Published online Oct 6, 2018. doi: 10.12998/wjcc.v6.i11.426
Peer-review started: May 24, 2018
First decision: July 3, 2018
Revised: July 16, 2018
Accepted: July 31, 2018
Article in press: August 2, 2018
Published online: October 6, 2018
To determine the distribution of rotavirus VP7 gene in hospitalized children in Yunnan, China.
A total of 366 stool specimens were collected from hospitalized children in hospitals in Yunnan Province from September 2010 to December 2013. The genomic RNA electropherotypes and the G genotypes of the rotaviruses were determined. A phylogenetic analysis of the VP7 gene was performed. Rotavirus isolation was performed, and characterized by plaque, minimum essential medium, and all genes sequence analysis. Quantification of antibodies for inactivated vaccine prepared with ZTR-68 was examined by enzyme-linked immunosorbent assay and microneutralization assay.
Group A human rotavirus was detected in 177 of 366 (48.4%) stool samples using a colloidal gold device assay. The temporal distribution of rotavirus cases showed significant correlation with the mean air temperature. Rotaviruses were isolated from 13% of the rotavirus-positive samples. The predominant genotype was G1 (43.5%), followed by G3 (21.7%), G9 (17.4%), G2 (4.3%), G4 (8.7%), and mixed (4.3%) among a total of 23 rotavirus isolates. A rotavirus strain was isolated from a rotavirus-positive stool sample of a 4-month-old child in The First People’s Hospital of Zhaotong (2010) for use as a candidate human inactivated rotavirus vaccine strain and for further research, and was designated ZTR-68. The genotype of 11 gene segments of strain ZTR-68 (RVA/Human-wt/CHN/ZTR-68/2010/G1P[8]) was characterized. The genotype constellation of strain ZTR-68 was identified as G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. The VP7 and VP4 genotypes of strain ZTR-68 were similar to Wa-like strains.
CONCLUSIONS
A high prevalence of the G1, G2, and G3 genotypes was detected from 2010 to 2012. However, a dominant prevalence of the G9 genotype was identified as the cause of gastroenteritis in children in Yunnan, China, in 2013. A candidate human inactivated rotavirus vaccine strain, designated ZTR-68 was isolated, characterized, and showed immunogenicity. Our data will be useful for the future formulation and development of a vaccine in China.
Core tip: A high prevalence of the G1, G2, and G3 genotypes was detected from 2010 to 2012. However, a dominant prevalence of the G9 genotype was identified as the cause of gastroenteritis in children in Yunnan, China, in 2013. A candidate human inactivated rotavirus vaccine strain, designated ZTR-68 was isolated, characterized, and showed immunogenicity. Our data will be useful for the future formulation and development of a new inactivated rotavirus vaccine in China.
- Citation: Wu JY, Zhou Y, Zhang GM, Mu GF, Yi S, Yin N, Xie YP, Lin XC, Li HJ, Sun MS. Isolation and characterization of a new candidate human inactivated rotavirus vaccine strain from hospitalized children in Yunnan, China: 2010-2013. World J Clin Cases 2018; 6(11): 426-440
- URL: https://www.wjgnet.com/2307-8960/full/v6/i11/426.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v6.i11.426
Rotavirus is a major cause of diarrhea in children aged below five years, and causes approximately 600000 deaths in both developed and developing countries each year[1,2]. It is estimated that approximately 35000 children die per year in China from rotavirus, which is the second largest number of rotavirus deaths in the world[3,4]. Rotavirus is a non-enveloped RNA virus, and the viral genome contains 11 segments of dsRNA. The surface structural proteins VP7 and VP4 define the virus’ G and P genotypes, respectively[5]. Globally, human infections have been mainly caused by five G types, which are G1-G4 and G9[6,7]. Together, the genome codes for six structural proteins and five nonstructural proteins. Rotaviruses are now classified into G-genotypes based on the relatedness of the genes encoding VP7[8,9]. Molecular sequencing of rotaviruses has also led to the development of a classification system. In this system, each internal gene is assigned a particular genotype based on established nucleotide identity cut-off percentages. Now, the acronym Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx is used to classify the VP7-VP4-VP6-VP1-VP2-VP3-NSP1-NSP2-NSP3-NSP4-NSP5/6-encoding segments[10].
In China, 70% of children deaths caused by rotavirus occur in rural areas each year, mostly among children aged below five years[11]. Many changes in China, such as economic development, improved access to medical care, improved sanitation, and the one child family rule, may all contribute to this decline in overall diarrheal deaths and in the number of rotavirus deaths as well. In western China, urbanization is ongoing, and the economy is still poor and developing. City Zhaotong, County Xiangyun and XishuangBanna are relatively impoverished districts in Yunnan province, located in southwestern China. In this study, stool samples from hospitalized children admitted for diarrhea were collected to determine the distribution of rotavirus. All hospitalized children were under five years old. The prevalence of rotavirus infection peaks during the winter season, when the temperature is low. We wanted to isolate and characterize a human virus for use as a candidate human inactivated rotavirus vaccine strain and for further research. A rotavirus strain was isolated from a rotavirus-positive stool sample of a 4-month-old child in The First People’s Hospital of Zhaotong (2010), which was designated ZTR-68.
Collection and use of human stool specimens was approved by the Ethics Committee of the Institute of Medical Biology (YISHENGLUNZI [2016] 3), and was provided following children stool samples with written informed consent.
Stool samples were collected from hospitalized children in Yunnan, China. Group A rotavirus antigens were detected using an enzyme immunoassay (colloidal gold device assay, Rotavirus Group A Diagnostic assay, Colloidal gold device, Beijing Wantai Biological Pharmacy Enterprise Co., Ltd.) according to the manufacturer’s instructions. These samples were collected from September 2010 through December 2013.
An extract of child stool sample was agitated in 20% (wt/vol) PBS, pH 7.0 ± 0.2. The suspension sample was centrifuged for clarification at 8000 g for 20 min. The extract was treated with trypsin (10 μg/mL) for 60 min and inoculated onto a monolayer of MA104 cells for 90 min. After washing, the monolayer was maintained in serum-free minimum essential medium (MEM, Institute of Medical Biology, IMBCAMS, Kunming, China) supplemented with trypsin (1.5 μg/mL) and streptomycin (50 μg/mL) for 4 d. A viral lysate was treated with the freeze-thawing procedure three times and centrifuged at 7700 g for 30 min before infected in MA104 cells. The rotavirus strain ZTR-68 was isolated from a 4-month-old child hospitalized in The First People’s Hospital of Zhaotong, Yunnan Province, China. This patient had acute diarrhea associated with a positive result of fecal rotavirus antigen detected by Rotavirus Group A Diagnostic assay (Colloidal gold device, Beijing Wantai Biological Pharmacy Enterprise Co., Ltd.). The virus isolation procedure was performed by previously described methods[12,13].
Rotavirus dsRNA was extracted from stools and infected cell cultures by using the MiniBEST Viral RNA Extraction Kit (TaKaRa Biotechnology, Dalian, China) in accordance with the manufacturer’s instructions. Rotavirus dsRNA was analyzed by 10% polyacrylamide gel electrophoresis (PAGE) analysis. For the analysis, 15 μL of RNA was electrophoresed, and the gels were stained with silver nitrate and photographed.
The rotavirus dsRNA was used as a template for reverse transcription polymerase chain reaction (RT-PCR) using the Prime Script® One Step RT-PCR Kit (TaKaRa Biotechnology, Dalian, China) in accordance with the manufacturer’s instructions. The rotavirus VP7 general primers GeneralF/GeneralR designed in this study and previously described primers, Beg9/End9[14,15] (Table 1) were used. RT-PCR amplification was carried out in a 50 μL reaction volume containing PrimeScript 1 Step Enzyme Mix, 2 × 1 Step Buffer, 10 μmol/L primers, and template RNA (less than 1 μg). Thermocycling was performed for 30 min at 50 °C, 2 min denaturation step at 94 °C, and 30 cycles of 30 s denaturation step at 94 °C, 30 s annealing step at 55 °C-60 °C, and a 1 min extension step at 72 °C for each assay. The RT-PCR products were analyzed by electrophoresis on a 1% agarose gel (Invitrogen, Spain) in Tris-borate buffer containing ethidium bromide and visualized under ultraviolet light. The 2000 bp DNA ladder marker (Fermentase) was used as a size marker to estimate the lengths of the products. The PCR amplicons were purified using a column-based purification kit (OMEGA, the United States) and sequenced with an automated DNA sequencer (ABI 3730XL, the United States).
The VP7 nucleotide and amino acid sequences were analyzed. The selected sequences were aligned with ClustalX[16]. The phylogenetic tree (neighbor-joining, maximum parsimony, maximum likelihood) was constructed using the MEGA 4 from dissimilar distances and pairwise comparisons with the Kimura 2-parameter model[17].
The virus was plaque assessed by previously described methods with modifications[12]. Virus stocks were activated with trypsin (10 μg/mL) for 60 min at 37 °C. The virus was diluted in MEM and 300 μL/well were plated on 6-well plates (Corning, the United States) with a monolayer of MA104. Then the inoculums were covered with 4 mL per well of MEM with 3.5% agarose. The agar was natural solidification, and then the plates were incubated at 37 °C for 4 d. Visualized plaques could be read after adding 2% neutral red in 1 mL MEM.
RT-PCR was performed for amplification of all structural and nonstructural genes using the primers listed in the supplementary data. Briefly, the extracted RNA genome of rotavirus was denatured at 96 °C for 5 min, and RT-PCR was carried out by using a Prime Script® One Step RT-PCR Kit (TAKARA, China). This involved an initial reverse transcription step of 30 min at 50 °C, 2 min denaturation step at 94 °C, and 30 cycles of 30 s denaturation step at 94 °C, 30 s annealing step at 55 °C-60 °C and 1 min extension step at 72 °C for each assay. PCR products were electrophoresed in 2% agarose gels containing ethidium bromide and visualized under UV. PCR amplicons were purified by a column-based purification kit (OMEGA, the United States) and sequenced with an automated DNA sequencer (ABI 3730XL, Uthe United States). Phylogenetic tree (neighbor-joining, maximum parsimony, maximum likelihood) was constructed using the MEGA 4 from dissimilar distances and pairwise comparisons with 1000 bootstrap replicates and the Kimura 2-parameter model. The genotype of 11 gene segments of strain ZTR-68 (RVA/Human-wt/CHN/ZTR-68/2010/G1P[8]) was characterized. The genotype constellation of strain ZTR-68 was identified as G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1.
The morphology of the rotavirus strain ZTR-68 stained by phosphotungstic acid negative staining was checked by electron microscopy (HITACHI H-600, Japan) to ensure that the virus contained triple-layered particles (TLP).
Rotavirus strain ZTR-68 was cultivated in MA104 cells and the virus was purified by isopycnic CsCl gradient centrifugation as described previously[18], Triple-layered particles and double-layered particles were collected separately. Both purified viruses were dialyzed in 10 mmol/L PBS pH 7.0 to remove CsCl. TLPs were inactivated with formalin at 400 μg/mL concentration at 37 °C for 120 h. The inactivated vaccine was stored at 4 °C before use. The vaccine was quantified for proteins by Lowry method[19] and rotavirus antigen was quantified by enzyme-linked immunosorbent assay (ELISA). Balb/c mice (18-22 g) were purchased from Institute of Medical Biology, CAMS (Kunming, China). Mice were separated in groups of 20, and vaccinated with 0 μg, 10 μg or 20 μg inactivated rotavirus vaccine (IRV) formulated with alum by injection into the abdominal cavity. Pre-bleed was taken before injection. Serums were collected 2 wk after injection. All experiments were approved by the Institutional Animal Care and Use Committee of Institute of Medical Biology, CAMS (Kunming, China) and conducted in accordance with the ethical guidelines for animal experiments and safety guidelines.
MA104 cell line was maintained in MEM with 10% fetal calf serum (GIBCO, the United States) in 6-well microtiter plates (Corning, the United States). The cell monolayers were incubated at 37 °C in an incubator containing 50 mL/L CO2 under humid conditions, and infected with different viruses (strain Wa, SA11, S2, ZTR-68) with virus titer in 104-105 PFU/mL, and incubated for 16 h. The plates were fixed in chilled acetone (-20 °C) for 15 min and air dried, then washed with 0.01 mol/L PBS buffer and blocked with 3% BSA for 1 h at 37 °C. Mouse anti rotavirus strain ZTR-68 20 μg inactivated vaccine antibody (1:500 dilution, 0.5 mL volume) was added to microtitre plate wells and incubated at 37 °C for 1 h. After washing with 0.01 mol/L PBS, goat anti mouse FITC conjugate (MILLIPORE, 1:1000) was added and incubated for 1 h. The slides were examined under fluorescent microscope (Nikon, Japan) indicative of rotavirus.
RV-specific IgG antibodies were detected in sera on days 0, 14, and 28. Goat anti rotavirus antibodies (Millipore, the United States) coated 96-well ELISA plates overnight at 4 °C. The plates were washed with PBS pH 7.0 and blocked with 5% BSA in PBS. The plates were washed and incubated with supernatant of rotavirus Wa infected MA104 cells (~ 105 PFU/well) for 1 h at 37 °C. Serially diluted mouse serum was added in each well and incubated for 1 h at 37 °C, , then washed. HRP-conjugated rabbit anti mouse antibody (Millipore, the United States, 1:1000) was added. TMB (Tiangen, China) substrate was added for detection and was stopped with 2mol/L H2SO4. OD value was determined with an EIA reader (BioTek, the United States). Cutoff value was twofold of negative well value, and the antibody titer was defined as the highest dilution OD greater than the cutoff value. Geometric mean titer was given. Microneutralization assay was performed to measure rotavirus neutralizing activity as described previously[20]. Serial diluted mouse serum was added to 96-well plates and incubated with 1000 PFU of Wa per well for 1 h at 37 °C. MA104 cells were prepared in 96-well plates as a monolayer and cultivated in MEM with 5 μg/mL trypsin (GIBCO, the United States) at 37 °C for 3 d. Then all 96-well plates of neutralization were transferred into 96-well plates MA104 cells grown in a monolayer and incubated at 37 °C for another 7 d. The plates were lysed by freeze/thaw three times. RV antigen was detected by ELISA method with HRP-labeled goat anti rotavirus antibody (Institute of Medical Biology, CAMS, Kunming, China). Cutoff value was twofold of cells well without virus value, neutralizing antibody titer was defined as the highest dilution that gave a lower than antigen cutoff value. Virus-only controls were performed to confirm the effectiveness of ELISA system. Graph analysis was performed using GraphPad Prism 5.02, and statistical analysis was performed using Excel 2016 with the T test (two tailed). P values lower than 0.05 were considered statistically significant.
The sequences were deposited in GenBank under the following accession numbers: KM247264-KM247286. ZTR-68 genomic sequences: JX509930-JX509940.
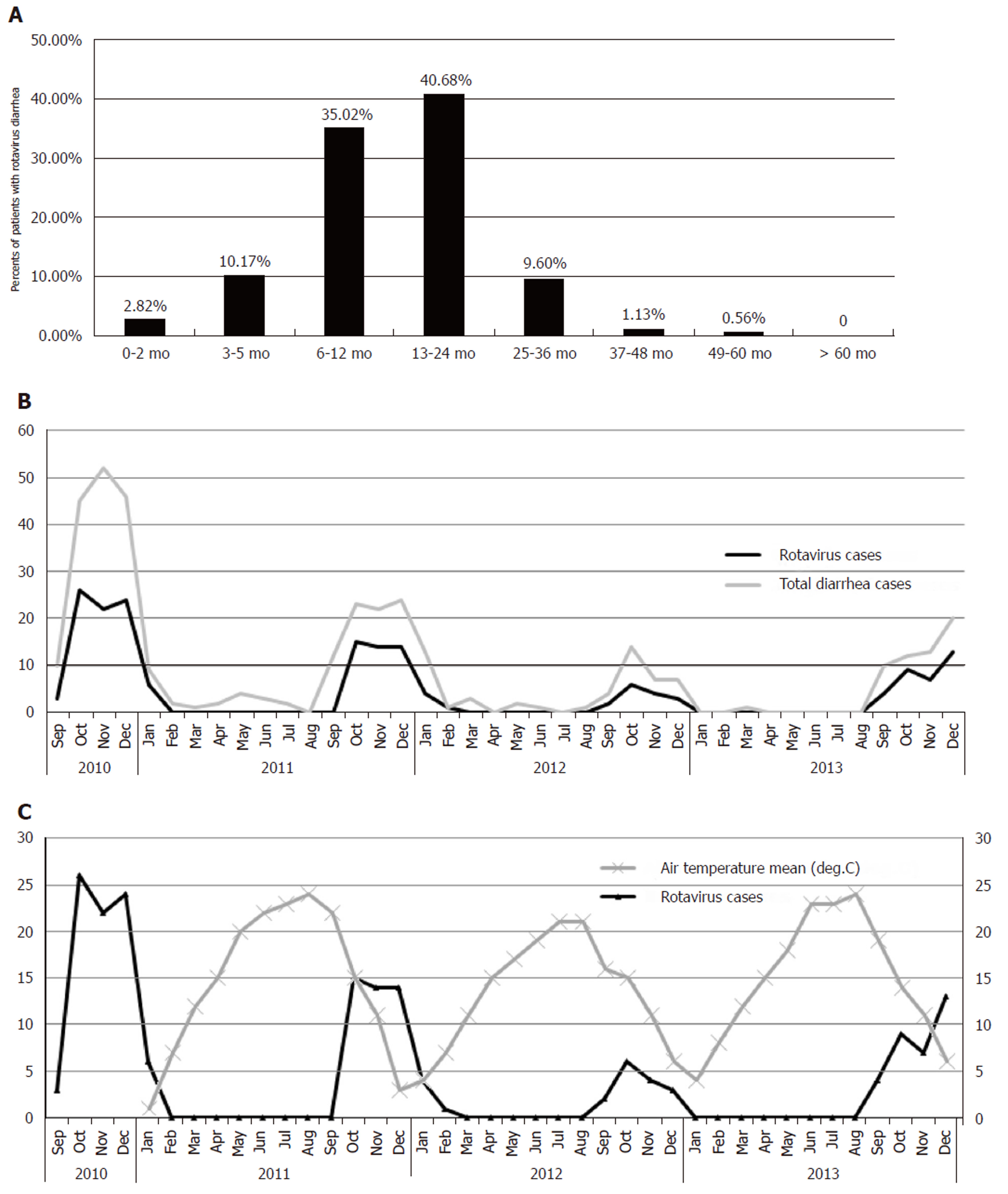
Group A human rotavirus was detected in 177 of 366 (48.4%) stool samples using the colloidal gold device assay (Table 2) and in 133 of 177 (75.14%) by dsRNA-PAGE between September 2010 and December 2013 (Figure 1A). On an average, the prevalence of rotavirus during each year was 48.4% and 51.2% during the peak season. The age of the 177 patients ranged from one and a half months to four and a half years old (Figure 2A), although the majority were less than 2 years old (n = 157, 88.69%). The temporal distribution of rotavirus cases showed a significant correlation with the mean air temperature. The number of rotavirus cases peaked during the fall and winter seasons when the temperature began to decrease (Figure 2B and 2C).
| During whole year | During September to February | |||
| Samples tested | Rotavirus positive | Samples tested | Rotavirus positive | |
| September 2010 to August 2011 | 176 | 81 (46.0) | 164 | 81 (49.4) |
| September 2011 to August 2012 | 102 | 48 (47.1) | 95 | 48 (50.5) |
| September 2012 to December 2013 | 88 | 48 (54.5) | 87 | 48 (55.2) |
| Total | 366 | 177 (48.4) | 346 | 177 (51.2) |
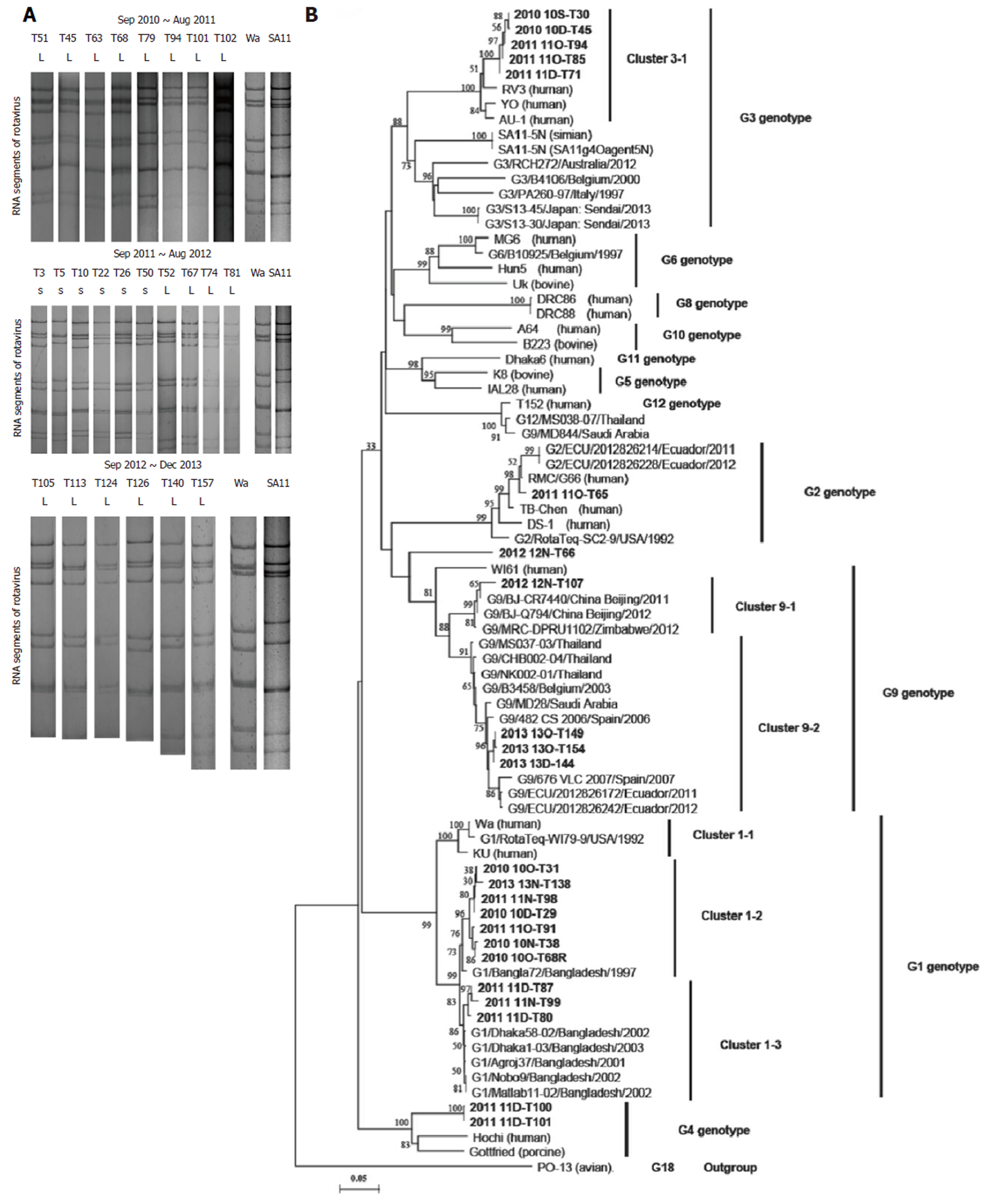
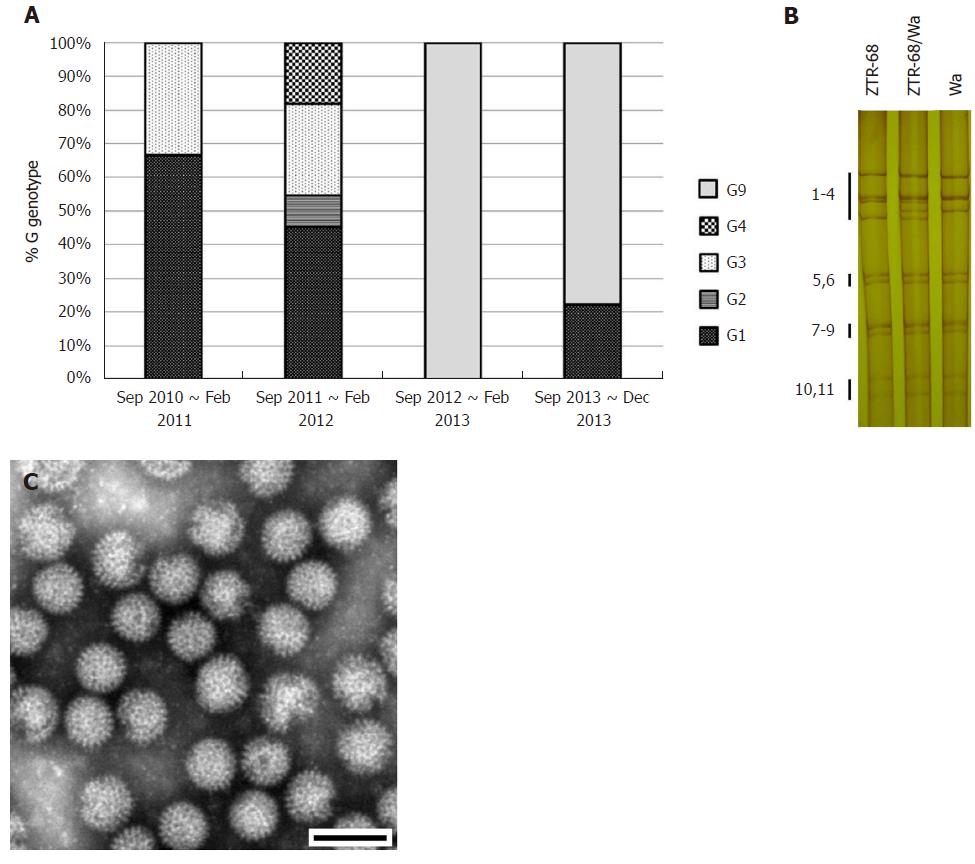
All 11 segments were visible in 133 samples, corresponding to 75.14% of the 177 rotavirus-positive samples and 36.33% of the total 366 stool samples. In total, 97 samples presented a long electropherotype, 36 presented a short electropherotype and none of the samples had a mixed pattern. The electropherotypes of the samples obtained from September 2011 to August 2012 presented a relatively higher level of diversity than the samples obtained from September 2010 to August 2011 and from September 2012 to December 2013. There was relative concordance between the electropherotypes and the distribution of G genotypes (Figure 1A and 3A).
We determined the VP7 nucleotide sequences for G1, G2, G3, G4, and G9 for the G genotype isolates obtained during the study period (Figure 1B, 3A). It was determined that phylogenetically, the G3 isolates clustered in one lineage, the G1 isolates clustered in two lineages, and the G2 and G4 isolates clustered in one lineage. Several of the G1 isolates maintained a similarity of 97.4% to 99.9% for 2010, 2011, and 2013 in cluster 1-2, and the others maintained a similarity of 98.5% to 99.2% for 2011 in cluster 1-3. The G3 isolates analyzed during this study showed 99.8% to 99.9% similarity for 2010 and 2011, and they composed one cluster with the strains RV3, YO, and AU-1. In the case of G9, one sequence in 2012 (12N-T107) was related to the rotavirus strains BJ-CR7440, BJ-Q794, and MRC-DPRU1102, which originated in China and Zimbabwe. Three sequences in 2013 showed high similarity (99.9%) with each other and were related to strains that originated in Spain (98.2%), Ecuador (97.9%-98.1%), Saudi Arabia (97.5%-97.9%), Belgium (98.6%-98.9%) and Thailand (98.3%-98.9%). One G2 isolate 11O-T65 was related to strains RMC/G66, TB-Chen, and DS-1, which originated in Ecuador. One mixed genotype isolate (12N-T66) was found, which contained a mixed G2 and G9 genotype and was phylogenetically located between the G2 cluster and the G9 cluster. In the case of G4, two 2011 sequences (11D-T100, 11D-T101) were related to the Hochi (85.5%) and Gottfried (86.5%-86.6%) strains. In the case of G1, seven isolates analyzed in one cluster, closed to Wa and KU cluster. One G1 isolate 10O-T68R was related to strains Wa and KU but not in one cluster. This isolate then cultured in MA104 cells and purified by plaque (Figure S3), and was designated as strain ZTR-68.
Positive rotavirus antigen was diagnosed in the diarrheal stool of a 4-month-old child who was infected naturally with rotavirus and developed severe, acute diarrhea sustained for four days and was hospitalized in The First People’s Hospital of Zhaotong, Yunnan Province, China. PAGE analysis of the rotavirus dsRNA segments from stool specimen revealed a typical 4-2-3-2 pattern of rotavirus, specific to the group A rotaviruses. (Figure 3B) The presence of virus-like particles, almost 70 nm in diameter was confirmed in this specimen by electron microscopy. (Figure 3C, Figure S2) The rotaviruses from isolate extract 10O-T68R were adapted to infect MA104 cells and purified by plague technique. ZTR-68 showed a typical CPE of rotavirus and virus titers grew to over 106.3 PFU/mL.
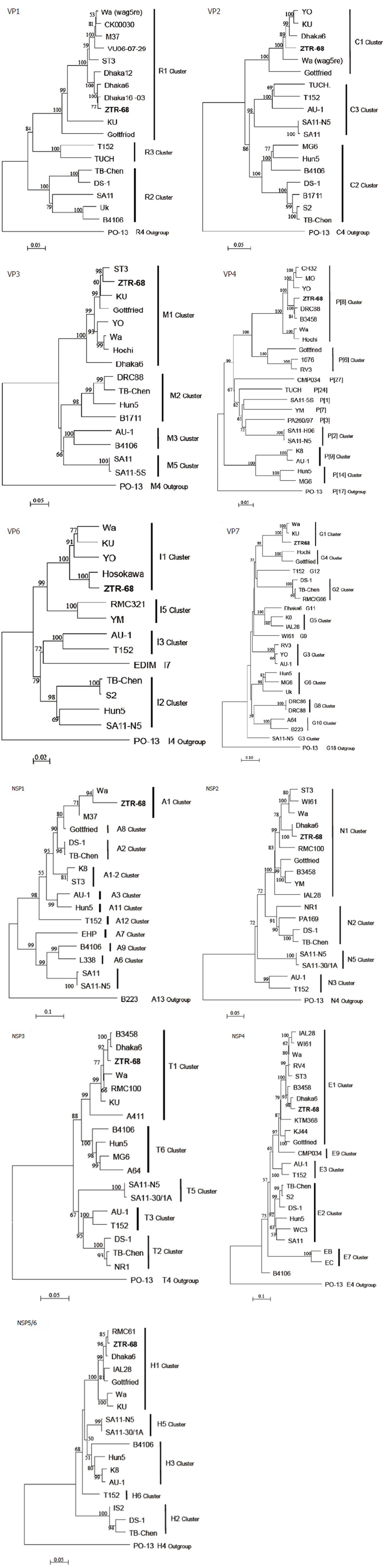
The ORF nucleotide sequences for the 11 genome segments of cell culture-adapted ZTR-68 strain were determined. The sequences deduced in this study are either identical or show a few changes from those that are already in GenBank for ZTR-68. Genotypes were assigned for each genome segment based on the nucleotide percent identity cut-off values defined by the RCWG and by submission to RotaC. Our analysis shows that ZTR-68 can be classified as a G1P[8] strain. Specifically, the gene segment 1 (VP1) of ZTR-68 showed 90.7%-99.2% nucleotide identity with the R1 genotype cluster. The sequence identity was found to be 98.6%-99.2%with strains Dhaka16-03, Dhaka6, and Dhaka12 of genotype R1 lineage. Percent nucleotide sequence identities with other genotypes were in the range of 77.6%-85.8%. Sequence analysis of the VP2 gene of rotavirus strain ZTR-68 showed genetic relatedness with the strains of the C1 genotype with 86.7%-95.2% nucleotide identity. The strain clustered with strains Wa, Dhaka6, YO, KU, and Gottfried. Percent nucleotide sequence identities with other genotypes (C2-C3) were in the range of 75.8%-77.5% (< the cut off value of 84%). Nucleotide sequence analysis of the VP3 gene of rotavirus strain ZTR-68 showed genetic relatedness with the strains of M1 genotype with 86.0%-95.2% nucleotide identity. The sequence identity was found to be 95.2%, 91.9%, and 86% with strains ST3, Wa, and Dhaka6, respectively. Percent nucleotide sequence identities with other genotypes (M2-M5) were in the range of 69.6%-71.6% (< the cut off value 81%). Sequence analysis of the VP4 gene of rotavirus strain ZTR-68 showed genetic relatedness with the strains of the P[8] genotype with 88.1%-97.3% nucleotide identity. The sequence identity was found to be 91.9%-97.1% with strains DRC88, B3458, MO, YO, and CH32 and 88.1%-89.1% with Wa and Hochi. Phylogenetically the strain ZTR-68 clustered with the strains of I1 genotype of the VP6 gene indicating 91.8%-97.7% nucleotide sequence identity and identified to be closer to strain Hosokawa. The nucleotide identity with other genotypes (I2-I7) ranged from 80.2% to 85.2%. Sequence analysis of the VP7 gene of rotavirus strain ZTR-68 showed genetic relatedness with the strains of G1 genotype with 91%-91.5% nucleotide identity. The sequence identity was found to be 91% and 91.5% with strains Wa and KU of genotype G1 lineage, respectively. Percent nucleotide sequence identities with other genotypes (G2-G10) were in the range of 53.7%-66.7% (< the cut off value 80%) (Figure 4).
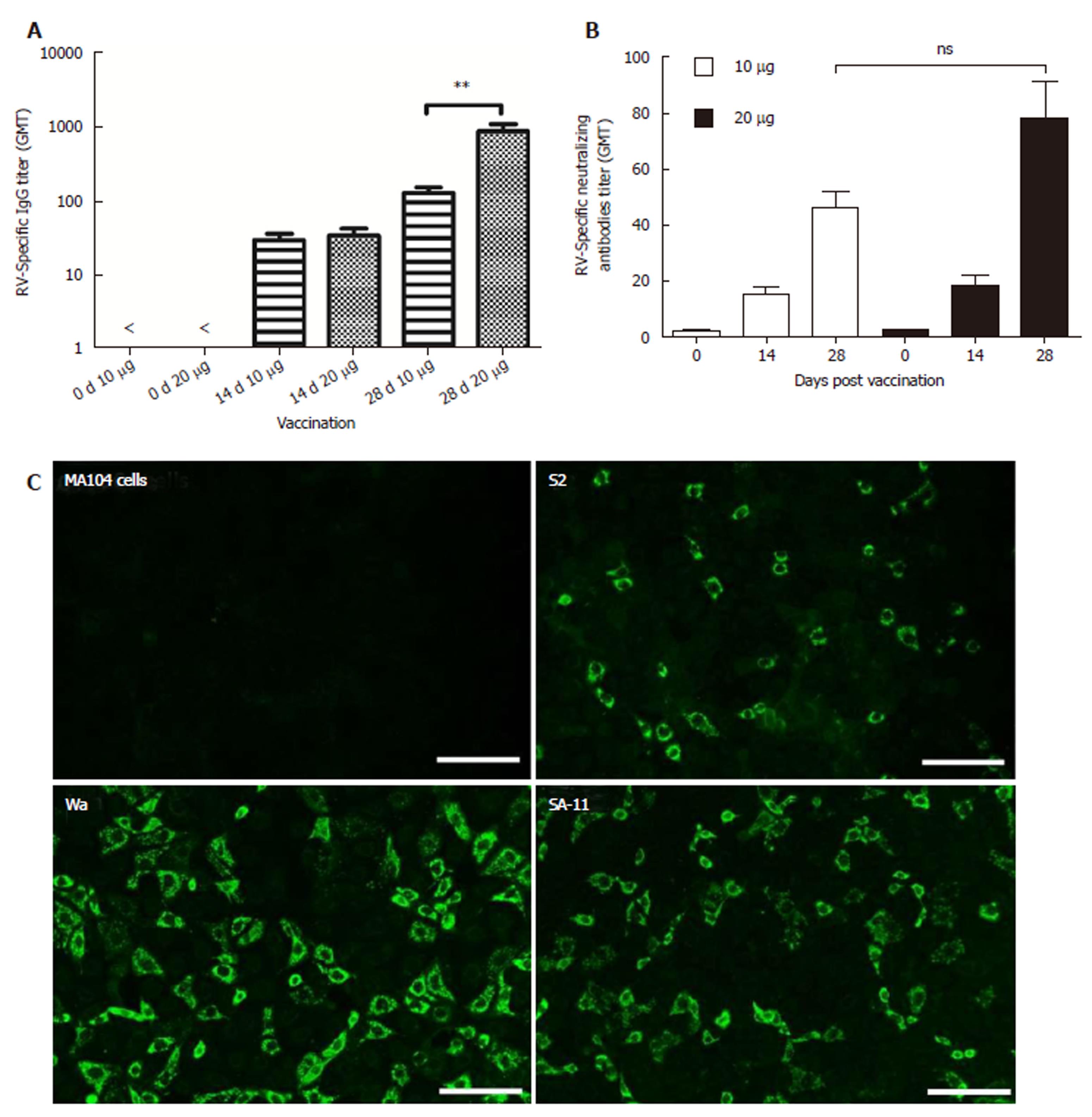
Immunogenicity of inactivated rotavirus vaccine prepared with ZTR-68 was examined by IgG ELISA and neutralization assay (Figure 5A and 5B). RV-specific IgG and neutralizing antibodies were detected after one dose vaccination, and its levels increased after two vaccinations. Ten micrograms of IRV induced lower levels of total IgG antibodies and neutralizing antibodies compared to 20 μg IRV. Both 10 μg and 20 μg of IRV induced increased IgG and neutralizing antibody levels. After the first vaccination (at 0 d), differences between 10 μg and 20 μg IRV were not statistically significant both in the IgG titer and neutralizing antibodies titer. In addition, a significant difference (P = 0.0043) was found in the IgG titer between 10 μg and 20 μg at 14 d and 28 d, but there was no significant difference (P = 0.0565) in the neutralizing antibodies titer. IRV ZTR-68 also could induce increased cross-typing anti-rotavirus antibodies anti-rotavirus strain Wa (G1P[8]), S2(G2P[4]), and SA11(G3P[2]) by the fluorescent antibody test (Figure 5C).
Rotavirus is recognized as a major public health problem for children in China[3,21-24]. Diarrhea remains the major common cause of Chinese children hospitalizations[11,25-28]. In Chinese rural areas, rotavirus caused about 33% of hospitalizations for severe pediatric gastroenteritis. The numbers of outpatient visits and all diarrhea episodes in the community are 28% and 7%, respectively[3,12,13,27,29,30]. The percentage of diarrhea patients hospitalized for rotavirus is consistent with findings in other Asian countries. In a 2004 study of rotavirus surveillance throughout Asia, 44%-53% of children hospitalized for gastroenteritis in Myanmar, Thailand, Indonesia, and Vietnam were detected as rotavirus-positive[31,32]. Our study demonstrated that the G1, G2, and G3 genotypes had a high prevalence from 2010 to 2011. However, the G9 genotype was the predominant cause of gastroenteritis in children in Yunnan, China, in 2012 and 2013. The most prevalent rotavirus G genotype shifted from G1–G4 and G9 to G1 + G9. Although the reason for this trend is unclear, the emergence of the G9 rotavirus has been documented in many studies over the last decade[33-35]. Previous studies have reported G3 as the most common strain in China[3,28], however, in this study, we found that G1 was predominant (43.3%), followed by G3 (21.7%), and G9 (17.4%). Notably, from 2010 to 2011, G1 + G3 were detected, while there was no G9 detected. However, from 2012 to 2013, G1 + G9 were predominant, and no G3 was detected. In this study, we observed an increase in the incidence of G9 rotavirus. In addition, G9P[4] rotavirus strains have been described as emerging in several Latin American countries[33,35]. The results of the phylogenetic sequence analyses demonstrating that some of the G9 rotavirus sequences detected in this study were closely related to sequences that originated in Ecuador may support this view. Some G9 rotavirus isolates were obtained during the surveillance, and full genomic sequencing and characterization will be performed in future studies.
Rotavirus diarrhea presents a serious health burden in China. A previous study suggested that every child in China experiences at least one episode of diarrhea due to rotavirus[3]. The use of rotavirus vaccines should be part of a comprehensive strategy to control diarrheal disease[18]. WHO/UNICEF recommend that all children receive solutions of low-osmolality oral rehydration salts to prevent and treat dehydration due to diarrhea. Rotarix[36] and RotaTeq are two approved vaccines used worldwide[37]. The Lanzhou Lamb Rotavirus Vaccine has already been licensed and used in China for several years. However, the immunization program for rotavirus is unpopular in the area of Yunnan. Our study may be useful for decisions regarding the need for rotavirus vaccines for use in rural China, such as Zhaotong, Xiangyun, and Xishuangbanna. It is assumed that G1 type rotavirus may be the most common strain in these areas. However, some issues should be considered, such as the reason for the G3 and G9 shift over time and the impact of the changes in the genetic diversity of rotavirus on the use of a rotavirus vaccine in China.
Whole-genome sequencing has shown that human RVs with the genotype constellation of G1/G3/G4/G9/G12-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 or G2-P[4]-I2-R2-C2-M2-A2-N2-T2-E2-H2 are globally dominant[7,38]. Based on all 11 rotavirus gene segments and the formation of RCWG classification system, rotavirus ZTR-68 has been designated G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 and was known as Wa-like rotavirus group A (RVA). Human Wa-like RVA strains are believed to have a common ancestor with porcine RVA strains[39]. The ZTR-68 and its plaque clone 3C were fully adapted to grow in MA-104 and Vero cell cultures (data not shown). ZTR-68 was identified as a Group A rotavirus, which was a Wa-like strain. The characterization of ZTR-68 G and P types are similar to Wa, but different in RNA electropherotype. It was interesting that our data demonstrated several rotavirus isolates with high similarity to ZTR-68 during the period of this study. It is assumed that G1P[8] type rotavirus may be the most common strain in the Zhaotong area. But some issues should be considered, such as the reason that the G3 and G9 type shifted as time went on, and the impact of the variation of genetic diversity of rotavirus for the use of rotavirus vaccine in China. This newly isolated human Group A rotavirus, ZTR-68 is being used to establish a new candidate rotavirus vaccine for use in studying the immune effectiveness.
Unfortunately, we were not able to acquire purified G3 and G9 strains after several passages in cell culture. Determining the circulation of various rotavirus strains in this area is of interest. However, no P types were identified during the genotype detection, which is another limitation of our study. The usefulness of next-generation DNA sequencing has previously been evaluated for the direct detection of bovine rotavirus from fecal samples[40]. More meticulous rotavirus surveillance should be conducted in Yunnan, China. In this study, a high prevalence of the G1, G2, and G3 genotypes was detected from 2010 to 2012. However, the G9 genotype was the predominant cause of gastroenteritis in children in Yunnan in 2013. Our data will be useful for future efforts to formulate and develop a vaccine in China.
Human rotavirus genotypes G1-G4 and G9 are major causes of acute gastroenteritis in children worldwide. Approximately 35000 children die per year in China due to rotavirus, which is the second highest rate of rotavirus deaths in the world.
In China, the elimination and control of the two kinds of diseases have experienced the process of alternating use of two kinds of vaccines, which proved to be feasible and effective. These successful experiences provide a theoretical basis and an approach to the development of an inactivated rotavirus vaccine. This newly isolated human Group A rotavirus, ZTR-68 is being used to establish a new candidate inactivated rotavirus vaccine for use in studying the immune effectiveness.
The aim of this study was to determine the distribution of rotavirus VP7 gene in hospitalized children in Yunnan, China. A new candidate human inactivated rotavirus vaccine strain was isolated and characterized.
A total of 366 stool specimens were collected from hospitalized children in hospitals in Yunnan Province from September 2010 to December 2013. The genomic RNA electropherotypes and the G genotypes of the rotaviruses were determined. A phylogenetic analysis of the VP7 genes was performed. Rotavirus isolation was performed, and characterized by plaque, EM, and all gene sequence analysis. The sequences were deposited in GenBank under the following accession numbers: KM247264 - KM247286. ZTR-68 genomic sequences: JX509930 - JX509940. Quantification of antibodies for inactivated vaccine prepared with ZTR-68 were examined by ELISA and microneutralization assay.
Group A human rotavirus was detected in 177 of 366 stool samples using a colloidal gold device assay. The temporal distribution of rotavirus cases showed significant correlation with the mean air temperature. Rotaviruses were isolated from 13% of the rotavirus-positive samples. The predominant genotype was G1 (43.5%), G3 (21.7%), G9 (17.4%), G2 (4.3%), G4 (8.7%), and mixed (4.3%). A rotavirus strain was isolated from a rotavirus-positive stool sample of a 4-month-old child in The First People’s Hospital of Zhaotong (2010) for use as a candidate human inactivated rotavirus vaccine strain and for further studies, which was designated ZTR-68. The genotype of 11 gene segments of strain ZTR-68 (RVA/Human-wt/CHN/ZTR-68/2010/G1P[8]) was characterized. The genotype constellation of strain ZTR-68 was identified as G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. The VP7 and VP4 genotypes of strain ZTR-68 was similar to Wa-like strains.
Group A human rotavirus was detected in 48.4% stool samples. A high prevalence of the G1, G2, and G3 genotypes was detected from 2010 to 2012. However, a dominant prevalence of the G9 genotype was identified as the cause of gastroenteritis in children in Yunnan, China, in 2013. A candidate human inactivated rotavirus vaccine strain, designated ZTR-68 was isolated, characterized, and showed immunogenicity.
Our data will be useful for the future formulation and development of a vaccine in China.
We are grateful to the members of the Pediatrics Department of the First People’s Hospital of Zhaotong City for introducing the patients and the collection of clinical samples.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, research and experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Islek A, Krishnan T S- Editor: Dou Y L- Editor: Filipodia E- Editor: Song H
| 1. | Bresee JS, Hummelman E, Nelson EA, Glass RI. Rotavirus in Asia: the value of surveillance for informing decisions about the introduction of new vaccines. J Infect Dis. 2005;192 Suppl 1:S1-S5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 2. | Parashar UD, Gibson CJ, Bresee JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12:304-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1010] [Cited by in F6Publishing: 1007] [Article Influence: 55.9] [Reference Citation Analysis (0)] |
| 3. | Orenstein EW, Fang ZY, Xu J, Liu C, Shen K, Qian Y, Jiang B, Kilgore PE, Glass RI. The epidemiology and burden of rotavirus in China: a review of the literature from 1983 to 2005. Vaccine. 2007;25:406-413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 52] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Parashar UD, Hummelman EG, Bresee JS, Miller MA, Glass RI. Global illness and deaths caused by rotavirus disease in children. Emerg Infect Dis. 2003;9:565-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1282] [Cited by in F6Publishing: 1237] [Article Influence: 58.9] [Reference Citation Analysis (0)] |
| 5. | Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, Franco MA, Greenberg HB, O’Ryan M, Kang G. Rotavirus infection. Nat Rev Dis Primers. 2017;3:17083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 324] [Article Influence: 46.3] [Reference Citation Analysis (0)] |
| 6. | Bányai K, László B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30 Suppl 1:A122-A130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 331] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 7. | Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29-56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 917] [Cited by in F6Publishing: 900] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 8. | Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gómara M, Maes P, Patton JT. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008;82:3204-3219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 729] [Cited by in F6Publishing: 702] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 9. | Rippinger CM, Patton JT, McDonald SM. Complete genome sequence analysis of candidate human rotavirus vaccine strains RV3 and 116E. Virology. 2010;405:201-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Bányai K, Estes MK, Gentsch JR, Iturriza-Gómara M, Kirkwood CD, Martella V. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008;153:1621-1629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 522] [Cited by in F6Publishing: 567] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 11. | Yee EL, Fang ZY, Liu N, Hadler SC, Liang X, Wang H, Zhu X, Jiang B, Parashar U, Widdowson MA. Importance and challenges of accurately counting rotavirus deaths in China, 2002. Vaccine. 2009;27 Suppl 5:F46-F49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Westerman LE, Jiang B, McClure HM, Snipes-Magaldi LJ, Griffin DD, Shin G, Gentsch JR, Glass RI. Isolation and characterization of a new simian rotavirus, YK-1. Virol J. 2006;3:40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Sato K, Inaba Y, Shinozaki T, Fujii R, Matumoto M. Isolation of human rotavirus in cell cultures: brief report. Arch Virol. 1981;69:155-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 164] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 14. | Cunliffe NA, Woods PA, Leite JP, Das BK, Ramachandran M, Bhan MK, Hart CA, Glass RI, Gentsch JR. Sequence analysis of NSP4 gene of human rotavirus allows classification into two main genetic groups. J Med Virol. 1997;53:41-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 15. | Pereira HG, Azeredo RS, Leite JP, Candeias JA, Rácz ML, Linhares AC, Gabbay YB, Trabulsi JR. Electrophoretic study of the genome of human rotaviruses from Rio de Janeiro, São Paulo and Pará, Brazil. J Hyg (Lond). 1983;90:117-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 86] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 16. | Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673-4680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47226] [Cited by in F6Publishing: 44069] [Article Influence: 1469.0] [Reference Citation Analysis (0)] |
| 17. | Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-1599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22436] [Cited by in F6Publishing: 19539] [Article Influence: 1149.4] [Reference Citation Analysis (0)] |
| 18. | Resch TK, Wang Y, Moon SS, Joyce J, Li S, Prausnitz M, Jiang B. Inactivated rotavirus vaccine by parenteral administration induces mucosal immunity in mice. Sci Rep. 2018;8:561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 19. | Waterborg JH, Matthews HR. The Lowry method for protein quantitation. Methods Mol Biol. 1994;32:1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 20. | Zhang B, Yi S, Ma Y, Zhang G, Zhang Y, Xie T, Li H, Sun M. Immunogenicity of a scalable inactivated rotavirus vaccine in mice. Hum Vaccin. 2011;7:248-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 21. | Dian Z, Wang B, Fan M, Dong S, Feng Y, Zhang AM, Liu L, Niu H, Li Y, Xia X. Completely genomic and evolutionary characteristics of human-dominant G9P[8] group A rotavirus strains in Yunnan, China. J Gen Virol. 2017;98:1163-1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | De La Cruz Hernández SI, Anaya Molina Y, Gómez Santiago F, Terán Vega HL, Monroy Leyva E, Méndez Pérez H, García Lozano H. Real-time RT-PCR, a necessary tool to support the diagnosis and surveillance of rotavirus in Mexico. Diagn Microbiol Infect Dis. 2018;90:272-276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 23. | Jing Z, Zhang X, Shi H, Chen J, Shi D, Dong H, Feng L. A G3P[13] porcine group A rotavirus emerging in China is a reassortant and a natural recombinant in the VP4 gene. Transbound Emerg Dis. 2018;65:e317-e328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Chang H, Zhang L, Ge Y, Cai J, Wang X, Huang Z, Guo J, Xu H, Gu Z, Chen H. A Hospital-based Case-control Study of Diarrhea in Children in Shanghai. Pediatr Infect Dis J. 2017;36:1057-1063. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 25. | Zheng J, Zheng H, Gupta RK, Li H, Shi H, Pan L, Gong S, Liang H. Interrelationship of rotavirus infection and Creatine Kinase-MB isoenzyme levels in children hospitalized with acute gastroenteritis in Guangzhou, China, 2012-2015. Sci Rep. 2017;7:7674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Shen J, Zhang BM, Zhu SG, Chen JJ. No direct correlation between rotavirus diarrhea and breast feeding: A meta-analysis. Pediatr Neonatol. 2018;59:129-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 27. | Wang XY, Du L, Von Seidlein L, Xu ZY, Zhang YL, Hao ZY, Han OP, Ma JC, Lee HJ, Ali M. Occurrence of shigellosis in the young and elderly in rural China: results of a 12-month population-based surveillance study. Am J Trop Med Hyg. 2005;73:416-422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 42] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Wang YH, Kobayashi N, Zhou X, Nagashima S, Zhu ZR, Peng JS, Liu MQ, Hu Q, Zhou DJ, Watanabe S. Phylogenetic analysis of rotaviruses with predominant G3 and emerging G9 genotypes from adults and children in Wuhan, China. J Med Virol. 2009;81:382-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 29. | Ma X, Li DD, Guo YQ, Xiang JY, Li XP, Duan ZJ. [Whole genome analysis of human group A rotavirus G9p[8] strains in Hebei lulong region, 2009-2011]. Bing Du Xue Bao. 2014;30:119-127. [PubMed] [Cited in This Article: ] |
| 30. | Ward RL, Knowlton DR, Pierce MJ. Efficiency of human rotavirus propagation in cell culture. J Clin Microbiol. 1984;19:748-753. [PubMed] [Cited in This Article: ] |
| 31. | Bresee J, Fang ZY, Wang B, Nelson EA, Tam J, Soenarto Y, Wilopo SA, Kilgore P, Kim JS, Kang JO. First report from the Asian Rotavirus Surveillance Network. Emerg Infect Dis. 2004;10:988-995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD; WHO-coordinated Global Rotavirus Surveillance Network. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 949] [Cited by in F6Publishing: 972] [Article Influence: 81.0] [Reference Citation Analysis (0)] |
| 33. | Linhares AC, Stupka JA, Ciapponi A, Bardach AE, Glujovsky D, Aruj PK, Mazzoni A, Rodriguez JA, Rearte A, Lanzieri TM. Burden and typing of rotavirus group A in Latin America and the Caribbean: systematic review and meta-analysis. Rev Med Virol. 2011;21:89-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 34. | Mitui MT, Chan PK, Nelson EA, Leung TF, Nishizono A, Ahmed K. Co-dominance of G1 and emerging G3 rotaviruses in Hong Kong: a three-year surveillance in three major hospitals. J Clin Virol. 2011;50:325-333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Quaye O, McDonald S, Esona MD, Lyde FC, Mijatovic-Rustempasic S, Roy S, Banegas DJ, Quiñonez YM, Chinchilla BL, Santiago FG. Rotavirus G9P[4] in 3 countries in Latin America, 2009-2010. Emerg Infect Dis. 2013;19:1332-1333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Ward RL, Bernstein DI. Rotarix: a rotavirus vaccine for the world. Clin Infect Dis. 2009;48:222-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 37. | Heaton PM, Ciarlet M. Vaccines: the pentavalent rotavirus vaccine: discovery to licensure and beyond. Clin Infect Dis. 2007;45:1618-1624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | McDonald SM, McKell AO, Rippinger CM, McAllen JK, Akopov A, Kirkness EF, Payne DC, Edwards KM, Chappell JD, Patton JT. Diversity and relationships of cocirculating modern human rotaviruses revealed using large-scale comparative genomics. J Virol. 2012;86:9148-9162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 39. | Matthijnssens J, Van Ranst M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr Opin Virol. 2012;2:426-433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 232] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 40. | Minami-Fukuda F, Nagai M, Takai H, Murakami T, Ozawa T, Tsuchiaka S, Okazaki S, Katayama Y, Oba M, Nishiura N. Detection of bovine group a rotavirus using rapid antigen detection kits, rt-PCR and next-generation DNA sequencing. J Vet Med Sci. 2013;75:1651-1655. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |