INTRODUCTION
Spinal cord injury (SCI) is one of the most destructive diseases of all traumatic diseases patients may encounter[1], and it has always been a challenging clinical disease, which exerts a considerable socio-economic impact on the patient’s family and health care system. Despite the improvement in medical care and rehabilitation, the outcome of SCI tissue regeneration treatment is still inadequate[2]. At present, SCI cannot be cured, and its treatment is limited to reducing secondary complications, and maximizing residual function through rehabilitation[3]. It can lead to lower limb paralysis, paraplegia, quadriplegia and other lifelong disabilities[4]. In addition, respiratory complications are common in patients with SCI[5]. The sequelae of SCI includes denervation atrophy and paralysis, glucose intolerance, skin and wound rupture, and depression[6]. SCI occurs worldwide, with an annual incidence of 15 of 40 cases, which is caused by factors including vehicle accidents, community violence, recreational activities and workplace injuries[7]. At this stage, scientists interpret SCI from various perspectives, and have achieved some success. Among them, the loss of downward control after SCI and the constant stimulation of a single synaptic pathway cause the intrinsic sensory impulse from muscles and tendons to enter the spinal cord, which leads to the exaggerated activity of alpha-motor neurons and the increase in reflex response[8]. At the same time, the down-regulation of microglia microRNA-128 may promote the development of neuropathic pain after SCI by activating P38[9]. In vivo experiments showed that over-expressed microRNA-136-5p promoted the production of inflammatory factors and chemokines in SCI rats, and inhibited the expression of A20 protein, inflammatory cell infiltration and SCI[10]. In addition, naringenin significantly inhibited SCI-induced neutrophil activation through microRNA-223[11]. On the other hand, microRNA-155 has been proved to be a new therapeutic target of SCI, which can overcome both internal and external neuronal barriers to repair SCI[12]. These findings deepen our understanding of the pathogenesis of SCI and provide guidance for further research.
Although there is a series of studies on SCI, the overall effect of these results is still elusive. In order to comprehensively and deeply examine the repair mechanism of astrocytes and non-astrocytes in SCI, we carried out a systematic modular analysis and exploration. In summary, our work describes in detail the relationship between multifactor-mediated dysfunction modules and astrocytes and non-astrocytes in SCI, and identifies potential therapeutic targets and related biological processes, which may help to understand and treat SCI. It provides abundant candidate resources for future experimental validation and drug relocation, and provides theoretical guidance for future research on SCI biology.
MATERIALS AND METHODS
Data resources
The NCBI Gene Expression Omnibus (GEO) database[13] includes a broad classification of high-throughput experimental data, which includes single-channel and dual-channel microarray-based determination of gene abundance, and experimental data on genomic DNA and protein molecules. In addition, it includes data from non-array-based high-throughput functional genomics and proteomics techniques. First, we collected a set of gene expression profiles of astrocytes and non-astrocytes in SCI repair from the GEO, the number of which is GSE7609[14]. The data set includes 11 astrocytes and 11 non-astrocytes under SCI. We then screened non-coding RNA (ncRNA)-RNA (protein) interaction pairs with a score ≥ 0.5 from the RAID v2.0 database[15], including 431937 interaction pairs involving 5431 ncRNAs. The RAID v2.0 database recruited more than 5.27 million RNA-related interactions, including more than 4 million RNA-RNA interactions and more than 1.2 million RNA-protein interactions. In addition, all human transcription factor target data, involving 2492 transcription factors (TF) and 9396 interaction pairs, were downloaded and used in the TRRUST V2 database[16].
Differentially expressed genes
The differential expression analysis of gene expression profile data in this study was implemented by R language Limma package[17-19]. Firstly, background correct function was used for background correction and standardization. Secondly, the normal between the array function quantile normalization method was used to filter out the control probe and the low expression probe. Then, differentially expressed genes in datasets were identified based on lmFit and eBayes functions, and default parameters were used.
Co-expression analysis
In order to explore the drivers of SCI repair, we analyzed the gene expression profiles of astrocytes and non-astrocytes and obtained the differential gene expression profiles of SCI repair. In addition, in order to explore the co-expression of differentially expressed genes in the repair of SCI, we used weighted gene co-expression network analysis[20] to analyze the matrix of differentially expressed genes in the repair of SCI, and to find the co-expression gene module. Firstly, the weighted value of the correlation coefficient, i.e., the N power of gene correlation coefficient, was used to calculate the correlation coefficient (Pearson Coefficient) between any two genes. The connection between genes in the network obeys scale-free networks, which makes the algorithm more biologically meaningful. Then, a hierarchical clustering tree was constructed by the correlation coefficient between genes. Different branches of the clustering tree represent different gene modules, and different colors represent different modules. According to the regulatory power of genes in each dysfunction module, we excavated the key genes that lead to the dysfunction module, and considered them as the key genes for the repair of SCI.
Functional and pathway enrichment
Exploring the functions and signaling pathways of gene involvement is often beneficial in studying the molecular mechanisms of diseases, and the enrichment of genes in dysfunctional modules is an effective means to explore the potential mechanisms of SCI repair. Therefore, we used R language Cluster Profiler package[21] to analyze the enrichment of GO function (P value Cutoff = 0.01, Q value Cutoff = 0.01) and KEGG pathway (P value Cutoff = 0.05, Q value Cutoff = 0.2). Cluster Profiler is a software package by Bioconductor, which can be adopted for statistical analysis and visualization of functional clustering of gene sets or gene clusters. In addition, we used BinGO[22] application by Cytoscape to analyze the integrated module network.
TF and ncRNA of regulatory dysfunction module
Gene transcription and post-transcriptional regulation are often driven by non-coding genes (ncRNA) and TF. Therefore, we scientifically predicted and tested its role in the relevant modules of SCI repair. Axonal regulators are defined as regulators that have significant regulatory effects on modules during the repair of SCI, including ncRNA and TF. We required that there were more than two control links between each regulator and each module, and that the significance of the enriched targets in each module based on hypergeometric test calculation had a P value lower than 0.01.
RESULTS
To determine the expression disorders of astrocytes and non-astrocytes in the repair mechanism of SCI
Biologists have carried out many experiments and studies on the pathogenesis of SCI, and thus identified the potential pathogenic genes involved. However, the complex molecular linkages and overall effects of these genes remain unclear. In order to analyze the changes in the repair mechanism of astrocytes and non-astrocytes during SCI repair, we analyzed the differential expression of astrocytes and non-astrocytes based on microarray data. The integration results showed that 4937 differentially expressed genes were identified. We believe that these differentially expressed genes are related to the repair mechanism of SCI.
Functional modules for identifying genes related to SCI repair
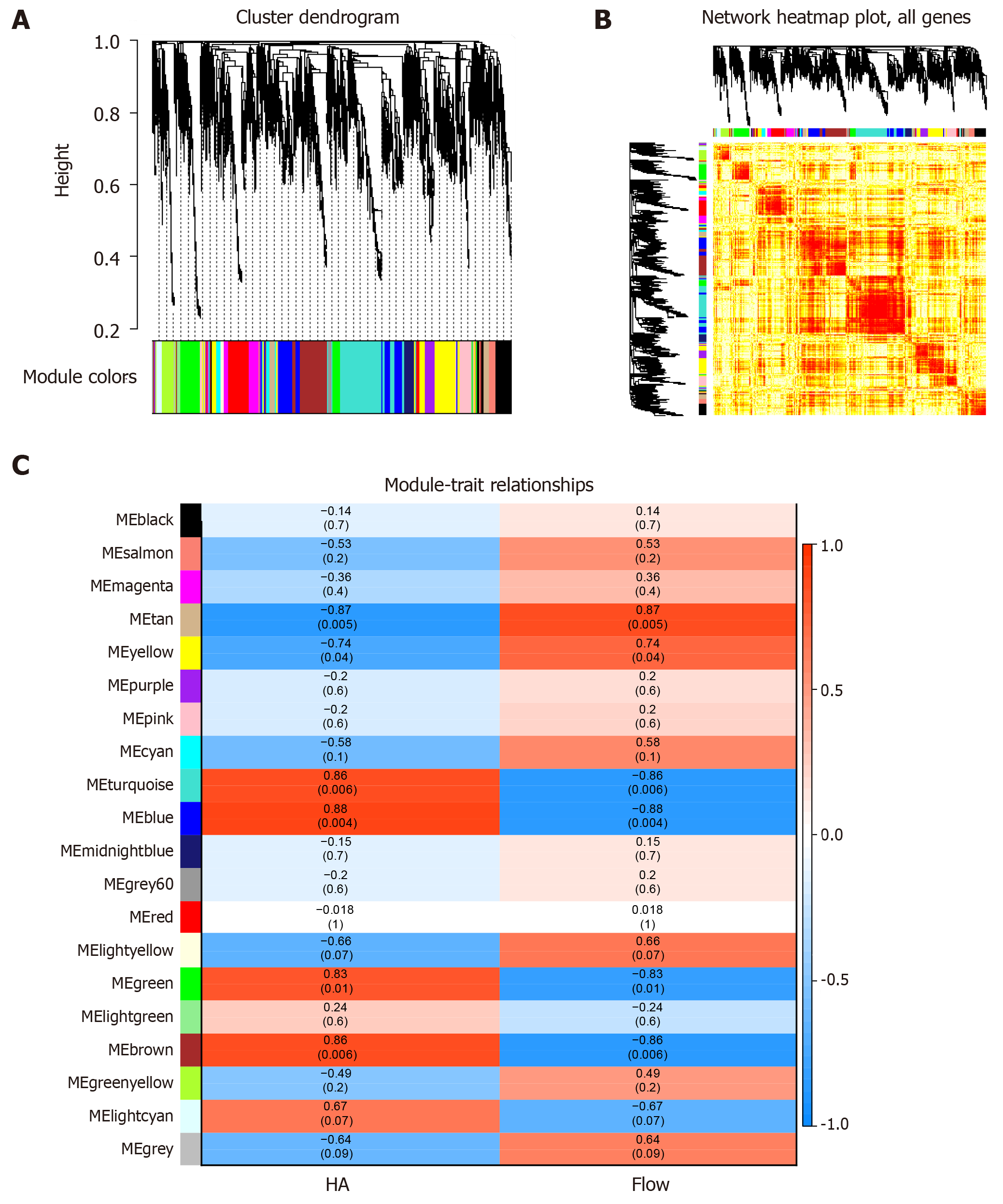
Firstly, the expression profile matrix was constructed based on 4937 differentially expressed genes and their interaction genes from SCI patients. Then, based on weighted gene co-expression network analysis, we observed significant grouping co-expression of these genes in disease samples. Modularization is a subsystem that deals with global complex systems and decomposes them into more detailed and orderly subsystems. Each subsystem has its own characteristics. For each element gene, a module is a set of genes with co-expression relationship, and the genes of the same module have the same expression behavior. On the other hand, each module also has certain interaction relationships. The overall effect of these interaction relationships represents the global characteristics and is the bridge for each element gene to play a role in the global network. Clustering the expression behavior of SCI in patients' samples into modules is helpful for us to observe the complex synergistic relationship between these genes from the perspective of expression behavior. Therefore, by identifying the co-expression group as a module, we obtained 19 functional impairment modules of SCI (Figure 1A, 1B). The key genes of each module were identified based on the functional impairment module, and the core genes with Sox13, Syt6 and so on were obtained. According to the correlation between module and phenotypic data, we can conclude that MEturquoise, MEblue and MEbrown are related to the repair mechanism of astrocytes in SCI, while MEtan is related to the repair mechanism of non-astrocytes in SCI (Figure 1C).
Figure 1 Synergistic expression of differentially expressed genes in patients with spinal cord injury.
A: Nineteen expression groups were identified as modules, and nineteen colors represented nineteen expression modules; B: The expression thermograms of all genes in the samples were clustered into 19 expression modules; C: Each row represents a module, and each column represents a phenotype. The color of each cell is mapped by the corresponding correlation coefficient. The values range from - 1 to 1, and the color changes from blue to white, then to red. The deeper the color, the stronger the correlation.
Functions and pathways of interested gene participation
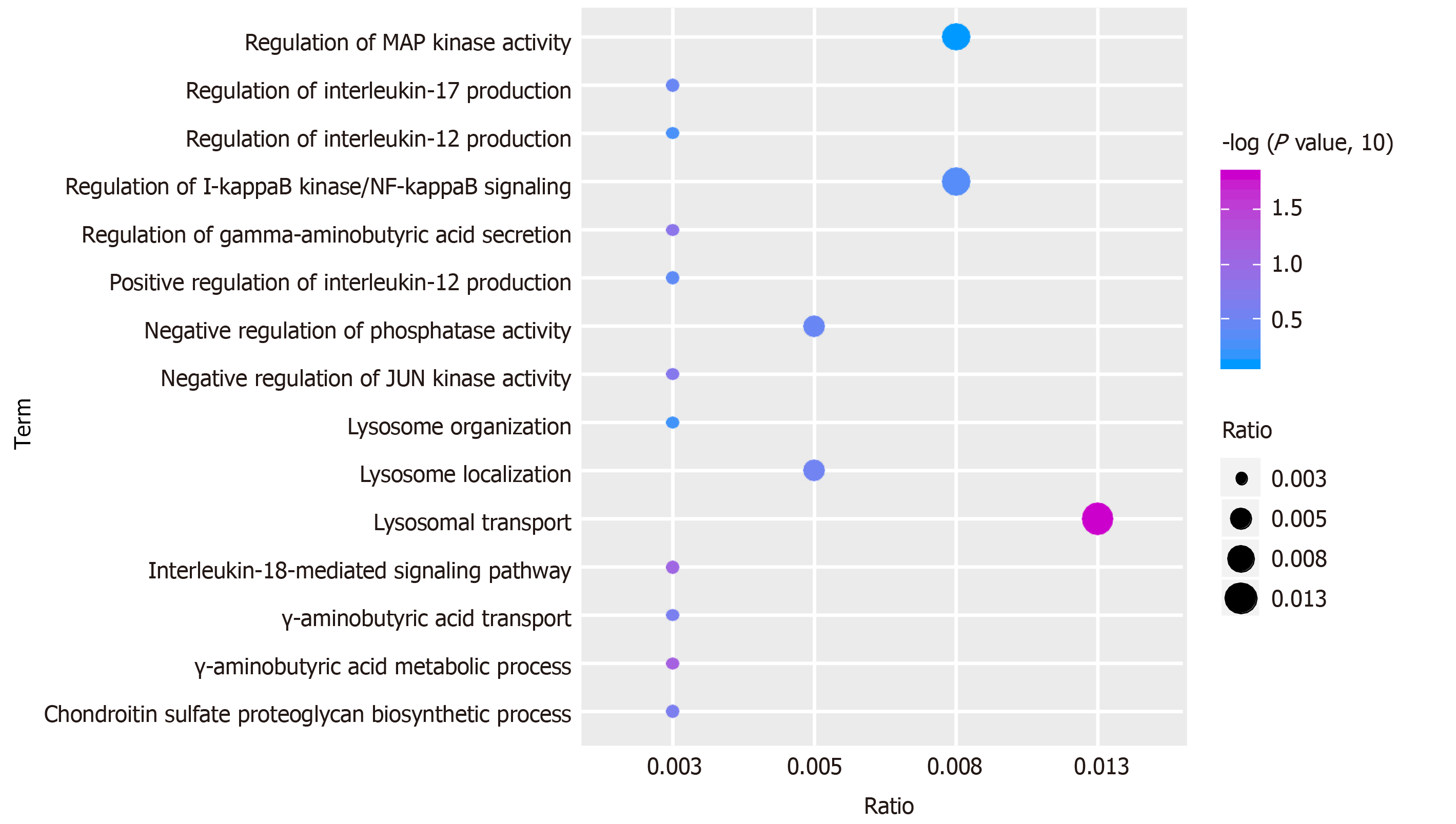
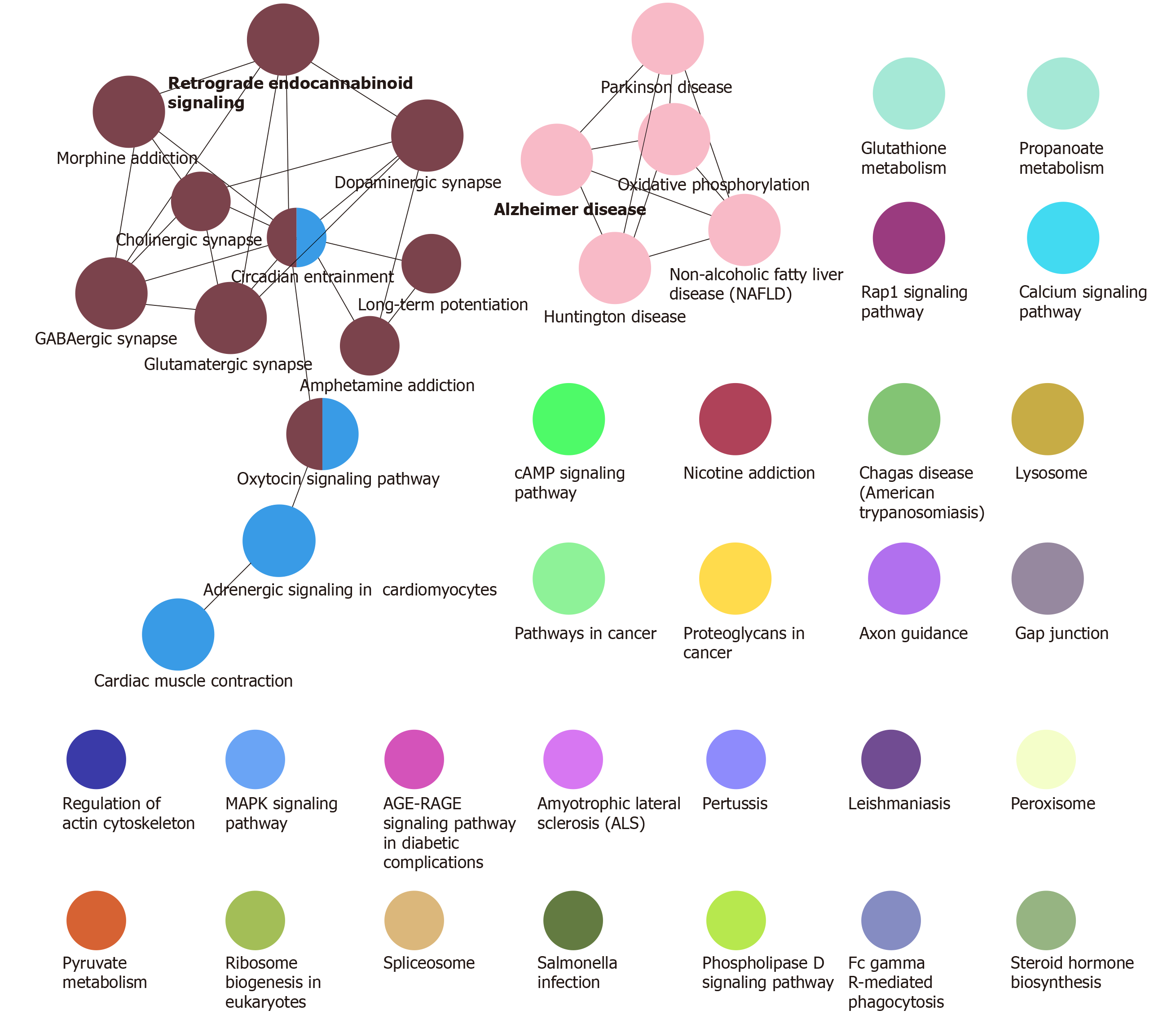
Function and pathway are important mediators of disease physiological response. Exploring the function and pathway of gene involvement in dysfunctional modules is not only helpful to determine the upstream and downstream relationship of the same pathway gene in the module, but also helpful to establish the molecular bridge between module and disease in system biology, and to deepen the understanding of the potential molecular mechanism of disease. We analyzed the enrichment of GO function and KEGG pathway in 19 modules, and obtained 59518 biological processes (Figure 2), 6988 cell components, 11522 molecular functions and 3532 KEGG pathways. It was found that these functions were mainly concentrated in reactive oxygen species, phosphoribose metabolism, regulation of T cell activation and T cell activation. On the other hand, the enrichment of KEGG pathway demonstrated that the differentially expressed genes of SCI were mainly involved in MAPK, PI3K-Akt, Ras and other signaling pathways. Looking back at the overall situation, we integrated 19 module networks and used BinGO for circuit analysis (Figure 3).
Figure 2 Functional and pathway enrichment analysis of modular genes.
GO functional enrichment analysis of module genes (excerpts). From blue to purple, the enrichment increased significantly. The larger the circle, the larger the proportion of module genes in GO functional entry genes.
Figure 3 Path analysis of the integrated module network.
TF and ncRNA driving modules related to SCI repair
From the perspective of systems biology and genetics, gene transcription and post-transcriptional regulation have been considered key regulators of disease occurrence and development, and TF and ncRNA are common regulators of expression and function. Although many biologists have paid attention to the regulation of single or several TF and ncRNA on the repair process of SCI, few studies have focused on their overall effects on dysfunctional mechanisms and their bridging role in their development. Therefore, in this study, we conducted pivot analysis of the common modules based on the targeting regulation relationship between TF and ncRNA to determine the key transcriptional regulators regulating the repair process of SCI. The predicted results showed that 173 ncRNAs involved 210 ncRNA-module regulatory pairs and 59 TF involved 66 TF-module target pairs. These results were introduced into Cytoscape to observe the regulation of regulatory factors in dysfunctional modules. In addition, the number of pivot regulatory modules was statistically analyzed, and the dysfunctional modules with the most regulation of ncRNA (miR-758-3p) and TF (Nfkb1, Sp1) were obtained. These TF and ncRNA may regulate the repair process of SCI by mediating dysfunctional modules. Therefore, we identified these potential regulators as dysfunctional molecules in the repair process of SCI. Finally, by constructing a comprehensive landscape of astrocytes and non-astrocytes in the repair mechanism of SCI, we found that the key gene Lrrtm2 of ncRNA RNA RNA RNA RNA micro344d-3p regulation module 3 and Rest of microRNA-302b-3p regulation module 19. Rest is a key gene as well as a TF.
DISCUSSION
SCI is a destructive acute nervous system disease, with loss of function and poor long-term prognosis, which is usually associated with loss of motor and sensory function and, sexual dysfunction[23,24]. At present, SCI is a major medical problem worldwide[25]. It has been found that astrocytes around lesions become reactive after traumatic injury of the central nervous system, including SCI, and usually undergo hypertrophy and elongation. These reactive astrocytes migrate to the heart and contribute to the tissue repair process[26]. In this study, we collected genes of astrocytes or non-astrocytes from SCI lesions in the NCBI Gene Expression Omnibus database. Based on their differentially expressed gene profiles, the functional modules of repair genes of SCI were analyzed in order to further understand the repair mechanism of astrocytes and non-astrocytes in SCI. At the module level, modules are significantly involved in reactive oxygen species, phosphoribose metabolism, T cell activation and T cell activation. In addition, they also participate in the signaling pathways of MAPK, PI3K-Akt, Ras, endocytosis and human T-cell leukemia virus 1 infection. Resveratrol is an antioxidant that has a protective effect in rat SCI by inhibiting the MAPK pathway. Some studies have also shown that through inhibition of EGFR/MAPK, microglia activation and related cytokines can be inhibited, and secondary injury related to neuroinflammation can be reduced, thus providing neuroprotection in SCI rats[27]. Fibroblast growth factor 10 derived from neurons and microglia/macrophages activates the signal transduction of fibroblast growth factor 2/PI3K/Akt and inhibits microglia/macrophages TLR4/NF-kappa B-dependent neuroinflammation to improve functional recovery after SCI[28]. There is evidence that activation of the PI3K/Akt/mTOR signaling pathway is associated with glial scar formation after SCI[29]. In addition, studies have shown that Ras/Raf/ERK1/2 signaling may be up-regulated in the injured spinal cord and participate in the recovery of SCI[30]. On the other hand, autophagy plays a key role in SCI, including traumatic SCI and ischemia-reperfusion SCI[31].
At the molecular level, 19 key genes such as Sox13 and Scrt2 were identified by co-expression analysis. These core genes are not only differentially expressed, but also play an important regulatory role in dysfunction modules. Sox13 is mainly expressed in neuroepithelial precursors, oligodendrocytes and astrocytes in developing mouse spinal cord. Sox13 is an important regulator of oligodendrocyte development[32]. Scratch 2 regulates neurogenesis and cell migration by antagonizing bHLH protein in the developing neocortex[33]. In addition, CD52 can regulate T cell activation through its intracellular signaling pathway or through the interaction between soluble CD52 and Siglec-10 expressed on T cells[34]. The effects of other key genes on the repair of SCI have not been found, but the results of our analysis show that its significant regulatory dysfunction module is the direction of further research in the future.
In addition, 173 ncRNAs were predicted to participate in the repair mechanism of astrocytes and non-astrocytes in SCI through mediation modules, and their abnormal expression in SCI was verified by differential analysis. According to the statistical analysis, we confirmed that microRNA-758-3p has significant effects on four dysfunctional modules and is the gene that regulates the most modules. However, miR-124-3p, miR-136-5p, miR-24-3p, miR-34c-5P and miR-449b regulate three modules. MiR-758-3p inhibits the proliferation, migration and invasion of hepatocellular carcinoma cells by targeting MDM2 and mTOR[35]. In addition, microRNA-758-3p, as a tumor suppressor, plays a key role in inhibiting the proliferation, migration and invasion of gastric cancer by targeting chromobox 5, suggesting its potential application in cancer therapy[36]. MiR-124-3p attenuates MPP-induced neuronal damage by targeting STAT3 in SH-SY5Y cells[37]. In addition, silencing of miR-136-5p significantly reduced the protein expression of miR-136-5p after overexpression, and improved inflammatory cell infiltration and SCI, which may be a new target for SCI treatment[38]. On the other hand, long non-coding RNA NEAT1 promotes glioma pathogenesis by regulating the miR-449b-5p/c-Met axis[39]. In addition to the fact that miR-136-5p is associated with the repair of SCI, no effect on SCI has been found in other ncRNA studies that significantly regulate dysfunction modules. However, our analysis showed that it affected the repair of SCI, which is one of the key research directions in the future. In addition, other ncRNAs that significantly regulate the dysfunction modules of SCI may also participate in the basic process of SCI repair, which can be used as candidates for further molecular experimental verification.
We then identified 59 TF differentially expressed in varying degrees and significantly regulated the repair dysfunction module of SCI. According to statistical analysis, Nfkb1 and Sp1 significantly regulated three modules, which may play an important role in the repair of SCI. Among them, the importance of Nfkb1 function can be seen in Nfkb1 mouse models with increased inflammation and susceptibility to certain forms of DNA damage, leading to cancer and rapid aging phenotype[40]. Also, NK cells activated by human IL-2 could not up-regulate the expression of NKp44, an activation marker, and showed decreased proliferation ability[41]. Some studies have shown that TF LEF1 and SP1 may play an important role in regulating cholesterol metabolism and injury response after SCI[42]. In addition, transcription factor SP1 has been shown to be up-regulated in SCI rats, and is predicted to be a potential transcription regulator of classical inflammatory response genes in rats[43]. Other TF that significantly regulate the dysfunction module of metastatic breast cancer may also participate in the basic process of asthma, which requires experimental confirmation. Finally, by constructing a comprehensive landscape of astrocytes and non-astrocytes in the repair mechanism of SCI, we found that ncRNA miR344d-3p regulated the key gene Lrrtm2 of module 3 and miR-302b-3p regulated the rest of module 19. The rest is both a transcription factor and a key gene. LRRTM2 interacts with PSD-95 and regulates the expression of the AMPA receptor. Lentivirus-mediated LRRTM2 knockdown in vivo decreases the intensity of the induced excitatory synaptic current[44]. IGF-1R is the direct target of microRNA-302b-3p. Overexpression of microRNA-302b-3p and silencing of IGF-1R decrease AKT phosphorylation[45]. In addition, down-regulation of N-acetylglucosaminotransferase GCNT3 by microRNA-302b-3p can reduce the proliferation, migration and invasion of non-small cell lung cancer (NSCLC)[46].
ARTICLE HIGHLIGHTS
Research background
Astrocytes are key regulators of inflammatory responses within the central nervous system following spinal cord injury (SCI).
Research motivation
The role and mechanism of astrocytes expression and its function in SCI are still unknown.
Research objectives
The aim of this study was to investigate the effect of astrocytes and non-astrocytes on SCI and to determine the mechanism of action in SCI repair.
Research methods
To identify the core genes that effected SCI at the molecular level using bioinformatics methods.
Research results
Among the four groups of differential results, miR-494, XIST and other core genes were found following further analysis.
Research conclusions
Our findings suggest a role for key genes and potential regulatory factors in regulating core dysfunction mechanisms following SCI.
Research perspectives
The key genes and potential regulatory factors play an important role in the occurrence and development of SCI. Significant genes in neurological tissue repair will be further discussed in future studies.