Published online Feb 28, 2021. doi: 10.35712/aig.v2.i1.1
Peer-review started: October 15, 2020
First decision: December 17, 2020
Revised: December 29, 2020
Accepted: February 12, 2021
Article in press: February 12, 2021
Published online: February 28, 2021
Hepatitis A virus (HAV) infection is still an important health issue worldwide. Although several effective HAV vaccines are available, it is difficult to perform universal vaccination in certain countries. Therefore, it may be better to develop antivirals against HAV for the prevention of severe hepatitis A. We found that several drugs potentially inhibit HAV internal ribosomal entry site-dependent translation and HAV replication. Artificial intelligence and machine learning could also support screening of anti-HAV drugs, using drug repositioning and drug rescue approaches.
Core Tip: In certain areas, it is difficult to perform universal hepatitis A virus (HAV) vaccination. We found that several drugs potentially inhibit HAV internal ribosomal entry sites-dependent translation and HAV replication. After the application of machine and deep learning, artificial intelligence identified effective anti-HAV drugs more quickly, using drug repositioning and drug rescue.
- Citation: Kanda T, Sasaki R, Masuzaki R, Moriyama M. Artificial intelligence and machine learning could support drug development for hepatitis A virus internal ribosomal entry sites. Artif Intell Gastroenterol 2021; 2(1): 1-9
- URL: https://www.wjgnet.com/2644-3236/full/v2/i1/1.htm
- DOI: https://dx.doi.org/10.35712/aig.v2.i1.1
Infection with hepatitis A virus (HAV) can lead to acute hepatitis, occasionally resulting in acute liver failure, which is associated with death or liver transplant
HAV infects humans through the fecal-oral route when HAV-contaminated foods and water are ingested[6]. Recently, hepatitis A has also been recognized as a sex-transmitted disease[7]. Several effective HAV vaccines are available, but they are relatively expensive, and in some countries, it is difficult to perform universal vaccination[4,5]. Therefore, to prevent severe hepatitis A, it may be better to develop antivirals against HAV[8].
Recently, information and communication technology, and artificial intelligence (AI) have played roles in daily clinical practice[9,10]. AI also plays an important role in drug discovery[11]. With the progress of machine learning methods and the accumulation of pharmacological data, AI has become a powerful data mining tool in the area of drug discovery, such as in silico screening, quantitative structure-activity relationship (QSAR) analysis, de novo drug design, and in silico evaluation of absorption, distribution, metabolism, excretion and toxicity[12].
Structure-based drug design is becoming an essential tool for faster, more cost-efficient drug discovery, compared to traditional methods[13]. The combination of AI and deep learning, which is a family of machine learning models that use artificial neural networks, may be a more powerful tool for drug discovery. The associations of machine learning, deep learning and AI are shown in Figure 1. Moreover, network-based in silico drug efficacy screening allows us to predict novel drug-disease associations, which may provide us with drug repositioning or drug rescue information[14]. In this minireview article, we discuss the recent involvement of AI in drug discovery and its application in the development of antivirals against HAV in the near future.
Translation of HAV protein is performed in a cap-independent manner under the control of the internal ribosomal entry site (IRES), which is mainly located at 5' untranslated region (5'UTR)[15]. It was reported that the HAV 5'UTR was more than 25-fold less active than the encephalomyocarditis virus IRES in producing translated proteins[16]. Thus, the relatively weaker activity of the HAV IRES may be due to a reduced affinity for several cellular translation factors[16]. Mutations within the HAV 5'UTR could enhance cap-independent translation in African green monkey kidney BS-C-1 cells[17]. Further studies are needed to identify specific mutations related to the severity of hepatitis A[18-20], although among HAV strains from HAV outbreaks in Korea and Japan, we did not identify specific mutations associated with severe hepatitis A in the HAV 5'UTR[21,22]. We also demonstrated that the inhibition of HAV IRES activity by small interfering RNAs (siRNAs) targeting HAV IRES could lead to the suppression of HAV replication[23]. Therefore, HAV IRES is an attractive target of antivirals against HAV.
HAV is a nonenveloped and enveloped positive-sense single-stranded RNA virus approximately 7.6 kb in length[24,25]. The HAV genome includes a 5′UTR, one open reading frame encoding structural (VP4, VP2, VP3, VP1 and 2A) and nonstructural proteins (2B, 2C, 3A, 3B, 3C and 3D) and a 3′UTR[26].
Among HAV proteins, HAV proteinase 3C suppressed HAV IRES-dependent translation[27]. Furthermore, HAV 3C cleaves the polypyrimidine tract-binding protein (PTB), which interacts with the HAV IRES[27,28]. Among host proteins, autoantigen La[27], glyceraldehyde-3-phosphate dehydrogenase[29], PTB[28], poly(C) binding protein 2[30], polyadenylate-binding protein-1[31], eukaryotic translation initiation factor 4E[32] and eukaryotic translation initiation factor 4E[33] are reported to interact with HAV IRES.
We demonstrated that siRNA against cellular cofactors for HAV IRES could inhibit HAV IRES-mediated translation[34]. The Janus kinase (JAK) inhibitors SD-1029 and AG490 could reduce La protein expression and inhibit HAV IRES-mediated translation as well as HAV replication[34]. The JAK2 inhibitor AZD1480 could reduce La expression and inhibit HAV IRES activity and HAV replication[35]. We also reported that the sirtuin inhibitor sirtinol[36] and broad-spectrum antivirals, such as amantadine[20,37,38], interferon-alpha[38] and interferon-lambda (interleukin-29)[39], could inhibit HAV IRES-mediated translation and HAV replication. Thus, in vitro drug screening with human hepatocytes revealed that several drugs inhibit HAV replication through the inhibition of HAV IRES activity.
Bioinformatics and cheminformatics are newer strategies to screen and design various drug candidates for HAV, as performed for severe acute respiratory syndrome coronavirus 2 in the coronavirus disease 2019-era[40]. Das et al[41] performed a genome-wide CRISPR screen and identified 39 candidate essential hepatovirus host factors, which form 4 clusters as follows: HAV IRES-mediated translation, chaperone activity, mitochondrial integrity and ganglioside synthesis. This strategy seems to result in the generation of more accurate approaches and techniques for HAV management.
HAV needs a HAV 3C protease to form its viral replication complex. X-ray structures were reported for HAV 3C protease with HAV 3C protease inhibitor N-benzyloxycarbonyl-l-serine-β-lactone (1a), resulting in a lead compound that was further developed to produce a potent inhibitor of HAV 3C protease through the alkylation of the sulfur atom at the active site Cys172[42]. Furthermore, soaking N-iodoacetyl-valine-phenylalanine-amide, which inhibited HAV 3C protease activity, into HAV 3C–1a crystals through the modification of His102 Nε2-alkylated protein could lead to the successful utilization of this new crystal form in the study of enzyme–inhibitor interactions in the proteolytic active site[42]. In general, antivirals are used after hepatitis virus infects the liver. It may be better to prevent infection rather than to treat HAV.
Koirala et al[43] also reported a 2.84-Å resolution crystal structure of HAV IRES domain V in complex with a synthetic antibody fragment - a crystallization chaperone. This is useful for drug repositioning to compare other picornaviral HAV structures with those of HAV.
AI and machine learning can contribute to drug development for viral infection by improving the speed and efficiency of repurposing and proposing new potent molecules to inhibit viral replication[40]. Both AI and machine learning can also be employed to make network-based predictions of drug-target interactions[44] or associations between gene expression and HAV infection[45]. This information is crucial to feed into AI and machine learning systems for the development of potent anti-HAV drugs. Although new drug discovery typically takes more than 10 years[46], this method may be useful for drug repositioning and drug rescue, which allows us to develop anti-HAV drugs more quickly. For example, the hepatitis C virus (HCV) NS5B polymerase inhibitor sofosbuvir and its derivatives could suppress HAV replication[47,48].
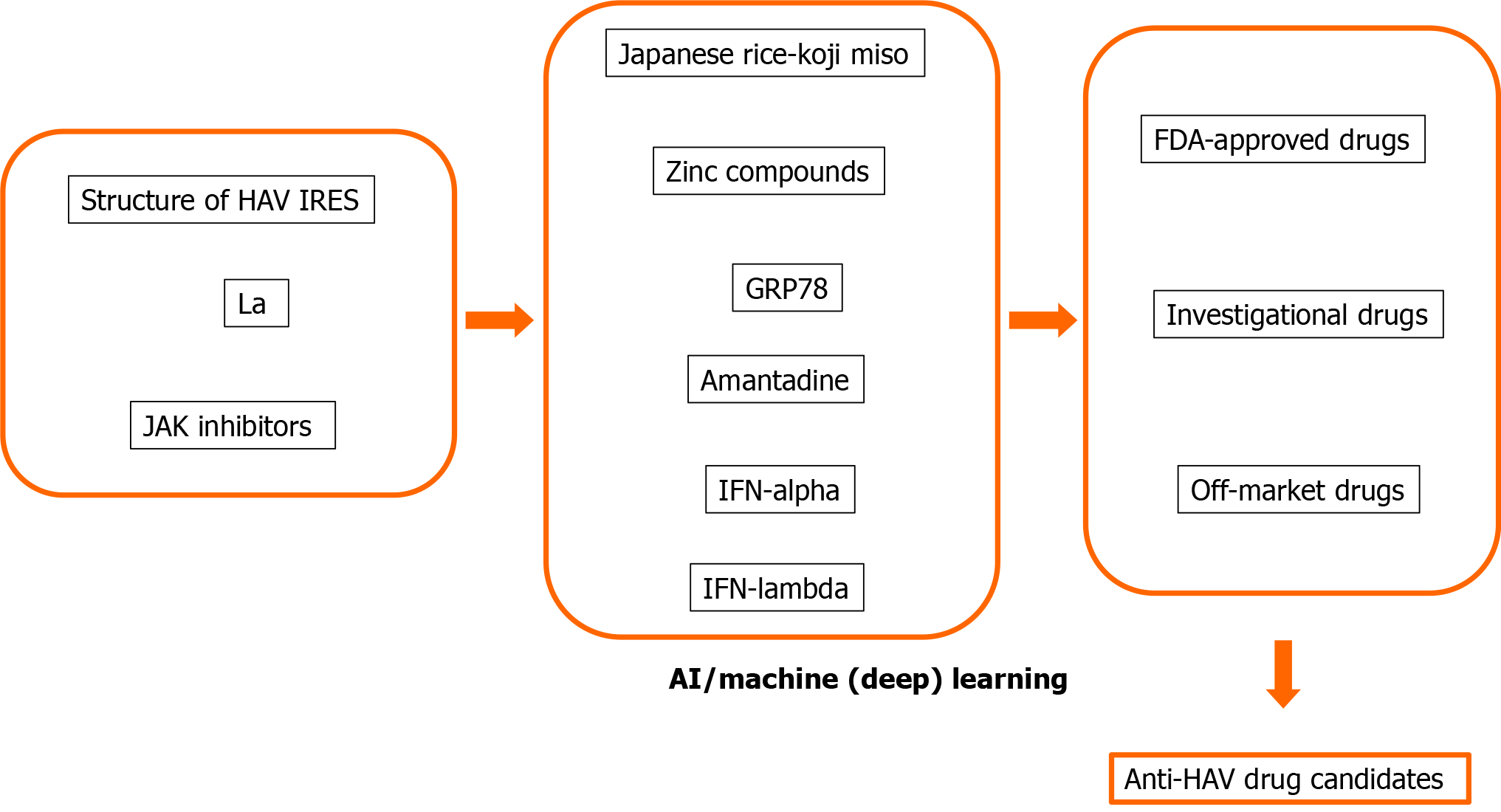
Many human proteins are involved in viral replication and pathogenesis[8,48]. The advantage of host-targeted antivirals is that the target is abundant. Another advantage is that they are less prone to resistance than those directly targeting the virus[8,49]. We and others also reported that host-targeted antivirals are useful for the suppression of HAV replication[8,34,35,50-53]. We would like to apply AI, machine learning and deep learning methods for drug repositioning and rescue to discover anti-HAV drug candidates (Figure 2). AI, machine learning and deep learning methods may also be useful for the avoidance of drug side effects.
Qureshi et al[54] developed virus-specific as well as general QSAR models and computed approximately 18000 chemical descriptors (1D, 2D and 3D), including geometric, constitutional, electrostatic, topological, hydrophobic and binary fingerprints, using PaDEL, an open-source software to calculate molecular descriptors and fingerprints[54]. They also employed SVMlight software (Freely available at http://svmlight.joachims.org) for machine learning. After attribute selection, there were 15 relevant descriptors for HBV. Arora et al[55] performed a QSAR study based on a series of anti-hepatitis B virus (HBV) agents, namely, a series of novel bis(Lamino acid) ester prodrugs of 9-[2-(phosphonomethoxy)ethyl]adenine, a similar series of compounds comprising 2-amino-6-arylthio-9-[2-(phosphonoethoxy)ethyl] purine bis(2,2,2-trifluoroethyl) esters, and a series of 1-isopropylsulfonyl-2-amine benzimidazoles. These systems may also be useful for the development of anti-HAV drugs.
Deep learning has been applied for the diagnosis and treatment of chronic hepatitis B. Compared with two-dimensional shared wave elastography and fibrosis biomarkers, deep learning radiomics of elastography is valuable and practical as a noninvasive accurate diagnosis of liver fibrosis in HBV-infected patients[56]. Analysis of the quasispecies pattern of HBV genomes by the combination of deep sequencing and machine learning is also useful for the prediction of hepatocellular carcinoma (HCC) and direct therapeutic strategies[57,58]. A valid systematic approach based on big data mining and genome-wide RNA-seq data may be imperative to further investigate the pathogenic mechanism and identify biomarkers for drug design[59].
Weidlich et al[60] developed SAR with advanced machine learning methods and performed in vitro antiviral assays, resulting in the identification of the candesartan cilexetil, which is used to treat hypertension, as an HCV NS5B inhibitor. Using a support vector machine (SVM), three classification models were built in HCV NS3 protease inhibitors[61] or HCV NS5B polymerase inhibitors[62]. Qin et al[63] reported that the combination of the best sub- and whole dataset SVM models can be used as reliable lead design tools for new NS3/4A protease inhibitors.
Wei et al[64] reported that the multiple QSAR method is useful in predicting chemical-protein interactions for the discovery of multitarget inhibitors for the treatment of HIV/HCV coinfection. This strategy may be useful for the treatment of the cooccurrence of HAV infection and chronic liver disease[65].
Combination information from yeast-based library screening, next-generation sequencing, and structure-based modeling in a supervised machine learning approach is useful for the comprehensive sequence-energetics-function mapping of the specificity landscape of the HCV NS3/4A protease, whose function-site-specific cleavages of the viral polyprotein are a key determinant of viral fitness[66]. Deep learning recurrent neural network models could be used to identify patients with HCV-related cirrhosis with a high risk of developing HCC for risk-based HCC outreach and surveillance strategies[67]. Deep learning should also be helpful for the development of antivirals.
We previously found that glucose-regulated protein 78 (GRP78) is an antiviral target for HAV (Table 1)[50-52]. Computational drug discovery using the structure of HAV and GRP78 may lead to the discovery of new anti-HAV drugs or drug repositioning and drug repurposing for anti-HAV drugs[68-71].
We found that several drugs potentially inhibit HAV IRES-dependent translation and HAV replication. Approaches that utilize AI, machine learning and deep learning methods could have the most promise in the discovery of new anti-HAV drugs. A systematic approach based on big data mining with AI is also useful for the development of anti-HAV drugs[71].
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Japan
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Raiter A, Ravaioli F S-Editor: Wang JL L-Editor: A P-Editor: Li JH
| 1. | Fujiwara K, Yokosuka O, Ehata T, Imazeki F, Saisho H, Miki M, Omata M. Frequent detection of hepatitis A viral RNA in serum during the early convalescent phase of acute hepatitis A. Hepatology. 1997;26:1634-1639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 2. | Navarro MED, Yao CC, Whiteley A, Movahedi B, Devuni D, Barry C, Zacharias I, Theodoropoulos NM, Bozorgzadeh A, Martins PN. Liver transplant evaluation for fulminant liver failure due to acute hepatitis A infection: Case series and literature review. Transpl Infect Dis. 2020: e13476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Agrawal A, Singh S, Kolhapure S, Hoet B, Arankalle V, Mitra M. Increasing Burden of Hepatitis A in Adolescents and Adults and the Need for Long-Term Protection: A Review from the Indian Subcontinent. Infect Dis Ther. 2019;8:483-497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 4. | Fukushima S, Kiyohara T, Ishii K, Nakano T, Hamada A. Immunogenicity of aluminum-adsorbed hepatitis A vaccine (Havrix®) administered as a third dose after primary doses of Japanese aluminum-free hepatitis A vaccine (Aimmugen®) for Japanese travelers to endemic countries. Vaccine. 2017;35:6412-6415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Yamamoto C, Ko K, Nagashima S, Harakawa T, Fujii T, Ohisa M, Katayama K, Takahashi K, Okamoto H, Tanaka J. Very low prevalence of anti-HAV in Japan: high potential for future outbreak. Sci Rep. 2019;9:1493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Vaughan G, Goncalves Rossi LM, Forbi JC, de Paula VS, Purdy MA, Xia G, Khudyakov YE. Hepatitis A virus: host interactions, molecular epidemiology and evolution. Infect Genet Evol. 2014;21:227-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 7. | Koibuchi T, Koga M, Kikuchi T, Horikomi T, Kawamura Y, Lim LA, Adachi E, Tsutsumi T, Yotsuyanagi H. Prevalence of Hepatitis A Immunity and Decision-tree Analysis Among Men Who Have Sex With Men and Are Living With Human Immunodeficiency Virus in Tokyo. Clin Infect Dis. 2020;71:473-479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 8. | Kassem AF, Batran RZ, Abbas EMH, Elseginy SA, Shaheen MNF, Elmahdy EM. New 4-phenylcoumarin derivatives as potent 3C protease inhibitors: Design, synthesis, anti-HAV effect and molecular modeling. Eur J Med Chem. 2019;168:447-460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 9. | Masuzaki R, Kanda T, Sasaki R, Matsumoto N, Nirei K, Ogawa M, Moriyama M. Application of artificial intelligence in hepatology: Minireview. Artif Intell Gastroenterol. 2020;1:5-11. [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Haga H, Sato H, Koseki A, Saito T, Okumoto K, Hoshikawa K, Katsumi T, Mizuno K, Nishina T, Ueno Y. A machine learning-based treatment prediction model using whole genome variants of hepatitis C virus. PLoS One. 2020;15:e0242028. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Hessler G, Baringhaus KH. Artificial Intelligence in Drug Design. Molecules. 2018;23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 12. | Zhong F, Xing J, Li X, Liu X, Fu Z, Xiong Z, Lu D, Wu X, Zhao J, Tan X, Li F, Luo X, Li Z, Chen K, Zheng M, Jiang H. Artificial intelligence in drug design. Sci China Life Sci. 2018;61:1191-1204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 13. | Batool M, Ahmad B, Choi S. A Structure-Based Drug Discovery Paradigm. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 234] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 14. | Guney E, Menche J, Vidal M, Barábasi AL. Network-based in silico drug efficacy screening. Nat Commun. 2016;7:10331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 278] [Cited by in F6Publishing: 297] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 15. | Glass MJ, Jia XY, Summers DF. Identification of the hepatitis A virus internal ribosome entry site: in vivo and in vitro analysis of bicistronic RNAs containing the HAV 5' noncoding region. Virology. 1993;193:842-852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Brown EA, Zajac AJ, Lemon SM. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5' nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J Virol. 1994;68:1066-1074. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 131] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 17. | Schultz DE, Honda M, Whetter LE, McKnight KL, Lemon SM. Mutations within the 5' nontranslated RNA of cell culture-adapted hepatitis A virus which enhance cap-independent translation in cultured African green monkey kidney cells. J Virol. 1996;70:1041-1049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 80] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 18. | Fujiwara K, Yokosuka O, Ehata T, Imazeki F, Saisho H. PCR-SSCP analysis of 5'-nontranslated region of hepatitis A viral RNA: comparison with clinicopathological features of hepatitis A. Dig Dis Sci. 2000;45:2422-2427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Fujiwara K, Yokosuka O, Fukai K, Imazeki F, Saisho H, Omata M. Analysis of full-length hepatitis A virus genome in sera from patients with fulminant and self-limited acute type A hepatitis. J Hepatol. 2001;35:112-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 77] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Fujiwara K, Yokosuka O, Ehata T, Saisho H, Saotome N, Suzuki K, Okita K, Kiyosawa K, Omata M. Association between severity of type A hepatitis and nucleotide variations in the 5' non-translated region of hepatitis A virus RNA: strains from fulminant hepatitis have fewer nucleotide substitutions. Gut. 2002;51:82-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Kanda T, Jeong SH, Imazeki F, Fujiwara K, Yokosuka O. Analysis of 5' nontranslated region of hepatitis A viral RNA genotype I from South Korea: comparison with disease severities. PLoS One. 2010;5:e15139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 22. | Wu S, Nakamoto S, Kanda T, Jiang X, Nakamura M, Miyamura T, Shirasawa H, Sugiura N, Takahashi-Nakaguchi A, Gonoi T, Yokosuka O. Ultra-deep sequencing analysis of the hepatitis A virus 5'-untranslated region among cases of the same outbreak from a single source. Int J Med Sci. 2014;11:60-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Kanda T, Zhang B, Kusov Y, Yokosuka O, Gauss-Müller V. Suppression of hepatitis A virus genome translation and replication by siRNAs targeting the internal ribosomal entry site. Biochem Biophys Res Commun. 2005;330:1217-1223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 486] [Cited by in F6Publishing: 522] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 25. | Hirai-Yuki A, Hensley L, Whitmire JK, Lemon SM. Biliary Secretion of Quasi-Enveloped Human Hepatitis A Virus. mBio. 2016;7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 26. | Debing Y, Neyts J, Thibaut HJ. Molecular biology and inhibitors of hepatitis A virus. Med Res Rev. 2014;34:895-917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 27. | Cordes S, Kusov Y, Heise T, Gauss-Müller V. La autoantigen suppresses IRES-dependent translation of the hepatitis A virus. Biochem Biophys Res Commun. 2008;368:1014-1019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 28. | Gosert R, Chang KH, Rijnbrand R, Yi M, Sangar DV, Lemon SM. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol Cell Biol. 2000;20:1583-1595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 109] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 29. | Schultz DE, Hardin CC, Lemon SM. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5'-nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134-14142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 109] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Graff J, Cha J, Blyn LB, Ehrenfeld E. Interaction of poly(rC) binding protein 2 with the 5' noncoding region of hepatitis A virus RNA and its effects on translation. J Virol. 1998;72:9668-9675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 71] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 31. | Zhang B, Morace G, Gauss-Müller V, Kusov Y. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Res. 2007;35:5975-5984. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Ali IK, McKendrick L, Morley SJ, Jackson RJ. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J Virol. 2001;75:7854-7863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Borman AM, Kean KM. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology. 1997;237:129-136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 92] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 34. | Jiang X, Kanda T, Wu S, Nakamoto S, Saito K, Shirasawa H, Kiyohara T, Ishii K, Wakita T, Okamoto H, Yokosuka O. Suppression of La antigen exerts potential antiviral effects against hepatitis A virus. PLoS One. 2014;9:e101993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 35. | Jiang X, Kanda T, Nakamoto S, Saito K, Nakamura M, Wu S, Haga Y, Sasaki R, Sakamoto N, Shirasawa H, Okamoto H, Yokosuka O. The JAK2 inhibitor AZD1480 inhibits hepatitis A virus replication in Huh7 cells. Biochem Biophys Res Commun. 2015;458:908-912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 36. | Kanda T, Sasaki R, Nakamoto S, Haga Y, Nakamura M, Shirasawa H, Okamoto H, Yokosuka O. The sirtuin inhibitor sirtinol inhibits hepatitis A virus (HAV) replication by inhibiting HAV internal ribosomal entry site activity. Biochem Biophys Res Commun. 2015;466:567-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 37. | Kanda T, Yokosuka O, Imazeki F, Fujiwara K, Nagao K, Saisho H. Amantadine inhibits hepatitis A virus internal ribosomal entry site-mediated translation in human hepatoma cells. Biochem Biophys Res Commun. 2005;331:621-629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Yang L, Kiyohara T, Kanda T, Imazeki F, Fujiwara K, Gauss-Müller V, Ishii K, Wakita T, Yokosuka O. Inhibitory effects on HAV IRES-mediated translation and replication by a combination of amantadine and interferon-alpha. Virol J. 2010;7:212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 39. | Kanda T, Wu S, Kiyohara T, Nakamoto S, Jiang X, Miyamura T, Imazeki F, Ishii K, Wakita T, Yokosuka O. Interleukin-29 suppresses hepatitis A and C viral internal ribosomal entry site-mediated translation. Viral Immunol. 2012;25:379-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 40. | Omolo CA, Soni N, Fasiku VO, Mackraj I, Govender T. Update on therapeutic approaches and emerging therapies for SARS-CoV-2 virus. Eur J Pharmacol. 2020;883:173348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 41. | Das A, Barrientos R, Shiota T, Madigan V, Misumi I, McKnight KL, Sun L, Li Z, Meganck RM, Li Y, Kaluzna E, Asokan A, Whitmire JK, Kapustina M, Zhang Q, Lemon SM. Gangliosides are essential endosomal receptors for quasi-enveloped and naked hepatitis A virus. Nat Microbiol. 2020;5:1069-1078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 42. | Yin J, Bergmann EM, Cherney MM, Lall MS, Jain RP, Vederas JC, James MN. Dual modes of modification of hepatitis A virus 3C protease by a serine-derived beta-lactone: selective crystallization and formation of a functional catalytic triad in the active site. J Mol Biol. 2005;354:854-871. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 43. | Koirala D, Shao Y, Koldobskaya Y, Fuller JR, Watkins AM, Shelke SA, Pilipenko EV, Das R, Rice PA, Piccirilli JA. A conserved RNA structural motif for organizing topology within picornaviral internal ribosome entry sites. Nat Commun. 2019;10:3629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | DiNunno NM, Goetschius DJ, Narayanan A, Majowicz SA, Moustafa I, Bator CM, Hafenstein SL, Jose J. Identification of a pocket factor that is critical to Zika virus assembly. Nat Commun. 2020;11:4953. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 45. | Sirota M, Dudley JT, Kim J, Chiang AP, Morgan AA, Sweet-Cordero A, Sage J, Butte AJ. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci Transl Med. 2011;3:96ra77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 556] [Cited by in F6Publishing: 540] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 46. | Gao K, Nguyen DD, Chen J, Wang R, Wei GW. Repositioning of 8565 Existing Drugs for COVID-19. J Phys Chem Lett. 2020;11:5373-5382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 47. | Jiang W, Muhammad F, Ma P, Liu X, Long G. Sofosbuvir inhibits hepatitis A virus replication in vitro assessed by a cell-based fluorescent reporter system. Antiviral Res. 2018;154:51-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 48. | Abu-Zaied MA, Hammad SF, Halaweish FT, Elgemeie GH. Sofosbuvir Thio-analogues: Synthesis and Antiviral Evaluation of the First Novel Pyridine- and Pyrimidine-Based Thioglycoside Phosphoramidates. ACS Omega. 2020;5:14645-14655. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Harrison C. Drug researchers pursue new lines of attack against COVID-19. Nat Biotechnol. 2020;38:659-662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 50. | Shubin AV, Demidyuk IV, Lunina NA, Komissarov AA, Roschina MP, Leonova OG, Kostrov SV. Protease 3C of hepatitis A virus induces vacuolization of lysosomal/endosomal organelles and caspase-independent cell death. BMC Cell Biol. 2015;16:4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 51. | Choi YS, Jung MK, Lee J, Choi SJ, Choi SH, Lee HW, Lee JJ, Kim HJ, Ahn SH, Lee DH, Kim W, Park SH, Huh JR, Kim HP, Park JY, Shin EC. Tumor Necrosis Factor-producing T-regulatory Cells Are Associated With Severe Liver Injury in Patients With Acute Hepatitis A. Gastroenterology. 2018;154:1047-1060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | Ogawa M, Kanda T, Suganami A, Nakamoto S, Win NN, Tamura Y, Nakamura M, Matsuoka S, Yokosuka O, Kato N, Ohara O, Okamoto H, Moriyama M, Shirasawa H. Antiviral activity of zinc sulfate against hepatitis A virus replication. Future Virol. 2019;14:399-406. [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 53. | Mo L, Zeng Z, Deng R, Li Z, Sun J, Hu N, Shi J, Hu Y. Hepatitis A virus-induced hsa-miR-146a-5p attenuates IFN-β signaling by targeting adaptor protein TRAF6. Arch Virol. 2021;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 54. | Qureshi A, Kaur G, Kumar M. AVCpred: an integrated web server for prediction and design of antiviral compounds. Chem Biol Drug Des. 2017;89:74-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 55. | Arora PK, Patil VM, Gupta SP. A QSAR study on some series of anti-hepatitis B virus (HBV) agents. Bioinformation. 2010;4:417-420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 56. | Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, Wu C, Liu C, Huang L, Jiang T, Meng F, Lu Y, Ai H, Xie XY, Yin LP, Liang P, Tian J, Zheng R. Deep learning Radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut. 2019;68:729-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 273] [Article Influence: 54.6] [Reference Citation Analysis (1)] |
| 57. | Mueller-Breckenridge AJ, Garcia-Alcalde F, Wildum S, Smits SL, de Man RA, van Campenhout MJH, Brouwer WP, Niu J, Young JAT, Najera I, Zhu L, Wu D, Racek T, Hundie GB, Lin Y, Boucher CA, van de Vijver D, Haagmans BL. Machine-learning based patient classification using Hepatitis B virus full-length genome quasispecies from Asian and European cohorts. Sci Rep. 2019;9:18892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 58. | Chen S, Zhang Z, Wang Y, Fang M, Zhou J, Li Y, Dai E, Feng Z, Wang H, Yang Z, Li Y, Huang X, Jia J, Li S, Huang C, Tong L, Xiao X, He Y, Duan Y, Zhu S, Gao C. Quasispecies pattern of hepatitis B virus predicts hepatocellular carcinoma using deep-sequencing and machine learning. J Infect Dis. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 59. | Chang S, Wang LH, Chen BS. Investigating Core Signaling Pathways of Hepatitis B Virus Pathogenesis for Biomarkers Identification and Drug Discovery via Systems Biology and Deep Learning Method. Biomedicines. 2020;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Weidlich IE, Filippov IV, Brown J, Kaushik-Basu N, Krishnan R, Nicklaus MC, Thorpe IF. Inhibitors for the hepatitis C virus RNA polymerase explored by SAR with advanced machine learning methods. Bioorg Med Chem. 2013;21:3127-3137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 61. | Wang M, Xuan S, Yan A, Yu C. Classification models of HCV NS3 protease inhibitors based on support vector machine (SVM). Comb Chem High Throughput Screen. 2015;18:24-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 62. | Wang M, Wang K, Yan A, Yu C. Classification of HCV NS5B polymerase inhibitors using support vector machine. Int J Mol Sci. 2012;13:4033-4047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 63. | Qin Z, Wang M, Yan A. QSAR studies of the bioactivity of hepatitis C virus (HCV) NS3/4A protease inhibitors by multiple linear regression (MLR) and support vector machine (SVM). Bioorg Med Chem Lett. 2017;27:2931-2938. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 64. | Wei Y, Li W, Du T, Hong Z, Lin J. Targeting HIV/HCV Coinfection Using a Machine Learning-Based Multiple Quantitative Structure-Activity Relationships (Multiple QSAR) Method. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 65. | Vento S, Garofano T, Renzini C, Cainelli F, Casali F, Ghironzi G, Ferraro T, Concia E. Fulminant hepatitis associated with hepatitis A virus superinfection in patients with chronic hepatitis C. N Engl J Med. 1998;338:286-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 535] [Cited by in F6Publishing: 439] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 66. | Pethe MA, Rubenstein AB, Khare SD. Data-driven supervised learning of a viral protease specificity landscape from deep sequencing and molecular simulations. Proc Natl Acad Sci USA. 2019;116:168-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 67. | Ioannou GN, Tang W, Beste LA, Tincopa MA, Su GL, Van T, Tapper EB, Singal AG, Zhu J, Waljee AK. Assessment of a Deep Learning Model to Predict Hepatocellular Carcinoma in Patients With Hepatitis C Cirrhosis. JAMA Netw Open. 2020;3:e2015626. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 68. | Wang L, Ma C, Wipf P, Liu H, Su W, Xie XQ. TargetHunter: an in silico target identification tool for predicting therapeutic potential of small organic molecules based on chemogenomic database. AAPS J. 2013;15:395-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 131] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 69. | Schuler J, Hudson ML, Schwartz D, Samudrala R. A Systematic Review of Computational Drug Discovery, Development, and Repurposing for Ebola Virus Disease Treatment. Molecules. 2017;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 70. | Zhou KY, Wang YX, Zhang S, Gachloo M, Kim JD, Luo Q, Cohen KB, Xia JB. GOF/LOF knowledge inference with tensor decomposition in support of high order link discovery for gene, mutation and disease. Math Biosci Eng. 2019;16:1376-1391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 71. | Im JH, Woo HT, Ha B, Jung J. Effectiveness of single-dose administration of inactivated hepatitis A virus vaccination in the Republic of Korea armed forces, 2013-2016. J Viral Hepat. 2020;27:537-539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |