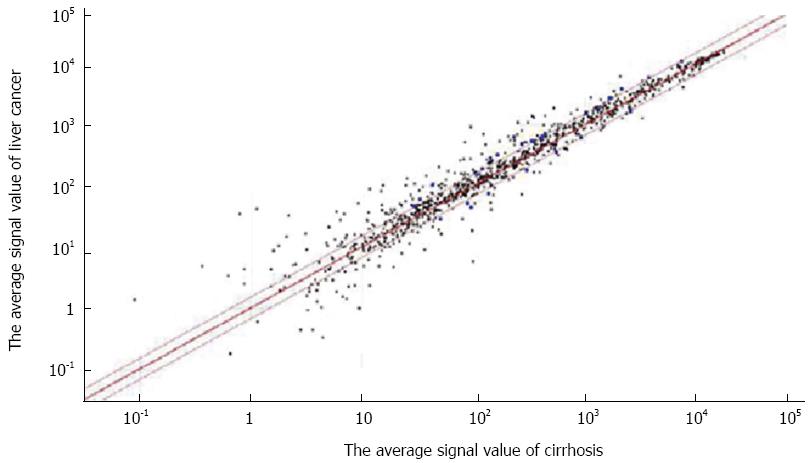
GenomeStudio (Illumina) was used to calculate the signal value of each point on the gene chip after scanning by the BeadArray Reader software (Illumina). The Mann-Whitney test showed significant differences in miRNA between each group, with a detection p-value of < 0.05. Illumina software was set up with Diff Score set to ± 13, meaning that P = 0.001. The scatter diagram shown in Figure 2 was drawn by the software, without the negative logarithm loop. The red line is the cut-off line: the upper parts were > 1.5-fold and the lower parts were < 0.67-fold.
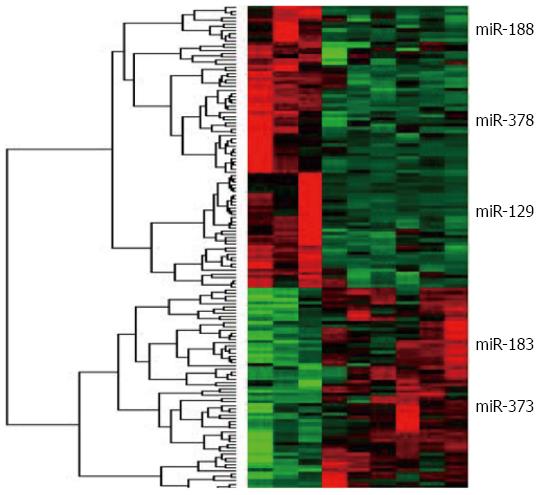
Cluster3.0 software was used to analyze the differences in miRNA between each group. MiRanda was used for miRNA target prediction (http://www.microrna.org) which is shown in Figure 3. Three groups showed significant changes in miRNA expression. The expression of five important miRNAs changed. miRNA-183 was the most up-regulated, followed by miRNA-373. miRNA-129 and miRNA-188 were greatly down-regulated and miRNA-378 was down-regulated a small amount.
The quantization map shows the expression difference in human serum between these five miRNAs. The radar chart in Figure 4 shows the changes in each miRNA in comparison to the baseline of the control group.
In the pentagon created by these five miRNAs, three groups showed significant deviation. The liver cancer group had a bigger deviation trend. The chart indicated that miRNA expression changes occurred in the hepatocirrhosis group, which increased in the liver cancer disease course and were irreversible.
Hepatocellular carcinoma, an aggressive malignancy with poor prognosis and one of the most common tumors in humans, has become a leading cause of cancer-related death in adults from Asia and Africa. Despite advances in the diagnosis and treatment of HCC, the prognosis of patients with HCC remains dismal. The poor prognosis of HCC has been associated with recurrence and metastasis[17-24]. Therefore, a better understanding of the molecular mechanisms involved in HCC development and metastasis is needed.
Early diagnosis is essential for cancer prevention and control. Previously, we found that a specific course of liver cancer disease had a different protein expression and different miRNA regulation. Recurrent chromosomal aberrations are often observed in HCC but little is known about the role of functional non-coding sequences, particularly miRNA, at the chromosomal breakpoints[14-16].
miRNAs are small non-coding RNAs that function as key regulators of gene expression at the post-transcriptional level. They play important roles in cell proliferation, differentiation and apoptosis.
A study from Anhui Medical University investigated the functions of miRNA-183 in HCC and discussed the construction of an artificial miRNA cluster expression vector[26]. The miRNA-183 expression profile from HCC tumor tissues and adjacent normal liver tissues were compared using real-time PCR. The results showed that miRNA-183 was significantly up-regulated (2 to 300 fold) in 68% of tumors. The author suggested that miRNA-183, which is up-regulated in HCC, repressed the expression of the tumor-suppressor PDCD4 post-transcriptionally and inhibited TGF-β1-induced apoptosis in human HCC cells. Therefore, miRNA-183 may play an important role in HCC development. Their artificial miRNA cluster efficiently expressed mature miRNA and miRNA procession suppressed the expression of the protein with miRNA host genes. Moreover, their artificial miRNA cluster on cell cycle arrest was more effective than single miRNAs.
Disturbance of miRNA expression and function results in tumors. Different miRNA have different roles as they regulate the expression of different target genes. miRNAs that originate from the same pre-miRNA transcript can have different functions, such as miRNA-371/miRNA-373. A study from Kunming University of Science and Technology showed that the molecular mechanisms of miRNA-373 related to the occurrence and development of tumors[27]. The real-time quantitative PCR results showed that there was a 6.684-fold decrease in miRNA-373 in BCSCs compared to MCF-7 cells and that EIF4A1 was a target gene of miRNA-373. However, our study found that more miRNA-373 was found in serum from patients with liver cancer and hepatocirrhosis, which may be due to differences in the cancer types.
The results from the bioinformatics prediction showed that miRNA-373 targeted genes involved in cell proliferation, apoptosis, cell cycle regulation, cell signal transduction, ontogenesis, tumor suppression and other closely related genes involved in tumor development and metastasis. These results indicate that some of the imbalance between the expression levels of key miRNAs might be important in breast and liver cancer occurrence.
A study from the National Engineering Center for Biochip in Shanghai investigated the diagnostic value of serum miRNA, including miRNA-129 in colorectal cancer (CRC)[28,29]. They identified 10 serum-specific miRNAs from patients with CRC. A set of serum-specific miRNAs, including miRNA-129, that are considered to be biomarkers for CRC detection were validated by RFQ-PCR. The area under the receiver operating characteristic curve for this set of serum miRNAs reached a maximum value of 0.914 with a sensitivity of 77.78% and a specific sensitivity of 100% for CRC. The set of serum-specific miRNA-129s may become a group of feasible and effective indices in the screening and early diagnosis of CRC, which we also found in our study.
A study from Central South University found that miRNA-188 was frequently down-regulated in HCC[30]. miRNA-188 expression was correlated with clinicopathological characteristics and the prognosis of HCC. miRNA-188 inhibited HCC cell proliferation and metastasis in vitro. Its direct downstream target was AAC11.
A study from Soochow University found that miRNA-378 may suppress the growth characteristics of HBV-related HCC by directly targeting the IGF 1R 3’-UTR and inhibiting its expression[31].
Additionally, it was reported that 22 miRNAs were often amplified or deleted in HCC and that miRNA-151 was correlated with intrahepatic metastasis of HCC, which we did not find in our study[32]. miRNA-151 was often expressed together with its host gene FAK, focal adhesion kinase, which significantly increased HCC cell migration and invasion in vitro and in vivo, mainly through miRNA-151-5p, but not through miRNA-151-3p. As such, more research into miRNA, tumor invasion and metastasis of HCC is needed.
Hepatocellular carcinoma (HCC) is one of the most common human malignancies worldwide, especially in East Asia and South Africa. MicroRNAs (miRNAs) are short non-coding RNA molecules, similar to siRNAs. Recent studies using DNA microarray techniques have identified unique gene expression profiles in hepatitis B and C virus-associated HCC. As such, a better understanding of the molecular mechanisms of miRNA expression changes involved in HCC is needed.
A study from Anhui Medical University investigated the functions of miRNA-183 in HCC and discussed the construction of an artificial miRNA cluster expression vector. A study from Kunming University of Science and Technology showed that the molecular mechanisms of miRNA-373 related to the occurrence and development of tumors. A study from the National Engineering Center for Biochip in Shanghai investigated the diagnostic value of serum miRNA, including miRNA-129, in colorectal cancer. A study from Central South University found that miRNA-188 was frequently down-regulated in HCC.
A better understanding of the molecular mechanisms involved in HCC development and metastasis is needed. Early diagnosis is essential for cancer prevention and control. Previously, the authors found that a specific course of liver cancer disease had different protein expression and different miRNA regulation. Cluster analysis and a radar chart were applied to the miRNA changes. By using a gene chip, large-scale integration detection could be made at one time which was more meaningful for the same specimen.
Early diagnosis is essential for cancer prevention and control. The conclusion from the present study is helpful for early diagnosis and intervention. Drug development might be a process change of miRNA expression.
Overall, this study is well designed and the manuscript is well written.
P- Reviewer: Maneesh D, Mulvihill SJ S- Editor: Yu J L- Editor: Roemmele A E- Editor: Ma S
| 1. | Wang X, Lin SX, Tao J, Wei XQ, Liu YT, Chen YM, Wu B. Study of liver cirrhosis over ten consecutive years in Southern China. World J Gastroenterol. 2014;20:13546-13555. [PubMed] [Cited in This Article: ] |
| 2. | Paul SB, Shalimar V, Gamanagatti SR, Sharma H, Dhamija E, Acharya SK. Incidence and risk factors of hepatocellular carcinoma in patients with hepatic venous outflow tract obstruction. Aliment Pharmacol Ther. 2015;41:961-971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 3. | Bogaerts E, Heindryckx F, Devisscher L, Paridaens A, Vandewynckel YP, Van den Bussche A, Verhelst X, Libbrecht L, van Grunsven LA, Geerts A. Time-dependent effect of hypoxia on tumor progression and liver progenitor cell markers in primary liver tumors. PLoS One. 2015;10:e0119555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Hong S, Jeong SH, Lee SS, Chung JW, Yang SW, Chung SM, Jang ES, Kim JW, Kim JH, Kim H. Prevalence and outcomes of extrahepatic primary malignancy associated with hepatocellular carcinoma in a Korean population. BMC Cancer. 2015;15:146. [PubMed] [Cited in This Article: ] |
| 5. | Kar P. Risk factors for hepatocellular carcinoma in India. J Clin Exp Hepatol. 2014;4:S34-S42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Li Y, Chen L, Chan TH, Guan XY. Hepatocellular carcinoma: transcriptome diversity regulated by RNA editing. Int J Biochem Cell Biol. 2013;45:1843-1848. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Liu L, An J, Liu J, Wen J, Zhai X, Liu Y, Pan S, Jiang J, Wen Y, Liu Z. Potentially functional genetic variants in microRNA processing genes and risk of HBV-related hepatocellular carcinoma. Mol Carcinog. 2013;52 Suppl 1:E148-E154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 8. | van Meer S, de Man RA, Siersema PD, van Erpecum KJ. Surveillance for hepatocellular carcinoma in chronic liver disease: evidence and controversies. World J Gastroenterol. 2013;19:6744-6756. [PubMed] [Cited in This Article: ] |
| 9. | Minami Y, Nishida N, Kudo M. Therapeutic response assessment of RFA for HCC: contrast-enhanced US, CT and MRI. World J Gastroenterol. 2014;20:4160-4166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 53] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 10. | Rou WS, Lee BS, Moon HS, Lee ES, Kim SH, Lee HY. Risk factors and therapeutic results of early local recurrence after transcatheter arterial chemoembolization. World J Gastroenterol. 2014;20:6995-7004. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 29] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Yang P, Markowitz GJ, Wang XF. The hepatitis B virus-associated tumor microenvironment in hepatocellular carcinoma. Natl Sci Rev. 2014;1:396-412. [PubMed] [Cited in This Article: ] |
| 12. | Chen CI, Kuan CF, Fang YA, Liu SH, Liu JC, Wu LL, Chang CJ, Yang HC, Hwang J, Miser JS. Cancer risk in HBV patients with statin and metformin use: a population-based cohort study. Medicine (Baltimore). 2015;94:e462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 61] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 13. | Deng Y, Xie M, Xie L, Wang J, Li T, He Y, Li R, Li S, Qin X. Association between polymorphism of the interleukin-13 gene and susceptibility to hepatocellular carcinoma in the Chinese population. PLoS One. 2015;10:e0116682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Tong Hv, Bock CT, Velavan TP. Genetic insights on host and hepatitis B virus in liver diseases. Mutat Res Rev Mutat Res. 2014;762:65-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 15. | Li CL, Yeh KH, Liu WH, Chen CL, Chen DS, Chen PJ, Yeh SH. Elevated p53 promotes the processing of miR-18a to decrease estrogen receptor-α in female hepatocellular carcinoma. Int J Cancer. 2015;136:761-770. [PubMed] [Cited in This Article: ] |
| 16. | Shimakawa Y, Lemoine M, Bottomley C, Njai HF, Ndow G, Jatta A, Tamba S, Bojang L, Taal M, Nyan O. Birth order and risk of hepatocellular carcinoma in chronic carriers of hepatitis B virus: a case-control study in The Gambia. Liver Int. 2015;35:2318-2326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Ji X, Zhang Q, Du Y, Liu W, Li Z, Hou X, Cao G. Somatic mutations, viral integration and epigenetic modification in the evolution of hepatitis B virus-induced hepatocellular carcinoma. Curr Genomics. 2014;15:469-480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Mao K, Zhang J, He C, Xu K, Liu J, Sun J, Wu G, Tan C, Zeng Y, Wang J. Restoration of miR-193b sensitizes Hepatitis B virus-associated hepatocellular carcinoma to sorafenib. Cancer Lett. 2014;352:245-252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 19. | Yu DL, Bao L. [Research on the relevance between the virulent genes differential expression and pathogenecity of Leptospira with microarray]. Sichuan Da Xue Xue Bao Yi Xue Ban. 2015;46:11-15. [PubMed] [Cited in This Article: ] |
| 20. | Zhang Y, Dai J, Deng H, Wan H, Liu M, Wang J, Li S, Li X, Tang H. miR-1228 promotes the proliferation and metastasis of hepatoma cells through a p53 forward feedback loop. Br J Cancer. 2015;112:365-374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 21. | Hu H, Ding X, Yang Y, Zhang H, Li H, Tong S, An X, Zhong Q, Liu X, Ma L. Changes in glucose-6-phosphate dehydrogenase expression results in altered behavior of HBV-associated liver cancer cells. Am J Physiol Gastrointest Liver Physiol. 2014;307:G611-G622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Kanda T, Jiang X, Yokosuka O. Androgen receptor signaling in hepatocellular carcinoma and pancreatic cancers. World J Gastroenterol. 2014;20:9229-9236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 39] [Reference Citation Analysis (0)] |
| 23. | Shi Y, Lv G, Chu Z, Piao L, Liu X, Wang T, Jiang Y, Zhang P. Identification of natural splice variants of SAMHD1 in virus-infected HCC. Oncol Rep. 2014;31:687-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Li J, Shi W, Gao Y, Yang B, Jing X, Shan S, Wang Y, Du Z. Analysis of microRNA expression profiles in human hepatitis B virus-related hepatocellular carcinoma. Clin Lab. 2013;59:1009-1015. [PubMed] [Cited in This Article: ] |
| 25. | Kondo Y, Kimura O, Shimosegawa T. Radiation therapy has been shown to be adaptable for various stages of hepatocellular carcinoma. World J Gastroenterol. 2015;21:94-101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 18] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 26. | Li JP. Study on the functions of miR-183 in HCC and construction of artificial miRNA cluster expression vector. Anhui Medical University Master Thesis. 2010;1-57. [Cited in This Article: ] |
| 27. | Liang M, Shi ZZ, Bai J. The study progress in relationship between miR373 and tumors. Zhongguo Shengmingkexue Gonggao. 2013;25:685-689. [Cited in This Article: ] |
| 28. | Liu HS, Ma Y, Xiao HS. The diagnostic value of serum microRNAs including miR-129-3p, miR-767-3p and miR-877 for colorectal cancer. Tumor. 2012;32:42-48. [Cited in This Article: ] |
| 29. | Liu XF. Identification of differentially expressed genes in the development and progression of human hepatocellular carcinoma by cDNA and tissue microarray techniques. Nanjing: Nanjing Medical University 2006; 1-93. [Cited in This Article: ] |
| 30. | Fang F. Study the growth, invasion and metastasis of hepatocellular carcinoma MiR-188-5p targeted inhibition AAC11. Changsha: Central South University 2011; 1-124. [Cited in This Article: ] |
| 31. | Li LH, Gao Q, Wang XY, Guo ZJ. [miR-378 suppresses HBV-related hepatocellular carcinoma tumor growth by directly targeting the insulin-like growth factor 1 receptor]. Zhonghua Ganzangbing Zazhi. 2013;21:609-613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 32. | Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, Ge C, Yao J, Chen T, Wan D. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390-399. [PubMed] [Cited in This Article: ] |