Published online May 14, 2010. doi: 10.3748/wjg.v16.i18.2283
Revised: February 4, 2010
Accepted: February 11, 2010
Published online: May 14, 2010
AIM: To elucidate the adherence and immunomodulatory properties of a probiotic strain Bifidobacterium lactis (B. lactis) HN019.
METHODS: Adhesion assays of B. lactis HN019 and Salmonella typhimurium (S. typhimurium) ATCC 14028 to INT-407 cells were carried out by detecting copies of species-specific genes with real-time polymerase chain reaction. Morphological study was further conducted by transmission electron microscopy. Interleukin-1β (IL-1β), interleukin-8, and tumor necrosis factor-α (TNF-α) gene expression were assessed while enzyme linked immunosorbent assay was used to detect IL-8 protein secretion.
RESULTS: The attachment of S. typhimurium ATCC 14028 to INT407 intestinal epithelial cells was inhibited significantly by B. lactis HN019. B. lactis HN019 could be internalized into the INT-407 cells and attenuated IL-8 mRNA level at both baseline and S. typhimurium-induced pro-inflammatory responses. IL-8 secretion was reduced while IL-1β and TNF-α mRNA expression level remained unchanged at baseline after treated with B. lactis HN019.
CONCLUSION: B. lactis HN019 does not up-regulate the intestinal epithelium expressed pro-inflammatory cytokine, it showed the potential to protect enterocytes from an acute inflammatory response induced by enteropathogen.
-
Citation: Liu C, Zhang ZY, Dong K, Guo XK. Adhesion and immunomodulatory effects of
Bifidobacterium lactis HN019 on intestinal epithelial cells INT-407. World J Gastroenterol 2010; 16(18): 2283-2290 - URL: https://www.wjgnet.com/1007-9327/full/v16/i18/2283.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i18.2283
The intestinal tract acts as a reservoir for the intestinal microbiota that exert both harmful and beneficial effects on human health; intestinal microbiota contains an extraordinarily complex variety of metabolically active bacteria and fungi which interact with the host’s epithelial cells and provide constant antigenic stimulation to the mucosal immune system[1]. The intestinal epithelium presents the first line of defense against invading or attaching bacteria. In addition to serving as a physical barrier to microbial penetration, the intestinal mucosa is the main interface between the immune system and the luminal environment[2]. Intestinal epithelial cells (IECs) appear as an essential link in communicating with the immune cells in the underlying mucosa and the microflora in the lumen via the expression of many mediators. These mediators included antibacterial peptides such as defensins[3], mucins (MUC3), and chemokines and cytokines such as interleukin (IL)-8.
Under certain pathological circumstances, such as enteropathogens infection, the intestinal epithelium releases the chemokine IL-8 and other pro-inflammatory molecules that provoke an acute inflammatory response[4]. Therefore, a prolonged infection can result in a high level and protracted IL-8 release by IECs. The final outcome is a considerable infiltration of neutrophils that may perpetuate inflammation and eventually lead to cell damage, epithelial barrier dysfunction and pathophysiologic change of diarrhea[5].
For the protection against enteropathogen infections, the possibility of using food supplements containing probiotic bacteria has been recently considered. Probiotics are a group of live micro-organisms administered in adequate amounts which confer a beneficial effect on the health of the host[6]. Most probiotics found are resident members of the intestinal flora and are of major economic importance to the food industry. Several health-related effects associated with the intake of probiotics have been reported in different animal models as well as in human studies[7]. This bacterial community plays a pivotal role in human nutrition and health by promoting the supply of nutrients, preventing pathogen colonization and shaping and maintaining normal mucosal immunity[8]. While the precise mechanistic basis of the beneficial effects of probiotics is still obscure and will most likely vary depending on the strain and species used, a number of mechanisms have been suggested[9]. Protecting the host from enteropathogen colonization (barrier effects) and immunomodulatory effects toward host immune response have been demonstrated in humans and laboratory animals[10,11].
The interaction between probiotic strains and the intestinal epithelium is a key determinant for cytokine production by enterocytes, and probably the initiating event in probiotic immunomodulatory activity, as it occurs prior to the encounter with the immune system cells. It has been reported that several strains of probiotics belonging to Bifidobacterium and Lactobacillus are highly relevant to the prevention of the invasion of tissues by enteropathogens[12]. Moreover, by inhibiting the production of IL-8 in enterocytes, these strains are also found to be effective in modulating the pro-inflammatory response in IECs challenged by enteropathogens such as Salmonella typhimurium (S. typhimurium); such induction is species and strain specific[13,14].
Since the immunomodulatory properties are strain-specific[15-17], for each probiotic strain, profiles of the cytokines secreted by lymphocytes, enterocytes and/or DCs that come into contact with the strain should be established. Strains belonging to the Bifidobacterium are the most widely used probiotic bacteria, and are included in many functional foods and dietary supplements.
We focused our study mainly on how IEC respond to widely used probiotic bacteria. The aim of this work was to study the cytokine pattern induced by the interaction of probiotics with intestinal epithelium cells. Furthermore, whether probiotic bacteria still function after inactivation was evaluated. This study may contribute to verify the pro- and anti-inflammatory properties of the strain in question and define specific clinical applications.
A globally consumed commercial probiotic strain Bifidobacterium lactis (B. lactis) HN019 was studied in the experiment. The B. lactis HN019 powder was a gift from Danisco. Live strain was inoculated in De Man, ROGOSA and SHARPE (MRS) broth (Difco) supplemented with 0.05% (w/v) cysteine hydrochloride at 37°C under anaerobic conditions for 24 h. The stationary-phase bacteria were harvested and Gram-stained to confirm purity. Heat-killed B. lactis HN019 were prepared by heating the bacteria at 80°C for 20 min. Effectiveness of the heat-killing method was confirmed by plating the treated bacteria on MRS agar at 37°C for 48 h. S. typhimurium ATCC 14028 was used as an enteropathogenic reference strain. S. typhimurium was cultured in Tryptone Soy broth (TSB) at 37°C with shaking overnight.
Bacteria number was estimated by measuring the absorbance at 600 nm, and correlating the absorbance value to a standard curve of colony-forming units (CFU) on MRS agar (Merck) estimated in preliminary experiments.
For storage, liquid cultures of B. lactis HN019 grown for 24 h were frozen in 30% glycerol at -80°C; vacuum freeze-drying method was also applied.
INT-407 cells which derived from human intestinal epithelium, were purchased from American Type Culture Collection (ATCC strain CCL-6). Cells were grown in a humidified incubator at 37°C under 5% CO2. INT-407 cells were cultured in Dulbcco’s Modifed Eagle Medium (DMEM) (GIBCO) containing 10% (v/v) heat-inactivated fetal bovine serum (Invitrogen), 50 U/mL penicillin, and 50 μg/mL streptomycin. The cells were routinely propagated in 25 cm2 tissue culture flasks until they approached 80%-90% confluences. Subsequently, the cells were subcultured and the medium was replaced every two to three days in the culturing process.
Each adhesion assay was conducted in triplicate over three to five successive passages of INT-407 cells. Briefly, INT-407 cells were seeded on sterile six-well plates and grown to 80% confluence. The monolayers of INT- 407 cells were rinsed three times with modified Hank’s Balanced Salt Solution (mHBSS) without calcium and magnesium. Late exponential cultures of B. lactis HN019 or/and S. typhimurium ATCC 14028 were adjusted at an optical density at 600 nm corresponding to 1 × 108 cells/mL. After rinsing, the bacterial cells were resuspended in DMEM and used for the adhesion assay. After incubation for 2 h and 6 h at 37°C in the atmosphere of 5% CO2 and 95% air, unattached bacteria were removed by washing the monolayers four times with sterile PBS. After detachment of the INT-407 cells from the plastic surface by incubation with 200 μL trypsin/EDTA per well (10 min, 37°C), the INT-407 cells and the adhesive bacteria were transferred into a 1.5 mL reaction tube. The wells were rinsed with 200 μL sterile PBS which was also transferred into the 1.5 mL-reaction tube.
Bacterial adhesion to INT-407 cells was evaluated by quantification of INT-407-bound bacteria with genus-specific primers via real-time polymerase chain reaction (PCR) as reported by Candela et al[18]. To quantify the bacterial cells by real-time PCR, the cell suspensions obtained from the adhesion assays were thawed at room temperature and, after mixing, an aliquot of 20 μL was transferred into a 0.2 mL-reaction tube and incubated for 10 min at room temperature with 3.8 μL of Trypsin inhibitor solution. Then the bacterial cells (Bifidobacterium and Salmonella) were specifically quantified by real-time PCR performed with the genus-specific primers listed in Table 1. pMD19 T-vector which carried genus-specific genes was constructed as internal standards.
Real-time PCR was performed in ABI 7500 instrument and SYBR Green I fluorophore was used to correlate the amount of PCR product with the fluorescent signal. Amplification was carried out in a 20 μL final volume containing 2 μL of cell suspension, 10 μL SYBR Premix Ex Taq (TAKARA), 0.2 μmol/L of each primer and 0.4 μL ROX Reference Dye II. The experimental protocol consisted of the following programs: starting preincubation at 95°C for 10s; amplification including 40 cycles of 95°C for 5 s, and 60°C for 34 s. Since the copy numbers of standard plasmids could be quantified, we amplified serial dilutions of the plasmids that represent bacteria number ranging from 1 × 107 to 1 × 103 CFU/μL.
The data reported represent mean values obtained in 3-5 independent experiments. Each experiment was performed in duplicate.
INT-407 cells treated with B. lactis HN019 for 2 h and 6 h as previously described were washed three times with PBS buffer solution. Formaldehyde (40%) and glutaraldehyde (10%) were added for fixation. Ultrastructure of the cells was examined under transmission electron microscopy. The micrographs were produced at 5800 × and 13 500 × magnification. Cells from the control group which had not been treated with bacteria were processed in the same way. Two samples from each group were analyzed.
Monolayer of INT-407 cells were washed in PBS and supplemented with DMEM plus 2% FBS without antibiotics overnight before infection. B. lactis HN019 and S. typhimurium ATCC 14028 was harvested in stationary-phase, centrifuged and resuspended in fresh DMEM medium. After heat-killing treatment, viable B. lactis HN019, S. typhimurium ATCC 14028 and inactivated B. lactis HN019 were added into six-well plates for co-culture with INT-407 cells at a bacterial to epithelial cell ratio of 10:1. Untreated cells with normal medium were used as negative control. The plates were kept in humidified atmosphere containing 5% CO2 for 2 h.
The ability of B. lactis to inhibit binding of S. typhimurium to INT-407 cells was also assessed. INT-407 cells (1 × 106) seeded in six-well plates in antibiotic-free medium were incubated for 24 h. After replenishing the medium, the following assays were performed. (1) Coincubation of B. lactis HN019 and S. typhimurium ATCC 14028 with INT-407 cells for 2 h; (2) INT-407 cells were pretreated with 10:1 B. lactis HN019 for 2 h prior to infection with 10:1 S. typhimurium ATCC 14028 for further 2 h; (3) INT-407 cells were pre-stimulated with 10:1 S. typhimurium ATCC 14028 for 2 h prior to retreatment with 10:1 B. lactis HN019 for further 2 h; and (4) Untreated INT-407 cells were used as baseline control.
Following bacterial treatment, cell culture supernatants were removed and washed three times with sterile D-Hank’s. INT-407 cells collected for RNA preparation were immediately lysed in TRIZOL Reagent (Invitrogen). Total RNA was isolated from the lysed cells according to the manufacturer’s instructions. Purification of RNA was performed by RNase-free DNase and standard phenol chloroform extraction method. The quantity of RNA was evaluated by spectrophotometric analysis and the quality of RNA samples was determined based on the A260:A280 ratio. All RNA samples were further analyzed by agarose gel electrophoresis to check for the integrity of 5S, 18S and 28S rRNA.
Total RNA (2 μg) was reverse-transcribed to yield cDNA using SuperScript III First-Strand Synthesis System (Invitrogen) as the manufacture’s protocol for the following experiments. Primers targeting four genes of interest were designed for detecting mRNA expression. Real-time RT-PCR was performed in an ABI 7500 system (Applied Biosystems) using SYBR Premix Ex Taq as mentioned above. Calculations were performed using β-actin as an internal standard. Primer sequences are shown in Table 2. All reactions were performed in triplicate. Relative gene expression data was determined by the 2-ΔΔCT method as previously described[19]. Fold changes > 2 or < -0.5 was defined as significantly changed between treated and untreated cells.
| Gene | Primer sequences (5’-3’) | Product size (bp) |
| IL-1β | ACTCACTTAAAGCCCGCCTG | 80 |
| TCAGAATGTGGGAGCGAATG | ||
| IL-8 | CAAGAGCCAGGAAGAAACCAC | 101 |
| TGCAGAAATCAGGAAGGCTG | ||
| TNF-α | TCTTCTCGAACCCCGAGTGA | 150 |
| CCTCTGATGGCACCACCAG | ||
| β-actin | TGTTACAGGAAGTCCCTTGCC | 101 |
| AATGCTATCACCTCCCCTGTG |
Subconfluent or confluent INT-407 cells were incubated with varying groups of bacteria: (1) Cells were incubated with B. lactis HN019 or heat-killed S. typhimurium ATCC 14028 at a bacterial to epithelial cell ratio of 10:1 for 12 h; (2) Confluent monolayers of INT-407 cells were stimulated with 10 ng/mL LPS (derived from S. typhimurium, Sigma) for 12 h; (3) INT-407 cells were co-incubated with B. lactis HN019 (multiplicity of infection = 10) and LPS (10 ng/mL) simultaneously for 12 h; (4) Pretreated with 10:1 B. lactis HN019 for 2 h prior to stimulation with 10 ng/mL LPS for further 12 h; (5) Pre-stimulated with 10 ng/mL LPS for 2 h, and then treated with 10:1 B. lactis HN019 for further 12 h; and (6) Untreated cells were used as negative control.
Cells from all treatments were washed three times with PBS. Immunoreactive IL-8 protein levels in cell-culture supernatants were quantified using an ELISA DuoSet kit (R & D Systems) according to the manufacturer’s protocol.
Values were given as mean ± SD of triplicate measurements. Statistical analyses were performed using unpaired two-tailed Student’s t test. P value < 0.05 was considered to be statistically significant.
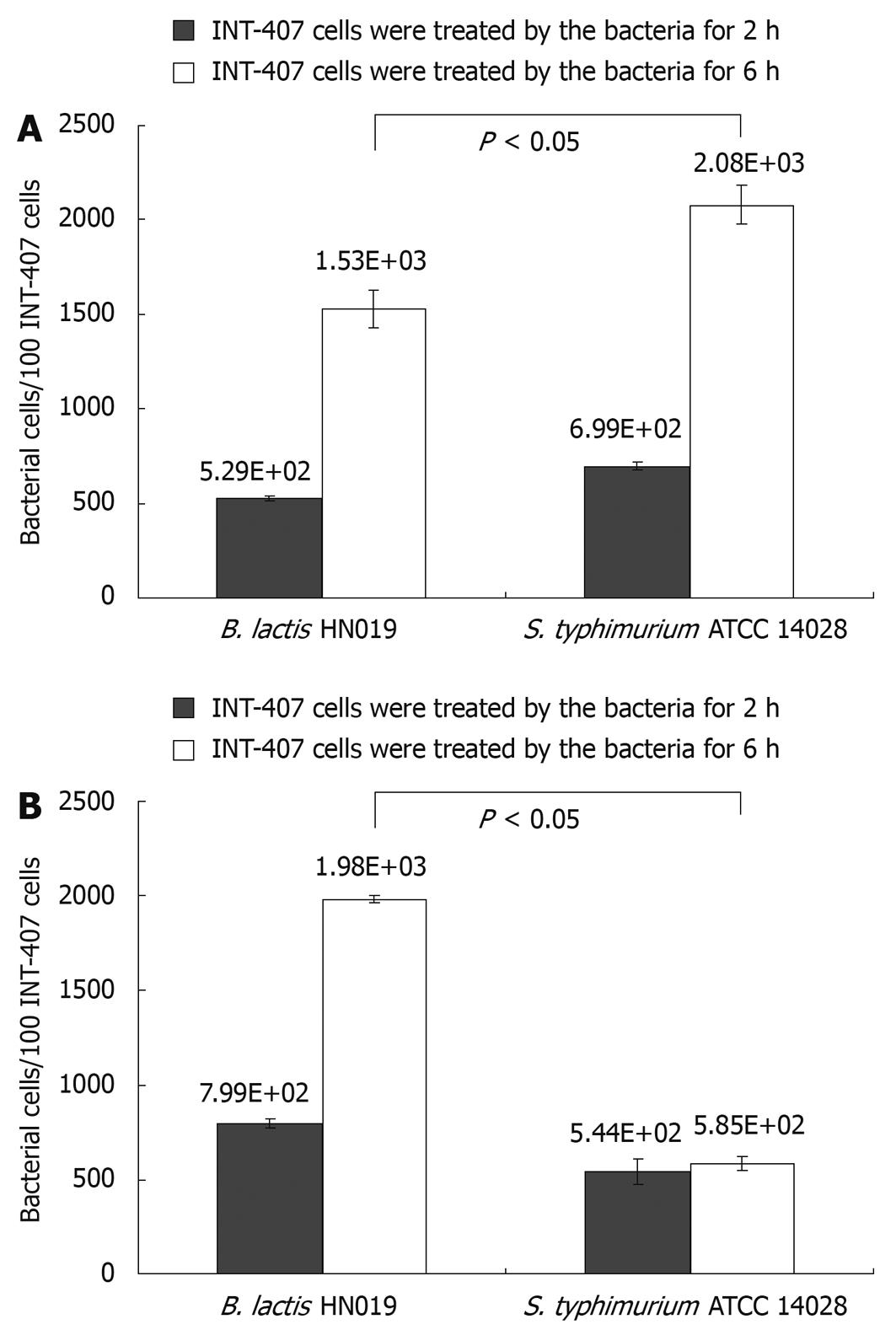
The B. lactis HN019 strains in this study were evaluated with regard to their ability to inhibit the attachment of pathogenic S. typhimurium ATCC 14028 to INT-407 cells. Adherence activity of B. lactis HN019 and/or S. typhimurium ATCC 14028 was evaluated by a real-time PCR as shown in Figure 1.
Candela et al[18] defined bacterial strains as non-adhesive with less than 5 bacterial cells adhered to one cell. Following this adhesion score, both B. lactis HN019 and S. typhimurium ATCC 14028 could effectively adhere to INT-407 cells. Moreover, the adhesive capacity of B. lactis HN019 was not higher than S. typhimurium ATCC 14028 when treated with either strain alone, showing the adhesion values of 5.29 × 102 and 6.99 × 102 bacterial cells/100 INT-407 cells after 2 h while 1.53 × 103 and 2.08 × 103 bacterial cells/100 INT-407 cells after 6 h.
Nevertheless, the adhesion values obtained in co-treated assays showed that the B. lactis HN019 could significantly decrease the attached number of S. typhimurium ATCC 14028 from 2.08 × 103 to 5.85 × 102 bacterial cells/100 INT-407 cells after 6 h treatment, and the number of attached B. lactis HN019 was not affected by the existence of S. typhimurium ATCC 14028.
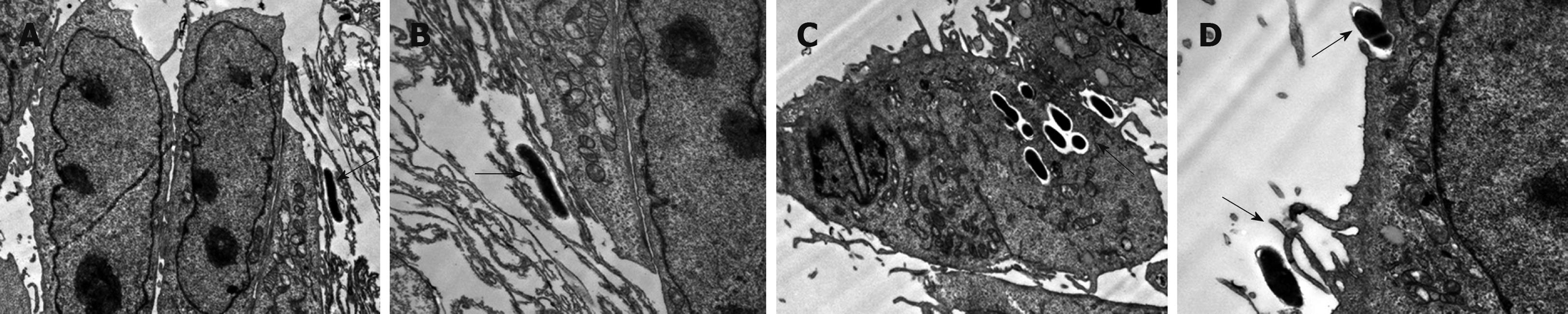
In these studies we observed that B. lactis HN019 was able to interact with the epithelial cells of small intestine (Figure 2), and it was interesting that the B. lactis HN019 could be internalized into the INT-407 cells after 6 h treatment.
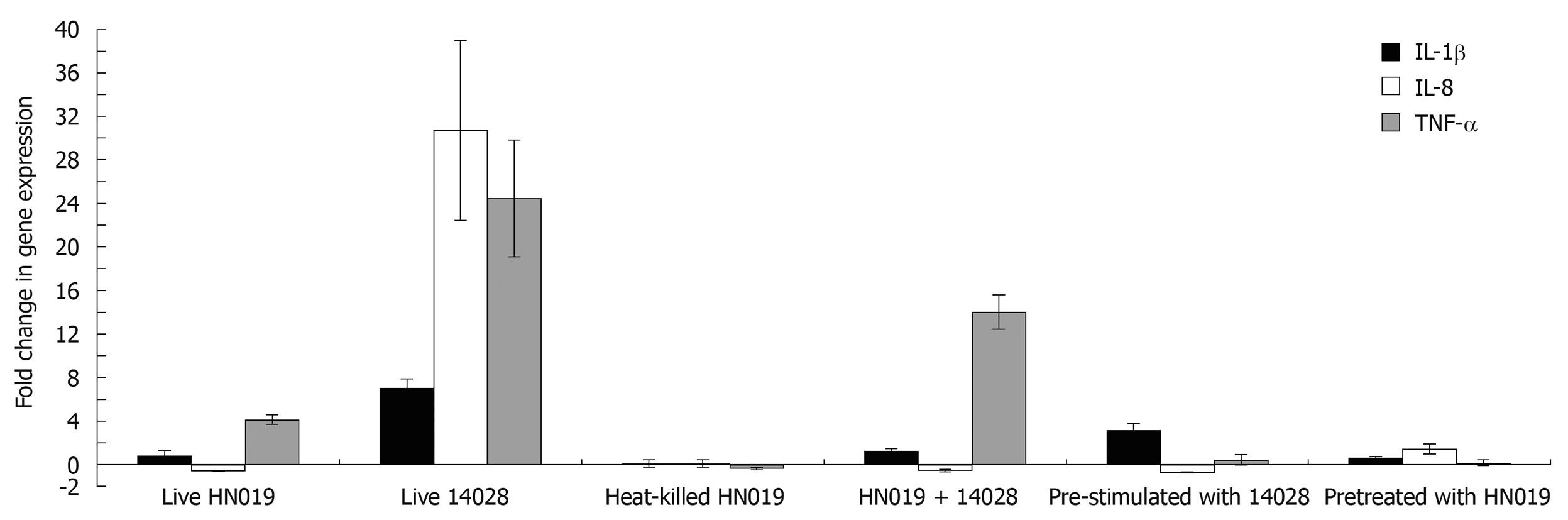
Inflammatory gene expression in bacteria-treated INT-407 cells was analyzed with real-time PCR. The results showed that live B. lactis HN019 had no effect on gene expression of IL-1β, but could up-regulate TNF-α expression and down-regulate IL-8 expression. Heat-killed B. lactis HN019 had no effect on gene expression of IL-1β, IL-8 and TNF-α. Pathogenic S. typhimurium ATCC 14028 could significantly up-regulate the mRNA level of pro-inflammatory cytokines. Although co-incubation with B. lactis HN019 and S. typhimurium ATCC 14028 could induce the gene expression of TNF-α and IL-1β, the extent is significantly lower than in those stimulated with S. typhimurium ATCC 14028 alone (Figure 3).
In bacterial interference assays, IL-1β, IL-8 and TNF-α mRNA expression was not changed at baseline after pretreated with B. lactis HN019, and addition with S. typhimurium ATCC 14028 also did not activate these pro-inflammatory cytokines expression. For groups pre-stimulated with S. typhimurium ATCC 14028, B. lactis HN019 lowered the S. typhimurium ATCC 14028 induced IL-1β, IL-8 and TNF-α levels from 8.73, 47.97 and 9.3 folds to 2.6, 0.89 and 0.37 folds as compared with the untreated group. The results are summarized in Figure 3.
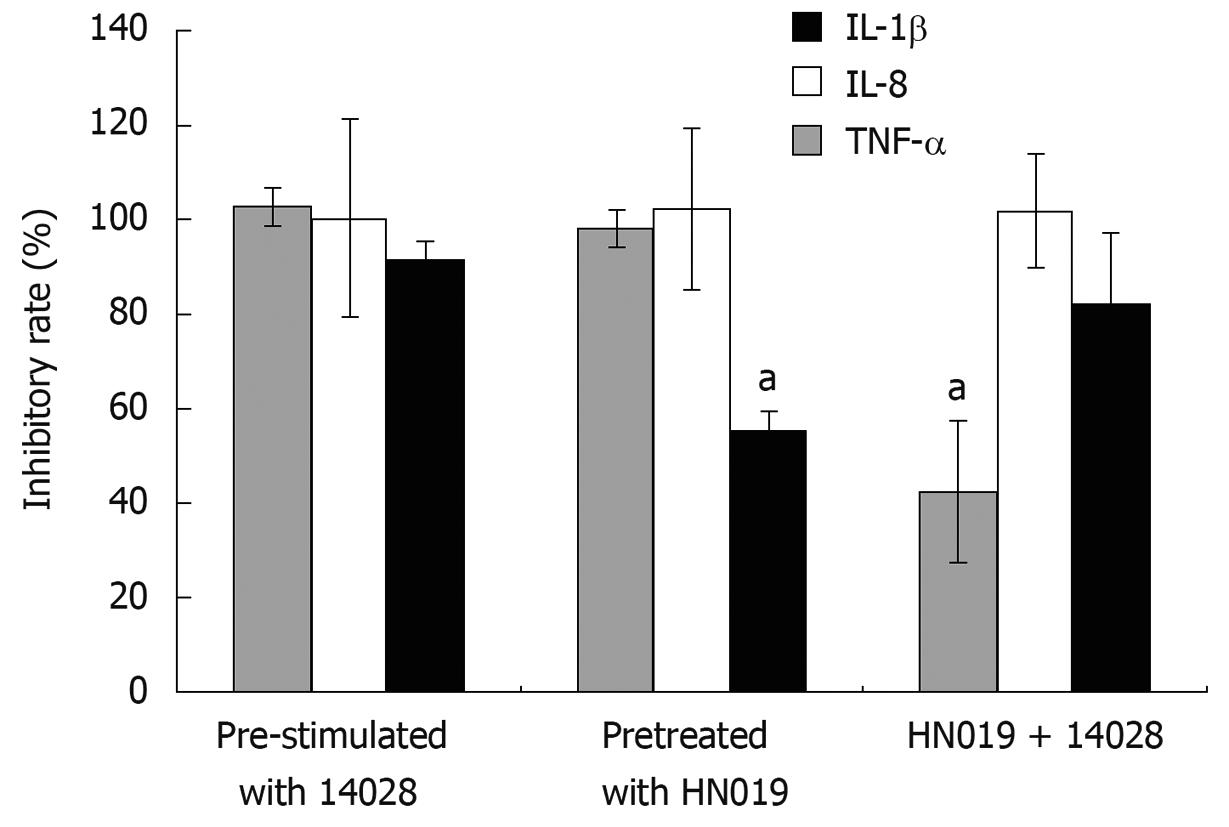
Through either incubating the INT-407 cells with B. lactis HN019 and S. typhimurium ATCC 14028 simultaneously or pre-treating with each strain respectively, the pro-inflammatory cytokines induced by S. typhimurium ATCC 14028 could be suppressed by B. lactis HN019. We therefore compared the inhibitory effects of these three groups and found that for IL-1β, the inhibition rate of B. lactis HN019 pretreated group was much lower than the other two groups. For IL-8, there was no significant difference among the three groups. For TNF-α, the inhibition rate of co-treated group was much lower than the other two groups (Figure 4).
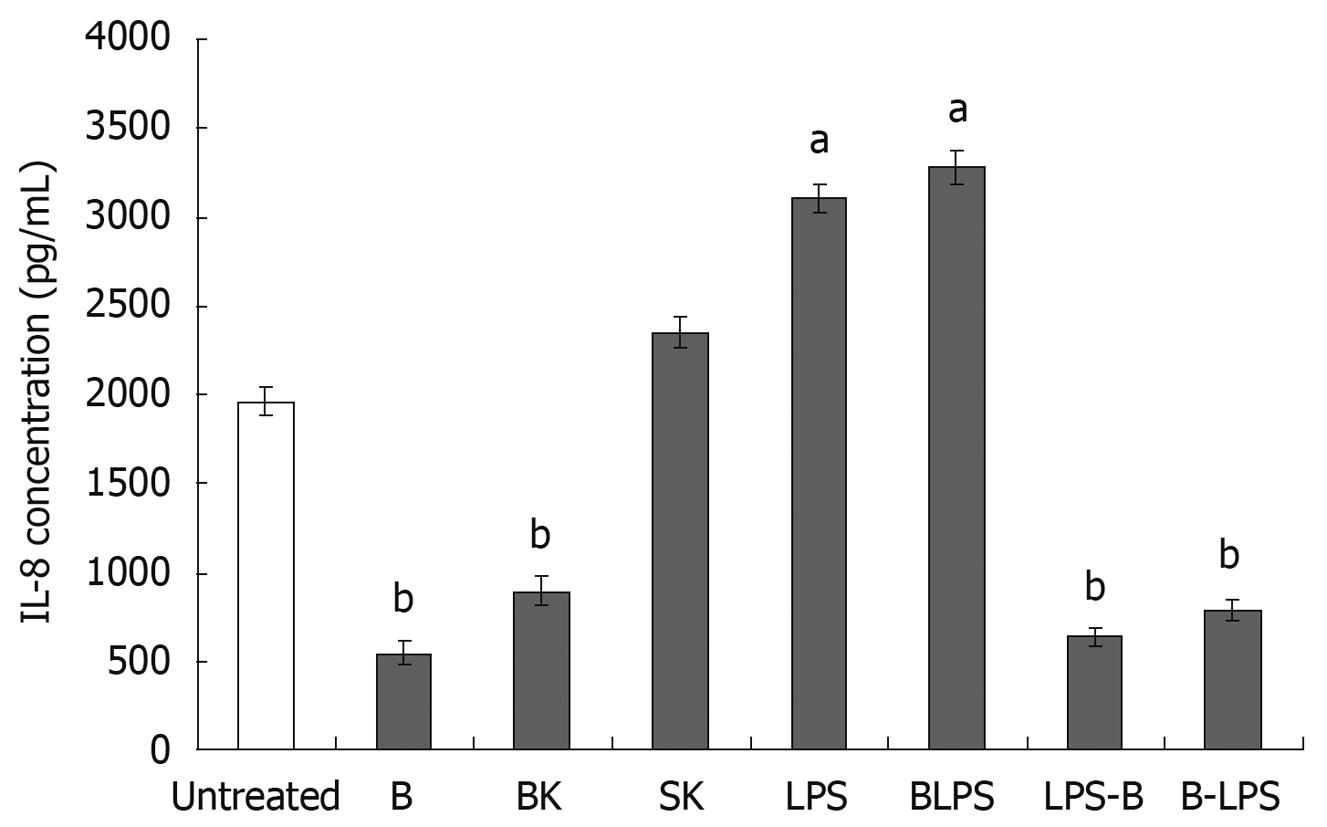
To assess the effects of probiotic strain on IL-8 secretion, INT-407 cells were incubated with live or heat-killed B. lactis and S. typhimurium and/or lipopolysaccharide. The experiments were performed 2-5 times in replicate; the results are demonstrated in Figure 5.
Firstly, we tested the effects of live B. lactis HN019, heat-killed B. lactis HN019 and S. typhimurium ATCC 14028, LPS derived from S. typhimurium on IL-8 secretion. The results showed that both live and dead B. lactis HN019 decreased IL-8 levels (844 ± 64 pg/mL and 897 ± 80 pg/mL) compared with the untreated group (1963 ± 77 pg/mL). Co-culturing with heat-killed S. typhimurium ATCC 14028 induced no change in IL-8 concentration. However, stimulation of INT-407 cells with LPS resulted in a significant increase of IL-8 production (3104 ± 80 pg/mL).
Secondly, in order to assess the anti-inflammatory properties of the probiotic strain, the IL-8 production was evaluated by incubating INT-407 cell monolayers with B. lactis HN019 in the presence of LPS. As presented in Figure 5, IL-8 protein level secreted by the cells noticeably increased after co-incubated with LPS and probiotic strain compared with untreated group. However, the probiotic strain could decrease the LPS-induced IL-8 expression, and the cells pretreated with probiotic strain could also remain at a low level IL-8 secretion even after re-stimulated with LPS as compared with untreated group. This suggests that this probiotic strain B. lactis HN019 exerted anti-inflammatory effects on the epithelium by down-regulating the secretion of IL-8.
The role of probiotic bacteria derived from normal intestinal microbiota in the physiology of the gastrointestinal tract is not completely understood. In particular, the mechanisms underlying the benefits from biotherapy with probiotics have not been sufficiently elucidated. B. lactis HN019 is a world-wide consumed probiotic strain which is associated with several health-related immunomodulatory effects: such as reducing the severity of pathogen infection and enhancing immunity in the elderly[20-22]. Nevertheless, the mechanism is yet to be known. This is the first study on the differential pro-inflammatory cytokine responses of intestinal epithelium under conditions employing live and heat-killed B. lactis HN019. Furthermore, the enteropathogen exclusion effect of B. lactis HN019 was not evaluated.
In the present study, we used a rapid, accurate and sensitive method for studying bacteria adhesion. Provided specific primers for bifidobacteria are known, our method allows the quantification of this organism’s cell number adhering to INT-407 cells which is used as a model in our investigation. A further important advantage of the real-time PCR based method is its efficacy in detecting and quantifying different bacterial genera and species simultaneously adhering to an epithelial cell monolayer. This method can be useful for probiotic strain selection. By this method, we found that B. lactis HN019 exerted a strong adhesive activity to human intestinal epithelial cells INT-407. The ability to adhere to the intestinal epithelium represents a significant prerequisite for the transient intestinal colonization of probiotic bacteria[23]. Bifidobacteria are normal gastrointestinal flora of many animals and have been shown to be adherent to a variety of epithelial cells in vitro[18,24,25]. However, adherence to INT-407 cells is seldom reported. This adherence to INT-407 cells may be mediated via fibronectin binding[26]. It is known that probiotic strains have the ability to counteract the pathogenic bacteria invasion[27,28]. In particular, the bifidobacteria strain tested is effective in inhibiting the adhesion of pathogenic S. typhimurium ATCC 14028 to INT-407 cells. This behavior is a fundamental prerequisite for probiotic bacteria to exert antagonistic activities against enteropathogens[29]. Such inhibitory effects of bifidobacteria can be explained by the mechanisms of competition for common adhesive sites or producing antibacterial lipophilic factor(s)[30]. In addition, it is reported that bifidobacteria could produce a proteinaceous factor that inhibits in vitro adherence of an enterotoxigenic E. coli strain to gangliotetraosylceramide molecules, which are physiological constituents of the mammalian intestinal epithelial surface[31].
Interactions of B. lactis HN019 and INT-407 cells observed under transmission electron microscope (TEM) indicated that the bacteria are able to contact with the epithelial cells of the small intestine. Moreover, the B. lactis HN019 could be internalized into INT-407 cells. It has been described that pathogens (bacteria, virus and parasites) can cross the mucosal barrier through different routes: transcellular route, paracellular route across cells for the tight junctions and via M cells[32]. The B. lactis used in this study was originally isolated from a human host. Therefore, it may be that this strain can internalize into epithelial cells because of its host specificity. We considered that such internalization may be of potential risks if the safety status of the internalized bacteria were not carefully assessed, especially the translocation capacity which correlate with its ability to induce infections[33].
The colonization of the intestinal tract by enteropathogenic bacteria induces the activation of the inflammatory cascade, resulting in the epithelial cell secretion of IL-8 and other pro-inflammatory molecules and in the consequent recruitment of neutrophils and other inflammatory cells. IL-8 amplifies an ongoing acute immune response and provides a set of signals for the activation of mucosal inflammatory responses in the earliest phases of microbial invasion. In some cases, a massive and prolonged infiltration of neutrophils may perpetuate inflammation and ultimately lead to cell damage, epithelial barrier dysfunction and pathophysiologic change of diarrhea. In the present study, INT-407 cells behaved like human enterocytes and could secrete IL-8 either constitutively or after stimulation with physiological agonists. Probiotic strains presented differences in their capacity to augment IL-8 expression, however, some strains seemed to rather decrease epithelial-cell production of IL-8[15-17,34-36]. Our results showed a robust IL-8 expression of INT-407 cells induced when exposed to S. typhimurium. However, live and heat-killed B. lactis HN019 not only functionally modulate the epithelium by inhibiting the constitutive mRNA level of IL-8 and attenuating S. typhimurium-induced IL-8 gene expression, but also protect the INT-407 cells from IL-8 protein production activated by LPS. The same results were discovered on TNF-α and IL-1β gene expression except for up-regulation of TNF-α expression by live B. lactis HN019, but the S. typhimurium could induce much higher TNF-α expression than live B. lactis HN019. High concentrations of IL-1β and TNF-α cause cachexia, tissue injury, disseminated intravascular coagulation and shock. Therefore, the control of secretion of these pro-inflammatory cytokines in vivo is very important. B. lactis HN019 has a beneficial effect in converting the infection-induced excessive cytokine expression and helping maintain an immunological balance. These data indicated that INT-407 displayed immunological quiescence when exposed to both live and dead B. lactis HN019. B. lactis HN019 can modulate epithelial responses to limit inflammatory signals and markedly inhibit the inflammatory response induced by pathogen.
We used three approaches to detect the influence of probiotic strain on the pathogenic strain, and found that although the B. lactis HN019 could reduce the expression of pro-inflammatory cytokines, the inhibitory effect was different depending on the treatment methods. Pre-stimulated INT-407 cells with S. typhimurium ATCC 14028 could strongly induce a high-level of pro-inflammatory cytokine expression and such up-regulation could be reversed to a normal level after re-treatment with probiotic B. lactis HN019. This treatment achieved the maximum inhibitory rate. These results indicated that B. lactis HN019 may be more effective as adjunctive therapeutic agent than as preventive agent in inflammatory diseases.
In conclusion, our studies revealed that as a probiotic strain, B. lactis HN019 could modulate immune system towards anti-inflammatory action and exclude enteropathogenic adhesion, thus contributing to the homeostasis of the intestinal epithelium. This finding will offer, in the near future, new therapeutic means to counteract the inflammatory disorders observed in human inflammatory bowel disease.
Bifidobacterium lactis (B. lactis) HN019 is a world-wide consumed probiotic strain and several health-related effects of the strain have been reported, especially the immunomodulatory effect. The immunomodulatory properties are strain-specific, and the interaction between probiotics and the intestinal epithelium is the initiating event in immunomodulatary activity. The authors focused their studies on how intestinal epithelial cells (IEC) respond to widely used probiotic bacteria to explore the mechanism of the immunomodulatory effects of B. lactis HN019.
Studies in probiotics are one of the hottest research fields at present. The interactions between bacteria and the host may reveal the mechanisms of bacteria on the host. This study may contribute to better applications of probiotic bacteria.
This is the first study on different pro-inflammatory cytokine responses of intestinal epithelium under conditions employing B. lactis HN019. The authors used a rapid, accurate and sensitive method for studying bacteria adhesion and found that B. lactis HN019 exerted a strong adhesive activity to human intestinal epithelium cells and the bifidobacteria strain tested was effective in inhibiting the adhesion of pathogen. The authors also indicated that B. lactis HN019 could modulate epithelial responses to limit inflammatory signals and markedly inhibit the inflammatory response induced by pathogen.
The study revealed that as a world-wide consumed probiotic strain, B. lactis HN019 could modulate immune system towards anti-inflammatory action and exclude enteropathogenic adhesion. These findings may help with better applications of the B. lactis HN019 and will contribute to offering new therapeutic means to counteract the inflammatory disorders observed in human inflammatory disease.
The manuscript by Liu et al describes the adherence and immuno-modulatory properties of B. lactis HN019 against INT-407 intestinal epithelial cells, and additionally reports the anti-S. typhimurium properties of this probiotic strain. This is an interesting manuscript which has been well executed.
Peer reviewers: Dr. William R Parker, PhD, Assistant Professor, Department of Surgery, Duke University Medical Center, Box 2605, Durham, NC 27710, United States; Catherine Greene, PhD, Senior Lecturer, Department of Medicine, Royal College of Surgeons in Ireland, Education and Research Centre, Beaumont Hospital, Dublin 9, Ireland
S- Editor Wang YR L- Editor Ma JY E- Editor Ma WH
| 1. | Tlaskalová-Hogenová H, Stepánková R, Hudcovic T, Tucková L, Cukrowska B, Lodinová-Zádníková R, Kozáková H, Rossmann P, Bártová J, Sokol D. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97-108. [Cited in This Article: ] |
| 2. | Wershil BK, Furuta GT. 4. Gastrointestinal mucosal immunity. J Allergy Clin Immunol. 2008;121:S380-S383; quiz S415. [Cited in This Article: ] |
| 3. | Takahashi A, Wada A, Ogushi K, Maeda K, Kawahara T, Mawatari K, Kurazono H, Moss J, Hirayama T, Nakaya Y. Production of beta-defensin-2 by human colonic epithelial cells induced by Salmonella enteritidis flagella filament structural protein. FEBS Lett. 2001;508:484-488. [Cited in This Article: ] |
| 4. | Macdonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307:1920-1925. [Cited in This Article: ] |
| 5. | Roselli M, Finamore A, Britti MS, Mengheri E. Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr. 2006;95:1177-1184. [Cited in This Article: ] |
| 6. | Heczko PB, Strus M, Kochan P. Critical evaluation of probiotic activity of lactic acid bacteria and their effects. J Physiol Pharmacol. 2006;57 Suppl 9:5-12. [Cited in This Article: ] |
| 7. | Roberfroid MB. Prebiotics and probiotics: are they functional foods? Am J Clin Nutr. 2000;71:1682S-1687S; discussion 1688S-1690S. [Cited in This Article: ] |
| 8. | Parvez S, Malik KA, Ah Kang S, Kim HY. Probiotics and their fermented food products are beneficial for health. J Appl Microbiol. 2006;100:1171-1185. [Cited in This Article: ] |
| 9. | Boirivant M, Strober W. The mechanism of action of probiotics. Curr Opin Gastroenterol. 2007;23:679-692. [Cited in This Article: ] |
| 10. | Nova E, Wärnberg J, Gómez-Martínez S, Díaz LE, Romeo J, Marcos A. Immunomodulatory effects of probiotics in different stages of life. Br J Nutr. 2007;98 Suppl 1:S90-S95. [Cited in This Article: ] |
| 11. | Resta-Lenert S, Barrett KE. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut. 2003;52:988-997. [Cited in This Article: ] |
| 12. | Fooks LJ, Gibson GR. Probiotics as modulators of the gut flora. Br J Nutr. 2002;88 Suppl 1:S39-S49. [Cited in This Article: ] |
| 13. | Ruiz PA, Hoffmann M, Szcesny S, Blaut M, Haller D. Innate mechanisms for Bifidobacterium lactis to activate transient pro-inflammatory host responses in intestinal epithelial cells after the colonization of germ-free rats. Immunology. 2005;115:441-450. [Cited in This Article: ] |
| 14. | Vinderola G, Matar C, Perdigon G. Role of intestinal epithelial cells in immune effects mediated by gram-positive probiotic bacteria: involvement of toll-like receptors. Clin Diagn Lab Immunol. 2005;12:1075-1084. [Cited in This Article: ] |
| 15. | Lammers KM, Helwig U, Swennen E, Rizzello F, Venturi A, Caramelli E, Kamm MA, Brigidi P, Gionchetti P, Campieri M. Effect of probiotic strains on interleukin 8 production by HT29/19A cells. Am J Gastroenterol. 2002;97:1182-1186. [Cited in This Article: ] |
| 16. | Otte JM, Podolsky DK. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am J Physiol Gastrointest Liver Physiol. 2004;286:G613-G626. [Cited in This Article: ] |
| 17. | McCracken VJ, Chun T, Baldeón ME, Ahrné S, Molin G, Mackie RI, Gaskins HR. TNF-alpha sensitizes HT-29 colonic epithelial cells to intestinal lactobacilli. Exp Biol Med (Maywood). 2002;227:665-670. [Cited in This Article: ] |
| 18. | Candela M, Seibold G, Vitali B, Lachenmaier S, Eikmanns BJ, Brigidi P. Real-time PCR quantification of bacterial adhesion to Caco-2 cells: competition between bifidobacteria and enteropathogens. Res Microbiol. 2005;156:887-895. [Cited in This Article: ] |
| 19. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [Cited in This Article: ] |
| 20. | Gill HS, Rutherfurd KJ, Cross ML, Gopal PK. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am J Clin Nutr. 2001;74:833-839. [Cited in This Article: ] |
| 21. | Shu Q, Lin H, Rutherfurd KJ, Fenwick SG, Prasad J, Gopal PK, Gill HS. Dietary Bifidobacterium lactis (HN019) enhances resistance to oral Salmonella typhimurium infection in mice. Microbiol Immunol. 2000;44:213-222. [Cited in This Article: ] |
| 22. | Shu Q, Gill HS. A dietary probiotic (Bifidobacterium lactis HN019) reduces the severity of Escherichia coli O157:H7 infection in mice. Med Microbiol Immunol. 2001;189:147-152. [Cited in This Article: ] |
| 23. | Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S. Quality assurance criteria for probiotic bacteria. Am J Clin Nutr. 2001;73:393S-398S. [Cited in This Article: ] |
| 24. | Riedel CU, Foata F, Goldstein DR, Blum S, Eikmanns BJ. Interaction of bifidobacteria with Caco-2 cells-adhesion and impact on expression profiles. Int J Food Microbiol. 2006;110:62-68. [Cited in This Article: ] |
| 25. | Collado MC, Gueimonde M, Hernández M, Sanz Y, Salminen S. Adhesion of selected Bifidobacterium strains to human intestinal mucus and the role of adhesion in enteropathogen exclusion. J Food Prot. 2005;68:2672-2678. [Cited in This Article: ] |
| 26. | Kapczynski DR, Meinersmann RJ, Lee MD. Adherence of Lactobacillus to intestinal 407 cells in culture correlates with fibronectin binding. Curr Microbiol. 2000;41:136-141. [Cited in This Article: ] |
| 27. | Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizzello F, Campieri M, Brigidi P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol. 2008;125:286-292. [Cited in This Article: ] |
| 28. | Gopal PK, Prasad J, Smart J, Gill HS. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. Int J Food Microbiol. 2001;67:207-216. [Cited in This Article: ] |
| 29. | Bernet MF, Brassart D, Neeser JR, Servin AL. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut. 1994;35:483-489. [Cited in This Article: ] |
| 30. | Liévin V, Peiffer I, Hudault S, Rochat F, Brassart D, Neeser JR, Servin AL. Bifidobacterium strains from resident infant human gastrointestinal microflora exert antimicrobial activity. Gut. 2000;47:646-652. [Cited in This Article: ] |
| 31. | Fujiwara S, Hashiba H, Hirota T, Forstner JF. Purification and characterization of a novel protein produced by Bifidobacterium longum SBT2928 that inhibits the binding of enterotoxigenic Escherichia coli Pb176 (CFA/II) to gangliotetraosylceramide. J Appl Microbiol. 1999;86:615-621. [Cited in This Article: ] |
| 32. | Hershberg RM, Mayer LF. Antigen processing and presentation by intestinal epithelial cells-polarity and complexity. Immunol Today. 2000;21:123-128. [Cited in This Article: ] |
| 33. | Ishibashi N, Yamazaki S. Probiotics and safety. Am J Clin Nutr. 2001;73:465S-470S. [Cited in This Article: ] |
| 34. | Ma D, Forsythe P, Bienenstock J. Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor alpha-induced interleukin-8 expression. Infect Immun. 2004;72:5308-5314. [Cited in This Article: ] |
| 35. | Bai AP, Ouyang Q, Zhang W, Wang CH, Li SF. Probiotics inhibit TNF-alpha-induced interleukin-8 secretion of HT29 cells. World J Gastroenterol. 2004;10:455-457. [Cited in This Article: ] |
| 36. | Wang S, Ng LH, Chow WL, Lee YK. Infant intestinal Enterococcus faecalis down-regulates inflammatory responses in human intestinal cell lines. World J Gastroenterol. 2008;14:1067-1076. [Cited in This Article: ] |
| 37. | Kok RG, de Waal A, Schut F, Welling GW, Weenk G, Hellingwerf KJ. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl Environ Microbiol. 1996;62:3668-3672. [Cited in This Article: ] |
| 38. | Trkov M, Avgustin G. An improved 16S rRNA based PCR method for the specific detection of Salmonella enterica. Int J Food Microbiol. 2003;80:67-75. [Cited in This Article: ] |