Published online Feb 27, 2016. doi: 10.5496/wjmg.v6.i1.1
Peer-review started: October 21, 2015
First decision: November 27, 2015
Revised: January 14, 2016
Accepted: February 16, 2016
Article in press: February 18, 2016
Published online: February 27, 2016
Atrial fibrillation (AF) is the most frequent arrhythmogenic syndrome in humans. With an estimate incidence of 1%-2% in the general population, AF raises up to almost 10%-12% in 80+ years. Thus, AF represents nowadays a highly prevalent medical problem generating a large economic burden. At the electrophysiological level, distinct mechanisms have been elucidated. Yet, despite its prevalence, the genetic and molecular culprits of this pandemic cardiac electrophysiological abnormality have remained largely obscure. Molecular genetics of AF familiar cases have demonstrated that single nucleotide mutations in distinct genes encoding for ion channels underlie the onset of AF, albeit such alterations only explain a minor subset of patients with AF. In recent years, analyses by means of genome-wide association studies have unraveled a more complex picture of the etiology of AF, pointing out to distinct cardiac-enriched transcription factors, as well as to other regulatory genes. Furthermore a new layer of regulatory mechanisms have emerged, i.e., post-transcriptional regulation mediated by non-coding RNA, which have been demonstrated to exert pivotal roles in cardiac electrophysiology. In this manuscript, we aim to provide a comprehensive review of the genetic regulatory networks that if impaired exert electrophysiological abnormalities that contribute to the onset, and subsequently, on self-perpetuation of AF.
Core tip: Atrial fibrillation (AF) is the most prevalent arrhythmogenic defect in the human population. Genetic factors such as mutations in distinct ion channel encoding genes have been described, yet representing less than 10% of all AF cases. Genome wide association studies have widened the genetic culprits contributing to AF. We provide herein a state-of-the art review on the genetic components underlying AF. Experimental evidences demonstrated that PITX2 plays a pivotal role regulating cellular, molecular and electrophysiological characteristics of the developing and adult heart that, if impaired, predispose to AF, leading to complex regulatory networks with transcriptional and post-transcriptional (microRNA) regulatory mechanisms.
- Citation: Franco D, Lozano-Velasco E, Aranega A. Gene regulatory networks in atrial fibrillation. World J Med Genet 2016; 6(1): 1-16
- URL: https://www.wjgnet.com/2220-3184/full/v6/i1/1.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v6.i1.1
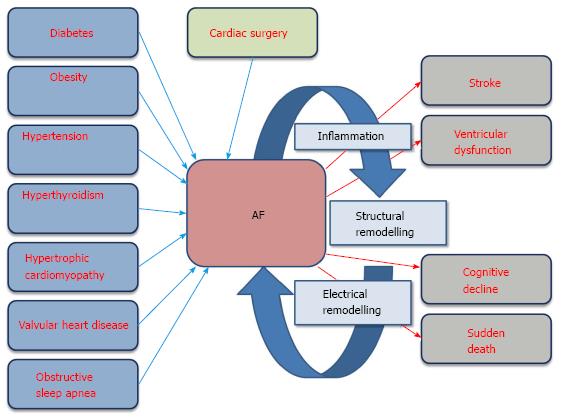
Atrial fibrillation (AF) is the most frequent arrhythmogenic syndrome in humans. With an estimate incidence of 1%-2% in the general population, AF raises up to almost 10%-12% in 80+ years[1-5]. Cardiovascular risk factors such diabetes, obesity, hypertension and hyperthyroidism promote AF[6-9]. In addition, the occurrence of AF can be also triggered by preceding cardiovascular diseases such as hypertrophic cardiomyopathy and valvular heart diseases[10,11] as well as it can be boosted by cardiac surgery, obstructive sleep apnea and inflammatory processes[12-15]. Importantly, besides the global risk factors for AF, it is well-established that the debut of an AF episode triggers subsequent and more severe AF episodes, leading to electrical and structural remodeling of the diseased heart, a condition quoted as “AF begets AF”[16,17]. Thus AF per se is a progressive disease. Electrical remodeling involves progressive changes in the cardiac electrical properties, possibly triggered by oxidative stress imbalance, leading to EADs, DADs and/or changes in the ADP configuration[18,19], culminating thus in rotor formation[20] (Figure 1). Structural remodeling involves atrial dilation, fibrosis and/or inflammation[21,22] which then indirectly promotes the generation of rotors and thus electrical re-entry circuitries[20] (Figure 1). Moreover, suffering from AF predisposes to additional health problems, ranging from bleeding and stroke[23-25], cognitive decline and dementia[26,27], ventricular dysfunction[28,29] and even sudden death[30] (Figure 1). Thus, these data support the notion that AF is an epidemic disease with large socio-economic burden worldwide.
While large epidemiological data support the previously mentioned predictive risk factors for AF, there are also unquestionable evidences that in a subset of AF patients no concurrent previous risk factors are diagnosed and thus AF occurs in an isolated manner, a condition dubbed lone AF[31-34]. These observations, in addition to seminal observations supporting that AF can also be familial[35], strengthen the notion of a genetic component on the onset of AF. In this context, two waves of genetic information have been gained over the last decades. Seminal studies on familiar AF cases followed by genetic linkage analyses, and subsequently by genetic screening of candidate genes identified a large number of point mutations in distinct genes encoding for proteins involved in cardiac electrophysiology[36-38]. These data, along with meticulous electrophysiological mapping analyses and in silico data modeling, provided an important cornerstone to understand AF pathophysiology[39-43]. However, genetic identification of candidate AF genes can only explained around 10%-15% of all AF patients. Thus, new approaches to untangle the genetic bases of AF were envisioned. Genome-wide association analyses (GWAS) lightened the discovery of new genes associated to AF. Seminal worked by Gubdjartsson et al[44] firstly identify common risk variants highly associated to the onset of lone AF in distinct large European and Chinese patient cohorts. Subsequent GWAS studies and meta-GWAS have further identified new candidate genes for AF pathophysiology[45-49], yet the functional link between these variants and the AF pathophysiology is a rather unexplored. A tortuous route to dissect their functional roles is envisioned, given that most of these risk variants are located rather apart from gene coding sequences. Thus, a first proposal based on these findings is that risk variants might affect gene regulatory networks within the vicinity and thus underscore AF onset. In addition, a novel layer of complexity in gene regulatory networks has emerged with the discovery of non-coding RNAs that can mediate post-transcriptional regulation. In this review, we aim to elaborate on the current state-of-the-art of the gene regulatory networks involved in AF pathophysiology.
The cardiac action potential is driven by inward and outward flow of distinct ion currents along the cardiomyocyte membrane. In atrial and ventricular cardiomyocytes, the configuration of the action potential is initiated by the upstroke depolarization of the sodium current, followed by a repolarization mediated by distinct potassium currents until the resting membrane potential is restored. A conduction-contraction coupling is modulated within each action potential by a complex regulatory network of calcium handling[50]. Interestingly, nodal conductive cells display a distinct upstroke configuration which is modulated by cation selective currents, yet repolarization is rather similar to working cardiomyocytes. A large number of mutations in the SCN5A gene, coding for the pore-forming subunits of the INa current, have been associated to AF, alone[51-56] or in combination to other concurrent cardiac pathologies[57-62]. Similarly, mutations in SCN1B, SCN2B, SCN3B and SCN4B ancillary subunits, respectively, have also been linked to AF[63-67]. At present, the functional interpretation of these findings suggest that impaired INa current can promote AF onset, yet electrophysiological evidences are only available for a subset of these mutations. Importantly, mutations in SCN5A are also linked to other cardiac arrhythmias such as long QT and Brugada[68-70] demonstrating a pivotal role for SNC5A on cardiac electrophysiology and illustrating that distinct phenotype might be acquired depending on subtles differences on the mutation location as well as on plausible modulatory accompanying proteins. In addition, mutations in the HCN4 cation channel, responsible for the cardiac action potential upstroke of conductive cells, have also been reported in AF[71].
Mutations in genes encoding for proteins controlling the repolarization phase of the cardiac action potential have also been associated to AF. In particular, KCNQ1[36,72-79], KCNA5[80] and KCNE2[38,81] mutations have been identified. In this setting, point mutations seem to shorten the repolarization phase providing an electrophysiological substrate for AF onset. Similarly, alteration of the resting membrane potential have been related to trigger AF onset and a large number of mutations in the gene coding for KCNJ2 have been reported[37,38,82,83]. Similarly as for the sodium channels, mutation in potassium channels have been associated to AF and concomitantly with other arrhythmogenic syndromes such as long[72] and short[84] QT, respectively.
Multiple lines of evidences have demonstrated the essential role of impaired calcium homeostasis as a triggering factor of AF[2,85-90]. In this context, mutations in RYR2 are associated to AF[91] and in conjunction with other cardiac pathophysiological conditions such as catecholaminergic polymorphic ventricular tachycardia[92,93]. Curiously, RYR2 mutations can also affect sodium channel expression[94]. Importantly, murine model of RYR2 mutation leads to AF, reinforcing the genetic evidences of impaired calcium homeostasis as a trigger of AF[95].
In addition, mutations in gap junction proteins such as GJA1 (connexin43)[96,97] and GJA5 (connexin40)[98-104], which are critical for the coordinated transmission of the electrical impulse among cardiomyocytes, have also been identified in association to AF. Seminal work described that mutation in connexin proteins were occurring in somatic cells (i.e., cardiomyocytes)[98], whereas more recently germline mutations have also been identified[98-104] and the concept of somatic cell mutation challenged[105]. In this setting, electrophysiological analyses of connexin mutations in the AF context demonstrated that cardiomyocyte electrical transmission is impaired[106-111] and thus can result in cardiac re-entry circuitries and then on AF.
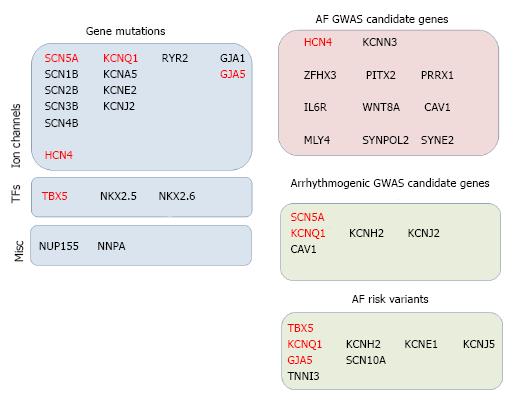
Furthermore, mutations in several other genes, besides those directly contributing to the electrophysiological properties of the heart, have also been identified in the AF setting. In particular, mutations in the transcription factors TBX5[112], NKX2.5[113-117] and NKX2.6[118] as well as in the nuclear pore component NUP155[119] and the atrial natriuretic factor (NNPA[120]). A summary of gene mutations associated to AF is provided on Figure 2. It is important to highlight in this context that mutation in NUP155 was found in patients with AF and early sudden death[118], while TBX5 and NKX2.5 are developmental transcription factors that play determinant roles during cardiogenesis and mutations in these genes have also been reported in distinct congenital heart diseases as well as GWAS candidate genes in other cardiac electrophysiological defects[121-128].
As previously said, classical genetic approaches provided an entry site to discover discrete genes involved in AF pathophysiology, but felt short to explain most of the diagnosed AF cases. Genome-wide association studies (GWAS) introduced a revolutionary genetic approach to understand AF, and cardiac arrhythmias at large[129,130]. Seminal work by Gudbjartsson et al[44] identified risk variant at 4q25 highly associated to lone AF. Soon thereafter these observations were corroborated in distinct studies worldwide[131-136], yet with some controversial findings[137,138]. Surprisingly, 4q25 risk variants are located in a gene desert, being the closest annotated gene a homeobox transcription factor, PITX2, around 150 kb downstream. Soon thereafter, experimental observations provided evidences that impaired Pitx2 function in animal models triggered increase atrial arrhythmias susceptibility[139,140]. However, how risk variants can influence PITX2 expression and/or function have only been partially revealed[141], since regulatory elements containing the risk variants can molecularly interact with PITX2 but also ENPEP promoter elements in a tissue- and developmental-specific manner[141]. Importantly, controversial findings in humans have been reported. Chinchilla et al[136] firstly described decreased PITX2 expression in AF patients, yet these claims were challenged since no correlation between risk variants and PITX2C expression was observed[142]. However, more recently a correlation with PITX2A expression has been reported, limiting their functional significance since data were obtained from right atrial biopsies[143]. Thus, further investigations are required to clarify these controversial findings.
Besides PITX2, additional GWAS have enlightened the putative role of other novel genes in AF pathophysiology, such as KCNN3[47], ZFHX3[45,48], IL6R[49] and MLY4[144]. In most cases, these findings have been corroborated in independent studies with distinct ethnicity cohorts as for ZFHX3[145,146], and IL6R[147], but to date no additional reports are available for KCNN3 and MYL4.
While arguments for the plausible involvement in AF have been provided in all cases, experimental evidences remain largely missing. KCNN3 loss-of-function mutant mice display no overt cardiac electrophysiological defects, yet overexpression leads to sudden death[148]. Similarly ZFHX3 function role in the heart is scare, yet recent findings suggests that atrial arrhythmias[149] and pacing[150] leads to significant impaired ZFHX3 expression. Importantly, risk variants associated to KCNN3, ZFHX3 and IL6R lie within intronic regions, respectively, and thus have no predictive functional consequences. Therefore, as in the cases of 4q25 risk variants (PITX2), a functional role on plausible regulatory elements is hypothesized, but functional evidences are yet missing.
In addition to these GWAS studies, at least two meta-GWAS studies have digged for additional risk variants associated to AF[46,151]. In this context, six new loci have been uncovered using such approach; CAV1, HCN4, SYNE2, SYNPOL2, PRRX1 and WTN8A. Replication analyses of the risk variants have been confirmed for SYNE2[152] and PRRX1[149], are controversial for CAV1[53,153,154] or have not been reported to date for HCN4 and SYNPOL2. Importantly, a 12-SNPs risk score have been developed that can individually identify the risk of AF and stroke[155].
While this approach broadens the plausible genetic bases of AF, it is also true that their contribution to explain AF pathophysiology would be directed linked to the statistical significance associated with those findings. It is important to say in this context that risk variants associated to PITX2 display highest significance values, KCNN3, ZFHX3 and IL6R are moderately significant and those revealed by meta-GWAS display the lowest, yet statistical significant, values. In a simplistic way, this could be transferred as PITX2 being contributing to larger spectrum of AF pathophysiology and thus being in the upper level of a hierarchical signaling pathway while CAV1, HCN4, SYNE2, SYNPOL2, PRRX1 and WTN8A will be rather discretely contributing to AF pathophysiology in a rather discrete, small subset of AF patients, and therefore being at the bottom of such hierarchy. We have recently provided experimental evidences supporting this notion, as detailed below.
GWAS studies have also been reported in other electrophysiological parameters such as PR interval[129,156-158], heart rate[157], QRS interval[157] as well as physiopathological conditions such as Brugada syndrome[159] and long QT[160-162]. Several of these cardiac arrhythmogenic conditions are sometimes directly associated to AF, such as for example impaired PR interval or Brugada syndrome. Importantly, such GWAS studies have identified multiple genes, some of which are shared with AF, such as CAV1 (PR interval[129,156-158]) while other have been previously involved in AF pahthophysiology, such as KCNQ1, KCNH2, SCN5A, KCNJ2 (long QT GWAS analyses[160-162]). In addition to GWAS studies, risk variants have been associated to AF using more discrete/small AF patient cohorts. Among them it is important to highlight risk variants at the following loci; KCNQ1[163-165], KCNH2[163], KCNE1[163], KCNJ5[166], GJA5[167] and TBX5[168]. In addition to these GWAS studies, several independent AF cohort studies have reported risk variants associated to AF in other loci, such as TNNI3[169] and SCN10A[170,171], yet the functional relevance of these findings remains unsolved. These data suggest that complex and intricate gene networks are likely to be operative between these regulatory genes resulting in AF (Figure 3) as well as other electrophysiological associated disorders.
Gene regulatory networks have been classically associated as hierarchical interactions between master genes, such as transcription factors, and structural genes, such as actin and myosin genes forming the sarcomere or genes encoding ion channels configuring therefore the cardiac action potential. In this setting, transcriptional regulation has been considered the master regulatory point, deciding whether to go or not to go, whereas translation and post-translational modifications are considered as minor regulatory nodes. Over the last decade, we have witnessed the discovery that large part of the so called “rubbish DNA” in fact codes for a large variety of non-coding RNAs species with highly relevant functional properties. In broad sense, non-coding RNAs can be sub-classified into long non-coding (lncRNAs) and small non-coding RNAs, exerting a distinct and variable functional roles[172]. Among small non-coding RNAs, microRNAs have emerged as the larger group, playing a pivotal role on post-transcriptional regulation of mRNA transcript expression[173]. MicroRNAs are 22-24 nt non-coding RNAs which can block translation or trigger transcript degradation by binding to complementary seed sequence in the 3’untranslated region (3’UTR) of nascent mRNA transcripts[173]. At present the hallmark of microRNA transcriptomics has been uncovered in the developing and adult heart in both normal and pathological conditions[174,175]. In the context of AF the microRNA fingerprint has been profiled in different settings[176-178], providing highly valuable biomarkers[179-183] as well as opening new avenues to dissect the functional role of these differentially expressed microRNAs in AF pathology. In addition distinct sets of microRNAs have also been reported to selectively modulate expression of distinct gene encoding for ion channels, opening thus new pathways to dissect their genetic contribution to AF.
As stated above, evidence on the functional role of mutiple genes have been provided by the identification and experimental testing of AF patient gene mutations. In this setting, a causative link can, in most cases, be established. However, GWAS analyses provide circumstantial evidences whereas functional linkage between the risk variants and coding genes, whether protective or deleterious, is not directly established. To date, an example of successful linkage have been reported[184] in the cardiovascular context, however in most cases, this is not yet established. Therefore, dissecting the functional role of GWAS candidate genes represents the first step for the analyses of their causative link.
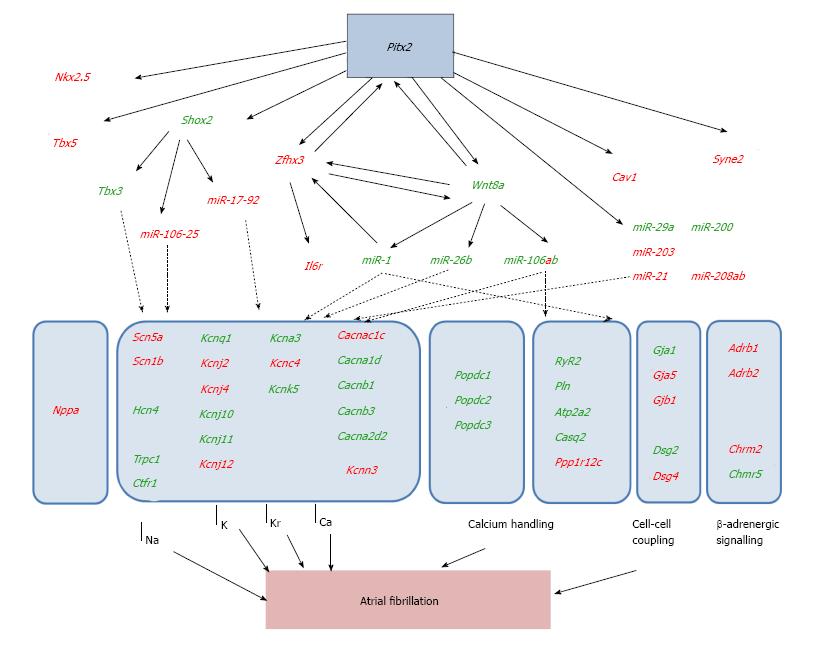
Large set of evidences suggest that multiple genetic pathways can contribute to the onset of AF. Over the last years we have carefully dissected the role of the homeobox transcription factor PITX2 in this context and we have made efforts to dissect the regulatory interaction of PITX2 with those genes previously reported to play a role in AF pathophysiology. Several studies have demonstrated that PITX2 can influence expression of distinct ion channels in the developing and adult hearts. Wang et al[138] firstly demonstrate that PITX2 haploinsufficiency is sufficient to increase AF susceptibility. These authors demonstrated that PITX2 haploinsufficiency led to developmental impaired expression of key transcription factors (SHOX2, TBX3) directing sinoatrial morphogenesis and thus impacting on the normal onset of the cardiac pacemaker structures. However, in this context, no basal AF occurs and no ECG defects were observed. In this line of thinking soon thereafter Kirchoff et al[139] demonstrate similar findings and furthermore reported that expression of multiple ion channels were impaired as well as cell-cell interacting proteins (Figure 3). However, none of these genes were reported to play a pivotal role on cardiac action potential and thus, molecular and electrophysiological links were still on its infancy. Using a distinct experimental approach, i.e., an atrial-specific PITX2 conditional transgenic mouse line, Chinchilla et al[135] revealed that ECG defects were observed at rest (no triggering needed), electrophysiological measurements were impaired, i.e., increased resting membrane potential and prolonged action potential duration, and furthermore using a candidate approach analyses, distinct ion channels were also impaired, particularly SCN5A (INa), KCNJ2 (IK1) and KCNJ12 (IK1). Thus, these data demonstrate for the first time that functional impairment of PITX2 leads to molecular defects which provoke electrophysiological alterations and thus increase rate of atrial arrhythmias. In line with these findings, Tao et al[185] using a PITX2 deletion conditional approach in the adult heart also demonstrated that basal ECG defects were observed, similar to those already reported by Chinchilla et al[135], and moreover they demonstrated that key determinants of the cardiac action potential were impaired, such as KCNQ1 (IKs), KCNH2 (IKr), CACNA1C (ICa), among others. Overall these data demonstrated that Pitx2 insufficiency can both promote and trigger molecular and electrophysiological alterations, namely affecting the depolarization, repolarization and resting membrane potential characteristics of the cardiac action potential. They also illustrate that alteration of a single transcription factor can influence the functional role of multiple ion channels previously reported to be associated to AF. Therefore it can be inferred that PITX2 is upstream of this hierarchical signaling pathway and its impairment would explain a large fraction of AF patients.
In addition to PITX2, other GWAS studies have provided evidence on the plausible role of distinct gene in AF pathophysiology, yet as previously said, their functional implication has been poorly documented. We investigated whether these genes would be under control of PITX2. Using a series of gain and loss-of-function approaches, Lozano-Velasco et al[149] demonstrated that PITX2 can modulate expression of ZFHX3, KCNN3 and IL6R. The role of ZFHX3 in atrial arrhythmias is basically unexplored. However, given its transcriptional capacity if might be plausible that it can modulate multiple genes previously related to AF. The role of KCNN3 in atrial arrhythmias is under debate, its plausible contribution to the configuration of the cardiac action potential, if any, seems to be residual. On the other hand, the involvement of IL6R could be related to inflammatory processes that, if impaired, are associated to AF as previously mentioned. We have noticed that IL6R expression in the atrial chambers is impaired in PITX2 deficient mice yet plasma levels of soluble IL6 and TFNα ligands are not altered. While these data support a tissue-specific involvement of IL6R, its links to AF remain to be elucidated.
We also investigated if PITX2 can influence the role of those genes related to AF by meta-GWAS analyses. Among them, previous evidences support that PITX2 can modulate expression of HCN4[138]. More recently, Lozano-Velasco et al[151] demonstrated that also CAV1, SYNE2 and WNT8A are modulated by PITX2. It is important nonetheless to highlight that risk variants associated to CAV1 and SYNE2 have also been reported in other cardiac electrophysiological disorders yet their functional role remains poorly characterized. Importantly, WNT signaling has been largely documented to play pivotal role during cardiac development and homeostasis[186,187], however WNT8 has not been reported to play fundamental roles therein[188].
Given the potential signaling properties of WNT8A and those transcriptional capacity exerted by ZFHX3 we explored in any of those pathways might be involved on the regulation of distinct ion channels previously reported to be modulated by PITX2 and related to AF. Gain- and loss-of-function approaches demonstrated that WNT8A but not ZFHX3 can modulate expression of calcium handling proteins[151]. Importantly, impaired expression of WNT8A is observed in PITX2 insufficiency models with basal ECG alterations but not in those with normal ECG recordings[151], supporting the notion that WNT signaling plays a fundamental role conferring susceptibility vs triggering capacity to the atrial chambers during AF onset.
In addition to gene regulatory mechanisms driven by PITX2, several lines of evidence support that PITX2-regulated microRNAs also contribute to AF pathophysiology. Chinchilla et al[135] already demonstrated that impaired miR-1 expression in PITX2 deficient mice underlie abnormal resting membrane potential configuration (by modulating KCNJ2 and KCNJ12 post-transcriptional regulation). Huang et al[153] has confirmed that PITX2 inhibits miR-1 expression, which negatively regulates ZFHX3. Wang et al[189] demonstrated that two distinct genomic microRNA clusters, namely miR-17-92 and miR-106b-25, as well as miR-335 and miR-423, are under control of PITX2. Genetic deletion of miR-17-92 and miR-106b-25 microRNA clusters, respectively display normal baseline electrophysiological parameters but were susceptible to pacing-induced AF. Cardiac-specific deletion (Nkx2.5Cre) of miR-17-92 cluster resulted in prolonged PR interval. Compound double mutant mice using conditional Nkx2.5Cre mediated deletion resulted in sinus node dysfunction, a condition reminiscent of sick sinus syndrome in humans. Molecular analyses revealed that SHOX2 and TBX3 expression were impaired. Furthermore, RYR2-mediated calcium-leak was reported in miR-106b-25 deficient mice[190] since several members of this cluster, namely miR-106b, miR-93 and miR-25 directly target the 3´UTR of Ryr2. More recently, Lozano-Velasco et al[149] demonstrate that a large number of microRNAs, previously reported to be associated to AF in patients, are also modulated by PITX2. In particular, miR-1, miR-26b, miR-29a, miR-30e, miR-106b, miR-133 and miR-200 were up-regulated in absence of PITX2 whereas miR-21, miR-106a, miR-203, miR-208a and miR-208b were down-regulated. Several of these microRNAs, such as miR-1, miR-133, miR-21, miR-106b and miR-26 have been previously reported to regulate calcium (CACNA1C[191]; RYR2[190]), sodium (SCN5A[192]), potassium (KCNJ2[193], KCNE1[194], KCNB2[194]), cation (HCN4[195]) channel subunits, respectively. Overall, these data demonstrate a highly complex gene regulatory networks leading to AF as summarized in Figure 3.
In this review we have highlighted the complexity of risk factors influencing the onset of AF, both clinically and genetically. Several lines of evidences demonstrated that PITX2 exerts a pivotal role on the genetic determinants of AF[136,139,140,149,185]. While it has been robustly demonstrated that PITX2 insufficiency predisposes to atrial arrhythmia genesis in experimental mouse models[136,139,140,149,185], discrepancies remain as whether PITX2 is impaired in AF risk variants human patient carriers[136,142,143]. Importantly some discrepancies also exist as whether predisposes or triggers AF[136,139]. Our recent findings suggest the notion that PITX2 insufficiency in the embryo predisposes whereas PITX2 insufficiency in the adult atrial chamber triggers AF. In this context, we have provided evidences that such switching mechanisms seems to be regulated by WNT signaling and the downstream activation/repression of key microRNAs[149,165], yet future experiments are needed to provide additional evidences on this front. In the last decade, our understanding of the downstream pathways controlled by PITX2 has advanced at a quick pace. Several studies have demonstrated that PITX2 controls signaling pathways regulation sinus node formation[189,196], including therein the pivotal role of a microRNA cluster[189,190]. These data shed light thus on the concurrence of AF with other cardiac physiopathological conditions such as sick sinus syndrome[190]. Similarly we and others have demonstrated the key role exerted by PITX2 regulating microRNAs impacting on calcium handling and thus on profibrillatory risk factors[136,153,189]. It remains nonetheless to be established if PITX2 also leads to impaired regulation of inflammatory processes and/or redox signaling which might impact on structural remodeling of the atrial chambers. To date, no evidences of atrial fibrosis have been reported in any of the PITX2 deficient mouse models[136,139,140,149,185], while some incipient evidences have been reported on the inflammatory link[149] but not on redox signaling. Thus, future lines of research should be envisioned to clarify this point.
In addition, whereas our understanding of the PITX2 downstream signaling pathways in the context of AF have progressively increased, scarce insights are currently available on the impact of AF clinically related risk factors on PITX2. A link between 4q25 risk variant carriers and increase left atrial volume has been recently reported[197] in AF patients, however it remains to be established this is modulated by PITX2, as suggested by previous evidence in PITX2 deficient mice in which atrial volume is already increased at fetal developmental stages[136]. A recent study reported that aging and hypertension, two well-established risk factors for AF, severely decreased PITX2 expression in a rat experimental model when both risk factors were combined[198]. We recently reported that Pitx2 is severely impaired in dilated cardiomyopathy patients as well as in an experimental heart failure pig model, shedding light into a possible connection between ventricular disfunction and AF[199,200]. Yet, it remains to be established if other clinically relevant AF risk factors, such as diabetes, obesity, hyperthyroidism, valvular heart disease and/or obstructive sleep apnea are also impairing PITX2 function and thus predisposing to AF. Overall these data demonstrate the pivotal role of PITX2 regulating multiple aspect that if impaired are pro-arrhythogenic and they also open new pathways to explore therapeutical approaches that could eventually lead to minime the burden of AF in the human population.
P- Reviewer: Kettering K, Liu T, Said SAM S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Hakim FA, Shen WK. Atrial fibrillation in the elderly: a review. Future Cardiol. 2014;10:745-758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Nattel S, Dobrev D. The multidimensional role of calcium in atrial fibrillation pathophysiology: mechanistic insights and therapeutic opportunities. Eur Heart J. 2012;33:1870-1877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 3. | Nalliah CJ, Sanders P, Kottkamp H, Kalman JM. The role of obesity in atrial fibrillation. Eur Heart J. 2015;pii:ehv486. [PubMed] [Cited in This Article: ] |
| 4. | Elahi MM, Flatman S, Matata BM. Tracing the origins of postoperative atrial fibrillation: the concept of oxidative stress-mediated myocardial injury phenomenon. Eur J Cardiovasc Prev Rehabil. 2008;15:735-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 5. | Zhang Q, Liu T, Ng CY, Li G. Diabetes mellitus and atrial remodeling: mechanisms and potential upstream therapies. Cardiovasc Ther. 2014;32:233-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 55] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 6. | Abed HS, Wittert GA. Obesity and atrial fibrillation. Obes Rev. 2013;14:929-938. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Vargas-Uricoechea H, Sierra-Torres CH. Thyroid hormones and the heart. Horm Mol Biol Clin Investig. 2014;18:15-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Goudis CA, Korantzopoulos P, Ntalas IV, Kallergis EM, Ketikoglou DG. Obesity and atrial fibrillation: A comprehensive review of the pathophysiological mechanisms and links. J Cardiol. 2015;66:361-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 92] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 9. | Goudis CA, Korantzopoulos P, Ntalas IV, Kallergis EM, Liu T, Ketikoglou DG. Diabetes mellitus and atrial fibrillation: Pathophysiological mechanisms and potential upstream therapies. Int J Cardiol. 2015;184:617-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 10. | De Caterina R, Camm AJ. What is ‘valvular’ atrial fibrillation? A reappraisal. Eur Heart J. 2014;35:3328-3335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 11. | Kumar KR, Mandleywala SN, Link MS. Atrial and ventricular arrhythmias in hypertrophic cardiomyopathy. Card Electrophysiol Clin. 2015;7:173-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 12. | Riber LP, Larsen TB, Christensen TD. Postoperative atrial fibrillation prophylaxis after lung surgery: systematic review and meta-analysis. Ann Thorac Surg. 2014;98:1989-1997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Qaddoura A, Kabali C, Drew D, van Oosten EM, Michael KA, Redfearn DP, Simpson CS, Baranchuk A. Obstructive sleep apnea as a predictor of atrial fibrillation after coronary artery bypass grafting: a systematic review and meta-analysis. Can J Cardiol. 2014;30:1516-1522. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 14. | Anumonwo JM, Kalifa J. Risk factors and genetics of atrial fibrillation. Cardiol Clin. 2014;32:485-494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Yadava M, Hughey AB, Crawford TC. Postoperative atrial fibrillation: incidence, mechanisms, and clinical correlates. Cardiol Clin. 2014;32:627-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 16. | Akoum N, Marrouche N. Assessment and impact of cardiac fibrosis on atrial fibrillation. Curr Cardiol Rep. 2014;16:518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 17. | Berenfeld O, Jalife J. Mechanisms of atrial fibrillation: rotors, ionic determinants, and excitation frequency. Cardiol Clin. 2014;32:495-506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Heijman J, Voigt N, Wehrens XH, Dobrev D. Calcium dysregulation in atrial fibrillation: the role of CaMKII. Front Pharmacol. 2014;5:30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Wolke C, Bukowska A, Goette A, Lendeckel U. Redox control of cardiac remodeling in atrial fibrillation. Biochim Biophys Acta. 2015;1850:1555-1565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 20. | Jalife J, Kaur K. Atrial remodeling, fibrosis, and atrial fibrillation. Trends Cardiovasc Med. 2015;25:475-484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 21. | Corradi D. Atrial fibrillation from the pathologist’s perspective. Cardiovasc Pathol. 2014;23:71-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 75] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 22. | Dzeshka MS, Lip GY, Snezhitskiy V, Shantsila E. Cardiac Fibrosis in Patients With Atrial Fibrillation: Mechanisms and Clinical Implications. J Am Coll Cardiol. 2015;66:943-959. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 336] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 23. | Senoo K, Lane D, Lip GY. Stroke and bleeding risk in atrial fibrillation. Korean Circ J. 2014;44:281-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Zimetbaum P, Waks JW, Ellis ER, Glotzer TV, Passman RS. Role of atrial fibrillation burden in assessing thromboembolic risk. Circ Arrhythm Electrophysiol. 2014;7:1223-1229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Hirsh BJ, Copeland-Halperin RS, Halperin JL. Fibrotic atrial cardiomyopathy, atrial fibrillation, and thromboembolism: mechanistic links and clinical inferences. J Am Coll Cardiol. 2015;65:2239-2251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 142] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 26. | Hui DS, Morley JE, Mikolajczak PC, Lee R. Atrial fibrillation: A major risk factor for cognitive decline. Am Heart J. 2015;169:448-456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 27. | Jacobs V, Cutler MJ, Day JD, Bunch TJ. Atrial fibrillation and dementia. Trends Cardiovasc Med. 2015;25:44-51. [PubMed] [Cited in This Article: ] |
| 28. | Luong C, Barnes ME, Tsang TS. Atrial fibrillation and heart failure: cause or effect? Curr Heart Fail Rep. 2014;11:463-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Wijesurendra RS, Casadei B. Atrial fibrillation: effects beyond the atrium? Cardiovasc Res. 2015;105:238-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 30. | Chen LY, Benditt DG, Alonso A. Atrial fibrillation and its association with sudden cardiac death. Circ J. 2014;78:2588-2593. [PubMed] [Cited in This Article: ] |
| 31. | Potpara TS, Lip GY. A brief history of ‘lone’ atrial fibrillation: from ‘a peculiar pulse irregularity’ to a modern public health concern. Curr Pharm Des. 2015;21:679-696. [PubMed] [Cited in This Article: ] |
| 32. | Andreasen L, Nielsen JB, Olesen MS. Genetic aspects of lone atrial fibrillation: what do we know? Curr Pharm Des. 2015;21:667-678. [PubMed] [Cited in This Article: ] |
| 33. | Tello-Montoliu A, Hernández-Romero D, Sanchez-Martínez M, Valdes M, Marín F. Lone atrial fibrillation - a diagnosis of exclusion. Curr Pharm Des. 2015;21:544-550. [PubMed] [Cited in This Article: ] |
| 34. | Andreasen L, Nielsen JB, Christophersen IE, Holst AG, Sajadieh A, Tveit A, Haunsø S, Svendsen JH, Schmitt N, Olesen MS. Genetic modifier of the QTc interval associated with early-onset atrial fibrillation. Can J Cardiol. 2013;29:1234-1240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 35. | Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, Brugada J, Girona J, Domingo A, Bachinski LL. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997;336:905-911. [PubMed] [Cited in This Article: ] |
| 36. | Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251-254. [PubMed] [Cited in This Article: ] |
| 37. | Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, Zhou Q, Yang Y, Liu Y, Liu B. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012-1019. [PubMed] [Cited in This Article: ] |
| 38. | Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, Liang B, Lin J, Liu Y, Liu B. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899-905. [PubMed] [Cited in This Article: ] |
| 39. | Berenfeld O, Zaitsev AV, Mironov SF, Pertsov AM, Jalife J. Frequency-dependent breakdown of wave propagation into fibrillatory conduction across the pectinate muscle network in the isolated sheep right atrium. Circ Res. 2002;90:1173-1180. [PubMed] [Cited in This Article: ] |
| 40. | Chen J, Mandapati R, Berenfeld O, Skanes AC, Gray RA, Jalife J. Dynamics of wavelets and their role in atrial fibrillation in the isolated sheep heart. Cardiovasc Res. 2000;48:220-232. [PubMed] [Cited in This Article: ] |
| 41. | Mandapati R, Skanes A, Chen J, Berenfeld O, Jalife J. Stable microreentrant sources as a mechanism of atrial fibrillation in the isolated sheep heart. Circulation. 2000;101:194-199. [PubMed] [Cited in This Article: ] |
| 42. | Skanes AC, Mandapati R, Berenfeld O, Davidenko JM, Jalife J. Spatiotemporal periodicity during atrial fibrillation in the isolated sheep heart. Circulation. 1998;98:1236-1248. [PubMed] [Cited in This Article: ] |
| 43. | Tanaka K, Zlochiver S, Vikstrom KL, Yamazaki M, Moreno J, Klos M, Zaitsev AV, Vaidyanathan R, Auerbach DS, Landas S. Spatial distribution of fibrosis governs fibrillation wave dynamics in the posterior left atrium during heart failure. Circ Res. 2007;101:839-847. [PubMed] [Cited in This Article: ] |
| 44. | Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353-357. [PubMed] [Cited in This Article: ] |
| 45. | Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879-881. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 301] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 46. | Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670-675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 452] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 47. | Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 363] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 48. | Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njølstad I, Nyrnes A. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876-878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 358] [Cited by in F6Publishing: 354] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 49. | Schnabel RB, Kerr KF, Lubitz SA, Alkylbekova EL, Marcus GM, Sinner MF, Magnani JW, Wolf PA, Deo R, Lloyd-Jones DM. Large-scale candidate gene analysis in whites and African Americans identifies IL6R polymorphism in relation to atrial fibrillation: the National Heart, Lung, and Blood Institute’s Candidate Gene Association Resource (CARe) project. Circ Cardiovasc Genet. 2011;4:557-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 50. | Amin AS, Tan HL, Wilde AA. Cardiac ion channels in health and disease. Heart Rhythm. 2010;7:117-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 51. | Amin AS, Bhuiyan ZA. SCN5A mutations in atrial fibrillation. Heart Rhythm. 2010;7:1870-1871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 52. | Blana A, Kaese S, Fortmüller L, Laakmann S, Damke D, van Bragt K, Eckstein J, Piccini I, Kirchhefer U, Nattel S. Knock-in gain-of-function sodium channel mutation prolongs atrial action potentials and alters atrial vulnerability. Heart Rhythm. 2010;7:1862-1869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 53. | Chen S, Wang C, Wang X, Xu C, Wu M, Wang P, Tu X, Wang QK. Significant Association Between CAV1 Variant rs3807989 on 7p31 and Atrial Fibrillation in a Chinese Han Population. J Am Heart Assoc. 2015;4:pii: e001980. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 54. | Laitinen-Forsblom PJ, Mäkynen P, Mäkynen H, Yli-Mäyry S, Virtanen V, Kontula K, Aalto-Setälä K. SCN5A mutation associated with cardiac conduction defect and atrial arrhythmias. J Cardiovasc Electrophysiol. 2006;17:480-485. [PubMed] [Cited in This Article: ] |
| 55. | Li Q, Huang H, Liu G, Lam K, Rutberg J, Green MS, Birnie DH, Lemery R, Chahine M, Gollob MH. Gain-of-function mutation of Nav1.5 in atrial fibrillation enhances cellular excitability and lowers the threshold for action potential firing. Biochem Biophys Res Commun. 2009;380:132-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 56. | Makiyama T, Akao M, Shizuta S, Doi T, Nishiyama K, Oka Y, Ohno S, Nishio Y, Tsuji K, Itoh H. A novel SCN5A gain-of-function mutation M1875T associated with familial atrial fibrillation. J Am Coll Cardiol. 2008;52:1326-1334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 57. | Benito B, Brugada R, Perich RM, Lizotte E, Cinca J, Mont L, Berruezo A, Tolosana JM, Freixa X, Brugada P. A mutation in the sodium channel is responsible for the association of long QT syndrome and familial atrial fibrillation. Heart Rhythm. 2008;5:1434-1440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 58. | Calloe K, Schmitt N, Grubb S, Pfeiffer R, David JP, Kanter R, Cordeiro JM, Antzelevitch C. Multiple arrhythmic syndromes in a newborn, owing to a novel mutation in SCN5A. Can J Physiol Pharmacol. 2011;89:723-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 59. | Dolz-Gaitón P, Núñez M, Núñez L, Barana A, Amorós I, Matamoros M, Pérez-Hernández M, González de la Fuente M, Alvarez-López M, Macías-Ruiz R. Functional characterization of a novel frameshift mutation in the C-terminus of the Nav1.5 channel underlying a Brugada syndrome with variable expression in a Spanish family. PLoS One. 2013;8:e81493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 60. | Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447-454. [PubMed] [Cited in This Article: ] |
| 61. | Ziyadeh-Isleem A, Clatot J, Duchatelet S, Gandjbakhch E, Denjoy I, Hidden-Lucet F, Hatem S, Deschênes I, Coulombe A, Neyroud N. A truncating SCN5A mutation combined with genetic variability causes sick sinus syndrome and early atrial fibrillation. Heart Rhythm. 2014;11:1015-1023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 62. | Rossenbacker T, Carroll SJ, Liu H, Kuipéri C, de Ravel TJ, Devriendt K, Carmeliet P, Kass RS, Heidbüchel H. Novel pore mutation in SCN5A manifests as a spectrum of phenotypes ranging from atrial flutter, conduction disease, and Brugada syndrome to sudden cardiac death. Heart Rhythm. 2004;1:610-615. [PubMed] [Cited in This Article: ] |
| 63. | Watanabe H, Darbar D, Kaiser DW, Jiramongkolchai K, Chopra S, Donahue BS, Kannankeril PJ, Roden DM. Mutations in sodium channel β1- and β2-subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2009;2:268-275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 172] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 64. | Wang P, Yang Q, Wu X, Yang Y, Shi L, Wang C, Wu G, Xia Y, Yang B, Zhang R. Functional dominant-negative mutation of sodium channel subunit gene SCN3B associated with atrial fibrillation in a Chinese GeneID population. Biochem Biophys Res Commun. 2010;398:98-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 62] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 65. | Olesen MS, Jespersen T, Nielsen JB, Liang B, Møller DV, Hedley P, Christiansen M, Varró A, Olesen SP, Haunsø S. Mutations in sodium channel β-subunit SCN3B are associated with early-onset lone atrial fibrillation. Cardiovasc Res. 2011;89:786-793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 66. | Li RG, Wang Q, Xu YJ, Zhang M, Qu XK, Liu X, Fang WY, Yang YQ. Mutations of the SCN4B-encoded sodium channel β4 subunit in familial atrial fibrillation. Int J Mol Med. 2013;32:144-150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 67. | Olesen MS, Holst AG, Svendsen JH, Haunsø S, Tfelt-Hansen J. SCN1Bb R214Q found in 3 patients: 1 with Brugada syndrome and 2 with lone atrial fibrillation. Heart Rhythm. 2012;9:770-773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 68. | Remme CA, Wilde AA, Bezzina CR. Cardiac sodium channel overlap syndromes: different faces of SCN5A mutations. Trends Cardiovasc Med. 2008;18:78-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 163] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 69. | Rook MB, Evers MM, Vos MA, Bierhuizen MF. Biology of cardiac sodium channel Nav1.5 expression. Cardiovasc Res. 2012;93:12-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 70. | Veerman CC, Wilde AA, Lodder EM. The cardiac sodium channel gene SCN5A and its gene product NaV1.5: Role in physiology and pathophysiology. Gene. 2015;573:177-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 71. | Macri V, Mahida SN, Zhang ML, Sinner MF, Dolmatova EV, Tucker NR, McLellan M, Shea MA, Milan DJ, Lunetta KL. A novel trafficking-defective HCN4 mutation is associated with early-onset atrial fibrillation. Heart Rhythm. 2014;11:1055-1062. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 72. | Hong K, Piper DR, Diaz-Valdecantos A, Brugada J, Oliva A, Burashnikov E, Santos-de-Soto J, Grueso-Montero J, Diaz-Enfante E, Brugada P. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc Res. 2005;68:433-440. [PubMed] [Cited in This Article: ] |
| 73. | Lundby A, Ravn LS, Svendsen JH, Olesen SP, Schmitt N. KCNQ1 mutation Q147R is associated with atrial fibrillation and prolonged QT interval. Heart Rhythm. 2007;4:1532-1541. [PubMed] [Cited in This Article: ] |
| 74. | Kharche S, Adeniran I, Stott J, Law P, Boyett MR, Hancox JC, Zhang H. Pro-arrhythmogenic effects of the S140G KCNQ1 mutation in human atrial fibrillation - insights from modelling. J Physiol. 2012;590:4501-4514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 75. | Das S, Makino S, Melman YF, Shea MA, Goyal SB, Rosenzweig A, Macrae CA, Ellinor PT. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm. 2009;6:1146-1153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 76. | El Harchi A, Zhang H, Hancox JC. The S140G KCNQ1 atrial fibrillation mutation affects ‘I(KS)’ profile during both atrial and ventricular action potentials. J Physiol Pharmacol. 2010;61:759-764. [PubMed] [Cited in This Article: ] |
| 77. | Bartos DC, Duchatelet S, Burgess DE, Klug D, Denjoy I, Peat R, Lupoglazoff JM, Fressart V, Berthet M, Ackerman MJ. R231C mutation in KCNQ1 causes long QT syndrome type 1 and familial atrial fibrillation. Heart Rhythm. 2011;8:48-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 78. | Hasegawa K, Ohno S, Ashihara T, Itoh H, Ding WG, Toyoda F, Makiyama T, Aoki H, Nakamura Y, Delisle BP. A novel KCNQ1 missense mutation identified in a patient with juvenile-onset atrial fibrillation causes constitutively open IKs channels. Heart Rhythm. 2014;11:67-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 79. | Ki CS, Jung CL, Kim HJ, Baek KH, Park SJ, On YK, Kim KS, Noh SJ, Youm JB, Kim JS, Cho H. A KCNQ1 mutation causes age-dependant bradycardia and persistent atrial fibrillation. Pflugers Arch. 2014;466:529-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 80. | Ravn LS, Aizawa Y, Pollevick GD, Hofman-Bang J, Cordeiro JM, Dixen U, Jensen G, Wu Y, Burashnikov E, Haunso S. Gain of function in IKs secondary to a mutation in KCNE5 associated with atrial fibrillation. Heart Rhythm. 2008;5:427-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 93] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 81. | Nielsen JB, Bentzen BH, Olesen MS, David JP, Olesen SP, Haunsø S, Svendsen JH, Schmitt N. Gain-of-function mutations in potassium channel subunit KCNE2 associated with early-onset lone atrial fibrillation. Biomark Med. 2014;8:557-570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 82. | Deo M, Ruan Y, Pandit SV, Shah K, Berenfeld O, Blaufox A, Cerrone M, Noujaim SF, Denegri M, Jalife J. KCNJ2 mutation in short QT syndrome 3 results in atrial fibrillation and ventricular proarrhythmia. Proc Natl Acad Sci USA. 2013;110:4291-4296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 83. | Kharche S, Garratt CJ, Boyett MR, Inada S, Holden AV, Hancox JC, Zhang H. Atrial proarrhythmia due to increased inward rectifier current (I(K1)) arising from KCNJ2 mutation--a simulation study. Prog Biophys Mol Biol. 2008;98:186-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 84. | Villafañe J, Fischbach P, Gebauer R. Short QT syndrome manifesting with neonatal atrial fibrillation and bradycardia. Cardiology. 2014;128:236-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 85. | Chelu MG, Sarma S, Sood S, Wang S, van Oort RJ, Skapura DG, Li N, Santonastasi M, Müller FU, Schmitz W. Calmodulin kinase II-mediated sarcoplasmic reticulum Ca2+ leak promotes atrial fibrillation in mice. J Clin Invest. 2009;119:1940-1951. [PubMed] [Cited in This Article: ] |
| 86. | Shan J, Xie W, Betzenhauser M, Reiken S, Chen BX, Wronska A, Marks AR. Calcium leak through ryanodine receptors leads to atrial fibrillation in 3 mouse models of catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2012;111:708-717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 87. | Li N, Wang T, Wang W, Cutler MJ, Wang Q, Voigt N, Rosenbaum DS, Dobrev D, Wehrens XH. Inhibition of CaMKII phosphorylation of RyR2 prevents induction of atrial fibrillation in FKBP12.6 knockout mice. Circ Res. 2012;110:465-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 88. | Zhang Y, Matthews GD, Lei M, Huang CL. Abnormal Ca(2+) homeostasis, atrial arrhythmogenesis, and sinus node dysfunction in murine hearts modeling RyR2 modification. Front Physiol. 2013;4:150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 89. | Beavers DL, Wang W, Ather S, Voigt N, Garbino A, Dixit SS, Landstrom AP, Li N, Wang Q, Olivotto I. Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization. J Am Coll Cardiol. 2013;62:2010-2019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 148] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 90. | Chiang DY, Li N, Wang Q, Alsina KM, Quick AP, Reynolds JO, Wang G, Skapura D, Voigt N, Dobrev D. Impaired local regulation of ryanodine receptor type 2 by protein phosphatase 1 promotes atrial fibrillation. Cardiovasc Res. 2014;103:178-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 91. | Di Pino A, Caruso E, Costanzo L, Guccione P. A novel RyR2 mutation in a 2-year-old baby presenting with atrial fibrillation, atrial flutter, and atrial ectopic tachycardia. Heart Rhythm. 2014;11:1480-1483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 92. | Zhabyeyev P, Hiess F, Wang R, Liu Y, Wayne Chen SR, Oudit GY. S4153R is a gain-of-function mutation in the cardiac Ca(2+) release channel ryanodine receptor associated with catecholaminergic polymorphic ventricular tachycardia and paroxysmal atrial fibrillation. Can J Cardiol. 2013;29:993-996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 93. | Kazemian P, Gollob MH, Pantano A, Oudit GY. A novel mutation in the RYR2 gene leading to catecholaminergic polymorphic ventricular tachycardia and paroxysmal atrial fibrillation: dose-dependent arrhythmia-event suppression by β-blocker therapy. Can J Cardiol. 2011;27:870.e7-870.e10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 94. | King JH, Wickramarachchi C, Kua K, Du Y, Jeevaratnam K, Matthews HR, Grace AA, Huang CL, Fraser JA. Loss of Nav1.5 expression and function in murine atria containing the RyR2-P2328S gain-of-function mutation. Cardiovasc Res. 2013;99:751-759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 95. | Zhang Y, Fraser JA, Jeevaratnam K, Hao X, Hothi SS, Grace AA, Lei M, Huang CL. Acute atrial arrhythmogenicity and altered Ca(2+) homeostasis in murine RyR2-P2328S hearts. Cardiovasc Res. 2011;89:794-804. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 96. | Thibodeau IL, Xu J, Li Q, Liu G, Lam K, Veinot JP, Birnie DH, Jones DL, Krahn AD, Lemery R. Paradigm of genetic mosaicism and lone atrial fibrillation: physiological characterization of a connexin 43-deletion mutant identified from atrial tissue. Circulation. 2010;122:236-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 97. | Tuomi JM, Tyml K, Jones DL. Atrial tachycardia/fibrillation in the connexin 43 G60S mutant (Oculodentodigital dysplasia) mouse. Am J Physiol Heart Circ Physiol. 2011;300:H1402-H1411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 98. | Delmar M, Makita N. Cardiac connexins, mutations and arrhythmias. Curr Opin Cardiol. 2012;27:236-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 99. | Gollob MH, Jones DL, Krahn AD, Danis L, Gong XQ, Shao Q, Liu X, Veinot JP, Tang AS, Stewart AF. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med. 2006;354:2677-2688. [PubMed] [Cited in This Article: ] |
| 100. | Yang YQ, Liu X, Zhang XL, Wang XH, Tan HW, Shi HF, Jiang WF, Fang WY. Novel connexin40 missense mutations in patients with familial atrial fibrillation. Europace. 2010;12:1421-1427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 101. | Yang YQ, Zhang XL, Wang XH, Tan HW, Shi HF, Jiang WF, Fang WY, Liu X. Connexin40 nonsense mutation in familial atrial fibrillation. Int J Mol Med. 2010;26:605-610. [PubMed] [Cited in This Article: ] |
| 102. | Christophersen IE, Holmegard HN, Jabbari J, Sajadieh A, Haunsø S, Tveit A, Svendsen JH, Olesen MS. Rare variants in GJA5 are associated with early-onset lone atrial fibrillation. Can J Cardiol. 2013;29:111-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 103. | Shi HF, Yang JF, Wang Q, Li RG, Xu YJ, Qu XK, Fang WY, Liu X, Yang YQ. Prevalence and spectrum of GJA5 mutations associated with lone atrial fibrillation. Mol Med Rep. 2013;7:767-774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 104. | Sun Y, Yang YQ, Gong XQ, Wang XH, Li RG, Tan HW, Liu X, Fang WY, Bai D. Novel germline GJA5/connexin40 mutations associated with lone atrial fibrillation impair gap junctional intercellular communication. Hum Mutat. 2013;34:603-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 105. | Roberts JD, Longoria J, Poon A, Gollob MH, Dewland TA, Kwok PY, Olgin JE, Deo RC, Marcus GM. Targeted deep sequencing reveals no definitive evidence for somatic mosaicism in atrial fibrillation. Circ Cardiovasc Genet. 2015;8:50-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 11] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 106. | Gemel J, Simon AR, Patel D, Xu Q, Matiukas A, Veenstra RD, Beyer EC. Degradation of a connexin40 mutant linked to atrial fibrillation is accelerated. J Mol Cell Cardiol. 2014;74:330-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 107. | Sun Y, Hills MD, Ye WG, Tong X, Bai D. Atrial fibrillation-linked germline GJA5/connexin40 mutants showed an increased hemichannel function. PLoS One. 2014;9:e95125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 108. | Sun Y, Tong X, Chen H, Huang T, Shao Q, Huang W, Laird DW, Bai D. An atrial-fibrillation-linked connexin40 mutant is retained in the endoplasmic reticulum and impairs the function of atrial gap-junction channels. Dis Model Mech. 2014;7:561-569. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 109. | Patel D, Gemel J, Xu Q, Simon AR, Lin X, Matiukas A, Beyer EC, Veenstra RD. Atrial fibrillation-associated connexin40 mutants make hemichannels and synergistically form gap junction channels with novel properties. FEBS Lett. 2014;588:1458-1464. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 110. | Bai D. Atrial fibrillation-linked GJA5/connexin40 mutants impaired gap junctions via different mechanisms. FEBS Lett. 2014;588:1238-1243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 111. | Lübkemeier I, Andrié R, Lickfett L, Bosen F, Stöckigt F, Dobrowolski R, Draffehn AM, Fregeac J, Schultze JL, Bukauskas FF. The Connexin40A96S mutation from a patient with atrial fibrillation causes decreased atrial conduction velocities and sustained episodes of induced atrial fibrillation in mice. J Mol Cell Cardiol. 2013;65:19-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 112. | Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, Lam J, Wilde AA, Lekanne Deprez RH, Moorman AF. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102:1433-1442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 122] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 113. | Gutierrez-Roelens I, De Roy L, Ovaert C, Sluysmans T, Devriendt K, Brunner HG, Vikkula M. A novel CSX/NKX2-5 mutation causes autosomal-dominant AV block: are atrial fibrillation and syncopes part of the phenotype? Eur J Hum Genet. 2006;14:1313-1316. [PubMed] [Cited in This Article: ] |
| 114. | Huang RT, Xue S, Xu YJ, Zhou M, Yang YQ. A novel NKX2.5 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2013;31:1119-1126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 115. | Xie WH, Chang C, Xu YJ, Li RG, Qu XK, Fang WY, Liu X, Yang YQ. Prevalence and spectrum of Nkx2.5 mutations associated with idiopathic atrial fibrillation. Clinics (Sao Paulo). 2013;68:777-784. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 116. | Yu H, Xu JH, Song HM, Zhao L, Xu WJ, Wang J, Li RG, Xu L, Jiang WF, Qiu XB. Mutational spectrum of the NKX2-5 gene in patients with lone atrial fibrillation. Int J Med Sci. 2014;11:554-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 117. | Yuan F, Qiu XB, Li RG, Qu XK, Wang J, Xu YJ, Liu X, Fang WY, Yang YQ, Liao DN. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int J Mol Med. 2015;35:478-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 118. | Wang J, Zhang DF, Sun YM, Li RG, Qiu XB, Qu XK, Liu X, Fang WY, Yang YQ. NKX2-6 mutation predisposes to familial atrial fibrillation. Int J Mol Med. 2014;34:1581-1590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 119. | Zhang X, Chen S, Yoo S, Chakrabarti S, Zhang T, Ke T, Oberti C, Yong SL, Fang F, Li L. Mutation in nuclear pore component NUP155 leads to atrial fibrillation and early sudden cardiac death. Cell. 2008;135:1017-1027. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 204] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 120. | Hua R, MacLeod SL, Polina I, Moghtadaei M, Jansen HJ, Bogachev O, O’Blenes SB, Sapp JL, Legare JF, Rose RA. Effects of Wild-Type and Mutant Forms of Atrial Natriuretic Peptide on Atrial Electrophysiology and Arrhythmogenesis. Circ Arrhythm Electrophysiol. 2015;8:1240-1254. [PubMed] [Cited in This Article: ] |
| 121. | Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis AR, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet. 1997;15:21-29. [PubMed] [Cited in This Article: ] |
| 122. | Costa MW, Guo G, Wolstein O, Vale M, Castro ML, Wang L, Otway R, Riek P, Cochrane N, Furtado M. Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ Cardiovasc Genet. 2013;6:238-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 123. | Qu XK, Qiu XB, Yuan F, Wang J, Zhao CM, Liu XY, Zhang XL, Li RG, Xu YJ, Hou XM. A novel NKX2.5 loss-of-function mutation associated with congenital bicuspid aortic valve. Am J Cardiol. 2014;114:1891-1895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 124. | Wang J, Mao JH, Ding KK, Xu WJ, Liu XY, Qiu XB, Li RG, Qu XK, Xu YJ, Huang RT. A novel NKX2.6 mutation associated with congenital ventricular septal defect. Pediatr Cardiol. 2015;36:646-656. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 125. | Lin Y, Guo X, Zhao B, Liu J, Da M, Wen Y, Hu Y, Ni B, Zhang K, Yang S. Association analysis identifies new risk loci for congenital heart disease in Chinese populations. Nat Commun. 2015;6:8082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 126. | Chowdhury R, Ashraf H, Melanson M, Tanada Y, Nguyen M, Silberbach M, Wakimoto H, Benson DW, Anderson RH, Kasahara H. Mouse Model of Human Congenital Heart Disease: Progressive Atrioventricular Block Induced by a Heterozygous Nkx2-5 Homeodomain Missense Mutation. Circ Arrhythm Electrophysiol. 2015;8:1255-1264. [PubMed] [Cited in This Article: ] |
| 127. | Zhou W, Zhao L, Jiang JQ, Jiang WF, Yang YQ, Qiu XB. A novel TBX5 loss-of-function mutation associated with sporadic dilated cardiomyopathy. Int J Mol Med. 2015;36:282-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 128. | Zhao L, Ni SH, Liu XY, Wei D, Yuan F, Xu L, Xin-Li RG, Qu XK, Xu YJ, Fang WY. Prevalence and spectrum of Nkx2.6 mutations in patients with congenital heart disease. Eur J Med Genet. 2014;57:579-586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 129. | Smith JG, Magnani JW, Palmer C, Meng YA, Soliman EZ, Musani SK, Kerr KF, Schnabel RB, Lubitz SA, Sotoodehnia N. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 2011;7:e1001304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 130. | den Hoed M, Eijgelsheim M, Esko T, Brundel BJ, Peal DS, Evans DM, Nolte IM, Segrè AV, Holm H, Handsaker RE. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet. 2013;45:621-631. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 228] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 131. | Parvez B, Shoemaker MB, Muhammad R, Richardson R, Jiang L, Blair MA, Roden DM, Darbar D. Common genetic polymorphism at 4q25 locus predicts atrial fibrillation recurrence after successful cardioversion. Heart Rhythm. 2013;10:849-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 132. | Mohanty S, Santangeli P, Bai R, Di Biase L, Mohanty P, Pump A, Natale A. Variant rs2200733 on chromosome 4q25 confers increased risk of atrial fibrillation: evidence from a meta-analysis. J Cardiovasc Electrophysiol. 2013;24:155-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 133. | Olesen MS, Holst AG, Jabbari J, Nielsen JB, Christophersen IE, Sajadieh A, Haunsø S, Svendsen JH. Genetic loci on chromosomes 4q25, 7p31, and 12p12 are associated with onset of lone atrial fibrillation before the age of 40 years. Can J Cardiol. 2012;28:191-195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 134. | Henningsen KM, Olesen MS, Haunsoe S, Svendsen JH. Association of rs2200733 at 4q25 with early onset of lone atrial fibrillation in young patients. Scand Cardiovasc J. 2011;45:324-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 135. | Kiliszek M, Franaszczyk M, Kozluk E, Lodzinski P, Piatkowska A, Broda G, Ploski R, Opolski G. Association between variants on chromosome 4q25, 16q22 and 1q21 and atrial fibrillation in the Polish population. PLoS One. 2011;6:e21790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 136. | Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpón E, Tamargo J, Cinca J, Hove-Madsen L, Aranega AE. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 137. | Sinner MF, Lubitz SA, Pfeufer A, Makino S, Beckmann BM, Lunetta KL, Steinbeck G, Perz S, Rahman R, Sonni A. Lack of replication in polymorphisms reported to be associated with atrial fibrillation. Heart Rhythm. 2011;8:403-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 138. | Lee KT, Yeh HY, Tung CP, Chu CS, Cheng KH, Tsai WC, Lu YH, Chang JG, Sheu SH, Lai WT. Association of RS2200733 but not RS10033464 on 4q25 with atrial fibrillation based on the recessive model in a Taiwanese population. Cardiology. 2010;116:151-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 139. | Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107:9753-9758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 224] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 140. | Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 210] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 141. | Aguirre LA, Alonso ME, Badía-Careaga C, Rollán I, Arias C, Fernández-Miñán A, López-Jiménez E, Aránega A, Gómez-Skarmeta JL, Franco D. Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP. BMC Biol. 2015;13:26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 142. | Gore-Panter SR, Hsu J, Hanna P, Gillinov AM, Pettersson G, Newton DW, Moravec CS, Van Wagoner DR, Chung MK, Barnard J. Atrial Fibrillation associated chromosome 4q25 variants are not associated with PITX2c expression in human adult left atrial appendages. PLoS One. 2014;9:e86245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 143. | Martin RI, Babaei MS, Choy MK, Owens WA, Chico TJ, Keenan D, Yonan N, Koref MS, Keavney BD. Genetic variants associated with risk of atrial fibrillation regulate expression of PITX2, CAV1, MYOZ1, C9orf3 and FANCC. J Mol Cell Cardiol. 2015;85:207-214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 144. | Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 544] [Cited by in F6Publishing: 497] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 145. | Li C, Wang F, Yang Y, Fu F, Xu C, Shi L, Li S, Xia Y, Wu G, Cheng X. Significant association of SNP rs2106261 in the ZFHX3 gene with atrial fibrillation in a Chinese Han GeneID population. Hum Genet. 2011;129:239-246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 146. | Zhai C, Cong H, Liu Y, Zhang Y, Liu X, Zhang H, Ren Z. Rs7193343 polymorphism in zinc finger homeobox 3 (ZFHX3) gene and atrial fibrillation: an updated meta-analysis of 10 case-control comparisons. BMC Cardiovasc Disord. 2015;15:58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 147. | Lin H, Sinner MF, Brody JA, Arking DE, Lunetta KL, Rienstra M, Lubitz SA, Magnani JW, Sotoodehnia N, McKnight B. Targeted sequencing in candidate genes for atrial fibrillation: the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Targeted Sequencing Study. Heart Rhythm. 2014;11:452-457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 148. | Mahida S, Mills RW, Tucker NR, Simonson B, Macri V, Lemoine MD, Das S, Milan DJ, Ellinor PT. Overexpression of KCNN3 results in sudden cardiac death. Cardiovasc Res. 2014;101:326-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 149. | Lozano-Velasco E, Hernández-Torres F, Daimi H, Serra SA, Herraiz A, Hove-Madsen L, Aránega A, Franco D. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc Res. 2016;109:55-66. [PubMed] [Cited in This Article: ] |
| 150. | Jiang Q, Ni B, Shi J, Han Z, Qi R, Xu W, Wang D, Wang DW, Chen M. Down-regulation of ATBF1 activates STAT3 signaling via PIAS3 in pacing-induced HL-1 atrial myocytes. Biochem Biophys Res Commun. 2014;449:278-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 151. | Kääb S, Darbar D, van Noord C, Dupuis J, Pfeufer A, Newton-Cheh C, Schnabel R, Makino S, Sinner MF, Kannankeril PJ. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813-819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 151] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 152. | Tsai CT, Hsieh CS, Chang SN, Chuang EY, Juang JM, Lin LY, Lai LP, Hwang JJ, Chiang FT, Lin JL. Next-generation sequencing of nine atrial fibrillation candidate genes identified novel de novo mutations in patients with extreme trait of atrial fibrillation. J Med Genet. 2015;52:28-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 153. | Huang Y, Wang C, Yao Y, Zuo X, Chen S, Xu C, Zhang H, Lu Q, Chang L, Wang F. Molecular Basis of Gene-Gene Interaction: Cyclic Cross-Regulation of Gene Expression and Post-GWAS Gene-Gene Interaction Involved in Atrial Fibrillation. PLoS Genet. 2015;11:e1005393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 154. | Li G, Zhang R, Gao L, Zhang S, Dong Y, Yin X, Chang D, Yang Y, Xia Y. Lack of association between rs3807989 in cav1 and atrial fibrillation. Int J Clin Exp Pathol. 2014;7:4339-4344. [PubMed] [Cited in This Article: ] |
| 155. | Tada H, Shiffman D, Smith JG, Sjögren M, Lubitz SA, Ellinor PT, Louie JZ, Catanese JJ, Engström G, Devlin JJ. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke. 2014;45:2856-2862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 156. | Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Müller M, Sotoodehnia N, Sinner MF. Genome-wide association study of PR interval. Nat Genet. 2010;42:153-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 346] [Cited by in F6Publishing: 329] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 157. | Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njølstad I. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 298] [Cited by in F6Publishing: 283] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 158. | Butler AM, Yin X, Evans DS, Nalls MA, Smith EN, Tanaka T, Li G, Buxbaum SG, Whitsel EA, Alonso A. Novel loci associated with PR interval in a genome-wide association study of 10 African American cohorts. Circ Cardiovasc Genet. 2012;5:639-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 159. | Bezzina CR, Barc J, Mizusawa Y, Remme CA, Gourraud JB, Simonet F, Verkerk AO, Schwartz PJ, Crotti L, Dagradi F. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1044-1049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 428] [Cited by in F6Publishing: 382] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 160. | Newton-Cheh C, Eijgelsheim M, Rice KM, de Bakker PI, Yin X, Estrada K, Bis JC, Marciante K, Rivadeneira F, Noseworthy PA. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 353] [Cited by in F6Publishing: 351] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 161. | Pfeufer A, Sanna S, Arking DE, Müller M, Gateva V, Fuchsberger C, Ehret GB, Orrú M, Pattaro C, Köttgen A. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 318] [Cited by in F6Publishing: 330] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 162. | Arking DE, Pulit SL, Crotti L, van der Harst P, Munroe PB, Koopmann TT, Sotoodehnia N, Rossin EJ, Morley M, Wang X. Genetic association study of QT interval highlights role for calcium signaling pathways in myocardial repolarization. Nat Genet. 2014;46:826-836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 223] [Cited by in F6Publishing: 228] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 163. | Li L, Shen C, Yao Z, Liang J, Huang C. Genetic variants of potassium voltage-gated channel genes (KCNQ1, KCNH2, and KCNE1) affected the risk of atrial fibrillation in elderly patients. Genet Test Mol Biomarkers. 2015;19:359-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 164. | Chu HM, Feng MJ, Li YG, Zhang YX, Ma JF, He B, Yu YB, Liu J, Chen XM. Polymorphisms but not mutations of the KCNQ1 gene are associated with lone atrial fibrillation in the Chinese Han population. ScientificWorldJournal. 2013;2013:373454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 165. | Zeng Z, Tan C, Teng S, Chen J, Su S, Zhou X, Wang F, Zhang S, Gu D, Makielski JC. The single nucleotide polymorphisms of I(Ks) potassium channel genes and their association with atrial fibrillation in a Chinese population. Cardiology. 2007;108:97-103. [PubMed] [Cited in This Article: ] |
| 166. | Jabbari J, Olesen MS, Holst AG, Nielsen JB, Haunso S, Svendsen JH. Common polymorphisms in KCNJ5 [corrected] are associated with early-onset lone atrial fibrillation in Caucasians. Cardiology. 2011;118:116-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 167. | Hauer RN, Groenewegen WA, Firouzi M, Ramanna H, Jongsma HJ. Cx40 polymorphism in human atrial fibrillation. Adv Cardiol. 2006;42:284-291. [PubMed] [Cited in This Article: ] |
| 168. | Zang X, Zhang S, Xia Y, Li S, Fu F, Li X, Wang F, Zhang R, Tian X, Gao L. SNP rs3825214 in TBX5 is associated with lone atrial fibrillation in Chinese Han population. PLoS One. 2013;8:e64966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 169. | Wang C, Wu M, Qian J, Li B, Tu X, Xu C, Li S, Chen S, Zhao Y, Huang Y. Identification of rare variants in TNNI3 with atrial fibrillation in a Chinese GeneID population. Mol Genet Genomics. 2015;291:79-92. [PubMed] [Cited in This Article: ] |
| 170. | Jabbari J, Olesen MS, Yuan L, Nielsen JB, Liang B, Macri V, Christophersen IE, Nielsen N, Sajadieh A, Ellinor PT. Common and rare variants in SCN10A modulate the risk of atrial fibrillation. Circ Cardiovasc Genet. 2015;8:64-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 171. | Delaney JT, Muhammad R, Shi Y, Schildcrout JS, Blair M, Short L, Roden DM, Darbar D. Common SCN10A variants modulate PR interval and heart rate response during atrial fibrillation. Europace. 2014;16:485-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 172. | Franco D, Aranega A. Post-transcriptional regulatory mechanisms. Clinic, Genetics and Molecular Pathways of Congenital Heart Diseases. In: Sperling S, Driscoll D, Kelly R, editors. Springer 2015; In press. [Cited in This Article: ] |
| 173. | Espinoza-Lewis RA, Wang DZ. MicroRNAs in heart development. Curr Top Dev Biol. 2012;100:279-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 174. | Zhou J, Dong X, Zhou Q, Wang H, Qian Y, Tian W, Ma D, Li X. microRNA expression profiling of heart tissue during fetal development. Int J Mol Med. 2014;33:1250-1260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 175. | Hu DL, Liu YQ, Chen FK, Sheng YH, Yang R, Kong XQ, Cao KJ, Zhang JS, Qian LM. Differential expression of microRNAs in cardiac myocytes compared to undifferentiated P19 cells. Int J Mol Med. 2011;28:59-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 176. | Synnergren J, Améen C, Lindahl A, Olsson B, Sartipy P. Expression of microRNAs and their target mRNAs in human stem cell-derived cardiomyocyte clusters and in heart tissue. Physiol Genomics. 2011;43:581-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 177. | Nishi H, Sakaguchi T, Miyagawa S, Yoshikawa Y, Fukushima S, Saito S, Ueno T, Kuratani T, Sawa Y. Impact of microRNA expression in human atrial tissue in patients with atrial fibrillation undergoing cardiac surgery. PLoS One. 2013;8:e73397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 178. | Wang J, Wang Y, Han J, Li Y, Xie C, Xie L, Shi J, Zhang J, Yang B, Chen D. Integrated analysis of microRNA and mRNA expression profiles in the left atrium of patients with nonvalvular paroxysmal atrial fibrillation: Role of miR-146b-5p in atrial fibrosis. Heart Rhythm. 2015;12:1018-1026. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 179. | Li M, Zhang J. Circulating MicroRNAs: Potential and Emerging Biomarkers for Diagnosis of Cardiovascular and Cerebrovascular Diseases. Biomed Res Int. 2015;2015:730535. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 180. | Orenes-Piñero E, Montoro-García S, Patel JV, Valdés M, Marín F, Lip GY. Role of microRNAs in cardiac remodelling: new insights and future perspectives. Int J Cardiol. 2013;167:1651-1659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 181. | Poudel P, Xu Y, Cui Z, Sharma D, Tian B, Paudel S. Atrial fibrillation: recent advances in understanding the role of microRNAs in atrial remodeling with an electrophysiological overview. Cardiology. 2015;131:58-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 182. | Gomes da Silva AM, Silbiger VN. miRNAs as biomarkers of atrial fibrillation. Biomarkers. 2014;19:631-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 183. | Liu Z, Zhou C, Liu Y, Wang S, Ye P, Miao X, Xia J. The expression levels of plasma micoRNAs in atrial fibrillation patients. PLoS One. 2012;7:e44906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 184. | van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJ, Klous P, McKean D, Muehlschlegel JD, Moosmann J, Toka O. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Invest. 2014;124:1844-1852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 136] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 185. | Tao Y, Zhang M, Li L, Bai Y, Zhou Y, Moon AM, Kaminski HJ, Martin JF. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet. 2014;7:23-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 83] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 186. | Nagy II, Railo A, Rapila R, Hast T, Sormunen R, Tavi P, Räsänen J, Vainio SJ. Wnt-11 signalling controls ventricular myocardium development by patterning N-cadherin and beta-catenin expression. Cardiovasc Res. 2010;85:100-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 187. | Pandur P, Läsche M, Eisenberg LM, Kühl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636-641. [PubMed] [Cited in This Article: ] |
| 188. | Martin A, Maher S, Summerhurst K, Davidson D, Murphy P. Differential deployment of paralogous Wnt genes in the mouse and chick embryo during development. Evol Dev. 2012;14:178-195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 189. | Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB, Tao Y, Chen Y, Wehrens XH, Martin JF. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc Natl Acad Sci USA. 2014;111:9181-9186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 190. | Chiang DY, Kongchan N, Beavers DL, Alsina KM, Voigt N, Neilson JR, Jakob H, Martin JF, Dobrev D, Wehrens XH. Loss of microRNA-106b-25 cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 expression and calcium release. Circ Arrhythm Electrophysiol. 2014;7:1214-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 84] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 191. | Barana A, Matamoros M, Dolz-Gaitón P, Pérez-Hernández M, Amorós I, Núñez M, Sacristán S, Pedraz Á, Pinto Á, Fernández-Avilés F. Chronic atrial fibrillation increases microRNA-21 in human atrial myocytes decreasing L-type calcium current. Circ Arrhythm Electrophysiol. 2014;7:861-868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 192. | Daimi H, Lozano-Velasco E, Haj Khelil A, Chibani JB, Barana A, Amorós I, González de la Fuente M, Caballero R, Aranega A, Franco D. Regulation of SCN5A by microRNAs: miR-219 modulates SCN5A transcript expression and the effects of flecainide intoxication in mice. Heart Rhythm. 2015;12:1333-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 193. | Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang N, Lin H, Xiao L, Maguy A, Qi XY. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Invest. 2013;123:1939-1951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 202] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 194. | Jia X, Zheng S, Xie X, Zhang Y, Wang W, Wang Z, Zhang Y, Wang J, Gao M, Hou Y. MicroRNA-1 accelerates the shortening of atrial effective refractory period by regulating KCNE1 and KCNB2 expression: an atrial tachypacing rabbit model. PLoS One. 2013;8:e85639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 195. | Li YD, Hong YF, Yusufuaji Y, Tang BP, Zhou XH, Xu GJ, Li JX, Sun L, Zhang JH, Xin Q. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol Med Rep. 2015;12:3243-3248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 196. | Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902-909. [PubMed] [Cited in This Article: ] |
| 197. | Mints Y, Yarmohammadi H, Khurram IM, Hoyt H, Hansford R, Zimmerman SL, Steinberg SJ, Judge DP, Tomaselli GF, Calkins H. Association of common variations on chromosome 4q25 and left atrial volume in patients with atrial fibrillation. Clin Med Insights Cardiol. 2015;9:39-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 198. | Scridon A, Fouilloux-Meugnier E, Loizon E, Rome S, Julien C, Barrès C, Chevalier P. Long-standing arterial hypertension is associated with Pitx2 down-regulation in a rat model of spontaneous atrial tachyarrhythmias. Europace. 2015;17:160-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 199. | Torrado M, Franco D, Hernández-Torres F, Crespo-Leiro MG, Iglesias-Gil C, Castro-Beiras A, Mikhailov AT. Pitx2c is reactivated in the failing myocardium and stimulates myf5 expression in cultured cardiomyocytes. PLoS One. 2014;9:e90561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 200. | Torrado M, Franco D, Lozano-Velasco E, Hernández-Torres F, Calviño R, Aldama G, Centeno A, Castro-Beiras A, Mikhailov A. A MicroRNA-Transcription Factor Blueprint for Early Atrial Arrhythmogenic Remodeling. Biomed Res Int. 2015;2015:263151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |