Published online Apr 28, 2021. doi: 10.35711/aimi.v2.i2.13
Peer-review started: March 4, 2021
First decision: March 14, 2021
Revised: March 30, 2021
Accepted: April 20, 2021
Article in press: April 20, 2021
Published online: April 28, 2021
Artificial intelligence (AI) is a computer science that tries to mimic human-like intelligence in machines that use computer software and algorithms to perform specific tasks without direct human input. Machine learning (ML) is a subunit of AI that uses data-driven algorithms that learn to imitate human behavior based on a previous example or experience. Deep learning is an ML technique that uses deep neural networks to create a model. The growth and sharing of data, increasing computing power, and developments in AI have initiated a transformation in healthcare. Advances in radiation oncology have produced a significant amount of data that must be integrated with computed tomography imaging, dosimetry, and imaging performed before each fraction. Of the many algorithms used in radiation oncology, has advantages and limitations with different computational power requirements. The aim of this review is to summarize the radiotherapy (RT) process in workflow order by identifying specific areas in which quality and efficiency can be improved by ML. The RT stage is divided into seven stages: patient evaluation, simulation, contouring, planning, quality control, treatment application, and patient follow-up. A systematic evaluation of the applicability, limitations, and advantages of AI algorithms has been done for each stage.
Core Tip: Beginning with the initial patient interview, artificial intelligence (AI) can help predict posttreatment disease prognosis and toxicity. Additionally, AI can assist in the automated segmentation of both the organs at risk and target volumes and the treatment planning process with advanced dose optimization. AI can optimize the quality control process and support increased safety, quality, and maintenance efficiency.
- Citation: Yakar M, Etiz D. Artificial intelligence in radiation oncology. Artif Intell Med Imaging 2021; 2(2): 13-31
- URL: https://www.wjgnet.com/2644-3260/full/v2/i2/13.htm
- DOI: https://dx.doi.org/10.35711/aimi.v2.i2.13
Artificial intelligence (AI) is a computer science branch that tries to imitate human-like intelligence in machines using computer software and algorithms without direct human input to perform certain tasks[1,2]. Machine learning (ML) is a subunit of AI that uses data-driven algorithms that learn to imitate human behavior based on previous example or experience[3]. Deep learning (DL) is an ML technique that uses deep neural networks to create a model. Increasing computing power and reduction of financial barriers led to the emergence of the domain of DL[4]. The growth and sharing of data, increasing computing power, and developments in AI have initiated a transformation in healthcare services. Advances in radiation oncology, clinical and dosimetric information from increasing cases, and computed tomography (CT) imaging before each fraction have resulted in the accumulation of a significant amount of information in big databases.
Evidence-based medicine is based on randomized controlled trials designed for large patient populations. However, the increasing number of clinical and biological parameters that need to be investigated makes it difficult to design studies[5]. New approaches are required for all patient populations. Clinicians should use all diagnostic tools, such as medical imaging, blood testing, and genetic testing, to decide on the appropriate combination of treatments (e.g., radiotherapy, chemotherapy, targeted therapy, and immunotherapy). There are a number of individual differences that are responsible for each patient's disease or associated with response to treatment and clinical outcome. The concept of personalized treatment is based on determining and using these factors for each patient[6]. Integrating such a large amount of heterogeneous of data and producing accurate models may present difficulties and subjective individual differences for the human brain from time to time.
Beginning with the initial patient interview, AI can help predict posttreatment disease prognosis and toxicity. Additionally, AI can assist in the automated segmentation of both the organs at risk and target volume and the treatment planning process, with advanced dose optimization. AI can optimize the quality control (QA) process and support increased safety, quality, and maintenance efficiency.
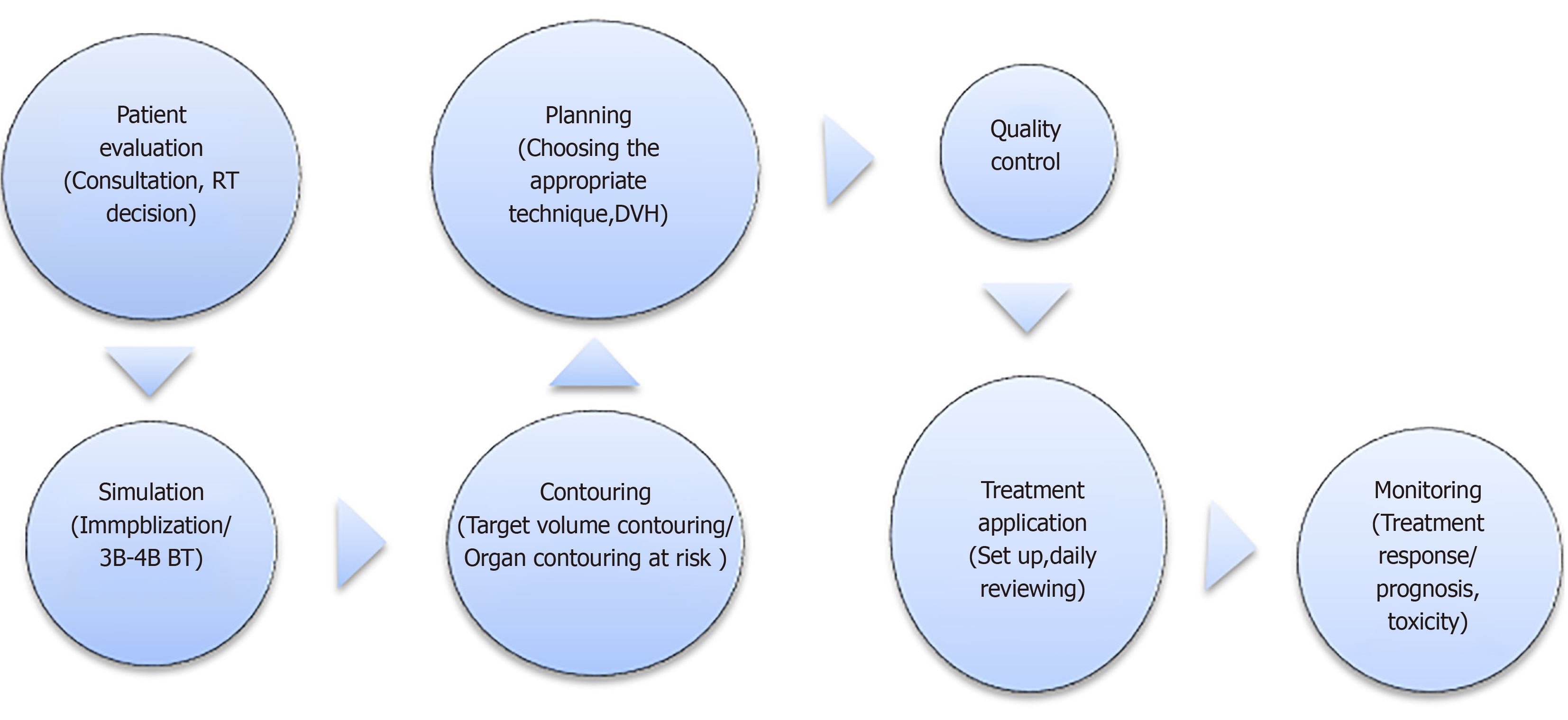
The aim of this review is to summarize the radiotherapy (RT) process in workflow order by identifying specific areas where quality and efficiency can be improved with AI. The RT stage is divided into seven stages: patient evaluation, simulation, contouring, planning, QA, treatment application, and patient follow-up, and the flow chart is given in Figure 1. A systematic evaluation of the applicability, limitations, and advantages of AI algorithms has been made to each stage.
Clinical radiation therapy workflow begins with patient assessment. This step typically includes a series of consultations including reviews of the radiation oncologist on the patient's symptoms, medical history, physical examination, pathological and genomic data, diagnostic studies of prognosis, comorbidities, and risk of toxicity from RT. The radiation oncologist then suggests a treatment plan based on the synthesis of these data. For clinicians involved in this process, the accumulation of big data beyond what people can quickly interpret is the biggest challenge[7]. AI-based methods that can be used in routine functioning may be important decision support tools for clinicians in the future. Such AI-based models have been reported to improve prognosis and predict treatment outcomes, but are not yet used in routine clinical practice[8].
The recent implementation of electronic health records has significantly increased the clinical documentation burden of physicians. The notes having constituted 34%-78% of physicians’ working days, for each hour a physician spends in direct contact with the patient, he spends an additional 2 h in front of the computer[9]. AI solutions have the potential to automate structured documentation. They can save time requirements that add to the documentation burden, reduce burnout, protect confidentiality, and organize medical data into searchable and available items[10]. In addition, an AI-supported electronic record system may have pre-consultation and disease pre-diagnosis power, by including a timeline and the outcomes of relevant tests, procedures, and treatments from various sources[10]. AI-based systems can record patient-doctor conversations and use speech recognition and natural language processing to create a coherent narrative. Such an AI-based system does not yet exist, significant technical advances for clinical and persuasive speech require the learning of hours of selected recording of patient speech[11]. According to patient demands AI systems can present information to the patient at low, medium, or high complexity levels.
The radiation oncologist should consider many factors during the evaluation of the patient and include consideration of their interactions when making treatment decisions. At this point, data-based forecasting models can guide the doctor and make the decision phase faster and more accurate. For example, when a patient diagnosed with lung cancer is being evaluated for stereotactic RT, the patient's respiratory functions, lung capacity, tumor size, proximity of the tumor to critical organs, comorbid diseases, and performance of the patient will affect both treatment response and toxicity. If modeling is made with these and similar factors, response and toxicity rates can be determined before starting treatment. In a case with a diagnosis of left breast cancer and treatment with breast-conserving surgery, modeling created with the patient and treatment characteristics can be predicted whether she can benefit from a breath-holding technique. Big data are needed to create these estimation models. The transition to the use of AI will also increase collaboration between centers in the data collection phase and make treatments more standardized. In addition, depending on the distribution of technology in the centers in the country, AI can direct patients to appropriate treatment centers. For example, it can direct pediatric cases requiring proton therapy to a specialized center, and cases requiring palliative treatment to a conventional center).
After the RT decision is made, a good simulation is required to choose the right treatment. Immobilization technique, scanning range, and the treatment area should be accurately determined. Preliminary preparations such as use of fiducial markers for simulation, full/empty bladder, whether an empty rectum is required, renal function tests, and fasting status should be carefully considered if intravenous contrast is to be applied. Accurate and good simulation is essential to obtain a high-quality, robust treatment plan for the patient. In clinical practice, it is not uncommon to repeat a CT during CT simulation because of deficiencies and inaccuracies such as insufficient scanning range, inadequate/incorrect immobilization technique, an inappropriate level of bladder/rectum content, and hardware-related artifacts[12]. There are many questions that can be answered with AI to improve overall workflow efficiency. For example, will the patient benefit from the use of an intravenous contrast agent? Which immobilization technique should be used? Is 4-dimensional (4D)-CT simulation necessary?
Depending on the location of the disease, this process can be very complex, and optimal patient immobilization is individual, so this process often requires the participation of a radiation oncologist and a medical physicist. For example, special care should be taken to assess potential interference between the immobilization device and treatment beam angles, or patient-specific problems that could cause collisions with the RT device. In simulation, CT is still used in many centers, but brain and prostate tumors can be seen better with magnetic resonance (MR). As a solution, efforts have been made to develop CT scans using MR data, also called synthetic CT (sCT) scans using the atlas-based, sparse coding-based, or learning-based methods. Convolutional neural networks (CNNs), which are less time consuming and more efficient AI-based method with fewer artifacts, increasingly used to convert MR data to sCT[13]. Therefore, in the future, sCT scans with AI-based methods may compensate for the need for CT scanning, as they can be created with electron density data faster and are more reliable for plan generation than MR. Compared with traditional sCT methods, DL methods can be fully automated. Training with MR-CT images has been improved by the use of cycle-consistent generative adversarial networks (GANs)[14]. GANs require a new DL algorithm using two networks, a generator network creates realistic images and a differential mesh distinguishes between real and created images[14]. Studies have reported that images created by sCT and DL are accurate enough for dose calculation[15,16]. The same method can be used for other image syntheses. For example, virtual 4D-MR images can be synthesized from 4D-BT in order to see liver tumors well in image-guided RT (IGRT)[17].
Simulation is one of the most important steps in RT because ant deficiencies or errors that occur are reflected in the entire treatment process. AI techniques can be used to increase the accuracy of the simulation, to personalize it according to the patient characteristics, and to better characterize the tumor, but more studies are needed for its routine clinical use.
Image registration is the process of spatially aligning two or more sets of images of the same region shot in different modalities at different times[17]. Commercially available automated image registration algorithms are typically designed to perform well only with modality-specific registration problems and require additional manual adjustments to achieve a clinically acceptable registration[7]. The two main registration methods used in RT are density based and have rigid registration. In a review of image registration Viergever et al[18] examined relevant developments between 1998 and 2016. They stated that DL approaches to registration can be novel game-changers in facilitating the implementation process and doing more, and they advocated the application of DL concepts to make it a routine integral part of the entire clinical imaging spectrum[18]. AI tools are also trained to determine the sequence of motion actions that result in optimal registration. These algorithms can provide better accuracy than various state-of-the-art registration methods and can be generalized to multiple display methods[19]. AI approaches have been shown to mitigate the effects of image artifacts like metal screws, guide wires, prostheses, and motion artifacts, which pose difficulties in both registration and segmentation[7].
In the standard workflow, the target volume and organs at risk (OARs) are manually contoured by the radiation oncologist in a cross-section. As a result, the process is long and has a high degree of variability as a result of individual differences[20]. Manual segmentation directly affects the quality of the treatment plan and dose distribution for OARs[21]. There have been some attempts at automatic segmentation. It is the most widely available atlas-based segmentation in clinical use. First, the target image is matched with one or more selected reference images. Then, the contours in the reference image are transferred to the target image[22]. Atlas-based methods depend on the choice of atlas and the accuracy of reference images[23]. AI can be used to minimize the differences between physicians and to shorten the duration of this step in RT planning.
Segmentation of at-risk organs: To protect at-risk organs and to correctly evaluate RT toxicity, the segmentation of OARs should be done correctly. To fully benefit from technological developments in RT planning and devices, at-risk organs must be identified correctly. In clinics with high patient density, this step can be rate limiting. In addition, there may be differences among the practitioners, and because of significant anatomical changes (e.g., edema, tumor response, weight loss, and others) during treatment, a new plan with new segmentation may be required. AI, particularly CNN, is a potential tool to reduce physician workload and define a standard segmentation. In recent years, DL methods have been widely used in medical applications such as organ segmentation in CNN, head-neck, lung, brain, and prostate cancers[24-27].
In a head and neck cancer study by Ibragimov et al[28] contouring the spinal cord, mandible, parotid glands, submandibular glands, larynx, pharynx, eyes, optic nerves, and optic chiasm was done in 50 patients by DL using CT images. They obtained dice similarity coefficients (DSCs) of between 37.4% (optic chiasm) and 89.5% (mandible). Compared with the contouring algorithm of current commercial software, contouring of the medulla spinalis, mandibular and parotid glands, larynx, pharynx, and eye globes, was better and that of the optic nerve, submandibular gland (SMG), and the optic chiasm was worse with DL. CT images were used in that study, and higher accuracy rates were achieved with MR image support[28]. In a study of head and neck organ segmentation in 200 patients with oropharyngeal squamous cell carcinoma, Chan et al[29] used CT for planning, with 160 cases used for training, 20 for internal validation, and 20 for testing. Mandibula, right and left parotid glands, oral cavity, brainstem, larynx, esophagus, right and left SMG, right and left temporomandibular joints were contoured. In a lifelong learning-based CNN (LL-CNN) comparison, manual contouring was used as the gold standard and DSC and root-mean-square error (RMSE) was used for accuracy. LL-CNN was then compared to 2D U-Net, 3D U-Net, single-task CNN (ST-CNN) and multitask CNN. Higher DSC and lower RMSE were obtained with LL-CNN compared with the other algorithms. The study found that LL-CNN had a better prediction accuracy than all alternative algorithms for the head and neck organs at risk[29]. In another study, Rooij et al[30] used CT images of 157 head and neck cancer patients, 142 for case training and 15 for testing. The right and left SMGs, right and left parotid glands, larynx, cricopharynx, pharyngeal constrictor muscle, upper esophageal sphincter, brain stem, oral cavity, and esophagus were contoured. With DL, contouring of the 11 OARs was < 10 s per patient. The mean DSC of seven of the 11 contoured organs ranged from 0.78 to 0.83, and the DSC values for the esophagus, brainstem, PMC and cricopharynx were 0.60, 0.64, 0.68 and 0.73, respectively[30]. The study found that for the head and neck OAR, DL-based segmentation was fast and performed well enough for treatment planning purposes for most organs and most patients.
OARs in the thorax area have also contoured for RT with AI[31-34]. Zhu et al[25] used CT images of 66 lung cancer cases, 30 cases for training and 36 cases for testing. CNN was used for segmentation, and compared with atlas-based automatic segmentation (ABAS). DSC, the mean surface distance (MSD), and 95% Hausdorff distance (95% HD) were used to evaluate the results. The MSD (mm) values for CNN and ABAS were 2.92 and 3.14 for the heart, 3.21 and 3.83 for the liver, 1.81 and 3.03 for ms, 2.65 and 2.67 for the esophagus, and 1.93 and 1.85 mm for the lungs. The 95% HD (mm) values for CNN and ABAS were 7.98 and 9.53 in the heart, 10.0 and 11.87 in the liver, 8.74 and 11.97 in ms, 9.25 and 9.45 in the esophagus, and 7.96 and 8.07 mm in the lungs[25]. According to the results of that study, CNN can be used in segmentation for RT of lung cancer. Zhang et al[33] compared CNN-based segmentation and ABAS and reported that CNN-based segmentation required 1.6 minutes per case and atlas-based contouring required 2.4 min (P < 0.001). Accuracy rates were measured by DSC and MSD and found that CNN-based segmentation was better than atlas-based segmentation for left lung and heart RT[33]. A study by Vu et al[34] that included 22411 CT images obtained from 168 cases reported training, validation, and test rates of 66%, 17% and 17%, respectively. CNN-based and atlas-based segmentation models were compared with verification by DSC and 95% HD. All differences were found to be statistically significant in favor of CNN-based segmentation[34].
Looking at other studies in the literature, Feng et al[32] evaluated 36 cases, with 24 used as training and 12 used as testing. The DSC obtained with 3D U-Net for medulla spinalis, right lung, left lung, heart, and esophagus were 0.89, 0.97, 0.97, 0.92, and 0.72, respectively. The corresponding MSDs were 0.66, 0.93, 0.58, 2.29 and 2.34 mm; and the 95% HDs were 1.89, 3.95, 2.10, 6.57, 8.71[32]. The conclusion was that because of the improved accuracy and low cost of OAR segmentation, DL has the potential to be clinically adopted in RT planning. Loap et al[31] performed AI-based heart segmentation with CT images obtained from 20 breast cancer cases. The performance of the model was evaluated by DSC, and this value was found to be 95% for the whole heart and 80% for the heart chambers[31].
Studies on OAR segmentation in the pelvic region have generally been done with cervical and prostate cancer[27,35]. The bladder, bone marrow, left femoral head, right femoral head, rectum, small intestine, and ms were contoured using CT images of 105 locally advanced cervical cancer cases. U-Net was used and the accuracy of the model was evaluated by DSC and 95% HD. The DSC of OARs ranged from 92% to 79%, with the best results in the bladder and the worst in the rectum. 95% HD values ranged between 5.09 and 1.39 mm[35]. Savenije et al[27] included 150 prostate cancer cases with MR imaging. DeepMedic and dense V-net were used in modeling. Bladder, rectum and femoral heads are contoured. The duration of DeepMedic, dense V-net, and atlas-based segmentation were 60 s, 4 s and 10-15 min, respectively. The accuracy of the DeepMedic algorithm that had been obtained in a feasibility study was confirmed the clinical setting in that study[27].
Additional evidence is available from a study by Ahn et al[36] who used CT images of 70 cases diagnosed with liver cancer, 45 for training, 15 for validation, and 19 for testing. The reference was accepted segmentation by three senior physicians. The model was created with deep CNN (DCNN). The accuracy rate was evaluated with 95% HD, DSC, volume overlap error (VOE), and relative volume difference (RVD). In ABAS, the DSCs were 0.92, 0.93, 0.86, 0.85, and 0.60 for the heart, liver, right kidney, left kidney and stomach. In the DCNN-based model, the values were 0.94, 0.93, 0.88, 0.86, and 0.73. The VOE% values in DCNN and atlas-based segmentation were 10.8 vs 15.17, 10.82 vs 13.52, 12.19 vs 17.51, 16.31 vs 25.63 and 37.53 vs 62.64. The RVD% values in DCNN and atlas-based segmentation were 5.17 vs 12.90, 1.86 vs 5.56, 4.53 vs 9.75, 2.45 vs 10.23 and 21.26 vs 50.6[36]. In that study, DL-based segmentation appeared to be more effective and efficient than atlas-based segmentation for most of the OAR in liver cancer RT.
Dolz et al[26] performed brainstem segmentation from the MR images of 14 brain cancer cases. A support vector machine (SVM) algorithm was used for the model, DSC, absolute volume difference (AVD) and percentage volume difference (pVD) between automatic and manual contours were used for the performance evaluation of the model. The mean values were, DSC 0.89-0.90, AVD 1.5 cm3 and pVD 3.99%[26]. The proposed approach has consistently shown similarity to manual segmentation and can be considered promising for adoption in clinical practice. Studies that investigated segmentation of OARs are summarized in Table 1.
| Ref. | Tumor site | Artificial intelligence technique | Patient number | Contouring | Results |
| Ibragimov et al[28], 2017 | Head-neck | CNN | 50 | Contoured with CT. OARs: (1) Ms; (2) Mandible; (3) Parotid; (4) SMG; (5) Larynx; (6) Pharynx; (7) Eyes; (8) Optic nerve; and (9) Optic chiasm | DSC: (1) Ms: 87%; (2) Mandible: 89.5%; (3) Right parotid gland: 77.9%; (4) Left parotid gland: 76.6%; (5) Left SMG: 69.7%; (6) Right SMG: 73%; (7) Larynx: 85.6%; (8) Pharynx: 69.3%; (9) Left eye glob: 63.9%; (10) Right eye glob: 64.5%; (11) Left optic nerve: 63.9%; (12) Right optic nerve: 64.5%; and (13) Optical chiasm: 37.4% |
| Chan et al[29], 2019 | Orafarenx | LL-CNN, 2D U-Net, 3D U-Net, ST-CNN, MT-CNN | 200 (160 training, 20 validation, 20 test) | Contoured with CT. OAR: (1) Mandible; (2) Right and left parotid gland; (3) Oral cavity; (4) Brain stem; (5) Larynx; (6) Esophagus; (7) Right and left SMG; and (8) Right and left TMJ | DSC (mm) for LL-CNN and RMSE: (1) Mandible: 0.91 and 0.66; (2) Right parotid gland: 0.86 and 1.67; (3) Left parotid gland: 0.85 and 1.86; (4) Oral cavity: 0.87 and 0.83; (5) Brain stem: 0.89 vs 0.96; (6) Larynx: 0.86 vs 1.34; (7) Esophagus: 0.86 vs 1.03; (8) Right SMG: 0.85 vs 1.24; (9) Left SMG: 0.84 vs 1.22; (10) Right TMJ: 0.87 vs 0.43; and (11) Left TMJ: 0.84 vs 0.47 |
| Rooij et al[30], 2019 | Head-neck | 3D U-Net | 157 (142 training, 15 tests) | Contoured with CT. OAR: (1) Right and left SMG; (2) Right and left parotid gland; (3) Larynx; (4) Cricopharynx,; (5) PCM; (6) UES; (7) Brain stem; (8) Oral cavity; and (9) Esophagus | DSC: (1) Right SMG: 0.81; (2) Left SMG: 0.82; (3) Right parotid gland: 0.83; (4) Left parotid gland: 0.83; (5) Larynx: 0.78; (6) Cricopharynx: 0.73; (7) PCM: 0.68; (8) UES: 0.81; (8) Brain stem: 0.64; (9) Oral cavity: 0.78; and (10) Esophagus: 0.60 |
| Zhu et al[25], 2017 | Lung | CNN | 66 (30 training, 36 tests) | Contoured with CT. OAR: (1) Heart; (2) Liver; (3) Ms; (4) Esophagus; and (5) Lung | MSD (mm) (CNN vs ABAS): (1) Heart: 2.92 vs 3.14; (2) Liver: 3.21 vs 3.83; (3) Ms: 1.81 vs 3.03; (4) Esophagus: 2.65 vs 2.67; and (5) Lung: 193 vs 1.85; 95% HD (mm) (CNN vs ABAS): (1) Heart: 7.98 vs 9.53; (2) Liver: 10.06 vs 11.87; (3) Ms: 8.74 vs 11.97; (4) Esophagus: 9.25 vs 9.45; and (5) Lung: 7.96 vs 8.07 |
| Zhang et al[33], 2020 | Lung | CNN | 200: training;50: validation 19: test | Contoured with CT. OAR: (1) Lungs; (2) Esophagus; (3) Heart; (4) Liver; and (5) Ms | DSC (CNN vs atlas based): (1) Left lung: 94.8% vs 93.2%; (2) Right lung: 94.3% vs 94.3%; (3) Heart: 89.3% vs 85.8%; (4) Ms: 82.1% vs 86.8%; (5) Liver: 93.7% vs 93.6%; and (6) Esophagus: 73.2% vs -; MSD (mm) (CNN vs atlas based): (1) Left lung: 1.10 vs 1.73; (2) Right lung: 2.23 vs 2.17; (3) Heart: 1.65 vs 3.66; (4) Ms: 0.87 vs 0.66; (5) Liver: 2.03 vs 2.11; and (6) Esophagus: 1.38 vs - |
| Vu et al[34], 2020 | Lung | 2D-CNN | 168 (66% training, 17% validation, 17% testing) | Contoured with CT. OAR: (1) Ms; (2) Lungs; (3) Heart; and (4) Esophagus | DSC (CNN vs atlas - based model): (1) Ms: 71% vs 67%; (2) Right lung: 96% vs 94%; (3) Left lung: 96% vs 94%; (4) Heart: 91% vs 85%; and (5) Esophagus: 63% vs 37%; 95% HD (mm) (CNN vs atlas - based model): (1) Ms: 9.5 vs 25.3; (2) Right lung: 5.1 vs 8.1; (3) Left lung: 4.0 vs 8.0; (4) Heart: 9.8 vs 15.8; and (5) Esophagus: 9.2 vs 20 |
| Feng et al[32], 2019 | Lung | 3D U-Net | 36 (24 training, 12 tests) | Contoured with CT. OAR: (1) Ms; (2) Right lung; (3) Left lung; (4) Heart; and (5) Esophagus | DSC: (1) Ms: 0.89; (2) Right lung: 0.97; (3) Left lung: 0.97; (4) Heart: 0.92; and (5) Esophagus: 0.72; 95% HD (mm): (1) Ms: 1.89; (2) Right lung: 3.95; (3) Left lung: 2.10; (4) Heart: 6.57; and (5) Esophagus: 8.71; MSD (mm): (1) Ms: 0.66; (2) Right lung: 0.93; (3) Left lung: 0.58; (4) Heart: 2.29; and (5) Esophagus 2.34 |
| Liu et al[35], 2019 | Cervix | 3D U-Net | 105 (77 training, 14 validation, 14 tests) | Contoured with CT. OAR: (1) Bladder; (2) Bone Marrow; (3) Left femoral head; (4) Right femoral head; (5) Rectum; (6) Small intestine; and (7) Ms | DSC: (1) Bladder: 0.92; (2) Bone Marrow: 0.86; (3) Left femoral head: 0.89; (4) Right femoral head: 0.89; (5) Rectum: 0.79; (6) Small intestine: 0.83; and (7) Ms: 0.82; 95% HD (mm): (1) Bladder: 5.09; (2) Bone marrow: 1.99; (3) Left femoral head: 1.39; (4) Right femoral head: 1.43; (5) Rectum: 5.94; (6) Small intestine: 5.21; and (7) Ms: 3.26 |
| Savenije et al[27], 2020 | Prostate | DeepMedic and Dense V-net | 48 (36 training, 16 tests) for feasibility study; 150 cases in total (97 train, 53 tests) | Contoured by MR. OAR: (1) Bladder; (2) Rectum; (3) Left femur; and (4) Right femur | DSC/95% HD (mm)/MSD (mm): (DeepMedic and dense V-net (feasibility study): (1) Bladder: 0.95/3.8/1.0; (2) Rectum: 0.85/8.3/2.1; (3) Left femur: 0.96/2.2/0.6; and (4) Right femur: 0.96/1.9/0.6; DSC/95% HD (mm)/MSD (mm): (Clinical application with DeepMedic): (1) Bladder: 0.96/2.5/0.6; (2) Rectum: 0.88/7.4/1.7; (3) Left femur: 0.97/1.6/0.5; and (4) Right femur: 0.97/1.5/0.5 |
| Ahn et al[36], 2019 | Liver | DCNN | 70 (45 training, 15 validation, 10 tests) | Contoured with CT. OAR: (1) Heart; (2) Liver; (3) Kidney; and (4) Stomach | DSC (DCNN vs atlas-based contouring): (1) Heart: 0.94 vs 0.92; (2) Liver: 0.93 vs 0.93; (3) Right kidney: 0.88 vs 0.86; (4) Left kidney: 0.86 vs 0.85; and (5) Stomach: 0.73 vs 0.60 |
Learning algorithms are trained to maximize measures of similarity between outcomes and examples given to them. Therefore, although they are increasingly skilled at imitating human-drawn contours, they are limited by the quality of their training samples. Until more concrete consensus definitions are specified for boundaries, machines cannot be more accurate than the human input taken as their clinically fundamental truth. Machine “accuracy” is only considered to be meaningful in the context of individuals and institutional protocols. More case numbers and multicenter studies are needed for the development and standardization of contouring models.
Target volume contouring: Target volume contouring is a labor-intensive step in the treatment planning flow in RT. Differences in manual contouring result from variability between contours, differences in radiation oncology education, or quality differences in imaging studies. Current automatic contouring methods aim to reduce manual workload and increase contour consistency, but still tend to require significant manual editing[37]. Recent studies have shown that DL-based automatic contouring of target volumes is promising, with greater accuracy and time savings compared with atlas-based methods.
The first reason for the necessity of computer-aided delineation is the variation between contours or even between contours of the same person at different times. Chao et al[38] reported that differences in defining CTVs from scratch among radiation oncologists is important, and the use of computer-aided methods reduces volumetric variation and improves geometric consistency[38]. The second reason is that it is time consuming. In a study by Chao et al[38], computer-assisted contouring provided 36%-29% time savings for experienced physicians and 38%-47% for less experienced physicians[38]. Ikushima et al[39] estimated the gross tumor volume (GTV) of 14 lung cancers. Six were solid, six were part-solid, and four had mixed ground-glass opacity (GGO) using AI. Image properties around the GTV contours were taught to the SVM algorithm during training, after which the algorithm was tested to generate GTV for each voxel. Diagnostic CT, planning CT and PET were used for image properties. The final GTV contour was determined using the optimum contour selection method. DSC was used for the performance of the algorithm and was determined to be 0.77 for 14 cases. The DSC values for solid, part-solid and mixed GGO were 0.83, 0.70 and 0.76, respectively[39]. In a study conducted by Cui et al[40], 192 cases of lung cancer (118 solid, 53 part-solid, and 21 pure GGO) with stereotactic body radiotherapy (SBRT) were contoured with dense V-networks using planning it. Of those, 147 cases were for training, 26 for validation, and 19 cases were for testing. Evaluation was performed with a DSC and HD 10-fold cross validation test. The 3D-DSC values were 0.838 ± 0.074, 0.822 ± 0.078, and 0.819 ± 0.059 for solid, part-solid, and GGO tumors respectively. The HD value of each inner group was 4.57 ± 2.44 mm[40]. The proposed approach has the potential to assist radiation oncologists in identifying GTVs for planning treatment of lung cancer SBRT.
Zhong et al[41] performed segmentation with 3-D DL and fully convolutional networks (DFCN) using both PET and BT images of 60 lung SBRT cases. Delineation was performed by three senior physicians. A simultaneous truth and performance level estimation algorithm was accepted as a reference, and DSC was used to evaluate DFCN performance. The mean DSCs were for 0.861 ± 0.037 for CT and 0.828 ± 0.087 for PET[41]. Kawata et al[42] used pixel-based MÖ techniques such as fuzzy-c-means clustering (FCM), artificial neural network (ANN), and SVM to evaluate the GVT of 16 lung cancer tumors (six solid, four GGO, six part-solid) for SBRT by AI using PET/CT. The performance of the algorithms was determined by DSC. The DSC values for FCM, ANN, and SVM were 0.79 ± 0.06, 0.76 ± 0.14 and 0.73 ± 0.14, respectively[42]. FCM had the highest accuracy rates of GTV contouring compared with the other algorithms.
There are also GTV and CTV contouring studies with AI in head and neck cancers[43-47]. In a study by Li et al43], tumor segmentation was performed in nasopharyngeal cancer by using CT images. The U-Net model was used, 302 cases were used for training, 100 for validation, and 100 for testing. In the U-Net model, DSC was found to be 65.8% for lymph nodes and 74.0% for tumor segmentation. Automatic delineation was calculated as 2.6 h per patient and manual delineation as 3 h[43]. This study found that DL increased the accuracy, consistency, and efficiency of tumor delineation and that additional physician input might be required for lymph node delineation. Multimodality medical images can be very useful for automated tumor segmentation as they provide complementary information that can make the segmentation of tumors more accurate. Ma et al[44] used multimodality CNN (M-CNN) based methods to investigate the segmentation of nasopharyngeal cancer using CT and MR images. M-CNN is designed to co-learn the segmentation of matched CT-MR images. Considering that each modality has certain distinctive features, it was planned to create a combined-CNN (C-CNN) using single-modality (S-CNN) and higher-layer features derived from M-CNN. Ninety CT and MR images were used, and positive predictive value (PPV), sensitivity (SE), DSC, and average symmetric surface distance (ASSD) were used to evaluate modalities. The PPV, SE, DSC and ASSD obtained by C-CNN, were 0.797 ± 0.109, 0.718 ± 0.121, 0.752 ± 0.043, and 1.062 ± 0.298 mm respectively. The included two main models, M-CNN and C-CNN, which can integrate complementary information from CT and MR images for tumor identification. The results in the clinical CT-MR dataset show that the proposed M-CNN can learn the correlations of two modalities and tumor segmentation together and perform better than using a single modality[44].
Zhao et al[45] used PET-CT and FCN to contour 30 nasopharyngeal cancer tumors. The mean DSC was 87.47% after threefold cross validation. Guo et al[46] performed GTV contouring with Dense Net and 3D U-Net using PET/CT and PET-CT in 250 head and neck cancer patients. DSC, MSD, and HD95 were calculated for each of the three imaging methods separately. For Dense Net, the DSC values were 0.73 for PET-CT, 0.67 for PET, and 0.32 for CT. The DSC for 3D-U-Net and PET-CT was 0.71. MSD, HD for Dense Net PET-BT were 2.88, 6.48 and 3.96 mm, respectively. For Dense Net PET, the MSD, and HD95 DC were 3.38, 8.29 and 5.56 mm, respectively. For 3D U-Net, the MSD, HD95, DC were 2.98, 7.57 and 4.40 mm respectively[46]. In a study using a deep deconvolutional neural network (DDNN), the GTVtumor, GTVlymph node, and CTV were determined from CT images of 230 nasopharyngeal cancer cases that were randomly allocated to 184 cases for training and 46 cases for testing. The DSC values were 80.9% for GTVtumor, 62.3% for GTVlymph node, and 82.6% for CTV[47].
AI-based contouring studies have also been performed in primary brain tumors and brain metastases[48-53]. In a study by Jeong et al[51], T1-weighted dynamic contrast-enhanced (DCE) perfusion MR images of 21 patients diagnosed with brain tumors were used for tumor segmentation. 3D mask region-based CNN (R-CNN) was used and algorithm performance was evaluated with DSC, HD, MSD, and center of mass distance. The values were 0.90 ± 0.04, 7.16 ± 5.78 mm, 0.45 ± 0.34 mm and 0.86 ± 0.91 mm, respectively[51]. The results support the feasibility of accurate localization and segmentation of brain tumors from DCE perfusion MRIs. Segmentation with 3D mask R-CNN in DCE perfusion imaging holds promise for future clinical use. Tang et al[53] described postoperative glioma segmentation of the CTV region using MR image information on CT. A deep feature fusion model (DFFM) guided by multisequence MR was used in CT images for postop glioma segmentation. DFFM is a multisequence MR-guided CNN that simultaneously learns deep features from CT and multisequence MR images and then combines the two deep features. In this study, 59 BT and MR (T1/T2-weighted FLAIR, T1-weighted contrast-enhanced, T2-weighted) data sets were used. The DSCs were 0.836 and 0.836[51]. Given the DSC rate, this algorithm can be used in the presegmentation stage to reduce the workload of the radiation oncologist.
Liver tumor segmentation with CT is difficult because the image contrast between liver tumors and healthy tissues is low, the boundary is blurred, and images of the liver tumor are complex, and vary in size, shape, and location. To solve these problems, Meng et al[54] performed liver tumor segmentation with 3D dual-path multiscale CNN (TDP-CNN). In the study, 81 CT images were used for training and 25 were used for the test. Tumor segmentation determined by an experienced radiologist was used as a reference. Performance evaluation was determined as DSC, HD average distance, and the values were 0.689, 7.69, and 1.07mm[54].
There are also studies using AI in pelvic tumor and CTV contouring[55-59]. In a prostate cancer study, MR images and the DeepLabV3 + method were used for with target volume segmentation. Volumetric DSC and surface DSC were used to evaluate performance, and these values were 0.83 ± 0.06 and 0.85 ± 0.11, respectively[56]. According to this model, the planning workflow can be accelerated with MR. Voxel-based ML was evaluated, and MR images of 78 cases were used in a study of tumor delineation in locally advanced cervical cancer. The model was trained according to the delineation of two radiologists, mean sensitivity was 94% and specificity was 52%[57]. CT images were used for CTV delineation in rectal cancer, and 218 randomly selected cases were used for training and 60 cases for validation. Deep dilated CNN (DDCNN) was used and the DSC for the model was 87.7%[59]. According to that study, the accuracy rate was high and effective in CTV segmentation in the DDCNN rectal cancer. Deep dilated residual network (DD-ResNet) was used in breast cancer CTV contouring, and the model was compared with DDCNN and DDNN. CT images of 800 breast cancer cases were used in the study, and the training/test rate was determined as 80%/20%. Mean DSC was used for segmentation accuracy. For the right and left breast, DD-ResNet was 0.91 and 0.91, 0.85 and 0.85, 0.88 and 0.87 for DDCNN and DDNN, respectively. HD values were 10.5 and 10.7 mm, 15.1 and 15.6 mm, and 13.5 and 14.1 mm, respectively. Mean segmentation times were 4, 21 and 15 s per patient[60]. The method proposed in the study contoured the CTV in a short time and with high accuracy. The studies of target volume segmentation are summarized in Table 2. More cases and multidisciplinary studies are needed to reduce the heterogeneity in tumor response and in GTV and CTV contouring, shorten the contouring step, and create standard delineations.
| Ref. | Tumor site | Artificial intelligence technique | Patient number | Contouring | Results |
| Ikushima et al[39], 2017 | Lung | SVM | 14 (solid: 6, GGO: 4, mixed GGO: 4) | GTV | DSC: (1) 0.777 for 14 cases; and (2) 0.763 for GGO, 0.701 for mixed GGO |
| Cui et al[40], 2021 | Lung | DVNs | 192 (solid: 118, part-solid:53, pure GGO: 21) | GTV | 3D-DSC: (1) Solid: 0.838 ± 0.074; (2) Part-solid: 0.822 ± 0.078; and (3) GGO: 0.819 ± 0.059 |
| Zhong et al[41], 2019 | Lung | 3D-DFCN | 60 | GTV | DSC: (1) CT: 0.861 ± 0.037; and (2) PET: 0.828 ± 0.087 |
| Kawata et al[42], 2017 | Lung | FCM, ANN, SVM | 16 (solid: 6, GGO:4, part-solid GGO:6) | GTV | DSC: (1) FCM-based framework:0.79 ± 0.06; (2) ANN-based framework: 0.76 ± 0.14; and (3) SVM-based framework: 0.73 ± 0.14 |
| Li et al[43], 2019 | Nasopharynx | U-Net | 502 | GTV | DSC: (1) Lymph nodes: 65.86%; (2) Primary tumor: 74.00%; HDs: (1) Lymph nodes: 32.10 mm; and (2) Primary tumor:12.85 mm |
| Zhao et al[45], 2019 | Nasopharynx | FCN | 30 | GTV | DSC: 87.47% |
| Guo et al[46], 2020 | Head and neck | Dense Net and 3D U-Net | 250 | GTV | DSC: (1) Dense Net with PET/CT: 0.73; (2) Dense Net with PET: 0.67; (3) Dense Net with CT: 0.32; and (4) 3D U-Net with PET/CT: 0.71; MSD: (1) Dense Net with PET/CT: 2.88; (2) Dense Net with PET: 3.38; (3) Dense Net with CT: -; and (4) 3D U-Net with PET/CT: 2.98; HD95: (1) Dense Net with PET/CT: 6.48; (2) Dense Net with PET: 8.29; (3) Dense Net with CT: -; and (4) 3D U-Net with PET/CT: 7.57 |
| Jeong et al[51], 2020 | Brain | 3D-R-CNN | 21 | GTV | DSC: 0.90 ± 0.04; HD: 7.16 ± 5.78 mm; MSD: 0.45 ± 0.34 mm; Center of mass distance: 0.86 ± 0.91 mm |
| Meng et al[54], 2020 | Liver | TDP-CNN | 106 | GTV | DSC: 0.689; HD: 7.69mm; Average distance: 1.07 mm |
| Elguindi et al[56], 2019 | Prostate | 2D-CNN, DeepLabV3 + | 50 | Prostate | Volumetric DSCL: 0.83 ± 0.06; Surface DSC: 0.85 ± 0.11 |
| Men et al[59], 2017 | Rectum | DDCNN | 278 | CTV | DSC: 87.7% |
RT planning process is quite complex. A mistake during planning can lead to life-threatening situations such as tumor incontinence or high doses of radiation to normal tissue. As technology advances, the margin given to the tumor also decreases, so even with a small margin of error, it is possible to miss the tumor geographically. After target volumes and OARs are defined, the planning process continues with the determination of dosimetric targets for targets and OARs, selection of an appropriate treatment technique [e.g., 3DCRT, intensity-adjusted RT (IMRT), volumetric modulated arc therapy (VMAT), protons], the achievement of planning goals, and evaluation and approval of the plan. Treatment planning, which is an RT design for each case, can be considered as both a science and an art.
Because of the complex mathematics and physics involved, RT planning includes computer-aided systems. During planning, humans interact many times with the computer-aided system, using their experience and skills to ensure the satisfactory quality of each plan. Planning is a very complex process. There are AI studies related to the planning steps of RT, such as dose calculation, dose distribution, dose-volume histogram (DVH), patient-specific dose calculation, IMRT area determination, beam angle determination, real-time tumor tracking, and replanning in adaptive RT[61-71].
The purpose of researching the dose calculation algorithm is to increase calculation accuracy while maximizing computational efficiency. In the study conducted by Zhu et al[61], it was aimed to calculate the 3D distribution of total energy release per unit mass and electron density based on CNN. Twelve sets of CT images were used for training, and a random beam configuration was created with a convolution/ superposition (CCCS) algorithm. 7500 samples were created for each single-energy photon model training set and 1500 samples for validation. Training included 0.5 MeV, 1 MeV, 2 MeV, 3 MeV, 4 MeV, 5 MeV, and 6 MeV monoenergetic photon models. To evaluate its usability under linear accelerator (Linac) conditions, 12 additional new CT images with different anatomical regions and 1512 samples were used for testing. For all anatomies, the mean value for the criterion of 3%/2mm, 95% lower confidence limit, and 95% upper confidence limit were 99.56%, 99.51%, and 99.61%, respectively. In that study, DL was investigated for CCCS dose calculation[61]. With DL, calculation accuracy can be improved and calculation efficiency can be increased, and the method can speed up dosing algorithms and also has great potential in adaptive RT.
In their study, Zhang et al[62] aimed to estimate voxel level doses by integrating the distance information between the planning target volume (PTV) and OAR as well as the image information into the DCNN. First, they created a four-channel feature map consisting of PTV image, OAR image, CT image, and distance image. A neural network was created and trained for dose estimation at the voxel level. Given that the shape and size of OARs are highly variable, dilated convolution was used to capture features from multiple scales. The network was evaluated by five-fold cross validation based on 98 clinically validated treatment plans. The voxel level mean absolute error values of the DCNN for PTV, left lung, right lung, heart, spinal cord and body were 2.1%, 4.6%, 4.0%, 5.1%, 6.0% and 3.4% respectively[62]. This method significantly improved the accuracy of the dose distribution estimated by the DCNN model. In their studies, Fan et al[64] aimed to develop a 3D dose estimation algorithm based on DL and create a treatment plan based on the dose distribution for IMRT. The DL model was trained to estimate a dose distribution based on patient-specific geometry and prescription dose. A total of 270 head and neck cancer cases, 195 in the training data set, 25 in the validation set, and 50 in the test set, were included in the study. All cases were treated with IMRT. The model input consisted of CT images and contours that defined the OAR and plot target volumes. The algorithm output was trained to estimate the dose distribution from the CT image slice. The resulting estimation model was used to estimate the patient dose distribution. An optimization target function was then created based on the estimated dose distributions for automatic plan generation. In the study, Differences between the prediction and the actual clinical plan in DVH for all OARs were not significant except for the brainstem, right, and left lens. Differences between PTVs (PTV70.4, PTV66, PTV60.8, PTV60, PTV56, PTV54, PTV51) in the estimated and the actual plan were significant only for PTV70.4[64]. In that study, optimization based on 3D dose distribution and an automatic RT planning system based on 3D dose estimation were developed. The model is a promising approach to realize automatic treatment planning in the future.
Ma et al[65] created a DVH prediction model that depended on support vector regression as the backbone of the ML model. A database containing VMAT plans of 63 prostate cancer cases was used, and a PTV plan was created for each patient. A correlative relationship between the OAR DVH (model input) of the PTV plan and the corresponding DVH (model output) of the clinical treatment plan was established with 53 training cases. The predictive model was tested with a validation group of ten cases. In the control of dosimetric endpoints for the training group, 52 of 53 bladder cases (98%) and 45 of 53 rectum cases were found to be within a 10% error limit. In the validation test group, 92% of the bladder cases and 96% of the rectum cases were within the 10% error limit. Eight of the ten validation plans (80%) were found to be within the 10% error margin for both rectum and bladder[65]. In that study, only the PTV plan was used for DVH estimation and an ML model was created based on new dosimetric characteristics. The framework had high accuracy for predicting the DVH for VMAT plans.
In lung cancer, as in other types of cancer, optimum selection of radiation beam directions is required to ensure effective coverage of the target volume by external RT and to prevent unnecessary doses to normal healthy tissues. IMRT planning is a lengthy process that requires the planner to iterate between selecting beam angles, setting dose-volume targets, and conducting IMRT optimization. The beam angle selection is made according to the planner's clinical experience. Mahdavi et al[67] planned to create a framework that used ML to automatically select treatment beam angles in thoracic cancers, intended to increase computational efficiency. They created an automatic beam selection model based on learning the relationship between beam angles and anatomical features. The plans of 149 cases who underwent clinically approved thoracic IMRT were used in the study. Twenty-seven cases were randomly selected and used to test the automated plan and the clinical plan. When the estimated and clinically used beam angles were compared, a good mean agreement was observed between the two (angular distance 16.8 ± 10◦, correlation 0.75 ± 0.2). The target volume of automated and clinical plans was found to be equivalent when evaluated in terms of winding and the OAR. The vast majority of plans (93%) were approved as clinically acceptable by three radiation oncologists[69].
Treatment planning is an important step in the RT workflow. It has become more sophisticated in the past few decades with the help of computer science, allowing planners to design highly complex RT plans to minimize damage of normal tissue while maintaining adequate tumor control. A need of individual patient plans has resulted in treatment planning becoming more labor-intensive and time consuming. Many algorithms have been developed to support those involved in RT planning. The algorithms have had a major impact on focusing on automating and/or optimizing the planning process and improving treatment planning efficiency and quality. Studies of treatment planning are summarized in Table 3.
| Ref. | Aim | Patient number | Artificial intelligence technique | Results |
| Zhu et al[61], 2020 | Calculating TERMA and ED | 24 | CNN | 3%/2 mm, 95% LCL, and 95% UCL to 99.56%, 99.51%, 99.61% |
| Zhang et al[62], 2020 | Making voxel level dose estimation by integrating the distance information between PTV and OAR | 98 | DCNN | MAEV: (1) PTV: 2.1%; (2) Left lung: 4.6%; (3) Right lung: 4.0%; (4) Heart: 5.1%; (5) Spinal cord: 6.0%; and (6) Body: 3.4% |
| Fan et al[64], 2019 | Developing a 3D dose estimation algorithm | 270 | Significant difference was found between the estimated and the actual plan in only PTV70.4 | |
| Ma et al[65], 2019 | Creating a DVH prediction model | 63 | SVR | The error limit of 10% for the bladder and rectum was 92% and 96% |
| Mahdavi et al[69], 2015 | Selecting treatment beam angles in thoracic cancers | 149 | ANN | The majority of plans (93%) were approved as clinically acceptable by three radiation oncologists |
Quality assurance (QA) is crucial in order to evaluate the RT plan and detect and report errors. Features of RT QA programs such as error detection, and prevention, and treatment device QA are very suitable for AI application[72-75]. Li et al[73] developed an application to estimate the performance of medical linear accelerators (Linacs) over time. Daily QA of RT in cancer treatment closely monitors Linac performance and is critical for the continuous improvement of patient safety and quality of care. Cumulative QA measures are valuable for understanding Linac behavior and enabling medical physicists to detect disturbances in output and take preventive action. Li et al[73] used a time series estimation model of ANNs and an autoregressive moving average to analyze 5-yr Linac QA data. Verification tests and other evaluations were made for all models and they reported that the ANN algorithm can be applied correctly and effectively in dosimetry and QA[73]. Valdes et al[72] developed AI applications to predict IMRT QA transition rates and automatically detect problems in the Linac imaging system. Carlson et al[76] developed an ML approach to predict multileaf collimator (MLC) position errors. Inconsistencies between planned and transmitted motions of multileaf collimators are a major source of error in dose distribution during RT. In their study, factors such as leaf movement parameters, leaf position and speed, leaf movement towards or away from the isocenter of the MLC were calculated from plan files of AI forecasting models. Position differences between synchronized DICOM-RT planning files and DynaLog files reported during QA delivery were used for training the models. To assess the effect on the patient, the DVH in the treated positions and the planned and anticipated DVHs were compared. In all cases, they found that the DVH parameters predicted for the OAR, especially around the treatment area, were closer to the DVHs in the treated position than to the planned DVH parameters[76].
The use of treatment plan features to predict patient-specific QA measurement results facilitate development of automated pretreatment validation workflows or provide a virtual assessment of treatment quality. Granville et al[77] trained a linear support vector classifier to classify the results of patient-specific VMAT QA measurements, using the complexity of the treatment plan and characteristics that define the Linac performance criteria. The “targets” in this model are simple classifications that represent the median dose difference between measured and expected dose distributions; median dose deviation was considered “hot” if > 1%, “cold” if < 1%, and “normal” if ± 1%. A total of 1620 patient-specific QA measurements were used for model development and testing’ and 75% of the data was used for model development and validation. The remaining 25% was used for the independent evaluation of model performance. Receiver operating characteristic (ROC) curve analysis was used to evaluate model performance. Of the ten variables that are considered important for prediction, half consist of treatment plan characteristics, and half are QA measures that characterize Linac performance. For this model, the micro-averaged area under the ROC curve was 0.93, and the macro-averaged area under the ROC curve was 0.88[77]. The study demonstrates the potential of using both treatment plan features and routine Linac QA results in the development of ML models for patient-specific VMAT QA measurements.
During radiation therapy, treatment may need to be adjusted to ensure that the plan is properly implemented. Need of adjustment may result from both online factors such as the patient's pretreatment position, and longer-term factors related to anatomical changes and response to treatment. Images taken before treatment should be aligned with the images in the planning CT and kept in alignment. Although many modern Linac devices currently have daily "cone-beam" CT (CBCT) using mega-voltage X-rays for treatment confirmation, but that imaging is not sufficient to distinguish soft tissue structures. However, those images are considered suitable for image-guided RT as they are used to adapt the treatment plans to the daily anatomy of the patient and to reduce intra-fractional shifts. When performing daily RT, the CBCT should be reviewed before each treatment. Two, or at least one experienced RT technician, are required for this procedure. When the RT technician sees an anatomical difference between the CBCT and the planning CT, she/he should inform the radiation oncologist and medical physicist. At that stage, it is necessary to decide whether to continue treatment with the difference or to require a new CBCT. Each of the steps delays patient treatment and causes a significant increase in the RT department workload. All this opens a path for the growth of AI in parallel with the training program in radiation oncology. In addition to the ability of existing staff to cope with the growing workload, innovations in modern technology and the ability to benefit from it are limited by access to adequate human resources[78]. In addition, AI replanning has been used to identify candidates for adaptive RT. Based on anatomical and dosimetric variations such as shrinkage of the tumor, weakening of the patient, or edema, classifiers and clustering algorithms have been developed to predict the patients who will benefit most from updated plans during fractionated RT[71,79]. However, it should also be kept in mind that the algorithm will mimic past protocols rather than determine the ideal time for replanning because AI learns from data about previous patients, their plans, and adaptive RT.
AI has the potential to change the way radiation oncologists follow definitive-treated patients. After surgery, the tumor may disappear during imaging, and tumor markers can quickly normalize. In contrast, imaging changes such as loss of contrast-enhancement, PET involvements or diffusion restriction, or size reduction, and the response of tumor markers after RT are gradual. Those characteristics are monitored regularly over time, and response assessments are made according to changes that are complemented by clinical experience and are considered indicative of therapeutic efficacy. Time is required for this assessment. However, if cases that will not respond to treatment can be predicted earlier, additional doses of RT or additional systemic treatments may be introduced earlier, which may improve oncological outcomes. In this context, early work in the field of radiology is promising. In radiology, quantitative features are extracted based on size and shape, image density, texture, relationships between voxels, and some characteristics to typify an image. AI algorithms can be used to correlate image-based features with biological observations or clinical outcomes[80-85]. The use of AI techniques for response and survival prediction in RT patients is a serious opportunity to further improve decision support systems and provide an objective assessment of the relative benefits of various treatment options for patients.
Cancer is the most common cause of death in developed countries, and it is estimated that the number of cases will increase further in aging populations[86,87]. Therefore, cancer research will continue to be the top priority for saving lives in the next decade. Prognosis studies have been conducted with AI on many types of cancer. The use of AI techniques for response and survival prediction in RT patients is a serious opportunity to further improve decision support systems and provide an objective assessment of the relative benefits of various treatment options for patients.
Six different ML algorithms were evaluated in a prognosis study with 72 cases of nasopharyngeal cancer. Age, weight loss, initial neutrophil/lymphocyte ratio, initial lactate dehydrogenase and hemoglobin values, RT time, tumor size, concurrent CT number, and T and N stage were determined as critical variables. The highest performing model among logistic regression, ANN, XGBoost, support-vector clustering, random forest, and Gaussian Naïve Bayes algorithms was determined as Gaussian Naïve Bayes, and the accuracy rate was found to be 88% (CI: 0.68-1)[88]. In a study using radionics obtained from clinical and PET-CT, prognosis was evaluated in 101 lung cancer cases, with 67% used for training and 33% validation and testing. The highest accuracy rate was achieved with an SVM algorithm that had an accuracy rate of 84%, a sensitivity of 86%, and a specificity of 82%[89]. In another study in which prognosis was predicted in prostate cancer, somatic gene mutations were evaluated and an accuracy rate of 66% was obtained with an SVM algorithm[90]. Post cystectomy bladder cancer prognosis was evaluated in 3503 cases using an SVM algorithm. Recurrence, 1-, 3, and 5-yr survival rates were estimated with sensitivity and specificity above 70%[91]. In a study including 75 gastric cancer patients, the accuracy of survival, distant metastasis, and peritoneal metastasis predictions were 81% for GNB, 86% for XGBoost, and 97% for Random Forest (97%)[92]. Pham et al[93] used AI to detect DNp73 expression associated with 5-yr overall survival and prognosis in 143 rectal cancer cases. Ten different CNN algorithms were used, and each immunochemical image was resized. For the algorithm, 90% of the images were used in training and 10% as test data. The accuracy of ten algorithms varied between 90% and 96%[93].
In oncological treatment, forecasting is crucial in the decision-making process because survival prediction is critical in making palliative vs curative treatment decisions. In addition, the estimation of remaining life expectancy can be an incentive for patients to live a fuller or more fulfilling life. It is also a question of which answer is sought by health insurance companies. Survival statistics assist oncologists in making treatment decisions. However, these are data from large and heterogeneous groups and are not well suited to predict what will happen to a specific patient. AI algorithms for the prediction of RT and chemotherapy oncological outcomes have attracted considerable attention recently. In cases diagnosed with cancer, predicting survival is critical for improving treatment and providing information to patients and clinicians. Considering the data set of rectal cancer patients with specific demographic, tumor, and treatment information, it is a crucial issue whether patient survival or recurrence can be predicted by any parameter. Today, many hospitals store medical records as digital data. By evaluating these large data sets using AI techniques, it may be possible to predict patient treatment outcomes, plan individualized patient treatment, improve corporate performance, and regulate health insurance premiums.
Although AI can take place at every step in radiation oncology, from patient consultation to patient monitoring, and can contribute to the clinician and the society, there are still many challenges and problems to be solved. Initially, Large data sets should be created for AI and then undergo continuing improvement. The development of estimation tools with a wide variety of variables and models limits the comparability of existing studies and the use of standards. Estimation algorithms can be standardized by sharing data between centers, data diversity, and establishing immense databases. In addition, models can be made clinically applicable by updating with entry of new data into the models. Today, the accuracy and quality of data are also of great importance, as no AI algorithm can fix problems in training data.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Turkey
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Lee KS S-Editor: Wang JL L-Editor: Filipodia P-Editor: Xing YX
| 1. | Meyer P, Noblet V, Mazzara C, Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med. 2018;98:126-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 144] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 2. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36149] [Cited by in F6Publishing: 17282] [Article Influence: 1920.2] [Reference Citation Analysis (0)] |
| 3. | Jarrett D, Stride E, Vallis K, Gooding MJ. Applications and limitations of machine learning in radiation oncology. Br J Radiol. 2019;92:20190001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 73] [Article Influence: 14.6] [Reference Citation Analysis (2)] |
| 4. | Boldrini L, Bibault JE, Masciocchi C, Shen Y, Bittner MI. Deep Learning: A Review for the Radiation Oncologist. Front Oncol. 2019;9:977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 5. | Chen C, He M, Zhu Y, Shi L, Wang X. Five critical elements to ensure the precision medicine. Cancer Metastasis Rev. 2015;34:313-318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 6. | Bibault JE, Giraud P, Burgun A. Big Data and machine learning in radiation oncology: State of the art and future prospects. Cancer Lett. 2016;382:110-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 171] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 7. | Huynh E, Hosny A, Guthier C, Bitterman DS, Petit SF, Haas-Kogan DA, Kann B, Aerts HJWL, Mak RH. Artificial intelligence in radiation oncology. Nat Rev Clin Oncol. 2020;17:771-781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 8. | Oberije C, De Ruysscher D, Houben R, van de Heuvel M, Uyterlinde W, Deasy JO, Belderbos J, Dingemans AM, Rimner A, Din S, Lambin P. A Validated Prediction Model for Overall Survival From Stage III Non-Small Cell Lung Cancer: Toward Survival Prediction for Individual Patients. Int J Radiat Oncol Biol Phys. 2015;92:935-944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 9. | Sinsky C, Colligan L, Li L, Prgomet M, Reynolds S, Goeders L, Westbrook J, Tutty M, Blike G. Allocation of Physician Time in Ambulatory Practice: A Time and Motion Study in 4 Specialties. Ann Intern Med. 2016;165:753-760. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 694] [Cited by in F6Publishing: 696] [Article Influence: 87.0] [Reference Citation Analysis (0)] |
| 10. | Lin SY, Shanafelt TD, Asch SM. Reimagining Clinical Documentation With Artificial Intelligence. Mayo Clin Proc. 2018;93:563-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 11. | Luh JY, Thompson RF, Lin S. Clinical Documentation and Patient Care Using Artificial Intelligence in Radiation Oncology. J Am Coll Radiol. 2019;16:1343-1346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Feng M, Valdes G, Dixit N, Solberg TD. Machine Learning in Radiation Oncology: Opportunities, Requirements, and Needs. Front Oncol. 2018;8:110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (1)] |
| 13. | Xiang L, Wang Q, Nie D, Zhang L, Jin X, Qiao Y, Shen D. Deep embedding convolutional neural network for synthesizing CT image from T1-Weighted MR image. Med Image Anal. 2018;47:31-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 14. | Adrian G, Konradsson E, Lempart M, Bäck S, Ceberg C, Petersson K. The FLASH effect depends on oxygen concentration. Br J Radiol. 2020;93:20190702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 108] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 15. | Maxim PG, Tantawi SG, Loo BW Jr. PHASER: A platform for clinical translation of FLASH cancer radiotherapy. Radiother Oncol. 2019;139:28-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 16. | Vozenin MC, De Fornel P, Petersson K, Favaudon V, Jaccard M, Germond JF, Petit B, Burki M, Ferrand G, Patin D, Bouchaab H, Ozsahin M, Bochud F, Bailat C, Devauchelle P, Bourhis J. The Advantage of FLASH Radiotherapy Confirmed in Mini-pig and Cat-cancer Patients. Clin Cancer Res. 2019;25:35-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 344] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 17. | Sheng K. Artificial intelligence in radiotherapy: a technological review. Front Med. 2020;14:431-449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Viergever MA, Maintz JBA, Klein S, Murphy K, Staring M, Pluim JPW. A survey of medical image registration - under review. Med Image Anal. 2016;33:140-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 113] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 19. | Wu G, Kim M, Wang Q, Munsell BC, Shen D. Scalable High-Performance Image Registration Framework by Unsupervised Deep Feature Representations Learning. IEEE Trans Biomed Eng. 2016;63:1505-1516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 20. | Roques TW. Patient selection and radiotherapy volume definition - can we improve the weakest links in the treatment chain? Clin Oncol (R Coll Radiol). 2014;26:353-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 21. | Weiss E, Hess CF. The impact of gross tumor volume (GTV) and clinical target volume (CTV) definition on the total accuracy in radiotherapy theoretical aspects and practical experiences. Strahlenther Onkol. 2003;179:21-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 129] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 22. | Sharp G, Fritscher KD, Pekar V, Peroni M, Shusharina N, Veeraraghavan H, Yang J. Vision 20/20: perspectives on automated image segmentation for radiotherapy. Med Phys. 2014;41:050902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 220] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 23. | Peressutti D, Schipaanboord B, van Soest J, Lustberg T, van Elmpt W, Kadir T, Dekker A, Gooding M. TU-AB-202-10: how effective are current atlas selection methods for atlas-based Auto-Contouring in radiotherapy planning? Medical Physics. 2016;43:3738-3739. [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Liang S, Tang F, Huang X, Yang K, Zhong T, Hu R, Liu S, Yuan X, Zhang Y. Deep-learning-based detection and segmentation of organs at risk in nasopharyngeal carcinoma computed tomographic images for radiotherapy planning. Eur Radiol. 2019;29:1961-1967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 25. | Zhu J, Zhang J, Qiu B, Liu Y, Liu X, Chen L. Comparison of the automatic segmentation of multiple organs at risk in CT images of lung cancer between deep convolutional neural network-based and atlas-based techniques. Acta Oncol. 2019;58:257-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Dolz J, Laprie A, Ken S, Leroy HA, Reyns N, Massoptier L, Vermandel M. Supervised machine learning-based classification scheme to segment the brainstem on MRI in multicenter brain tumor treatment context. Int J Comput Assist Radiol Surg. 2016;11:43-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Savenije MHF, Maspero M, Sikkes GG, van der Voort van Zyp JRN, T J Kotte AN, Bol GH, T van den Berg CA. Clinical implementation of MRI-based organs-at-risk auto-segmentation with convolutional networks for prostate radiotherapy. Radiat Oncol. 2020;15:104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 28. | Ibragimov B, Xing L. Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44:547-557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 308] [Cited by in F6Publishing: 306] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 29. | Chan JW, Kearney V, Haaf S, Wu S, Bogdanov M, Reddick M, Dixit N, Sudhyadhom A, Chen J, Yom SS, Solberg TD. A convolutional neural network algorithm for automatic segmentation of head and neck organs at risk using deep lifelong learning. Med Phys. 2019;46:2204-2213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 30. | van Rooij W, Dahele M, Ribeiro Brandao H, Delaney AR, Slotman BJ, Verbakel WF. Deep Learning-Based Delineation of Head and Neck Organs at Risk: Geometric and Dosimetric Evaluation. Int J Radiat Oncol Biol Phys. 2019;104:677-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 31. | Loap P, Tkatchenko N, Kirova Y. Evaluation of a delineation software for cardiac atlas-based autosegmentation: An example of the use of artificial intelligence in modern radiotherapy. Cancer Radiother. 2020;24:826-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Feng X, Qing K, Tustison NJ, Meyer CH, Chen Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3D images. Med Phys. 2019;46:2169-2180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 33. | Zhang T, Yang Y, Wang J, Men K, Wang X, Deng L, Bi N. Comparison between atlas and convolutional neural network based automatic segmentation of multiple organs at risk in non-small cell lung cancer. Medicine (Baltimore). 2020;99:e21800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Vu CC, Siddiqui ZA, Zamdborg L, Thompson AB, Quinn TJ, Castillo E, Guerrero TM. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology - use of non-domain transfer learning. J Appl Clin Med Phys. 2020;21:108-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Liu Z, Liu X, Xiao B, Wang S, Miao Z, Sun Y, Zhang F. Segmentation of organs-at-risk in cervical cancer CT images with a convolutional neural network. Phys Med. 2020;69:184-191. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 36. | Ahn SH, Yeo AU, Kim KH, Kim C, Goh Y, Cho S, Lee SB, Lim YK, Kim H, Shin D, Kim T, Kim TH, Youn SH, Oh ES, Jeong JH. Comparative clinical evaluation of atlas and deep-learning-based auto-segmentation of organ structures in liver cancer. Radiat Oncol. 2019;14:213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 37. | La Macchia M, Fellin F, Amichetti M, Cianchetti M, Gianolini S, Paola V, Lomax AJ, Widesott L. Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat Oncol. 2012;7:160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 38. | Chao KS, Bhide S, Chen H, Asper J, Bush S, Franklin G, Kavadi V, Liengswangwong V, Gordon W, Raben A, Strasser J, Koprowski C, Frank S, Chronowski G, Ahamad A, Malyapa R, Zhang L, Dong L. Reduce in variation and improve efficiency of target volume delineation by a computer-assisted system using a deformable image registration approach. Int J Radiat Oncol Biol Phys. 2007;68:1512-1521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 94] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 39. | Ikushima K, Arimura H, Jin Z, Yabu-Uchi H, Kuwazuru J, Shioyama Y, Sasaki T, Honda H, Sasaki M. Computer-assisted framework for machine-learning-based delineation of GTV regions on datasets of planning CT and PET/CT images. J Radiat Res. 2017;58:123-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 40. | Cui Y, Arimura H, Nakano R, Yoshitake T, Shioyama Y, Yabuuchi H. Automated approach for segmenting gross tumor volumes for lung cancer stereotactic body radiation therapy using CT-based dense V-networks. J Radiat Res. 2021;62:346-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 41. | Zhong Z, Kim Y, Plichta K, Allen BG, Zhou L, Buatti J, Wu X. Simultaneous cosegmentation of tumors in PET-CT images using deep fully convolutional networks. Med Phys. 2019;46:619-633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 42. | Kawata Y, Arimura H, Ikushima K, Jin Z, Morita K, Tokunaga C, Yabu-Uchi H, Shioyama Y, Sasaki T, Honda H, Sasaki M. Impact of pixel-based machine-learning techniques on automated frameworks for delineation of gross tumor volume regions for stereotactic body radiation therapy. Phys Med. 2017;42:141-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 43. | Li S, Xiao J, He L, Peng X, Yuan X. The Tumor Target Segmentation of Nasopharyngeal Cancer in CT Images Based on Deep Learning Methods. Technol Cancer Res Treat. 2019;18:1533033819884561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 44. | Ma Z, Zhou S, Wu X, Zhang H, Yan W, Sun S, Zhou J. Nasopharyngeal carcinoma segmentation based on enhanced convolutional neural networks using multi-modal metric learning. Phys Med Biol. 2019;64:025005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 45. | Zhao L, Lu Z, Jiang J, Zhou Y, Wu Y, Feng Q. Automatic Nasopharyngeal Carcinoma Segmentation Using Fully Convolutional Networks with Auxiliary Paths on Dual-Modality PET-CT Images. J Digit Imaging. 2019;32:462-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 46. | Guo Z, Guo N, Gong K, Zhong S, Li Q. Gross tumor volume segmentation for head and neck cancer radiotherapy using deep dense multi-modality network. Phys Med Biol. 2019;64:205015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 47. | Men K, Chen X, Zhang Y, Zhang T, Dai J, Yi J, Li Y. Deep Deconvolutional Neural Network for Target Segmentation of Nasopharyngeal Cancer in Planning Computed Tomography Images. Front Oncol. 2017;7:315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 121] [Article Influence: 17.3] [Reference Citation Analysis (1)] |
| 48. | Agn M, Munck Af Rosenschöld P, Puonti O, Lundemann MJ, Mancini L, Papadaki A, Thust S, Ashburner J, Law I, Van Leemput K. A modality-adaptive method for segmenting brain tumors and organs-at-risk in radiation therapy planning. Med Image Anal. 2019;54:220-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 49. | Estienne T, Lerousseau M, Vakalopoulou M, Alvarez Andres E, Battistella E, Carré A, Chandra S, Christodoulidis S, Sahasrabudhe M, Sun R, Robert C, Talbot H, Paragios N, Deutsch E. Deep Learning-Based Concurrent Brain Registration and Tumor Segmentation. Front Comput Neurosci. 2020;14:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 50. | Liu Y, Stojadinovic S, Hrycushko B, Wardak Z, Lau S, Lu W, Yan Y, Jiang SB, Zhen X, Timmerman R, Nedzi L, Gu X. A deep convolutional neural network-based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS One. 2017;12:e0185844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 51. | Jeong J, Lei Y, Kahn S, Liu T, Curran WJ, Shu HK, Mao H, Yang X. Brain tumor segmentation using 3D Mask R-CNN for dynamic susceptibility contrast enhanced perfusion imaging. Phys Med Biol. 2020;65:185009. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 52. | Charron O, Lallement A, Jarnet D, Noblet V, Clavier JB, Meyer P. Automatic detection and segmentation of brain metastases on multimodal MR images with a deep convolutional neural network. Comput Biol Med. 2018;95:43-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 139] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 53. | Tang F, Liang S, Zhong T, Huang X, Deng X, Zhang Y, Zhou L. Postoperative glioma segmentation in CT image using deep feature fusion model guided by multi-sequence MRIs. Eur Radiol. 2020;30:823-832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 54. | Meng L, Tian Y, Bu S. Liver tumor segmentation based on 3D convolutional neural network with dual scale. J Appl Clin Med Phys. 2020;21:144-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 55. | Eppenhof KAJ, Maspero M, Savenije MHF, de Boer JCJ, van der Voort van Zyp JRN, Raaymakers BW, Raaijmakers AJE, Veta M, van den Berg CAT, Pluim JPW. Fast contour propagation for MR-guided prostate radiotherapy using convolutional neural networks. Med Phys. 2020;47:1238-1248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 56. | Elguindi S, Zelefsky MJ, Jiang J, Veeraraghavan H, Deasy JO, Hunt MA, Tyagi N. Deep learning-based auto-segmentation of targets and organs-at-risk for magnetic resonance imaging only planning of prostate radiotherapy. Phys Imaging Radiat Oncol. 2019;12:80-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 57. | Torheim T, Malinen E, Hole KH, Lund KV, Indahl UG, Lyng H, Kvaal K, Futsaether CM. Autodelineation of cervical cancers using multiparametric magnetic resonance imaging and machine learning. Acta Oncol. 2017;56:806-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 58. | Karimi D, Zeng Q, Mathur P, Avinash A, Mahdavi S, Spadinger I, Abolmaesumi P, Salcudean SE. Accurate and robust deep learning-based segmentation of the prostate clinical target volume in ultrasound images. Med Image Anal. 2019;57:186-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 59. | Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44:6377-6389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 187] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 60. | Men K, Zhang T, Chen X, Chen B, Tang Y, Wang S, Li Y, Dai J. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med. 2018;50:13-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 86] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 61. | Zhu J, Liu X, Chen L. A preliminary study of a photon dose calculation algorithm using a convolutional neural network. Phys Med Biol. 2020;65:20NT02. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 62. | Zhang J, Liu S, Yan H, Li T, Mao R, Liu J. Predicting voxel-level dose distributions for esophageal radiotherapy using densely connected network with dilated convolutions. Phys Med Biol. 2020;65:205013. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 63. | Liu Z, Fan J, Li M, Yan H, Hu Z, Huang P, Tian Y, Miao J, Dai J. A deep learning method for prediction of three-dimensional dose distribution of helical tomotherapy. Med Phys. 2019;46:1972-1983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 59] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 64. | Fan J, Wang J, Chen Z, Hu C, Zhang Z, Hu W. Automatic treatment planning based on three-dimensional dose distribution predicted from deep learning technique. Med Phys. 2019;46:370-381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 188] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 65. | Ma M, Kovalchuk N, Buyyounouski MK, Xing L, Yang Y. Dosimetric features-driven machine learning model for DVH prediction in VMAT treatment planning. Med Phys. 2019;46:857-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 66. | Valdes G, Simone CB 2nd, Chen J, Lin A, Yom SS, Pattison AJ, Carpenter CM, Solberg TD. Clinical decision support of radiotherapy treatment planning: A data-driven machine learning strategy for patient-specific dosimetric decision making. Radiother Oncol. 2017;125:392-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 67. | Mahdavi SR, Tavakol A, Sanei M, Molana SH, Arbabi F, Rostami A, Barimani S. Use of artificial neural network for pretreatment verification of intensity modulation radiation therapy fields. Br J Radiol. 2019;92:20190355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 68. | Yan H, Dai JR. Intelligence-guided beam angle optimization in treatment planning of intensity-modulated radiation therapy. Phys Med. 2016;32:1292-1301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 69. | Amit G, Purdie TG, Levinshtein A, Hope AJ, Lindsay P, Marshall A, Jaffray DA, Pekar V. Automatic learning-based beam angle selection for thoracic IMRT. Med Phys. 2015;42:1992-2005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 70. | Sakata Y, Hirai R, Kobuna K, Tanizawa A, Mori S. A machine learning-based real-time tumor tracking system for fluoroscopic gating of lung radiotherapy. Phys Med Biol. 2020;65:085014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 71. | Guidi G, Maffei N, Meduri B, D'Angelo E, Mistretta GM, Ceroni P, Ciarmatori A, Bernabei A, Maggi S, Cardinali M, Morabito VE, Rosica F, Malara S, Savini A, Orlandi G, D'Ugo C, Bunkheila F, Bono M, Lappi S, Blasi C, Lohr F, Costi T. A machine learning tool for re-planning and adaptive RT: A multicenter cohort investigation. Phys Med. 2016;32:1659-1666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 72. | Valdes G, Morin O, Valenciaga Y, Kirby N, Pouliot J, Chuang C. Use of TrueBeam developer mode for imaging QA. J Appl Clin Med Phys. 2015;16:322-333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 73. | Li Q, Chan MF. Predictive time-series modeling using artificial neural networks for Linac beam symmetry: an empirical study. Ann N Y Acad Sci. 2017;1387:84-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 74. | Valdes G, Scheuermann R, Hung CY, Olszanski A, Bellerive M, Solberg TD. A mathematical framework for virtual IMRT QA using machine learning. Med Phys. 2016;43:4323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 121] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 75. | Valdes G, Chan MF, Lim SB, Scheuermann R, Deasy JO, Solberg TD. IMRT QA using machine learning: A multi-institutional validation. J Appl Clin Med Phys. 2017;18:279-284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 76. | Carlson JN, Park JM, Park SY, Park JI, Choi Y, Ye SJ. A machine learning approach to the accurate prediction of multi-leaf collimator positional errors. Phys Med Biol. 2016;61:2514-2531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 77. | Granville DA, Sutherland JG, Belec JG, La Russa DJ. Predicting VMAT patient-specific QA results using a support vector classifier trained on treatment plan characteristics and linac QC metrics. Phys Med Biol. 2019;64:095017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 78. | Boon IS, Au Yong TPT, Boon CS. Assessing the Role of Artificial Intelligence (AI) in Clinical Oncology: Utility of Machine Learning in Radiotherapy Target Volume Delineation. Medicines (Basel). 2018;5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 79. | Guidi G, Maffei N, Vecchi C, Gottardi G, Ciarmatori A, Mistretta GM, Mazzeo E, Giacobazzi P, Lohr F, Costi T. Expert system classifier for adaptive radiation therapy in prostate cancer. Australas Phys Eng Sci Med. 2017;40:337-348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 80. | Tseng HH, Luo Y, Cui S, Chien JT, Ten Haken RK, Naqa IE. Deep reinforcement learning for automated radiation adaptation in lung cancer. Med Phys. 2017;44:6690-6705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 81. | Varfalvy N, Piron O, Cyr MF, Dagnault A, Archambault L. Classification of changes occurring in lung patient during radiotherapy using relative γ analysis and hidden Markov models. Med Phys. 2017;44:5043-5050. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 82. | Oakden-Rayner L, Carneiro G, Bessen T, Nascimento JC, Bradley AP, Palmer LJ. Precision Radiology: Predicting longevity using feature engineering and deep learning methods in a radiomics framework. Sci Rep. 2017;7:1648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 83. | Lao J, Chen Y, Li ZC, Li Q, Zhang J, Liu J, Zhai G. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme. Sci Rep. 2017;7:10353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 318] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 84. | Li Z, Wang Y, Yu J, Guo Y, Cao W. Deep Learning based Radiomics (DLR) and its usage in noninvasive IDH1 prediction for low grade glioma. Sci Rep. 2017;7:5467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 173] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 85. | Cha KH, Hadjiiski L, Chan HP, Weizer AZ, Alva A, Cohan RH, Caoili EM, Paramagul C, Samala RK. Bladder Cancer Treatment Response Assessment in CT using Radiomics with Deep-Learning. Sci Rep. 2017;7:8738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 86. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13300] [Cited by in F6Publishing: 14452] [Article Influence: 2890.4] [Reference Citation Analysis (2)] |
| 87. | DeSantis CE, Miller KD, Dale W, Mohile SG, Cohen HJ, Leach CR, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for adults aged 85 years and older, 2019. CA Cancer J Clin. 2019;69:452-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 182] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 88. | Akcay M, Etiz D, Celik O, Ozen A. Evaluation of Prognosis in Nasopharyngeal Cancer Using Machine Learning. Technol Cancer Res Treat. 2020;19:1533033820909829. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 89. | Sepheri S, Upadhaya T, Desseroit MC, Visvikis D, Le Rest CC, Hatt M. Comparison of machine learning algorithms for building prognostic models in non-small cell lung cancer using clinical and radiomics features from 18F-FDG PET/CT images. J Nucl Med. 2018;59:328. [Cited in This Article: ] |
| 90. | Zhang S, Xu Y, Hui X, Yang F, Hu Y, Shao J, Liang H, Wang Y. Improvement in prediction of prostate cancer prognosis with somatic mutational signatures. J Cancer. 2017;8:3261-3267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 91. | Hasnain Z, Mason J, Gill K, Miranda G, Gill IS, Kuhn P, Newton PK. Machine learning models for predicting post-cystectomy recurrence and survival in bladder cancer patients. PLoS One. 2019;14:e0210976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 92. | Akcay M, Etiz D, Celik O. Prediction of Survival and Recurrence Patterns by Machine Learning in Gastric Cancer Cases Undergoing Radiation Therapy and Chemotherapy. Adv Radiat Oncol. 2020;5:1179-1187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 93. | Pham TD, Fan C, Zhang H, Sun XF. Artificial intelligence-based 5-year survival prediction and prognosis of DNp73 expression in rectal cancer patients. Clin Transl Med. 2020;10:e159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |