Published online Nov 28, 2014. doi: 10.3748/wjg.v20.i44.16498
Revised: June 26, 2014
Accepted: August 13, 2014
Published online: November 28, 2014
Gut microbes comprise a high density, biologically active community that lies at the interface of an animal with its nutritional environment. Consequently their activity profoundly influences many aspects of the physiology and metabolism of the host animal. A range of microbial structural components and metabolites directly interact with host intestinal cells and tissues to influence nutrient uptake and epithelial health. Endocrine, neuronal and lymphoid cells in the gut also integrate signals from these microbial factors to influence systemic responses. Dysregulation of these host-microbe interactions is now recognised as a major risk factor in the development of metabolic dysfunction. This is a two-way process and understanding the factors that tip host-microbiome homeostasis over to dysbiosis requires greater appreciation of the host feedbacks that contribute to regulation of microbial community composition. To date, numerous studies have employed taxonomic profiling approaches to explore the links between microbial composition and host outcomes (especially obesity and its comorbidities), but inconsistent host-microbe associations have been reported. Available data indicates multiple factors have contributed to discrepancies between studies. These include the high level of functional redundancy in host-microbiome interactions combined with individual variation in microbiome composition; differences in study design, diet composition and host system between studies; and inherent limitations to the resolution of rRNA-based community profiling. Accounting for these factors allows for recognition of the common microbial and host factors driving community composition and development of dysbiosis on high fat diets. New therapeutic intervention options are now emerging.
Core tip: The development of dysbiosis is driven by multiple factors. These include selective pressures imposed on the microbial community by the diet composition and feedback effects that involve either diet-host interaction or diet-microbiome-host interaction. The role of microbial signals in dysbiosis is well established but the involvement of host feedback mechanisms in aberrant host-microbial interactions is an under-appreciated part of disease progression. New opportunities to intervene in diseases of dysbiosis can result from targeting these distinct processes. These include stimulation of the host ability to self-regulate and blocking of deleterious host responses.
- Citation: Ha CW, Lam YY, Holmes AJ. Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World J Gastroenterol 2014; 20(44): 16498-16517
- URL: https://www.wjgnet.com/1007-9327/full/v20/i44/16498.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i44.16498
The gastrointestinal tract of animals typically harbours a large resident community of microorganisms that we will term the microbiome. The main function of the gut is to enable harvesting of nutrients from the external environment, however, animals live in a dynamic environment where their energy demands, exposure to foreign microorganisms and their access to nutrients are continually changing. Consequently gut functions also include containment of microbial activity to the intestinal lumen and integration of sensory perception of the intestinal environment with behavioural and physiological responses. Put simply, the gut is a major site for endocrine, immune and neural signalling in addition to digestion and nutrient absorption.
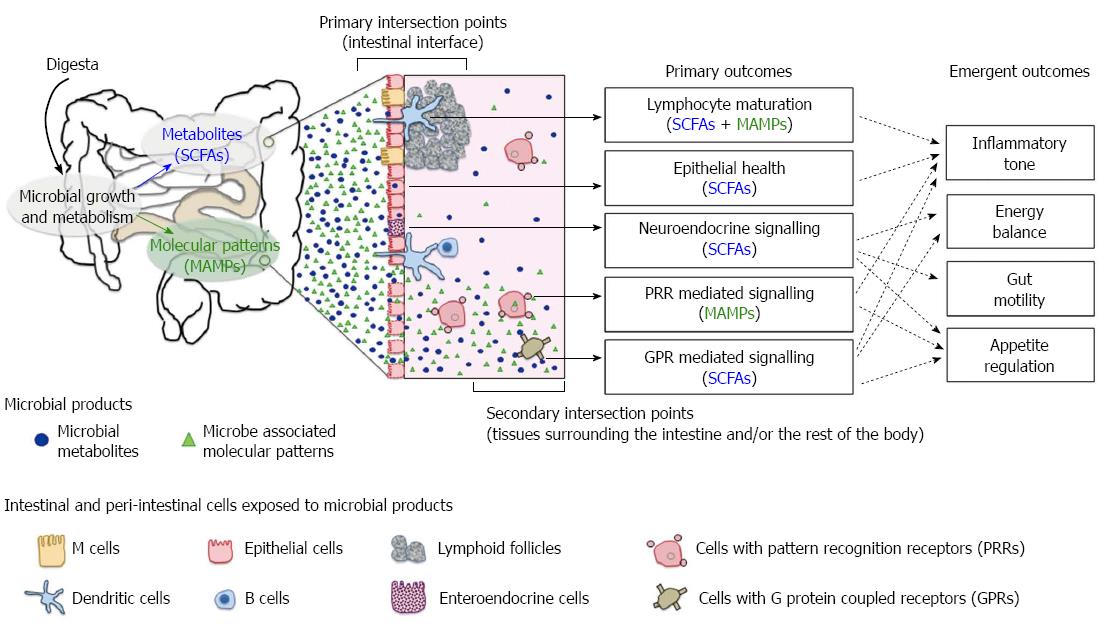
Many aspects of host physiology are strongly shaped by the presence and activities of the gut microbiome. The primary axis of host-microbiome interaction is in the intestinal tissues where microbial growth in the lumen contributes to the digestion of ingested food and directly shapes the chemical milieu of the gut. Host cells in the intestines are highly exposed to microbial activity, and microbial influence ranges from stimulation of receptors on those cells, to supply of energy sources to epithelial cells and triggering of developmental pathways in intestinal tissues[1,2] (Figure 1). Although the primary interaction with microbes is at the intestinal epithelium, their influence is projected beyond the gut through secondary host-microbiome interactions, which occur externally to the epithelium. Some of these influences such as nutrient uptake and systemic inflammation, result from translocation of or “escape” of microbial products[3,4]. Others such as appetite regulation, gut motility, energy balance and immune tone, result from the integration of multiple signals from the gut environment and bidirectional communication along the gut-brain axis[5,6]. Accordingly, it is now widely recognised that differences in microbial composition and activity result in effects of fundamental importance to health.
The breadth of potential influence of the microbiome means mechanisms that serve to regulate the microbial interface with host systems are critical for health. This view gives rise to the concept of dysbiosis: Disease states that result from dysregulated host-microbe interactions. Dysbiosis contributes to the underlying pathophysiology of a wide range of diseases, including obesity[7], diabetes[4,8], inflammatory bowel diseases[9], non-alcoholic fatty liver diseases[10,11] and cardiovascular diseases[12,13]. With awareness of the importance of dysbiosis in multiple diseases, attention has focused on how to define the microbe involvement in different diseases. The objectives here encompass the following: Identification of microbiota signatures (or biomarkers) that help define different dysbiosis states, ideally at the pre-clinical stage. Identification of the triggers of dysregulated host-microbe interactions that ultimately lead to disease. Development of intervention strategies based around restoration of normal host-microbiome interactions. Underpinning all these objectives is the need to understand the dynamics of gut microbial community composition. This review focuses on mechanisms that drive the changes in microbial community composition that ultimately lead to shifts in host-microbiome interactions.
Comparative studies on germ-free (GF) and conventionally raised (CONV) animals have been instrumental in establishing that the gut microbiome has influence on the physiological, immunological and nutritional state of its host. Such studies have consistently shown that GF animals are characterised by reduced intestinal vasculature[1], undeveloped gut-associated lymphoid tissue[14] and alterations in nutrition and energy metabolism[15], all of which are largely restored by reintroduction of gut bacteria. Collectively there is compelling evidence that the gut microbiome can influence postnatal development of gut tissues and the physiological state of animals.
The effects of microbes are interdependent with effects of diet or the host genotype. For instance, GF and CONV comparisons are not precisely recapitulated in different animal models[16], and there are also characteristic variations in microbiome composition between species[17]. Some of these variations almost certainly reflect genetically encoded differences in life history (carnivores vs herbivores) or gut structure (ruminants vs monogastrics). Others will reflect more subtle tissue specific differences, for example, the organisation of gut-associated lymphoid tissue in dogs and rodents are distinct[18]. Collectively these points serve to illustrate a broader issue. Host-microbiome interaction involves effects of the microbiome on the host, as well as effects of the host on the microbiome and these both occur within the context of environmental effects on the system (especially the nutritional environment). Studies that have addressed the influence of microbiome on differences between GF and CONV against defined genetic and diet differences in animals highlight the importance of this tripartite interaction[9,19].
The importance of variation in host diet and genotype has been observed through GF-CONV comparisons across different strains and species of inbred rodents. In a seminal paper Bäckhed, Gordon et al[15] raised the prospect that gut microbiota represent an environmental factor in obesity. They showed that GF C57BL/6 mice had less fat deposition than CONV counterparts despite higher food consumption. Moreover, the faecal caloric content of GF mice was significantly higher than that of CONV counterparts. These findings led to the conclusion that gut microbiota promote energy harvesting and fat storage, and the hypothesis that GF animals are protected from obesity[15,20]. In contrast to this mouse model, GF Fischer 344 rats displayed similar body weight and adiposity relative to CONV in two out of three experimental cohorts, and differences in daily food intake between the GF and CONV groups were insignificant[21]. Although this suggests different animal species may respond differently, it is important to note that these studies used standard rodent chow from different suppliers and almost certainly the diets were compositionally distinct[15,21].
Intersection between diet and genotype can also influence the phenotype of GF and CONV animals. The significance of this issue is highlighted in a report comparing the effect of three different diets on GF and CONV C3H mice[22]. There was no difference in weight gain between GF and CONV groups under low fat diet, but GF C3H mice actually showed significantly higher weight gain on a high fat diet (HFD) compared to CONV. Previous reports of obesity resistance on HFD in GF C57BL/6 mice had used a formulation with similar macronutrient balance but distinct sources of carbohydrates and fat[20]. When the two versions of high fat formulation were directly compared, GF and CONV C3H had comparable body fat content on the HFD with low sugar formulation but GF C3H mice was obesity resistant on the HFD with high sugar[22]. In summary, GF-CONV comparisons in different animal/diet models consistently show differences in energy harvest (faecal caloric content), energy storage (weight and body fat) and energy expenditure. Typically the effect of microbial presence is to increase adiposity, however, this does vary between experimental models and even between cohorts in the same model system. The major identifiable variables are animal species/strain and diet composition which differ between experimental cohorts.
Further exploration of the importance of microbiome composition has provided robust evidence supporting a causal link between gut microbiome composition and host outcomes. Specifically, some phenotypic traits of CONV animals can be recapitulated by conventionalisation of GF animals through microbiome transplantation[11,23-25]. When GF mouse models are conventionalised with gut microbiota from either obese or lean mice, metabolic profiles and physiological attributes of the recipients reflect their donors[23,24]. Evidently emergent properties of the total microbial community can drive differences in metabolic and physiological phenotypes. Precisely which microbes or how many are needed is unclear. For example, monocolonisation of GF mice with Enterobacter cloacae (a member of Proteobacteria isolated from an obese human) induced obesity and systemic insulin resistance in mice on HFD, while GF mice on HFD did not exhibit the same disease phenotypes[26].
In conclusion, host metabolic health is strongly influenced by the gut microbiome. The influence of gut microbes is dependent on microbiome composition and is interactive with the effects of diet and host genotype. The mechanisms of microbial influence stem from microbial activity in the intestinal tract, but are projected to the body system via multiple integrated pathways. The complexities of these interactions mean that although variations in microbial community composition can lead to different outcomes, associations may be diet or system-specific.
Broadly speaking microbiome association studies have two objectives: (1) To identify links with specific disease states[27]; and (2) To identify features of a healthy microbiome that may be a target in the restoration of health[28]. Although there have been many reports of microbiome associations with obesity or metabolic health indicators in cross-sectional studies[29,30], experimentally controlled treatments in humans[31,32] and animal models (Table 1), consistent patterns across studies are hard to discern. As discussed above the influence of the microbiome on host health is interdependent with diet and the host system. As such the apparent lack of consistent associations is likely to reflect the confounding effects of diet, host genotype and host epigenetic state. Since HFDs in Table 1 are not of the same formulation, some of the discrepancies observed almost certainly reflect variations in diet. Differences will also reflect some inherent limitations of taxon-based description of the gut microbial community.
| Type of high fat diet and duration | Detection method | Key microbial features1 | Observation and proposed mechanism for microbial outcomes | Reported host phenotype | Observation and proposed mechanism for host outcomes | Ref. | |||||
| F:B | Firmicutes | Bacteroidetes | Proteobacteria | Actinobacteria | Other | ||||||
| HF/HS2 for 8 wk | Fecal 454 [V4] | ↑ F:B | ↑ unclassified Lachnospiraceae, unclassified Ruminococcaceae, Turicibacter, Dorea, Roseburia | ↓ Barnesiella, unclassified Porphyromonadaceae | ↑ Bifidobacterium | ↑ Akkermansia | Host genetype influences gut microbiota plasticity in response to diet. | ↑ Body fat percent | [33] | ||
| ↓ Oscillibacter | |||||||||||
| HF/HS3 for 8 wk | Cecal full length 16S sequencing, shotgun sequencing and transcriptomics | ↑ F:B | ↑ Mollicutes/ Erysipelotrichaceae | ↓ Microbial diversity | Altered substrate availability ↑ microbes with the capacity to import and degrade sugars found in diet and/or host mucosa. | Weight gain, | Increased energy harvest. Specific microbes facilitate the transfer of calories from the diet to the host in the form of SCFAs. | [24] | |||
| ↑ Genes for PTS system | ↑ Body fat percent | ||||||||||
| ↑ SCFAs concentration | |||||||||||
| HF/HS4 for 12 wk | Colonic tissue 454 [V1-2], qPCR and DGGE [V3-5] | ↓ F:B | ↑ mucin-degrading Ruminococcus torques | ↑ Bacteroides-Prevotella spp | ↑ Proteobacteria | ↓ 16S rRNA gene copies | Diet type and host genotype ↑ bacteria with the ability to bind to glycosylated proteins and colonise mucosal surfaces. | Leaky gut | Diet-induced microbial changes at gut mucosa may aggravate inflammation in genetically susceptible host. | [34] | |
| HFD5 for 8 wk | Fecal 454 [V4] at baseline, Week 4 and Week 8 | Progressive ↑ F:B | Progressive ↓ Proteobacteria | Fecal energy and SCFAs fluctuate overtime, varied patterns in cecum and stool | Dietary factors determine microbial composition. Microbial community may adapt to HFD overtime | Weight gain and ↑ fat mass | Microbes may promote obesity via LPS or SCFA modulation of host gene expression rather than energy harvesting. | [35] | |||
| HFD5 for 20 wk | Fecal 454 [V4] and qPCR | ↑ F:B | ↑ Lactobacillus | ↓ Bacteroides | Weight gain, IR, fatty liver, adipose, and systemic inflammation | Antibiotic improves metabolic abnormalities. Gut microbiota modulates inflammatory responses. | [36] | ||||
| HFD5 for 21 wk | Fecal 454 [V1-2] and shotgun sequencing | ↑ F:B | ↑ Clostridiaceae | ↓ Bacteroidaceae, Prevotellaceae and Rickenellaceae | ↑ Desulfovibrionaceae | ↑ genes for ABC transporters, two-component system and cell motility↓ metabolic genes | Altered substrate availability ↑ microbes with the capacity to enhance nutrient uptake in an environment of limiting substrates | Weight gain | [37] | ||
| HFD6 for 8 to 12 wk | Fecal 454 [V6-8] | ↑ F:B | ↑ Oscillibacter, Blautia | ↓ Barnesiella, Parabacteroides | Weight gain, leaky gut, IR, adipose, gut and liver inflammation | Gut bacteria modulate gut barrier integrity. Leaky gut coupled with aberrant microbiota drive metabolic dysfunction. | [38] | ||||
| ↓ Lactobacillus | |||||||||||
| HFD7 for 12 wk | Cecal MiSeq [V4], metaproteome, metabolomics | ↓ Ruminococcaceae | ↑ Rikenellaceae | ↑ proteins for amino acid metabolism and transport and cell motility | Altered substrate availability shifts the composition and/or activity of microbiota, which favours amino acid metabolism | Weight gain, hyperglycemia | [39] | ||||
| ↑ Erysipelotrichales | No difference in microbial richness | ||||||||||
| HFD8 for 8 wk | Fecal 454 [V1-3], culture | ↑ F:B | ↑ Ruminococcaceae | ↑ Rikenellaceae | ↑ Enterobacteriaceae | ↓ Bifidobacterium | ↑ LPS | Weight gain, hyperglycemia, adipose, systemic and gut inflammation | Leaky gut and LPS induce pro-inflammatory cascade and accelerate obesity development | [40] | |
| ↓ Clostridiales | ↓ Bacteroidaceae | No difference microbial diversity | |||||||||
| HFD8 for 12 wk | Fecal 454 [V3] at every 2-4 wk | Progressive ↑ F:B | ↑ Lachnospiraceae, Ruminococcaceae, Lactococcus | ↑ selected OTUs in Bacteroides, Alistipes | Progressive ↑ Desulfovibrionaceae | ↓ Bifidobacterium | ↓ microbial diversity | Age-related effects and/or altered substrate availability | Weight gain, ↑ fat mass, IGT | ↑ antigen load (LPS) and H2S production may lead to chronic inflammation and leaky gut | [41] |
| ↓ Barnesiella | ↑ LPS binding protein | ||||||||||
| HFD8 for 25 wk | Fecal 454 [V3], DGGE [V3] and T-RFLP | Lineages in Mollicutes/ Erysipelotrichaceae responded differentially | ↑ Desulfovibrionaceae | ↓ Bifidobacteriaceae | Altered substrate availability and host genetics have differential impact on gut microbial profile | Weight gain, IGT | ↓ gut barrier protecting members, ↑ LPS and H2S production promote leaky gut and trigger inflammation | [42] | |||
| HFD8 for more than 35 wk | Cecal 454 [V1-2] | ↓ F:B | ↑ unclassified Lachnospiraceae, Lactococcus, Unclassified Ruminococcaceae, Roseburia | ↑ Bacteroides | ↑ Mucispirillum | Leptin may affect microbial composition by modulating mucin production in the intestine | Weight gain, ↑ leptin, adipose inflammation | Association between gut bacteria and body fat may be mediated by adipokines and inflammation | [43] | ||
| ↓ Allobaculum | ↓ Akkermansia | ||||||||||
| HFD8 for life, gut microbiota at week 62 is described here | Fecal 454 [V3] | ↑ selected OTUs in Allobaculum, Ruminococcaceae, Papillibacter, Lactococcus | ↑ selected OTUs in Rikenella, Alistipes | ↑ Bilophila | ↑ Mucispirillum | Altered substrate availability. Low plant polysaccharides may alter the balance of gut barrier protecting bacteria, butyrate producers and pathobionts | Weight gain, IGT, fatty liver, ↓ liver function, | ↑ antigen load (mainly LPS) may contribute to metabolic abnormalities | [44,45] | ||
| ↓ selected OTUs in Allobaculum | ↓ selected OTUs in Bacteroidiales, Prophyromonadaceae. | ↑ LPS binding protein | |||||||||
| HFD9 for 4 wk | Cecal FISH | ↓ Eubacterium rectale/ Closiridium coccoides | ↓ Bacteroides-like species | ↓ Bifidobacterium | ↑ LPS | Weight gain, IR, fatty liver, systemic and adipose inflammation | Dietary fat modulates LPS level in plasma and ↓ gut barrier protecting bacteria, which trigger inflammation and the onset of diabetes and obesity | [4] | |||
| HFDs10 with different sources of fat (safflower oil, milk fat or lard) for 24 d | Cecal 454 [V2-4] | ↑ F:B in lard HFD | ↑ Bilophila in milk fat HFD | ↓ microbial diversity in milk fat and safflower oil HFDs | Altered substrate availability. Milk-derived saturated fat ↑ the pool of sulphated bile acid, an antimicrobial but a growth substrate for Bilophila | Gut inflammation in genetically susceptible host | H2S or secondary bile acids from pathobiont may damage gut barrier and drive pro-inflammatory responses | [9] | |||
| ↓ F:B in other HFDs | |||||||||||
| HFDs10 with different sources of fat (safflower oil, milk fat or lard) for 4 wk | Fecal Illumina [V3-4] | ↑ F:B in all HFDs | ↑ Proteobacteria in milk fat and safflower oil HFDs | ↑ Actinobacteria | ↑ Tenericutes in lard HFD | Altered substrate availability. Dietary fat source modulates gut microbial profile | Weight gain (highest in milk fat), adipose inflammation (highest in safflower) | Diet induced alterations in the gut microbiota influence localised inflammation | [46] | ||
| HFDs11 with different sources of fat (palm, olive or safflower oil) for 8 wk | Fecal MITChip (microarray) | ↑ F:B in palm oil HFD only | ↑ Bacilli, Clostridium cluster XI, XVII, and XVIII in palm oil HFD only | ↓ microbial diversity in palm oil HFD only | Saturated fat diet leads to an overflow of dietary fat in the gut which may have an antimicrobial effect on microbiota | Weight gain (highest in palm oil), IR, fatty liver | [47] | ||||
Community profiling has two key requirements. These are the ability to recognise biologically distinct units and the capacity to effectively sample all such units in a community. The size and diversity of microbial communities mean that it is essential to meet these requirements with high throughput approaches. The limitations of the species concept in bacteriology, combined with poor cultivability of bacteria meant that historically this has been impossible. Advances in sequencing technologies and analysis programs over the past decade have made effective sampling possible for the first time. However, recognition of biologically meaningful taxonomic units is still limited.
The most widely used marker for community profiling is the 16S ribosomal RNA (rRNA) gene. Sample sizes of thousands to even millions of sequence reads are now readily obtained. A feature of the 16S rRNA is that it is a very flexible phylogenetic marker and taxonomic units can be readily made at a variety of scales. Generally defining taxonomic units at coarse scale (e.g., phylum; about 80% 16S rRNA identity) simplifies the analytical task of comparing units but at the expense of explanatory power. Variation in the gut microbiome is readily observable at this scale[48]. Many studies have reported an association between the ratio of the two dominant gut phyla, Bacteroidetes and Firmicutes, with obesity in cross-sectional studies and in experimental treatments[24,29,49]. However numerous exceptions have also been reported[50-52], and a recent exhaustive meta-analysis of human microbiome project data found no consistent relationship between the Bacteroidetes:Firmicutes ratio and obesity[53]. An almost certain contributing factor is that such coarse taxonomic units are less biologically meaningful than fine scale units.
There are some attributes of the gut microbiome that one can reasonably predict from the taxonomic profiles at phylum scale. For instance, Firmicutes and Bacteroidetes have fundamental differences in cell envelope composition, and polysaccharide foraging strategy[54]. However, detailed predictions of microbial functions and/or properties based on phylum classification alone are unrealistic. At finer scales of classification the biological homogeneity of taxa increases and more consistent patterns are observable. For example, it has been proposed that human gut microbiome variation occurs in three predominant variants termed enterotypes, which are recognisable through co-occurrence patterns defined by the genera Bacteroides, Prevotella and Ruminococcus[52]. Recently this concept has been intensively explored, highlighting that observation of specific patterns of association is subject to analytical and classification approaches[55], particularly how sequences are clustered into operational taxonomic units (OTUs) and how OTU-based distances between communities are calculated. This effect of analytical approach is likely to exist wherever community profiling does not (or cannot) classify into ecologically homogeneous units (ecotypes).
The inability to recognise ecotypes is an inherent limitation of 16S rRNA sequencing based approaches. Closely related species can have differential responses to specific nutrient sources and have divergent ecological roles[42,56,57]. Perhaps the most striking illustration of this issue derives from a study conducted by Li et al[58], where they used community fingerprinting and metabolomics to test for associations between Clostridia and urinary metabolites in humans. Distinct populations in the fingerprinting analysis that had mutually exclusive associations to different sets of urinary metabolites were classified to Faecalibacterium prausnitzii (F. prausnitzii). This indicates that strains of F. prausnitzii inseparable by rRNA-based classification had distinct metabolic impacts in the gut system. Hence, it is not surprising that even microbiome associations reported at the finest scales possible with rRNA-based classification are often contradictory between different studies. For instance, F. prausnitzii was found to be over-represented in obese subjects in comparison to the lean counterparts[59], which suggests high proportion of F. prausnitzii within the gut community is an indicator of poor health outcomes. Yet, other investigations have reported that healthy individuals carry more F. prausnitzii than patients with type 2 diabetes[30] or chronic inflammation[60]. Another example is the association of Akkermansia muciniphilia (A. muciniphilia) with health in some animal studies[61], other studies have noted an increased proportion of A. muciniphilia in obesity[33] and type 2 diabetes[30], or a role in exacerbating gut inflammation[62].
In summary, consideration of diet, host system and great care in methodological approaches to community profiling is necessary to identify consistent associations between microbes and metabolic health. The main limitation from a methodological perspective is linkage of relevant ecological properties of the microbial group to the taxonomic marker. An alternate approach to this is to profile the gut system and its resident bacteria from a functional perspective.
In effect functional profiling is delineation of taxonomic units based on a biochemical property. It is generally accepted that there is a high level of functional redundancy in the gut microbiome. This means bacteria from different taxonomic groups may contribute to the same ecological process (belong to the same guild) and they can substitute for one another. For instance, many gut bacteria can produce butyrate, a short chain fatty acid (SCFA) with widespread health implications, but the bacteria that carry out this function are phylogenetically diverse[63]. Associations between rRNA-based taxa and host outcomes that are critically dependent on butyrate availability are likely to be inconsistent because different members of the butyrate-producer guild may be dominant under different diets or host systems. Thus functional redundancy is almost certainly a contributor to the wide variation in associations of microbiome response and host outcomes to HFDs summarised in Table 1.
If the diet-microbiome-host outcomes listed in Table 1 are cross-examined from the perspective of microbial metabolites or microbe-associated molecular patterns (MAMPs) that are likely to be common features of ecological guilds, a more encouraging picture of associations between microbiome and metabolic health starts to emerge. Inferred or measured changes of microbial metabolite such as elevated total SCFA, elevated serum lipopolysaccharides (LPS) and hydrogen sulphide (H2S) production are recurrently observed. In the case of LPS and H2S these are also associated to taxa that are recognisable by rRNA-based classification, such as Enterobacteriaceae and Desulfovibrionaceae from the phylum Proteobacteria.
Metagenomic analysis provides a global dataset for functional profiling whereby multiple guilds can be looked at simultaneously. Such analyses have reported differences in the total level of carbohydrate degradation genes in the metagenomes of obese vs lean microbiomes raising the prospect that energy harvesting may be predictable from metagenome signatures[64], but more specific signatures have also been reported. Aside from microbial metabolites, MAMPs also stimulate host responses. Consistent with this, metagenome studies have found enrichment of microbial genes that encode cell motility[37] as well as an increase in flagellin proteins[65] associated with the obese state.
In summary, small scale single-cohort, rRNA-based studies of diet-microbiome-host interactions in response to HFD typically identify associations. Cursory comparisons of such studies reveal a confusing picture, however more detailed consideration of common ecological or physiological features reveals common patterns. Microbial structural motifs and metabolites with robust associations to HFD formulations and disease states have been seen and are regarded as the mechanistic links between gut microbiome and systemic complications. It is noteworthy that these MAMPs and microbial metabolites are present in the intestinal lumen but their systemic loads are known to increase during a HFD challenge[4,66-68] and in various aspects of metabolic disorders[4,51,69]. This raises the question of feedback processes that may further shape microbial community structure and the progression into dysbiosis.
Multiple host mechanisms are involved in restricting microbial growth and activity to the intestinal lumen (Figure 2). These processes may act against the gut microbiome in a generalised manner or target specific bacteria with distinct properties. Host secretions in the gut can function as environmental stressors that regulate bacterial growth. The primary role of bile acids is to facilitate dietary fat absorption but their amphipathic properties also disrupt bacterial membrane integrity and result in antibacterial activity[70]. When rats are fed with diet supplemented with bile acids, their gut communities are characterised by a reduction in Bacteroidetes and enrichment in Clostridia and Erysipelotrichi[71]. Intriguingly, this compositional change mirrors the patterns reported in HFD studies[24,37,38]. Higher amounts of bile acids are also linked to lower caecal concentrations of butyrate[71], a metabolite produced by subsets of gut bacteria. This finding suggests bile acids either select against the proliferation of butyrate producing bacteria or inhibit the metabolic pathways leading to butyrate synthesis. Collectively, bile acids have a contributing role in determining microbial composition and the products released by the gut microbiome.
At the intestinal interface, host-derived molecules work in synergy to exclude microbial colonisation along the gut epithelium and modulate the microbial composition in the vicinity. Secretory immunoglobulin A (IgA) is known to control bacterial migration patterns by sequestering the movement of motile organisms, thereby preventing their penetration across the gut epithelium[72]. Antimicrobial peptides such as defensins and RegIIIγ also influence microbial composition[73,74]. Mice expressing human α-defensin genes had marked depletion of segmented filamentous bacteria and less interleukin 17-producing T cells in the lamina propria than those with α-defensin deficiency[75]. RegIIIγ, on the other hand, generally selects against Gram positive bacteria, as LPS on Gram negative bacteria inhibit RegIIIγ activity[74,76]. Host secretions can also shape the gut microbiome by providing an ecological niche for specific bacteria. For instance, mucin, a glycosylated protein covering the intestinal epithelium, is a specific growth substrate for many commensal gut microbes, including Ruminococcus[77], Bacteroides[78] and Akkermansia[79]. In the event of gut inflammation, byproducts of immune responses may alter the gut microbiome by favouring the growth of selected organisms. For instance, host cells release reactive oxygen and nitrogen species into the lumen, which react to form nitrate[80-82]. It has been shown that Escherichia coli uses exogenous nitrate as electron acceptors for anaerobic respiration, giving it a competitive advantage over fermentative organisms[83].
While host secretions play an important role in determining the gut community structure, external factors such as host feeding behaviour are equally influential (Figure 2). A main driver of microbial change is the macronutrient intake of the host, in particular the type of carbohydrate ingested[57,84]. Changes in intake are likely to influence the gut microbiota composition or their nutrient acquisition strategies[85]. For instance, experiments in monocolonised mice have found that Bacteroides thetaiotaomicron responded to depletion of dietary polysaccharides by upregulating a set of genes adapted to degradation of host mucus glycans[78]. Similarly, Rumincoccus gnavus switches on different sets of carbohydrate-utilising enzymes in response to the availability of carbon sources (monosaccharides vs mucin) in the environment[86]. Escherichia coli can also adapt to nutrient changes in the environment by altering porin-mediated outer membrane permeability, broadening nutritional acquisition capacity[87], but at the expense of reduced resistance against bile[88]. Increase in the amount of fermentable polysaccharides changes intestinal transit rate, which modulates the membership of the gut community[89]. Faster transit rate may flush out slow growing organisms and those without the ability to adhere to the mucosal lining of epithelial cells. Altered microbial composition and associated metabolites, in turn, feedback to gut motility[89,90], which strongly influences nutrient absorption in the gut[91,92]. Additionally, high consumption of dietary saturated fat enhances the secretion and taurine conjugation of bile acids[9,93,94], which provides a strong selection pressure on the gut commensals due to its antibacterial activity. However, influx of taurocholic acid presents an additional source of sulphated compounds for bile tolerant, sulphate/sulphite-reducing bacteria (SRBs) to utilise in anaerobic respiration[9], thereby promoting their expansion in the gut community. Changes in diet can alter microbial composition in the matter of days[95,96]. If the altered state persists over time, it will result in a different repertoire of microbial products accumulating in the gut system[97].
A number of pattern recognition receptors (PRRs) on host cells, such as toll-like receptors (TLR4 and TLR5) and nucleotide-binding oligomerisation domain receptors (NOD1 and NOD2) are specialised for detection of MAMPs such as LPS, peptidoglycan (PGN) and flagellin. The structure and/or the extent to which MAMPs are released from bacterial cells can vary between species. Thus modification in community composition, or MAMPs expression, can promote changes in the host system. However MAMPs profile alone cannot determine host outcomes, specific host receptors and loss of gut barrier function are required to potentiate metabolic dysfunction. Localisation and expression of PRRs differ between cell types[98], this may explain the divergent outcomes of each MAMP/PRR interaction.
A wide range of gut bacteria have the capacity to produce flagella, including members of the phyla Firmicutes[99] and Proteobacteria[72]. Flagellin proteins derived from motile organisms are detected by TLR5, which is selectively expressed at a higher level in the cecum and proximal colon[100]. TLR5 are present on the basolateral surface of intestinal epithelial cells, apical surface of epithelial cells associated lymphoid follicles and mucosal dendritic cells[98,100]. TLR5 detection of flagellin is known to induce the secretion of anti-flagellin IgA, which quenches the motility of various Proteobacteria and Firmicutes species[72]. This restriction of microbial migration is a normal host response. When flagellin gains access into the intestinal mucosa, it triggers pro-inflammatory responses and increases the risk of chronic inflammation[101].
Aside from localised responses in the gut, flagellin activation is linked to regulation of physiological processes beyond the gut system. Mice lacking TLR5 had higher food consumption, and developed obesity, dyslipidemia, insulin resistance and hypertension in comparison to wild type (WT)[102]. While some of these phenotypes can be explained by increased dietary intake, food restriction in TLR5 knockout (KO) mice was only effective in preventing obesity but not insulin resistance. Remarkably, antibiotic treatment of TLR5 KO mice normalised food intake and ameliorated metabolic defects, while transplantation of TLR5 KO gut microbiota into WT recipients recapitulated metabolic dysfunction[102]. These results suggest that appropriate flagellin/TLR5 signalling cascade have a beneficial role in host feeding behaviour and thus, promote metabolic health.
LPS is a component of the outer membrane of most Gram negative bacteria, including Bacteroidetes and Proteobacteria. Chemical properties of LPS vary between species, which lead to differential capacity in activating the TLR4 signalling cascade[103]. It is thought that species from Proteobacteria exert a stronger immunostimulatory effect than Bacteroides[104]. In comparison to TLR5, TLR4 expression in intestinal epithelial cells is relatively low[105] and they are localised in the basolateral compartment[98]. Under normal circumstances, only small amounts of LPS pass through the gut epithelium and reach the bloodstream[4]. Consumption of HFD, however, is associated with reduced expression of tight junction proteins in the gut epithelium[106]. Loss of tight junction integrity increases the paracellular space in the epithelium and facilitates the leakage of luminal contents, including LPS, into adjacent tissues and the circulatory system[106]. Dietary fat is also believed to enhance chylomicron absorption of LPS from the intestinal lumen or enterocytes, which are then exported into the circulatory system[107,108]. Once LPS escapes from the intestinal lumen it can be recognised by cells with TLR4 in the peri-intestinal region or in insulin-targeting tissues, such as adipose tissue, liver, skeletal muscle and pancreas[109]. Activation of TLR4 induces the release of pro-inflammatory cytokines, which drives helper T cell (THelper) expansion and impairs insulin signalling[109,110]. In summary, LPS is an immunostimulatory agent but its exposure to TLR4 expressing cells and the capacity to drive dysbiosis is dependent on physiological properties of the host system such as intestinal permeability.
Physiological consequences of LPS/TLR4 signalling are demonstrated in mice with CD14 or TLR4 deficiencies. During HFD treatment or LPS infusion, both KO mouse models are protected from the hallmark features of metabolic dysfunction observed in the WT counterparts, including obesity, insulin resistance and inflammation[4,111]. These results indicate that TLR4 agonists, such as LPS, can influence health. Yet, TLR4 is also stimulated by non microbial structures, such as saturated fatty acids[112]. Systemic lipid infusion can trigger the TLR4 inflammatory cascade in adipose tissue and give rise to insulin resistance[113]. One might argue the activation of TLR4 cascade and associated metabolic defects is due to an excess of dietary lipid from HFD, rather than a consequence driven by a microbiota-derived compound. However, detoxification of LPS by intestinal alkaline phosphatase[114], reduced microbial load after antibiotic administration[106,115] or altered microbial profile after prebiotics treatment[61,116] can all lower plasma LPS. All these are thought to be concomitant with improved gut barrier function and/or restoration of metabolic health[106,114-116]. Since broad (antibiotics) and selective (prebiotics) alterations in the gut microbiota lead to improvements of metabolic parameters during HFD, these findings are in agreement that the availability of LPS has a fundamental role in driving metabolic outcomes.
NOD1 and NOD2 are sensors of PGN, but each receptor has a different substrate preference. NOD1 preferentially binds to a structural variant commonly found in Gram negative bacteria[117], while NOD2 detects a common motif of gram positive and gram negative organisms[118]. Similar to TLR4, NOD1 activation is implicated in the development of insulin resistance. Administration of NOD1 agonist to adipocytes upregulates the expression of pro-inflammatory cytokine TNF-α and chemokine MCP-1 in a dose dependent manner, which affects insulin signalling and decreases insulin-mediated glucose uptake[119]. Mice lacking NOD1 are protected from HFD-induced glucose intolerance and translocation of intact Gram negative bacteria from the gut lumen to mesenteric adipose tissue (MAT) and blood, compared to the WT[120]. The authors also demonstrated that bacterial translocation to MAT and the associated inflammation preceded glucose intolerance, suggesting NOD1 interaction with Gram negative gut bacteria drives the pathophysiology associated with HFD.
Apart from NOD1 signalling, NOD2 activation in the skeletal muscle also influences insulin action and glucose homeostasis. Tamrakar et al[121] have shown that a NOD2 agonist significantly reduced insulin-stimulated glucose uptake in rat skeletal muscle cell line, whereas NOD1 activation had minimal effect. However, interference with the NOD2 cascade does not necessarily protect the host from dysbiosis. Malfunctions in NOD2 signalling in patients with Crohn’s disease or in NOD2 KO mice, are linked to dysregulation of microbial containment, resulting in bacterial translocation to intestinal surface and aberrant stimulation of mucosal immune system[122,123]. Taken together, these findings demonstrate the diverse outcomes of host-microbial immune signalling. The net response is strongly dependent on the target site and is possibly linked to the ratio of Gram negative to Gram positive organisms as different PGN ligands lead to divergent downstream response.
SCFAs, such as acetate, propionate and butyrate, are arguably the most influential microbial metabolites in the context of health and disease. Both community composition and the available fermentable substrates influence the net SCFA profile[54,124,125]. As a consequence SCFA profile is an emergent property of the community and it is difficult to predict from taxon-based analysis. The majority of SCFA production is utilised locally by the gut epithelial cells but significant amounts are also transported across the epithelium to distant tissues via the circulatory system. Butyrate is metabolised in the gut epithelium and is the key energy source for colonocytes[126]. Propionate and acetate are metabolised as substrates for energy metabolism and lipid synthesis in the liver and other peripheral tissues[127]. Absorption of SCFAs accounts for 6%-9% of the total energy intake for humans and can contribute up to 44% in other animals[128,129]. In addition to their role as an energy substrate, SCFAs are signalling molecules in modulating neuroendocrine and anti-inflammatory responses at various sites.
G protein coupled receptors, GPR41 and GPR43, are the primary mediators of SCFA signalling. Butyrate and propionate have high stimulatory effect towards GPR41, while butyrate, propionate and acetate all show similar activity towards GPR43[130]. Evidence from KO models has led to the proposal that SCFA signalling via GPRs modulates energy balance, with WT mice having higher fat deposition than GPR41 KO[131]. The GPR41 KO is also characterised by a reduced expression of intestinal peptide YY (PYY), an enteroendocrine L cell hormone that in WT animals inhibits gut motility, potentially increasing the time for energy harvest and absorption[131]. Similarly, GPR43 KO mice are resistant to HFD-induced obesity, insulin insensitivity, and dyslipidemia[132], and there is supporting evidence that acetate and propionate promote adipogenesis through GPR43[133].
Other gut hormones are also influenced by SCFA signals. Glucagon-like peptide 1 (GLP-1) secreted by enteroendocrine cells has a range of effects that encompass promotion of satiety and glucose homeostasis[134], and its release can be stimulated by oral administration of butyrate[135]. Supplementation of butyrate to HFD fed mice reduced food intake and improved glucose control compared to HFD mice without the treatment[135], these phenotypic differences might be driven by differential secretion of GLP-1. Consistent with this observation, mice with impaired GPR43 signalling had reduced GLP-1 secretion, concomitant with glucose intolerance[136]. In adipocytes, SCFA activation of GPR41 induce the expression and production of leptin[137], a hormone that regulates feeding behaviour, metabolic rate and immune response.
Interactions via the gut-brain axis are also involved in the coordination of metabolic homeostasis. Propionate produced in the gut can activate GPR41 in the nerve fibres of the portal vein, which resulted in upregulation of genes required in intestinal synthesis of glucose, or intestinal gluconeogenesis (IGN)[138]. The IGN-derived glucose contributes to reduced appetite, improved glucose control and decreased hepatic glucose production, concomitant with lower body weight[138,139]. These emergent outcomes of propionate-induced IGN are mediated by the portal nervous system as denervation can abolish these effects[138,139].
It is evident that SCFA interactions with GPRs and subsequent neuroendocrine signalling affect a wide range of physiological functions, and the emergent outcomes are contingent on the type and location of the receptors as well as the agonists. As a consequence variation in microbial community composition that alters the SCFA profile can drive host responses via signalling pathways. The range of pathways triggered is influenced by other factors such as gut barrier function and SCFA translocation that impact which tissues are exposed to SCFA. The host responses, including appetite and intestinal motility, have potential to feedback to gut community composition.
The actions of SCFAs extend beyond energy balance and endocrine function, they are also involved in shaping immune regulation and possibly the progression of autoimmune diseases. In models of colitis, arthritis and asthma, GF mice and CONV GPR43 KO mice showed increased production of inflammatory mediators and enhanced recruitment of immune cells. Notably, exacerbated inflammation in GF mice was attenuated by acetate supplementation, supporting SCFA/GPR43 signalling resolves inflammatory responses[140]. However, other studies have proposed that SCFA mediated GPR43 signalling also has a role in potentiating tissue destruction[141,142].
Despite the competing views on the role of SCFAs/GPR signalling in inflammatory outcomes, SCFAs have emerged as the key microbial signal in modulating the balance of pro-inflammatory THelper and anti-inflammatory T regulatory cells (TReg). Atarashi et al[143] have shown that SCFA-producing species from Clostridium clusters IV and XIVa had greater capacity in expanding the population of colonic TReg than Bacteroides fragilis, which releases polysaccharide A (PSA) to promote immune homeostasis. More importantly, SCFAs on their own can modulate TReg responses and increase the expression of anti-inflammatory cytokine interleukin-10, which dampens pro-inflammatory responses and reduces the proliferation of effector CD4+ T cells[144]. Diets which promote SCFA production or administration of butyrate alone are able to recapitulate these effects[145,146]. Butyrate can also down regulate the expression of pro-inflammatory mediators in intestinal macrophages, such as nitric oxide, interleukin-6, and interleukin-12 by histone deacetylase inhibition, a mechanism independent of GPR activation[147].
These host-microbial immune feedbacks in the gut are proposed to have a role in the pathophysiology of autoimmune diseases in genetically susceptible individuals, such as type 1 diabetes (T1D). T1D is characterised by T cell mediated destruction of pancreatic β cells and deficiencies in TReg numbers or function[148,149]. Given the link between butyrate and T cell homeostasis, gut microbiota might be an environmental risk factor in T1D. High throughput sequencing studies have shown that the T1D gut is depleted in butyrate producing bacteria and a key gene involved in butyrate synthesis[8]. Butyrate depletion is linked to increased intestinal permeability, which precedes the clinical onset of T1D[150,151]. In individuals who are genetically susceptible to T1D, an aberrant gut microbiota with reduced butyrate production is predicted to increase the risk of the following events: increased intestinal permeability, leakage of MAMPs, subclinical intestinal inflammation, homeostatic imbalance of T cells and ultimately autoimmunity in pancreas[152,153].
In conclusion the widespread effects of SCFAs mean that factors altering their concentration and profile have multiple interacting consequences for the host and microbiome. SCFA are primary metabolites of microbial growth. Consequently the SCFA profile of the gut will be especially responsive to diet as changes in microbial nutrient supply can alter both community composition and their metabolic activity. These SCFA changes can lead to changes in gut barrier integrity, energy metabolism and inflammatory responses. All these may impact on host health, but also can feedback to impact microbial community structure. SCFAs are key factors in the interaction between gut microbiome and the host.
While butyrate fortifies the structural integrity of gut epithelium, other microbial metabolites, such as H2S, are implicated in impaired epithelial function. H2S is produced when sulphated compounds are utilised as terminal electron acceptor in anaerobic respiration. Most gut bacteria with this capability belong to the Desulfovibrionaceae family[154]. H2S is known to interfere with energy metabolism in the gut epithelium[155], ultimately leading to cell death, concomitant with gut inflammation[156]. In vitro studies of intestinal epithelial cells have demonstrated that H2S influences the expression of genes linked to cell cycle progression and stimulates both inflammatory and DNA repair responses[157,158]. Collectively, there is robust evidence that H2S has deleterious effects on the gut epithelium. A recurrent feature of HFD studies, especially those in which diet formulations have a high proportion of saturated fat, is an increase in Desulfovibrionaceae and gut inflammation (Table 1). Again the inferred loss of gut barrier function and associated changes in host-microbiome interaction have the potential to drive feedback responses in the microbial community.
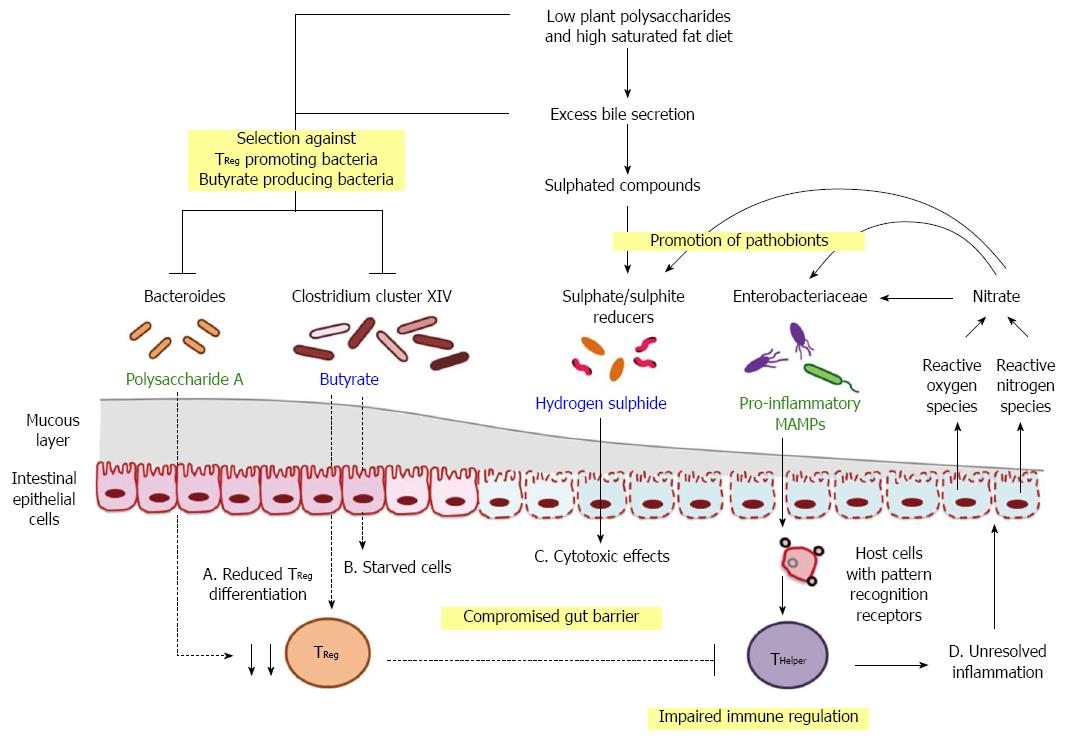
The interplay between diet, gut microbiome and host health has been the subject of numerous studies, and mechanisms that tip homeostasis to dysbiosis are starting to emerge. Nutrient competition is a major driver of community dynamics. Available evidence indicates that access to inorganic electron acceptors such as nitrate and sulphate occupies a special place in determining the outcome of nutrient competition between pathobionts and commensals at the epithelial interface[9,82]. The availability of these is tightly linked to inflammation and cell damage[9,82]. We postulate that microbes whose competitive advantage is dependent on anaerobic respiration adopt a pro-inflammatory life history strategy (which results in increased nitrate) and that their competitors promote mucosal homeostasis (which limits nitrate). Obesity and diet can skew the outcome of these opposing strategies by altering the “tipping point” at which inflammatory processes lead to elevated gut nitrate (Figure 3).
The effect of obesity, or more specifically MAT, is due to their potential to amplify the host response to metabolites that escape the intestine. Adipose tissue macrophages stimulated by MAMPs such as LPS switch to a pro-inflammatory state and increase the production of pro-inflammatory cytokines[159]. Pro-inflammatory cytokines can “escape” from the adipose tissue and promote inflammation and insulin resistance in other tissues[160].
The effects of diet are multiple but can be summarised as driving microbial changes that alter gut barrier function and immune tone. Diets that are depleted in fermentable polysaccharides are associated with lower levels of SCFA production. This state increases the risk of epithelial cell starvation (due to low butyrate levels) and reduces the numbers of TReg cells. Both host responses have the effect of increasing the potential for inflammation. Epithelial cell starvation and/or inflammation can both increase the availability of inorganic electron acceptors in the lumen that supports expansion of pro-inflammatory pathobionts, many of which are Proteobacteria. At this point the potential for positive feedback exists since the LPS of Proteobacteria is strongly pro-inflammatory. Diets that are also high in saturated fat exacerbate this basic model. Dietary fat results in increased bile secretion which has been observed to select against key groups of fermentative bacteria. Fat types that specifically promote taurocholate may exacerbate the inflammatory processes since they are strongly linked to expansion of SRBs and production of H2S. Collectively these two aspects of diet composition, levels of fermentable polysaccharide and saturated fat, can operate in synergy to reduce the fitness of bacteria that promote mucosal function via butyrate production and enhance the competitiveness of bacteria that drive inflammation via LPS.
In this conceptual framework there are two independent host feedback pathways, bile secretion and nitrate production, that facilitate the enrichment of pathobionts and drive pro-inflammatory responses. Host feedbacks to the gut microbiome may be an important determinant in disease progression, which warrants further investigation. Furthermore, there may be more than one type of commensal or pathobiont that influence disease states, especially when alternate microbial groups fulfil similar ecological functions within the gut community. Although Bilophila was the leading SRB pathobiont in the initial saturated fat/taurocholic acid/inflammation model[9], the above mechanism is applicable to other SRBs that produce H2S, such as Desulfovibrio in the Desulfovibrionaceae family and other representatives within the Clostridia class[154,161]. Similarly, several SRBs in the Desulfovibrionaceae family and other Proteobacteria have the capacity to utilise nitrate[162] and thus, Enterobacteriaceae such as E. coli may not be the only organisms with increased fitness during inflammation.
With many mechanistic links between gut community dynamics and host health are now established, microbiome-based applications for preventing and attenuating the progression of gut-related diseases are emerging. Potential therapeutic strategies may be in the form of restoring function or blocking feedback at specific nodes of the host-microbial network. If pro-inflammatory tone at the intestinal interface is the predominant driver of disease states, improving TReg ability to suppress THelper actions may ameliorate local and systemic complications associated with aberrant immune responses. Prebiotics with fermentable dietary carbohydrates are known to promote the proliferation of organisms that produce butyrate and PSA[163,164]. Stimulation of TReg differentiation by these beneficial microbial signals may help resolve inflammation.
Aside from rational modifications in diet composition, a change in feeding cycle, e.g., intermittent fasting, has been shown to have metabolic benefits[165]. Since periodic fasting will change nutrient availability to gut microbes and potentially interrupt host feedbacks to the gut microbiome, this may also help reverse dysbiosis. However, these postulated links require further investigations for validation. In conclusion, integration of metagenomics, metabolomics and taxonomic profiling has provided important insights into the functions of gut microbiome and the role of host-microbial crosstalk in dysbiosis. Our emerging understanding of interplay between nutrition, gut microbial dynamics and host responses will further the development of effective interventions on pathophysiology of lifestyle diseases.
P- Reviewer: Bernardo WM, Hu JZ, Mandi Y S- Editor: Ma YJ L- Editor: A E- Editor: Zhang DN
| 1. | Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA. 2002;99:15451-15455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 771] [Cited by in F6Publishing: 722] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 2. | Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27:201-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 388] [Cited by in F6Publishing: 456] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 3. | den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54:2325-2340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2408] [Cited by in F6Publishing: 2688] [Article Influence: 244.4] [Reference Citation Analysis (2)] |
| 4. | Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761-1772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4095] [Cited by in F6Publishing: 4129] [Article Influence: 242.9] [Reference Citation Analysis (0)] |
| 5. | Holzer P, Reichmann F, Farzi A. Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides. 2012;46:261-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 315] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 6. | Inoue D, Tsujimoto G, Kimura I. Regulation of Energy Homeostasis by GPR41. Front Endocrinol (Lausanne). 2014;5:81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 66] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 7. | de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol. 2010;299:G440-G448. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 628] [Cited by in F6Publishing: 633] [Article Influence: 45.2] [Reference Citation Analysis (0)] |
| 8. | Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, Casella G, Drew JC, Ilonen J, Knip M. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6:e25792. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 496] [Cited by in F6Publishing: 543] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 9. | Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1405] [Cited by in F6Publishing: 1265] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 10. | Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976-986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 430] [Cited by in F6Publishing: 514] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 11. | Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, Martin P, Philippe C, Walker F, Bado A. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787-1794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 622] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 12. | Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3327] [Cited by in F6Publishing: 3627] [Article Influence: 279.0] [Reference Citation Analysis (0)] |
| 13. | Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576-585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2783] [Cited by in F6Publishing: 2831] [Article Influence: 257.4] [Reference Citation Analysis (0)] |
| 14. | Rhee KJ, Sethupathi P, Driks A, Lanning DK, Knight KL. Role of commensal bacteria in development of gut-associated lymphoid tissues and preimmune antibody repertoire. J Immunol. 2004;172:1118-1124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 271] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 15. | Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718-15723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3967] [Cited by in F6Publishing: 4117] [Article Influence: 205.9] [Reference Citation Analysis (4)] |
| 16. | Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19:59-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 506] [Cited by in F6Publishing: 516] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 17. | Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776-788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1176] [Cited by in F6Publishing: 1045] [Article Influence: 65.3] [Reference Citation Analysis (0)] |
| 18. | Cesta MF. Normal structure, function, and histology of mucosa-associated lymphoid tissue. Toxicol Pathol. 2006;34:599-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 175] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 19. | Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 833] [Cited by in F6Publishing: 937] [Article Influence: 85.2] [Reference Citation Analysis (0)] |
| 20. | Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979-984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1778] [Cited by in F6Publishing: 1757] [Article Influence: 103.4] [Reference Citation Analysis (0)] |
| 21. | Swartz TD, Sakar Y, Duca FA, Covasa M. Preserved adiposity in the Fischer 344 rat devoid of gut microbiota. FASEB J. 2013;27:1701-1710. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 22. | Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104:919-929. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 297] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 23. | Duca FA, Sakar Y, Lepage P, Devime F, Langelier B, Doré J, Covasa M. Replication of obesity and associated signaling pathways through transfer of microbiota from obese-prone rats. Diabetes. 2014;63:1624-1636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 24. | Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2001] [Cited by in F6Publishing: 2044] [Article Influence: 127.8] [Reference Citation Analysis (0)] |
| 25. | Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2004] [Cited by in F6Publishing: 2039] [Article Influence: 145.6] [Reference Citation Analysis (0)] |
| 26. | Fei N, Zhao L. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J. 2013;7:880-884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 422] [Cited by in F6Publishing: 481] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 27. | Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3051] [Cited by in F6Publishing: 3198] [Article Influence: 266.5] [Reference Citation Analysis (0)] |
| 28. | Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4:137rv5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 188] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 29. | Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022-1023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5789] [Cited by in F6Publishing: 5918] [Article Influence: 348.1] [Reference Citation Analysis (0)] |
| 30. | Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3971] [Cited by in F6Publishing: 4291] [Article Influence: 357.6] [Reference Citation Analysis (0)] |
| 31. | Nadal I, Santacruz A, Marcos A, Warnberg J, Garagorri JM, Moreno LA, Martin-Matillas M, Campoy C, Martí A, Moleres A. Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes (Lond). 2009;33:758-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 234] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 32. | Santacruz A, Marcos A, Wärnberg J, Martí A, Martin-Matillas M, Campoy C, Moreno LA, Veiga O, Redondo-Figuero C, Garagorri JM. Interplay between weight loss and gut microbiota composition in overweight adolescents. Obesity (Silver Spring). 2009;17:1906-1915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 329] [Cited by in F6Publishing: 330] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 33. | Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17:141-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 399] [Cited by in F6Publishing: 386] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 34. | Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, Darfeuille-Michaud A, Barnich N. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 346] [Cited by in F6Publishing: 344] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 35. | Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635-1642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 647] [Cited by in F6Publishing: 650] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 36. | Murphy EF, Cotter PD, Hogan A, O’Sullivan O, Joyce A, Fouhy F, Clarke SF, Marques TM, O’Toole PW, Stanton C. Divergent metabolic outcomes arising from targeted manipulation of the gut microbiota in diet-induced obesity. Gut. 2013;62:220-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 188] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 37. | Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137:1716-24.e1-2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1057] [Cited by in F6Publishing: 1077] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 38. | Lam YY, Ha CW, Campbell CR, Mitchell AJ, Dinudom A, Oscarsson J, Cook DI, Hunt NH, Caterson ID, Holmes AJ. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One. 2012;7:e34233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 377] [Cited by in F6Publishing: 425] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 39. | Daniel H, Moghaddas Gholami A, Berry D, Desmarchelier C, Hahne H, Loh G, Mondot S, Lepage P, Rothballer M, Walker A. High-fat diet alters gut microbiota physiology in mice. ISME J. 2014;8:295-308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 468] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 40. | Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7:e47713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 666] [Cited by in F6Publishing: 775] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 41. | Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012;6:1848-1857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 301] [Cited by in F6Publishing: 333] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 42. | Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 651] [Cited by in F6Publishing: 768] [Article Influence: 51.2] [Reference Citation Analysis (1)] |
| 43. | Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring). 2012;20:738-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 301] [Cited by in F6Publishing: 299] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 44. | Zhang C, Li S, Yang L, Huang P, Li W, Wang S, Zhao G, Zhang M, Pang X, Yan Z. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat Commun. 2013;4:2163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 334] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 45. | Zhou B, Yang L, Li S, Huang J, Chen H, Hou L, Wang J, Green CD, Yan Z, Huang X. Midlife gene expressions identify modulators of aging through dietary interventions. Proc Natl Acad Sci USA. 2012;109:E1201-E1209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 46. | Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. JPEN J Parenter Enteral Nutr. 2013;37:746-754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 47. | de Wit N, Derrien M, Bosch-Vermeulen H, Oosterink E, Keshtkar S, Duval C, de Vogel-van den Bosch J, Kleerebezem M, Müller M, van der Meer R. Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am J Physiol Gastrointest Liver Physiol. 2012;303:G589-G599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 283] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 48. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8203] [Cited by in F6Publishing: 7225] [Article Influence: 602.1] [Reference Citation Analysis (2)] |
| 49. | Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070-11075. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4639] [Cited by in F6Publishing: 4164] [Article Influence: 219.2] [Reference Citation Analysis (1)] |
| 50. | Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond). 2008;32:1720-1724. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 817] [Cited by in F6Publishing: 813] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 51. | Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring). 2010;18:190-195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1670] [Cited by in F6Publishing: 1642] [Article Influence: 117.3] [Reference Citation Analysis (0)] |
| 52. | Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM. Enterotypes of the human gut microbiome. Nature. 2011;473:174-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4310] [Cited by in F6Publishing: 4470] [Article Influence: 343.8] [Reference Citation Analysis (0)] |
| 53. | Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One. 2014;9:e84689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 310] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 54. | Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859-5864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 475] [Cited by in F6Publishing: 505] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 55. | Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A guide to enterotypes across the human body: meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol. 2013;9:e1002863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 373] [Cited by in F6Publishing: 347] [Article Influence: 31.5] [Reference Citation Analysis (1)] |
| 56. | Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol. 2004;11:686-690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 57. | Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, Brown D, Stares MD, Scott P, Bergerat A. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1101] [Cited by in F6Publishing: 1106] [Article Influence: 79.0] [Reference Citation Analysis (0)] |
| 58. | Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117-2122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 808] [Cited by in F6Publishing: 774] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 59. | Balamurugan R, George G, Kabeerdoss J, Hepsiba J, Chandragunasekaran AM, Ramakrishna BS. Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br J Nutr. 2010;103:335-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 60. | Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, Mariat D, Corthier G, Doré J, Henegar C. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59:3049-3057. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 898] [Cited by in F6Publishing: 831] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 61. | Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066-9071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2639] [Cited by in F6Publishing: 2900] [Article Influence: 263.6] [Reference Citation Analysis (0)] |
| 62. | Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8:e74963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 315] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 63. | Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1125] [Cited by in F6Publishing: 1267] [Article Influence: 84.5] [Reference Citation Analysis (0)] |
| 64. | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-1031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7820] [Cited by in F6Publishing: 8095] [Article Influence: 476.2] [Reference Citation Analysis (1)] |
| 65. | Ferrer M, Ruiz A, Lanza F, Haange SB, Oberbach A, Till H, Bargiela R, Campoy C, Segura MT, Richter M. Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ Microbiol. 2013;15:211-226. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 160] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 66. | Pendyala S, Walker JM, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology. 2012;142:1100-1101.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 341] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 67. | Clemente-Postigo M, Queipo-Ortuño MI, Murri M, Boto-Ordoñez M, Perez-Martinez P, Andres-Lacueva C, Cardona F, Tinahones FJ. Endotoxin increase after fat overload is related to postprandial hypertriglyceridemia in morbidly obese patients. J Lipid Res. 2012;53:973-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 68. | Amar J, Burcelin R, Ruidavets JB, Cani PD, Fauvel J, Alessi MC, Chamontin B, Ferriéres J. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr. 2008;87:1219-1223. [PubMed] [Cited in This Article: ] |
| 69. | Harte AL, da Silva NF, Creely SJ, McGee KC, Billyard T, Youssef-Elabd EM, Tripathi G, Ashour E, Abdalla MS, Sharada HM. Elevated endotoxin levels in non-alcoholic fatty liver disease. J Inflamm (Lond). 2010;7:15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 270] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 70. | Kurdi P, Kawanishi K, Mizutani K, Yokota A. Mechanism of growth inhibition by free bile acids in lactobacilli and bifidobacteria. J Bacteriol. 2006;188:1979-1986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 226] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 71. | Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773-1781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 632] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 72. | Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe. 2013;14:571-581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 257] [Cited by in F6Publishing: 255] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 73. | Chamaillard M, Dessein R. Defensins couple dysbiosis to primary immunodeficiency in Crohn’s disease. World J Gastroenterol. 2011;17:567-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 14] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 74. | Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 983] [Cited by in F6Publishing: 978] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 75. | Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 835] [Cited by in F6Publishing: 859] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 76. | Mukherjee S, Zheng H, Derebe MG, Callenberg KM, Partch CL, Rollins D, Propheter DC, Rizo J, Grabe M, Jiang QX. Antibacterial membrane attack by a pore-forming intestinal C-type lectin. Nature. 2014;505:103-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 216] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 77. | Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420-2428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 863] [Cited by in F6Publishing: 923] [Article Influence: 65.9] [Reference Citation Analysis (0)] |
| 78. | Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955-1959. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 821] [Cited by in F6Publishing: 815] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 79. | Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54:1469-1476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1124] [Cited by in F6Publishing: 1261] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 80. | Kolios G, Valatas V, Ward SG. Nitric oxide in inflammatory bowel disease: a universal messenger in an unsolved puzzle. Immunology. 2004;113:427-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 366] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 81. | Reinders CA, Jonkers D, Janson EA, Stockbrügger RW, Stobberingh EE, Hellström PM, Lundberg JO. Rectal nitric oxide and fecal calprotectin in inflammatory bowel disease. Scand J Gastroenterol. 2007;42:1151-1157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708-711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 602] [Cited by in F6Publishing: 643] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 83. | Spees AM, Lopez CA, Kingsbury DD, Winter SE, Bäumler AJ. Colonization resistance: battle of the bugs or Ménage à Trois with the host? PLoS Pathog. 2013;9:e1003730. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 84. | Licht TR, Hansen M, Poulsen M, Dragsted LO. Dietary carbohydrate source influences molecular fingerprints of the rat faecal microbiota. BMC Microbiol. 2006;6:98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 85. | Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 838] [Cited by in F6Publishing: 906] [Article Influence: 75.5] [Reference Citation Analysis (0)] |
| 86. | Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8:e76341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 197] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 87. | Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 138] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 88. | De Paepe M, Gaboriau-Routhiau V, Rainteau D, Rakotobe S, Taddei F, Cerf-Bensussan N. Trade-off between bile resistance and nutritional competence drives Escherichia coli diversification in the mouse gut. PLoS Genet. 2011;7:e1002107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 89. | Kashyap PC, Marcobal A, Ursell LK, Larauche M, Duboc H, Earle KA, Sonnenburg ED, Ferreyra JA, Higginbottom SK, Million M. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144:967-977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 318] [Cited by in F6Publishing: 311] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 90. | Wichmann A, Allahyar A, Greiner TU, Plovier H, Lundén GÖ, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Bäckhed F. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe. 2013;14:582-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 261] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 91. | Chapman RW, Sillery JK, Graham MM, Saunders DR. Absorption of starch by healthy ileostomates: effect of transit time and of carbohydrate load. Am J Clin Nutr. 1985;41:1244-1248. [PubMed] [Cited in This Article: ] |
| 92. | Holgate AM, Read NW. Relationship between small bowel transit time and absorption of a solid meal. Influence of metoclopramide, magnesium sulfate, and lactulose. Dig Dis Sci. 1983;28:812-819. [PubMed] [Cited in This Article: ] |
| 93. | Reddy BS. Diet and excretion of bile acids. Cancer Res. 1981;41:3766-3768. [PubMed] [Cited in This Article: ] |
| 94. | Lindstedt S, Avigan J, Goodman DS, Sjövall J, Steinberg D. The effect of dietary fat on the turnover of cholic acid and on the composition of the biliary bile acids in man. J Clin Invest. 1965;44:1754-1765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 50] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 95. | Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4098] [Cited by in F6Publishing: 4107] [Article Influence: 315.9] [Reference Citation Analysis (0)] |
| 96. | Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94:58-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 801] [Cited by in F6Publishing: 782] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 97. | Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, Fischbach MA, Sonnenburg JL. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933-1943. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 239] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 98. | Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat Rev Immunol. 2010;10:131-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 856] [Cited by in F6Publishing: 866] [Article Influence: 61.9] [Reference Citation Analysis (0)] |
| 99. | Lozupone C, Faust K, Raes J, Faith JJ, Frank DN, Zaneveld J, Gordon JI, Knight R. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles of human gut symbionts. Genome Res. 2012;22:1974-1984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 106] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 100. | Feng T, Cong Y, Alexander K, Elson CO. Regulation of Toll-like receptor 5 gene expression and function on mucosal dendritic cells. PLoS One. 2012;7:e35918. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 101. | Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, González A. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 345] [Cited by in F6Publishing: 369] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 102. | Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1496] [Cited by in F6Publishing: 1501] [Article Influence: 107.2] [Reference Citation Analysis (0)] |
| 103. | Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect Immun. 2011;79:203-210. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 104. | Lindberg AA, Weintraub A, Zähringer U, Rietschel ET. Structure-activity relationships in lipopolysaccharides of Bacteroides fragilis. Rev Infect Dis. 1990;12 Suppl 2:S133-S141. [PubMed] [Cited in This Article: ] |
| 105. | Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010-7017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 942] [Cited by in F6Publishing: 917] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 106. | Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470-1481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3224] [Cited by in F6Publishing: 3232] [Article Influence: 202.0] [Reference Citation Analysis (0)] |
| 107. | Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J Lipid Res. 2009;50:90-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 437] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 108. | Laugerette F, Vors C, Géloën A, Chauvin MA, Soulage C, Lambert-Porcheron S, Peretti N, Alligier M, Burcelin R, Laville M. Emulsified lipids increase endotoxemia: possible role in early postprandial low-grade inflammation. J Nutr Biochem. 2011;22:53-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 190] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 109. | Kim JJ, Sears DD. TLR4 and Insulin Resistance. Gastroenterol Res Pract. 2010;2010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 110. | McAleer JP, Vella AT. Understanding how lipopolysaccharide impacts CD4 T-cell immunity. Crit Rev Immunol. 2008;28:281-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 111. | Saberi M, Woods NB, de Luca C, Schenk S, Lu JC, Bandyopadhyay G, Verma IM, Olefsky JM. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab. 2009;10:419-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 338] [Cited by in F6Publishing: 341] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 112. | Lee JY, Zhao L, Hwang DH. Modulation of pattern recognition receptor-mediated inflammation and risk of chronic diseases by dietary fatty acids. Nutr Rev. 2010;68:38-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 113. | Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116:3015-3025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2565] [Cited by in F6Publishing: 2597] [Article Influence: 144.3] [Reference Citation Analysis (0)] |
| 114. | Kaliannan K, Hamarneh SR, Economopoulos KP, Nasrin Alam S, Moaven O, Patel P, Malo NS, Ray M, Abtahi SM, Muhammad N. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc Natl Acad Sci USA. 2013;110:7003-7008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 115. | Carvalho BM, Guadagnini D, Tsukumo DM, Schenka AA, Latuf-Filho P, Vassallo J, Dias JC, Kubota LT, Carvalheira JB, Saad MJ. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia. 2012;55:2823-2834. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 206] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 116. | Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, Possemiers S, Van Holle A, François P, de Vos WM. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775-2786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 755] [Cited by in F6Publishing: 750] [Article Influence: 57.7] [Reference Citation Analysis (0)] |
| 117. | Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584-1587. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1178] [Cited by in F6Publishing: 1121] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 118. | Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869-8872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1755] [Cited by in F6Publishing: 1701] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 119. | Zhao L, Hu P, Zhou Y, Purohit J, Hwang D. NOD1 activation induces proinflammatory gene expression and insulin resistance in 3T3-L1 adipocytes. Am J Physiol Endocrinol Metab. 2011;301:E587-E598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 120. | Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermúdez-Humarán LG, Smirnova N, Bergé M, Sulpice T, Lahtinen S. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3:559-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 560] [Cited by in F6Publishing: 569] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 121. | Tamrakar AK, Schertzer JD, Chiu TT, Foley KP, Bilan PJ, Philpott DJ, Klip A. NOD2 activation induces muscle cell-autonomous innate immune responses and insulin resistance. Endocrinology. 2010;151:5624-5637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 122. | Rehman A, Sina C, Gavrilova O, Häsler R, Ott S, Baines JF, Schreiber S, Rosenstiel P. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60:1354-1362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 231] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 123. | Kosovac K, Brenmoehl J, Holler E, Falk W, Schoelmerich J, Hausmann M, Rogler G. Association of the NOD2 genotype with bacterial translocation via altered cell-cell contacts in Crohn’s disease patients. Inflamm Bowel Dis. 2010;16:1311-1321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 124. | Macfarlane S, Macfarlane GT. Regulation of short-chain fatty acid production. Proc Nutr Soc. 2003;62:67-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1150] [Cited by in F6Publishing: 1141] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 125. | Salonen A, Lahti L, Salojärvi J, Holtrop G, Korpela K, Duncan SH, Date P, Farquharson F, Johnstone AM, Lobley GE. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218-2230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 375] [Cited by in F6Publishing: 419] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 126. | Fitch MD, Fleming SE. Metabolism of short-chain fatty acids by rat colonic mucosa in vivo. Am J Physiol. 1999;277:G31-G40. [PubMed] [Cited in This Article: ] |
| 127. | Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40:235-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1742] [Cited by in F6Publishing: 1709] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 128. | McNeil NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984;39:338-342. [PubMed] [Cited in This Article: ] |
| 129. | Hume ID. Fermentation in the hindgut of mammals. Gastrointestinal Microbiology. New York, USA: Chapman & Hall 1997; 84-108. [Cited in This Article: ] |
| 130. | Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312-11319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1583] [Cited by in F6Publishing: 1595] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 131. | Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767-16772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1035] [Cited by in F6Publishing: 1070] [Article Influence: 66.9] [Reference Citation Analysis (0)] |
| 132. | Bjursell M, Admyre T, Göransson M, Marley AE, Smith DM, Oscarsson J, Bohlooly-Y M. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am J Physiol Endocrinol Metab. 2011;300:E211-E220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 206] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 133. | Hong YH, Nishimura Y, Hishikawa D, Tsuzuki H, Miyahara H, Gotoh C, Choi KC, Feng DD, Chen C, Lee HG. Acetate and propionate short chain fatty acids stimulate adipogenesis via GPCR43. Endocrinology. 2005;146:5092-5099. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 423] [Cited by in F6Publishing: 431] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 134. | Doyle ME, Egan JM. Mechanisms of action of glucagon-like peptide 1 in the pancreas. Pharmacol Ther. 2007;113:546-593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 497] [Cited by in F6Publishing: 465] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 135. | Lin HV, Frassetto A, Kowalik EJ, Nawrocki AR, Lu MM, Kosinski JR, Hubert JA, Szeto D, Yao X, Forrest G. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7:e35240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 760] [Cited by in F6Publishing: 830] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 136. | Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61:364-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1524] [Cited by in F6Publishing: 1411] [Article Influence: 117.6] [Reference Citation Analysis (0)] |
| 137. | Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, Yanagisawa M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci USA. 2004;101:1045-1050. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 484] [Cited by in F6Publishing: 502] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 138. | De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156:84-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1214] [Cited by in F6Publishing: 1416] [Article Influence: 141.6] [Reference Citation Analysis (0)] |
| 139. | Mithieux G, Misery P, Magnan C, Pillot B, Gautier-Stein A, Bernard C, Rajas F, Zitoun C. Portal sensing of intestinal gluconeogenesis is a mechanistic link in the diminution of food intake induced by diet protein. Cell Metab. 2005;2:321-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 139] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 140. | Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282-1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2007] [Cited by in F6Publishing: 2169] [Article Influence: 144.6] [Reference Citation Analysis (0)] |
| 141. | Sina C, Gavrilova O, Förster M, Till A, Derer S, Hildebrand F, Raabe B, Chalaris A, Scheller J, Rehmann A. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol. 2009;183:7514-7522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 271] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 142. | Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396-406.e1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 576] [Cited by in F6Publishing: 640] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 143. | Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2568] [Cited by in F6Publishing: 2647] [Article Influence: 189.1] [Reference Citation Analysis (0)] |
| 144. | Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2937] [Cited by in F6Publishing: 3373] [Article Influence: 306.6] [Reference Citation Analysis (0)] |
| 145. | Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451-455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2516] [Cited by in F6Publishing: 2923] [Article Influence: 265.7] [Reference Citation Analysis (0)] |
| 146. | Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446-450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2951] [Cited by in F6Publishing: 3272] [Article Influence: 297.5] [Reference Citation Analysis (0)] |
| 147. | Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA. 2014;111:2247-2252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1079] [Cited by in F6Publishing: 1258] [Article Influence: 125.8] [Reference Citation Analysis (0)] |
| 148. | Kukreja A, Cost G, Marker J, Zhang C, Sun Z, Lin-Su K, Ten S, Sanz M, Exley M, Wilson B. Multiple immuno-regulatory defects in type-1 diabetes. J Clin Invest. 2002;109:131-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 211] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 149. | Lindley S, Dayan CM, Bishop A, Roep BO, Peakman M, Tree TI. Defective suppressor function in CD4(+)CD25(+) T-cells from patients with type 1 diabetes. Diabetes. 2005;54:92-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 616] [Cited by in F6Publishing: 623] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 150. | Bosi E, Molteni L, Radaelli MG, Folini L, Fermo I, Bazzigaluppi E, Piemonti L, Pastore MR, Paroni R. Increased intestinal permeability precedes clinical onset of type 1 diabetes. Diabetologia. 2006;49:2824-2827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 279] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 151. | Secondulfo M, Iafusco D, Carratù R, deMagistris L, Sapone A, Generoso M, Mezzogiomo A, Sasso FC, Cartenì M, De Rosa R. Ultrastructural mucosal alterations and increased intestinal permeability in non-celiac, type I diabetic patients. Dig Liver Dis. 2004;36:35-45. [PubMed] [Cited in This Article: ] |
| 152. | Sorini C, Falcone M. Shaping the (auto)immune response in the gut: the role of intestinal immune regulation in the prevention of type 1 diabetes. Am J Clin Exp Immunol. 2013;2:156-171. [PubMed] [Cited in This Article: ] |
| 153. | Vaarala O. Human intestinal microbiota and type 1 diabetes. Curr Diab Rep. 2013;13:601-607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 154. | Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J Bacteriol. 1998;180:2975-2982. [PubMed] [Cited in This Article: ] |
| 155. | Babidge W, Millard S, Roediger W. Sulfides impair short chain fatty acid beta-oxidation at acyl-CoA dehydrogenase level in colonocytes: implications for ulcerative colitis. Mol Cell Biochem. 1998;181:117-124. [PubMed] [Cited in This Article: ] |
| 156. | Den Hond E, Hiele M, Evenepoel P, Peeters M, Ghoos Y, Rutgeerts P. In vivo butyrate metabolism and colonic permeability in extensive ulcerative colitis. Gastroenterology. 1998;115:584-590. [PubMed] [Cited in This Article: ] |
| 157. | Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ Mol Mutagen. 2010;51:304-314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 158. | Attene-Ramos MS, Wagner ED, Plewa MJ, Gaskins HR. Evidence that hydrogen sulfide is a genotoxic agent. Mol Cancer Res. 2006;4:9-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 229] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 159. | Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4154] [Cited by in F6Publishing: 4458] [Article Influence: 234.6] [Reference Citation Analysis (0)] |
| 160. | Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363-374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1082] [Cited by in F6Publishing: 1096] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 161. | Nava GM, Carbonero F, Croix JA, Greenberg E, Gaskins HR. Abundance and diversity of mucosa-associated hydrogenotrophic microbes in the healthy human colon. ISME J. 2012;6:57-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 127] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 162. | Warren YA, Citron DM, Merriam CV, Goldstein EJ. Biochemical differentiation and comparison of Desulfovibrio species and other phenotypically similar genera. J Clin Microbiol. 2005;43:4041-4045. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 70] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 163. | Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101-1111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 395] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 164. | Rios-Covian D, Arboleya S, Hernandez-Barranco AM, Alvarez-Buylla JR, Ruas-Madiedo P, Gueimonde M, de los Reyes-Gavilan CG. Interactions between Bifidobacterium and Bacteroides species in cofermentations are affected by carbon sources, including exopolysaccharides produced by bifidobacteria. Appl Environ Microbiol. 2013;79:7518-7524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 165. | Hatori M, Vollmers C, Zarrinpar A, DiTacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012;15:848-860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1232] [Cited by in F6Publishing: 1281] [Article Influence: 106.8] [Reference Citation Analysis (0)] |