Published online Nov 16, 2022. doi: 10.12998/wjcc.v10.i32.11671
Peer-review started: June 12, 2022
First decision: July 29, 2022
Revised: September 4, 2022
Accepted: October 11, 2022
Article in press: October 11, 2022
Published online: November 16, 2022
Gut microbiota imbalances play an important role in inflammatory bowel disease (IBD), but no single pathogenic microorganism critical to IBD that is specific to the IBD terminal ileum mucosa or can invade intestinal epithelial cells has been found. Invasive Escherichia coli (E. coli) adhesion to macrophages is considered to be closely related to the pathogenesis of inflammatory bowel disease. Further study of the specific biological characteristics of adherent invasive E. coli (AIEC) may contribute to a further understanding of IBD pathogenesis. This review explores the relationship between AIEC and the intestinal immune system, discusses the prevalence and relevance of AIEC in Crohn's disease and ulcerative colitis patients, and describes the relationship between AIEC and the disease site, activity, and postoperative recurrence. Finally, we highlight potential therapeutic strategies to attenuate AIEC colonization in the intestinal mucosa, including the use of phage therapy, antibiotics, and anti-adhesion molecules. These strategies may open up new avenues for the prevention and treatment of IBD in the future.
Core Tip: Gut microbiota imbalances play an important role in inflammatory bowel disease (IBD), but no single pathogenic microorganism critical to IBD, which is specific to the IBD terminal ileum mucosa or can invade intestinal epithelial cells, has been found. Invasive Escherichia coli (E. coli) adhesion to macrophages is considered to be closely related to the pathogenesis of IBD. Further study of the specific biological characteristics of adherent invasive E. coli (AIEC) may contribute to a further understanding of IBD pathogenesis. This review explores the relationship between AIEC and the intestinal immune system, discusses the prevalence and relevance of AIEC in Crohn's disease and ulcerative colitis patients, and describes the relationship between AIEC and the disease site, activity, and postoperative recurrence. Finally, we highlight potential therapeutic strategies to attenuate AIEC colonization in the intestinal mucosa, including the use of phage therapy, antibiotics, and anti-adhesion molecules. These strategies may open up new avenues for the prevention and treatment of IBD in the future.
- Citation: Zheng L, Duan SL, Dai YC, Wu SC. Role of adherent invasive Escherichia coli in pathogenesis of inflammatory bowel disease. World J Clin Cases 2022; 10(32): 11671-11689
- URL: https://www.wjgnet.com/2307-8960/full/v10/i32/11671.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i32.11671
Inflammatory bowel disease (IBD) is a chronic disease with spontaneous pathogenesis. The interaction between the intestinal flora and intestinal immune system is an important part of IBD pathogenesis, and adherent invasive Escherichia coli (E. coli) (AIEC) is attracting increasing attention as a potential IBD pathogen. Studies have shown that the colonization rate of AIEC in the intestinal mucosa is higher in IBD than in other digestive diseases, and colonization in the intestinal mucosa is related to the incidence of Crohn's disease (CD) in children, indicating that this pattern of colonization may aggravate CD activity and increase the severity of recurring postoperative CD as identified by endoscopy[1]. Animal experiments have shown that AIEC infection in mice caused chronic intestinal inflammation similar to the gross and microscopic pathological changes observed in IBD, indicating that AIEC caused intestinal fibrosis, a common complication in IBD[2]. Therefore, an exploration of the relationship between AIEC and intestinal mucosal immunity is important to advance the understanding of IBD pathogenesis and to identify new therapeutic targets. In this paper, we review details on the special biological characteristics of AIEC and the potential pathogenesis of IBD.
AIEC is one of the most concerning possible pathogens in IBD. The cause of E. coli overgrowth in IBD patients is not clear, but it may be related to the increase in reactive nitrogen, which allows rapid digestion, providing favorable conditions for E. coli proliferation[3]. AIEC, a unique pathogenic species, is believed to be related to human IBD and is considered to be one of the most active pathogenic bacteria.
AIEC attached to differentiated Caco-2 cells and undifferentiated I-407 intestinal epithelial cells (IECs) and survived attachment to J774-A1 macrophages, in which they replicated[4]. AIEC bound and invaded IECs through microtubule polymerization and actin recruitment, thereby inducing inflammatory cytokine secretion. When surviving and replicating in macrophages, AIEC induced tumor necrosis factor-α (TNF-α) secretion and promoted granuloma formation in vitro[5]. AIEC induced interleukin-1β (IL-1β) production through an NLRP3-dependent mechanism, and macrophages cleared AIEC through an NLRP3-independent mechanism. The aggressiveness of E. coli in IECs and secretion of inflammatory factors such as IL-1β may promote CD and ulcerative colitis (UC)[6]. Studies have shown that AIEC induced granulomatous lesions similar to those formed in early epithelioid granuloma in vitro. AIEC has type I pili and therefore binds to carcinoembryonic antigen-associated cell adhesion factor 6 (CEACAM6), a host adhesion receptor[7]. CEACAM6 is overexpressed in ileal tissue in patients with CD but not in normal individuals. In vitro experiments showed that the upregulation of CEACAM6 expression in IECs was caused by AIEC infection and the simultaneous stimulation of the proinflammatory cytokines interferon-γ (IFN-γ) and TNF-α[8]. In addition, AIEC crossed the mucosal barrier into lymphocytes through long pili.
AIEC LF82, isolated from the ileum of patients with CD, intensified inflammatory mucosal immune responses; for example, it triggered the upregulated expression of Toll-like receptor 5 (TLR5) and international powered access federation flagellin receptor. In CD, CEACAM6 is a receptor for AIEC in the intestinal mucosa, and increased CEACAM6 expression on the apical membrane of ileum cells promoted the abnormal colonization of AIEC in the ileum mucosa[9]. Furthermore, AIEC LF82 type I pili binding to CEACAM6 and TLR5 induced hiF-1A production and activated the classic NF-κB pathway, resulting in clinical colitis symptoms[10]. To mimic the high level of CEACAM6 expression in CD, transgenic CEABAC10-expressing mice were used to establish models expressing human CEACAMs. Experiments with this model demonstrated that AIEC LF82 induced clinical colitis symptoms in the transgenic mice by binding to the type I pili of CEACAM6[11].
AIEC NRG857c has been reported to colonize the intestinal mucosa of mice treated with stre
E. coli is a Gram-negative aerobic bacterium that can colonize within a few hours of initial replication. This species is important to the intestinal microecology and plays a key role in maintaining intestinal homeostasis[13]. Generally, it does not cause disease. However, some of E. coli strains are intestinal pathogens because they show certain pathogenic characteristics (e.g., outer membrane proteins), and five categories have been identified: Pathogenic E. coli, enterotoxigenic E. coli, enteroinvasive E. coli, enterohemorrhagic E. coli, enteroaggregative E. coli and enteroaggregative E. coli[14].
The aforementioned pathogenic bacteria do not specifically adhere to or invade the epithelial cells of IBD patients. However, bacteria without any of the aforementioned pathogenic characteristics can adhere to the surface of epithelial cells and enter the cytoplasm to infect eukaryotic cells[15]. Moreover, these invasive E. coli replicate within epithelial cells and macrophages. Therefore, these bacteria have been separately classified and defined as “AIEC strains”, and these strains have been reported to be involved in the pathogenesis of IBD in many countries.
It is currently unclear whether AIEC causes intestinal inflammation and leads to IBD or whether AIEC successfully colonizes the mucosa of patients with IBD and is thus an aggravating factor. Factors involved in AIEC damage to the intestinal mucosa have been identified. For example, deletions in traditional pathogenic genes play important roles in the pathogenesis of AIEC infection, such as mucosal immune abnormalities (e.g., in CD) or intestinal barrier dysfunction (e.g., in UC)[16]. AIEC virulence genes promote cell motility, and capsule and lipopolysaccharide (LPS) gene expression; increase serum resistance and iron uptake; and promote epithelial cell adhesion, invasion, and biofilm formation[17]. AIEC strains contain bacterial components and pathways that regulate these key pathogenic determinants. For example, lipoprotein, histone-like protein, ribonucleotide reductase NrdR, the flagellar transcriptional regulator FlhD2C2, sigma factor FliA, and the second messenger cyclodimer GMP are involved in interactions with AIEC with epithelial cells, while the sigma pathway is involved in biofilm formation[18]. In addition, the ability to form biofilm on epithelial cells and long polar fimbriae are pathogenic features of AIEC.
The toxicity of AIEC is related to its survival and replication in macrophages. For example, the LF82 strain does not enter the cytoplasm; in contrast, it replicates in the acidic environment of mature phagosomes, where oxidative stress-inducing compounds, proteolytic enzymes, and antibacterial compounds reside[19]. In macrophages, survival and replication of AIEC are mediated by combined actions of the stress protein HtrA, mercaptyl-disulfide REDOX enzyme DsbA, RNA-connexin Hfq, and FAD-dependent REDOX enzyme IbeA, which play roles in reactive oxygen species production[20]. In AIEC infection, TNF-α and other proinflammatory cytokines are released, further enhancing the imbalance and proliferation of AIEC without inducing host cell death. AIEC can evade host immune responses by inhibiting IFN-γ-mediated signal transduction and activation of transcription-1 phosphorylation in IECs, thereby preventing appropriate antimicrobial responses[21].
AIEC can colonize, invade, and then survive in the intestinal mucosa by affecting the function of intestinal immune cells. Moreover, the abnormal function of immune cells leads to an excessive inflammatory response to other intestinal symbiotic bacteria in the intestinal mucosa, resulting in intestinal inflammation.
Dendritic cells, macrophages, innate lymphocytes, and neutrophils complement the physical barrier formed by IECs and are the first line of defense of the innate immune system in the human intestinal mucosa. The intestinal barrier controls mucosal absorption and protects the mucosa from endotoxins, invasive microorganisms, and antigens. IECs connect to each other to form a physical barrier, and cell-to-cell contact is regulated by various proteins, including adhesion and tight junctions[22]. The epithelium, which is a barrier throughout the intestinal mucus layer, is not only involved in the synthesis of antimicrobial peptides but is also an integral part of natural immunity. An effective intestinal mucosal barrier is crucial to prevent exposure to the external environment. Therefore, dysfunction of the barrier system can increase intestinal permeability, an important characteristic in IBD[23]. Barrier disorders include a reduction in intestinal mucus layer thickness and composition, changes in tightly connected complexes, and disruptions to antimicrobial peptide synthesis. The presence of host intestinal barrier defects of genetic or environmental origin in the intestinal barrier may affect AIEC ability to colonize and be translocated into the intestinal tract[24]. Host defects in many patients with CD are associated with enhanced pathogenicity of AIEC LF82. In the context of inflammation, abnormal expression of IEC-specific receptors CEACAM6 and Gp96 reduces the ability of epithelial cells to resist adhesion and invasion by intestinal bacteria. Furthermore, defects in autophagy are associated with the function and expression of NOD2, ATG16L1, and IRGM, impairing the ability of host cells to resist external infections. Bacterial adhesion and colonization of IECs are considered to be key steps in the pathogenesis of IBD, which is evident before bacterial spread into the submucosa. Evidence suggests that AIEC can alter epithelial barrier function by replacing and redistributing the zO-1 protein, an essential component of tight apical junctions[25]. The reduced integrity of the barrier system makes it easier for AIEC to cross the epithelial barrier, increasing the pathogenicity of AIEC. Intestinal barrier damage and inflammatory responses can also exacerbate inflammatory symptoms in the gut[26].
The small intestine is covered with a thin layer of mucus, and the colon and stomach are covered with two layers of mucus. The mucus layer of the small intestine is similar to the thinner mucus layer in the colon (50 to 200 μm) and is closely connected to the intestinal epithelium, reinforcing the protection conferred by the epithelial layer[27]. This mucus layer provides the substrate for maintaining antimicrobial peptides, including alpha-defensins secreted by Paneth cells, which create a barrier between microbes and intestinal mucosal tissue[28]. The inner mucosa blocks direct contact between the epithelial cells and gut microbes, and the outer mucosa is colonized by many symbiotic bacteria that are difficult to eliminate. In IBD, decreased mucus production and antimicrobial peptide secretion provide favorable conditions for symbiotic bacteria to become opportunistic pathogens[29]. Studies have shown that the VAT-AIEC protease secreted by AIEC can promote the degradation of mucin and decrease the viscosity of mucus, thereby facilitating the colonization and invasion of AIEC into the intestinal mucosa. IBD patients reported present with greater colonization of Ruminococcus torques and Ruminococcus gnavus, which have mucus-dissolving abilities, in their intestines[30]. As a result, the overall number of mucosa-associated bacteria in the intestines of IBD patients is increased. These abundant bacteria can degrade human secretory MUC2, enhancing the adhesive and invasive abilities of opportunistic pathogens in contact with IECs.
In the transgenic CEABAC10 mouse model, dietary components rich in fat and sugar reduced the expression of mucin 2, Klf4, and Tff3, as well as the thickness of the mucus layer and the number of goblet cells in the colonic mucosa, thereby altering barrier function[31]. In these mouse models, AIEC showed a greater ability to colonize the intestinal mucosa and trigger intestinal inflammation due to changes in barrier function and increased TNF-α secretion[32]. In addition, some AIEC strains have been shown to resist the effects of antimicrobial peptides, which allows invasive bacteria to survive in the inner mucin layer. A better understanding of the molecular mechanisms underlying increased intestinal permeability in patients with CD may lead to the development of new drugs that target AIEC and prevent it from undermining the integrity of the intestinal barrier[33].
Peyer patches (PPs) play important roles in mucosal immunity and can cause intestinal epithelial disorders. The interaction between PPs and intestinal microbiota and species diversity have gradually become research hotspots. Some pathogens have evolved ways to cross the epithelial apical barrier. Studies have shown that certain microorganisms, especially invasive pathogenic bacteria, can access specific microfold (M) cells as the main entry point to invade the host, cross the intestinal barrier, and cause disease[34]. For example, Yersinia enterocolitica and Yersinia pseudotuberculosis cross the intestinal epithelial barrier by adhering to follicle-associated epithelial M cells. Aggregated pyloric lymph nodes may be the initial site of inflammation and play an important role in the early stage of CD[35]. Recent studies have shown that AIEC can leverage their LPF to interact with pyloric lymph nodes and thus gain access to M cells. In addition, the prevalence of an AIEC strain expressing LPF was higher in CD than in a control group. After transport through the epithelium, AIEC can enter immune cells, especially macrophages and dendritic cells; these immune cells release immunomodulators, such as TNF-α, that promote functional changes in the mucosal barrier and reduce mucosal permeability[36].
The intestinal mucosa contains a large number of effector lymphocytes, including Ig-secreting CD4+ effector T cells, such as IFN-γ-producing Th1 cells, IL-17-producing Th17 cells, Foxp3 regulatory T cells (Tregs), CD8+ T cells, and intraepithelial lymphocytes (mainly γδ T cells). Recent studies have shown that naive lymphocytes share the same functional characteristics as T cells[37].
The intestinal immune system must simultaneously sense whether the immune-tolerant microbiome and host defenses against microbial invasion are caused by pathogens or opportunistic pathogens. The inflammatory response is controlled by Fox P3+ Tregs originating in the thymus or from locally differentiated primary CD4+ T cells. Notably, innate immune lymphocytes also play major roles in the regulation of effector T-cell responses to symbiotic bacteria[38]. Under stable conditions, a mucosal firewall consisting of a mucus layer, an epithelial barrier, IgA, and regulatory cells controls microbial-derived and food-borne antigens and limits inappropriate immune responses. Therefore, microbiological antigens do not normally activate systemic immunity[39]. Any disruption to this mucosal firewall (even local or temporary breach), such as that caused by acute infection, epithelial damage, or increased permeability, causes microbial spread and can transform symbiotic bacteria into opportunistic pathogens[40].
The microbiome has been identified as a major activator and regulator of the mucosal immune system through the direct interactions of the microorganisms with epithelial cells or immune cells. In addition, the microbiota stimulates the regulatory response mainly through the production of active metabolites[41]. Studies have shown that feeding mice probiotics, such as Lactobacillus reuteri, increases the frequency of Tregs. Bacterial fermentation products, namely, short-chain fatty acids (such as butyrate and acetate produced by Bacteroides or Clostridium), inhibit inflammation, and the differentiation and function of colonic Tregs depend on GPR43 facilitation, which protect against colitis. Other types of microbial products, such as bacterial DNA, can lead to Th17 cell activation in a steady state. RORγ T+ cells and CD4+ T cells, which secrete IL-17, are generally thought to be harmful to the host because of their ability to induce recurrence of inflammation and autoimmune diseases[42]. In addition, the Th17 cell response protects against intestinal fungal and bacterial pathogens such as Acidobacteria. Thus, Th17 cells are involved in disease pathogenesis only in specific contexts, such as when the host is susceptible to autoimmune attack or when the immune system is overactivated by a large number of infectious bacteria[43]. Therefore, the gut microbiome plays a dual role in immune regulation. In the intestinal mucosa, the microbiota plays a role by stimulating the acquired immune system and promoting the formation of different T-cell subsets. The normal intestinal microbiota is involved in immune homeostasis. Notably, Tregs in the intestinal mucosa show specific responses to microbial-derived antigens, and any disruption to microbiome components affects the Treg content[44]. Studies have shown that an antibody group changed with changes to the microbiome, with the antibody composition adjusting in response to the latest microbial composition[45].
IBD is considered to be a result of an inappropriate response of the acquired immune system to microbial-derived antigens because, in most cases of IBD-related genetic diversity, the mucosal immune system response to microorganisms is affected. It had been widely believed that the Th1 cell response was the main cause of CD. This idea was supported by experiments showing that Th1 cells produce TNF-α and IFN-γ, and IFN-γ can activate macrophages to produce additional TNF-α, leading to the apoptosis of epithelial cells and inducing CD[46]. Moreover, experiments have shown that many Th17 cells infiltrate the inflammatory gastrointestinal mucosa of IBD patients, and the upregulation of Th17 cell-secreted cytokine expression (e.g., IL-21 and IL-22) can promote the Th1 cell response, thus exacerbating the inflammatory response of IBD. Therefore, Th17 cells are now also recognized as important in the pathogenesis of IBD[47].
Furthermore, acquired functional mutation of the IL23R gene has been reported to predispose humans to CD and UC, while IL-17 secreted by peripheral blood mononuclear cells of patients is related to the severity of UC but not CD, suggesting that IL-23 plays an important role in the pathogenesis of IBD, while Th17 cells play a different role in these two diseases[48]. Notably, IL-23 not only maintained the Th17 cell response but also promoted the secretion of IL-17 and IFN-γ by naive immune lym
Although the exact nature of antigen stimulation of the immune system remains unclear, evidence of B- and T-cell immunoactivation by inducible microbiota suggests that IBD is not caused by specific pathogenic antigens but by an intolerant microbiota in the gut. Antibodies targeting Saccharomyces cerevisiae, the E. coli membrane protein Omp C, and flagellin have been found in patients with IBD, and these antibodies have been associated with IBD development[50]. In addition, these antibodies have been detected in family members of IBD patients (who show the potential to acquire IBD). Notably, although no correlation between AIEC colonization and the anti-Omp C antibody titer in CD has been reported, both measures have been identified in the ileum and can cause ileum injury[51]. For example, relevant experiments have shown that the concentration of anti-Omp C antibodies is greatly increased in IBD patients. Moreover, same antibodies against the gut microbiota have been also found in healthy subjects who experience brief disruption of gut barrier function, indicating that specific microbiome systemic immunity is not the direct cause of IBD[52].
AIEC formed colonies in wild type and TLR5-knockout (KO) mice for only a brief period, but chronic colitis persisted in the TLR5-KO mice for months longer, suggesting that AIEC plays a role in colitis etiology. In contrast, inflammatory environments are conducive to AIEC proliferation and a decrease in the diversity of gut microbes[53]. With the development of a fecal sample bank, the origin of AIEC will be eventually identified. According to knowledge obtained to date, in contrast to an acquired infectious agent, AIEC can only cause disease in a specific host or environmental context. Notably, studies have shown that host factors play major roles in promoting intestinal bacterial infection, especially AIEC infection. For example, increased expression of CEACAM6, a specific bacterial adhesion factor, in the brush margin of the ileum led to the colonization of pathogenic AIEC in the ileum of CD patients[54].
AIEC adhered to CHI3LI through the Chi A protein domain, thereby promoting the pathogenic effect of AIEC. The outer membrane protein Omp A interacted with the endoplasmic reticulum (ER) stress response glycoprotein Gp96, which is overexpressed in the apical membrane of ileal epithelial cells in CD[55]. Given that ER stress is often associated with inflammatory responses, AIEC may also leverage ER stress to enhance its adhesiveness to the intestinal epithelium in CD. AIEC delays apoptosis of infected macrophages by increasing the protein mercaptosylation and degrading the apoptotic enzyme Caspase-3, enabling AIEC persistence in patients with CD. AIEC also regulates the ubiquitin– proteasome system and downregulates the NF-κB regulator CYLD, leading to degradation of Iκ-α and activation of NF-κB[56]. This feature is conducive to the intracellular replication of AIEC LF82, playing a very important role in the pathogenicity of AIEC. Studies have shown that abnormal autophagy contributes to the persistence of AIEC in the intestines of patients with IBD. Indeed, alteration in ATG16L1, IRGM, and NOD2 expression was beneficial to the replication of AIEC in macrophages, thus increasing the secretion of IL-6 and TNF-α[57]. In contrast, the numbers of AIEC cells and proinflammatory cytokines released by macrophages were significantly reduced under pharmacologically induced autophagy. Notably, autophagy was blocked at the autophagolysosome stage, and this inhibitory effect was accompanied by AIEC infection of PLB-985 neutrophil-like cells, which enabled intracellular bacterial survival and increased the Il-8 secretion level[58]. In addition, the microRNAs miR106B and miR93 reduced the expression of ATG16L1 and prevented the elimination of intracellular bacteria by regulating autophagy[59]. This self-phagocytic regulation appears to be disrupted in the colonic mucosa of patients with CD. AIEC infection upregulated the expression of miR30C and miR130A in T84 cells and mouse intestinal cells, resulting in reduced expression of ATG5 and ATG16L1, inhibition of autophagy, an increase in the number of AIEC in cells, and intensification of the inflammatory response[60].
Multiple deficiencies in the innate immune response associated with CD contribute to AIEC-induced inflammatory responses. Risk polymorphisms associated with autophagy lead to impaired innate immune system sensing and processing of intracellular bacteria, leading to inflammation. Autophagy is induced in AIEC-infected host cells and is required to inhibit AIEC intracellular replication[61].
Nucleotide-binding oligomerization domain-containing 2 (NOD2/CARD15, on chromosome 16) was the first CD susceptibility gene discovered. NOD2/CARD15-encoded proteins are expressed in monocytes, macrophages, dendritic cells, epithelial cells, and Paneth cells. This protein contains two N-terminal caspase-activating and recruitment domains (CARDs), a central nucleotide-binding domain (NBD) and C-terminal leucine-rich-repeat region (LRR). NOD2/CARD15 is mainly involved in the recognition and inflammatory response of various pathogenic microorganisms in cells, and it can recognize muramyl dipeptide (MDP), the cleavage product of peptidoglycan in the bacterial cell wall[62]. It is an important bridge between innate immunity and acquired immunity. One study suggested that NOD2/CARD15 regulated the release of defensins and antimicrobial peptides by intestinal Paneth cells, thus playing a role in maintaining the normal barrier function of the intestinal mucosa. High expression of CD147 promoted Listeria invasion of epithelial cells, while NOD2/CARD15 formed a complex with CD147, which reduce the level of free CD147, ultimately regulating intestinal tract stability. NOD2/CARD15 was identified through the Kront's method in 2001, and three single-nucleotide polymorphisms (SNPs) in this gene have been associated with IBD[63]. Studies have shown that NOD2/CARD15 mutations are promoters of IBD development. NOD2/CARD15 gene mutations or functional defects led to the impairment of the intestinal immune barrier, resulting in individual intestinal mutations related to the susceptibility to a variety of pathogenic bacteria, accompanied by reduced defensins and antimicrobial peptide levels, increased intestinal mucosal permeability, weakened intestinal bactericidal activity, easy bacterial invasion of epithelial cells, etc. Therefore, NOD2/CARD15 deletion in mice caused typical intestinal pathological outcomes and increased mouse susceptibility to Listeria. With further development of immunological, microbiological, and molecular biology techniques, details on the function of NOD2 in the molecular system through which intracellular bacteria are recognized will be further revealed, and certain factors interacting with NOD and their mechanisms of action will be gradually discovered, possibly revealing the whole picture of this functional system[64]. Advances in methodology will also lay a foundation for in-depth research into the contributions of NOD2 in the pathogenesis of IBD. The discovery that NOD2/CARD15 gene mutation or functional defects led to an impaired intestinal immune barrier was the first finding to link genetic susceptibility to CD, intestinal microecology, and the body's natural immunity in CD and was a major breakthrough in CD research. Moreover, later studies confirmed that the NOD2/CARD15 gene contains three SNPs (Arg702Trp, Gly908Arg, and Leu1007fsins C) and is only a susceptibility gene in Caucasians. Due to regional and ethnic differences, the NOD2/CARD15 mutation sites involved in CD among Chinese patients may be different from those identified in the West[65].
Autophagy-related 16-like gene (ATG16L1) is located on chromosome 2Q37.1. This gene is mainly expressed in IECs, lymphocytes, and macrophages and can encode proteins involved in the processing of intracellular bacterial autophagosome metabolism[66]. The autophagy proteins ATG5 and ATG12 form compounds that play an important role in bacterial clearance from cells. Studies based on a large sample size confirmed that the SNP rs2241880 allele of the ATG16L1 gene is associated with CD susceptibility in European people[67]. This SNP is located in exon 9, which is approximately 103 bp in length. The codon of the 47th allele encodes a mutation in which a threonine residue is replaced by an alanine residue (ACT is mutated to GCT). Mutation of the ATG16L1 gene leads to the invasion of a small number of bacteria and inhibited intracellular responses that reduce the killing effect of macrophages, resulting in the survival of the invading intracellular bacteria, activating the immune response to the expression of a variety of cytokines, and causing tissue damage and chronic intestinal inflammatory reactions. In addition, murine norovirus strains have been found to cause a CD-like condition in mice with ATG16L1 mutations[68]. This pattern of viral cosusceptible gene interaction depends on TNF-α and interferon-γ.
When AIEC cells adhere to and colonize IECs, the bacteria begin to invade the epithelium. CD-associated AIEC has been found to invade several human epithelial cell lines in vitro, including HEP-2 cells and the INT 407, CACO-2, and HCT-8 IEC cell lines. AIEC survived and replicated successfully in the cytoplasm of host cells[69]. The main virulence factors in CD-related AIEC are type 1 pili, which induce membrane extension; flagella, which endow bacteria with mobility; effector molecules, which enable AIEC to interact with host cell outer membrane vesicles; and Omp C, which regulates bacterial virulence factors through sigma signaling. Invading AIEC can cross the intestinal barrier and penetrate deep into tissue, where it interacts with macrophages to continuously activate immune cells. In addition, patients with CD are more sensitive to AIEC infection, possibly due to genetic defects in CD-susceptible people[70].
It is currently believed that the process by which E. coli induces CD is as follows: Through type I pili, the bacteria bind to the mucosal surface of cells with high expression of the human antigen-related cell adhesion molecule CEACAM6 and further adhere to and invade the intestinal epithelium[71]. The lysis of endocytic vesicles damages the intestinal epithelium, and the bacteria enter cells via pinocytosis. The internalized bacteria survive and replicate in lamina propria macrophages, potentially causing persistent inflammation. In addition, E. coli destroys and passes through the epithelium and penetrates deep into tissue, continuously activating macrophages and inducing the secretion of TNF-α in large quantities, resulting in granuloma formation[72].
AIEC strains are highly correlated with CD ileum mucosa; for example, pathogenic strains were isolated from ileum specimens of 36.4% of CD patients and 6% of control individuals. In colon samples, AIEC strains accounted for 3.7% of CD, 0% of UC, and 1.9% of control sample bacteria[73]. The AIEC strain was first identified in recurrent CD soon after surgery, and therefore, the AIEC strain may play a role in initiating inflammation and may not be merely a secondary invader. The presence of AIEC strains in the colon and ileum of healthy individuals ranged from 0% to 16% and from 6% to 19%[74], respectively, suggesting that AIEC strains are associated with CD-related factors, such as host genetic susceptibility and environmental stimuli, to induce colitis. Bacterial translocation and mucosal damage are rarely observed in healthy individuals.
Few studies have reported on the correlation between age of CD onset and AIEC. AIEC is less common in children with IBD than in adults with IBD. Two strains of AIEC were identified from a total of 85 lactose-fermented specimen isolated from 34 children with IBD (24 patients with CD and 10 patients with UC) and 23 healthy children[75]. In a recent study, 616 E. coli strains were isolated from the ileal mucosa biopsy samples obtained from 4 children with CD and 4 healthy children, indicating that the relative abundance of the AIEC strain was higher in the CD patients. Therefore, the studies of AIEC prevalence in children and newly diagnosed IBD patients demonstrate the importance of AIEC in the early diagnosis of IBD. However, the incidence of CD and AIEC in elderly people has not been reported[76].
AIEC strains are more easily found in the CD ileum than in the CD colon. Specifically, Martinez-medina et al reported a higher incidence of AIEC in the ileum than in the colon in CD[77]. The genetic susceptibility and immune response of the intestinal flora reportedly varied with the site of disease, which means that the composition and distribution of mucosal flora may vary on the basis of the disease phenotype. Most studies fail to report information on the loci of the CD-affected areas, which is needed to determine the role played by AIEC in disease phenotype acquisition[78]. Dysbiosis of the gut microbiome may be caused by changes in the oxygen content of the inflamed gastrointestinal tract, leading to a decrease in probiotics and an overgrowth of proinflammatory bacteria. In the most recently diagnosed cases of untreated pediatric CD, testing of gastrointestinal samples from multiple locations revealed an increase in the number of bacteria in the Enterobacteriaceae, Veronococcaceae, Fusobacteriaceae, and Pasteurelaceae and a decrease in the number of bacteria in the orders Bacteroides, Clostridium, and Erysiphales[79].
Studies have shown that E. coli infection plays a decisive role in disease flare-up and severity, but extensive research on AIEC is lacking. An analysis of the microbiota associated with the rectal mucosa can contribute to a CD diagnosis in the early stages of the disease[80]. Specifically, the composition of the fungal community differs in inflammatory and noninflammatory areas of the intestine. Therefore, studies into intestinal fungal flora can be used to assess disease flare-ups in CD. Biopsy samples of the right colon obtained from 34 adult IBD patients (23 with CD and 11 with UC) were found to contain E. coli types B2 and D, which were associated with inflammation[81]. One study based on fluorescence in situ hybridization confirmed that the number of E. coli in the epithelium and lamina propria of patients with active CD was significantly higher than that of patients with inactive CD[82]. Similarly, other studies have found higher E. coli abundance in patients with active CD than in those in remission. Some scholars have indicated that the mucosal abundance of E. coli is related to the severity of inflammation identified by endoscopy. With E. coli, however, this association has not been found in cells[83].
A number of studies have linked AIEC to ileal recurrence after CD surgery. Studies have shown that 65% of E. coli strains were recovered from patients with chronic inflammatory lesions removed from the ileum, and 100% of the E. coli strains were recovered from recurrent biopsy samples obtained soon after surgery[84]. The number of E. coli colonies in the ileum after early relapse was significantly higher than that in healthy people. These studies suggest that E. coli may play an important role in initiating inflammatory processes in CD. Nevertheless, studies have found the AIEC strain in 22% of patients without recurrence, as determined by endoscopy, suggesting that other factors were needed to initiate CD recurrence. Recent data from 170 patients with surgically treated CD suggested that the AIEC in the surgical site was associated with recurrence 6 mo after endoscopic surgery[85]. These results suggest that targeting AIEC colonization may be a potential adjuvant strategy for preventing postoperative recurrence.
A recent study showed that the bacterial community on the mucosal surface of the human gut can be classified into five highly conserved modules, with two modules presenting unique metabolic functions and a correlation with CD[86]. Based on an analysis of the microbial component modules, a holistic view of the microbial ecology related to CD data for an individual can be obtained. A reduction in Faecalibacterium prausnitzii in patients with ileal mucosa resection has been associated with recurrence of CD[87]. As a beneficial bacterium, Clostridium plasmodium can stimulate peripheral blood monocytes in vitro to induce the secretion of the anti-inflammatory factor IL-10 and inhibit the release of the proinflammatory factor IL-12. The number of C. plasmodium in the feces of UC patients was reported to be decreased. Due to the different proportions of C. plasmodium and E. coli in ileal and colonic CD, this ratio can be considered a promising biomarker for differential diagnoses and personalized treatments[88].
AIEC is closely related to CD, but a causal relationship between intestinal colonization of AIEC and CD remains unclear. Clinical trials targeting AIEC are ongoing, with results expected. Recent studies have shown that AIEC can not only induce intestinal inflammation in CD but can also promote the colonization of AIEC in the intestinal tract, thereby increasing the severity of intestinal lesions[89].
The levels of proinflammatory factors in CD with AIEC adhesion were significantly increased, especially the transcriptional levels of TNF-α, IFN-γ, and IL-8, which were closely related to the involvement of AIEC in the immune response in CD[90]. First, adhesion of AIEC to IECs can activate the NF-κB signaling pathway through phosphorylation of IκB-α and nuclear translocation of NF-κBp65, resulting in the release of downstream proinflammatory factors (TNF-α, IL-1β, IL-2, IL-6, IL-8, etc.) and causing intestinal inflammation. AIEC also degrades IκB-α by downregulating the expression of the NF-κB regulatory factor CYLD, leading to abnormalities in the ubiquitin-proteosome system, which is involved in the breakdown of almost all proteins in the body[91]. Second, AIEC flagella can induce secretion of the proinflammatory factor IL-8 and chemokine CCL20, leading to the recruitment of macrophages and dendritic cells to the site of infection and resulting in local infiltration of inflammatory cells. However, macrophages can express TNF-α and IFN-γ after phagocytosis of AIEC, and these proinflammatory factors in turn stimulate the expression of CEACAM6 in IECs, thus enhancing the colonization of AIEC[92].
Intestinal inflammation in patients with CD provides favorable conditions for the specific colonization of AIEC. Studies have shown that engraftment of AIEC through adhesion is realized only in DSS-induced colitis, and broad-spectrum antibiotics destroy the original intestinal flora in mice or induce immune deficiency, such as IL-10 loss, in knockout mice[93]. In CD, the concentration of the intestinal mucosa containing AIEC is far higher than that in the area of inflammation intestinal mucosa. An anomalous inflammatory state can lead to intestinal flora AIEC multiplication and subsequent chronic inflammation-induced injury. An increase in the AIEC engraftment rate in inflamed intestine increases the area of long-term inflammation in the intestines, possibly because IECs are closely connected, and therefore, when the connections between abnormal structures are disrupted, pores form[94]. In this case, the Claudin2 protein content increases significantly, and the content of pore-closing proteins decreases significantly. As a result, the permeability of the intestinal epithelium is transformed from "closed" to "leaky". The increase in the permeability of the intestinal epithelium enables AIEC to penetrate the intestinal mechanical barrier, and then, with its antigen exposed to immune cells, the invading AIEC triggers an immune response, resulting in inflammation[95]. Moreover, as mentioned above, inflammatory factors can stimulate the high expression of CEACAM6 in IECs, and CEACAM6 is the most important adhesion molecule receptor involved in AIEC adhesion.
In addition to affecting the immune response of the body and causing intestinal inflammation, AIEC directly induces a certain level of pathogenicity. In studies with mice in which the flagella receptor gene TRL5 was lacking, specific AIEC infecting strains were evident. The results showed that shortly after infection, the AIEC strains were not effectively cleared by the TRL5-KO mice compared with the clearance by the normal mice, significantly increasing the inflammation index and IL-6 levels in the mutant mice[96]. In addition, the proinflammatory LPS and flagella protein levels increased significantly. Other studies have shown that streptomycin-treated mice infected with an AIEC strain exhibit histological features similar to those of CD (mural inflammation and fibrotic changes in the intestinal cavity), demonstrating that AIEC infection induces an immunological response similar to that in CD[97]. In addition, AIEC colonization was found in the healthy intestinal mucosa of CD patients, suggesting that AIEC infection may not depend on a dysregulated inflammatory response or intestinal barrier injury.
The mechanism of AIEC action in the pathogenesis of UC is still unclear. Studies have suggested that the absence of autophagy may facilitate AIEC persistence in the gut. After AIEC infection, autophagy is blocked at the autophagolysosomal stage, during which vacuoles containing pathogens are destroyed and become targets for autophagy clearance. In patients with active UC, reduced levels of chromosomal associated protein-D3 (CAP-D3) are thought to be critical for the reduced autophagy capacity and inhibited intracellular bacterial clearance. AIEC may participate in the regulation of CAP-D3 expression-related amino acid transporters[98].
The prevalence of AIEC in UC is lower than that in CD patients. Although some studies have shown a high prevalence of E. coli in UC patients, few studies have identified AIEC bacteria in the colon of UC patients. Compared with CD patients, the prevalence of AIEC in adults and children with UC is low, ranging from 0% to 10%[99]. A number of studies have shown that E. coli abundance is increased in active UC lesions compared with inactive UC lesions. E. coli in phylogenetic group B2 with at least one adherence-related gene has been associated with increased disease flare-ups in UC patients. Other studies have shown differences in the adhesion or invasion of fecal strains obtained from UC patients during a flare-up and UC patients with inactive disease[100].
Some emulsifiers and food stabilizers provide favorable conditions for AIEC colonization. AIEC LF82 is more likely to generate unique biofilms in the presence of maltodextrin (MDX)[101]. MDX is an amyloid polysaccharide that promotes bacterial adhesion to human IECs and facilitates the expression of type I pili, which is essential for the formation of AIEC biofilms and cell adhesion. However, CEACAM6 is expressed independently, suggesting that AIEC adhesion to intestinal cells involves a unique mechanism[102]. In addition, bacteria isolated from patients with ileal CD showed a higher prevalence of MAL X genes. Since MDX is a ubiquitous dietary component, this finding suggests that MDX-rich diets are more conducive to intestinal colonization with E. coli, leading to dysbiosis in the body. Polysorbate 80, an emulsifier commonly used in processed foods, promotes the passage of AIEC HM605 through M cells and IECs. Experiments with animal models have shown that dietary components rich in fat and sugar can induce dysregulation of the body's flora and induce low levels of inflammation. Studies have shown that bacterial dysbiosis and low-level inflammation in susceptible individuals increase the AIEC colonization rate, exacerbating inflammatory responses and epithelial barrier disruption[103].
AIEC is considered to be a pathobiont, although many AIEC strain genomes encode virulence genes, and pathogenicity of the prototype strain LF82 has been reported[104]. This pathophysiology is supported by clinical data showing that IBD patients have underlying genetic mutations and that inflammation may be caused by a distinct microbiome not an infectious pathogen. Analysis of the AIEC genome revealed that the presence of specific genes may be associated with bacterial virulence, while disease-causing adaptive mutations in many other genes may be associated with the pathogenicity of AIEC in susceptible hosts[105]. For example, the Omp A protein in LF82 interacts with the host molecule Gp96 to allow bacteria to adhere to IECs. Recently, acquired mutations of certain strains were considered to be markers of the evolution of pathogenic adaptability of the bacteria, and the emergence of Fim H mutants in AIEC strains enables AIEC to achieve greater cell adhesion[106]. Hence, AIEC evolved into a disease-causing adaptive strain with mutant Fim H, which improved the ability of AIEC to colonize the intestine and induce inflammation in genetically susceptible hosts. A SNP analysis of FIM H can be performed to detect virulence of E. coli in IBD patients and can also be used in diagnostic or epidemiological investigations[107]. Interestingly, the protease methyldopa degraded the type I pili of AIEC and prevented AIEC from binding to mannosylated host receptors. Decreased methyldopa expression in patients with CD is associated with the severity of inflammation, suggesting that lack of protective methyldopa allows greater AIEC colonization in the intestinal mucosa.
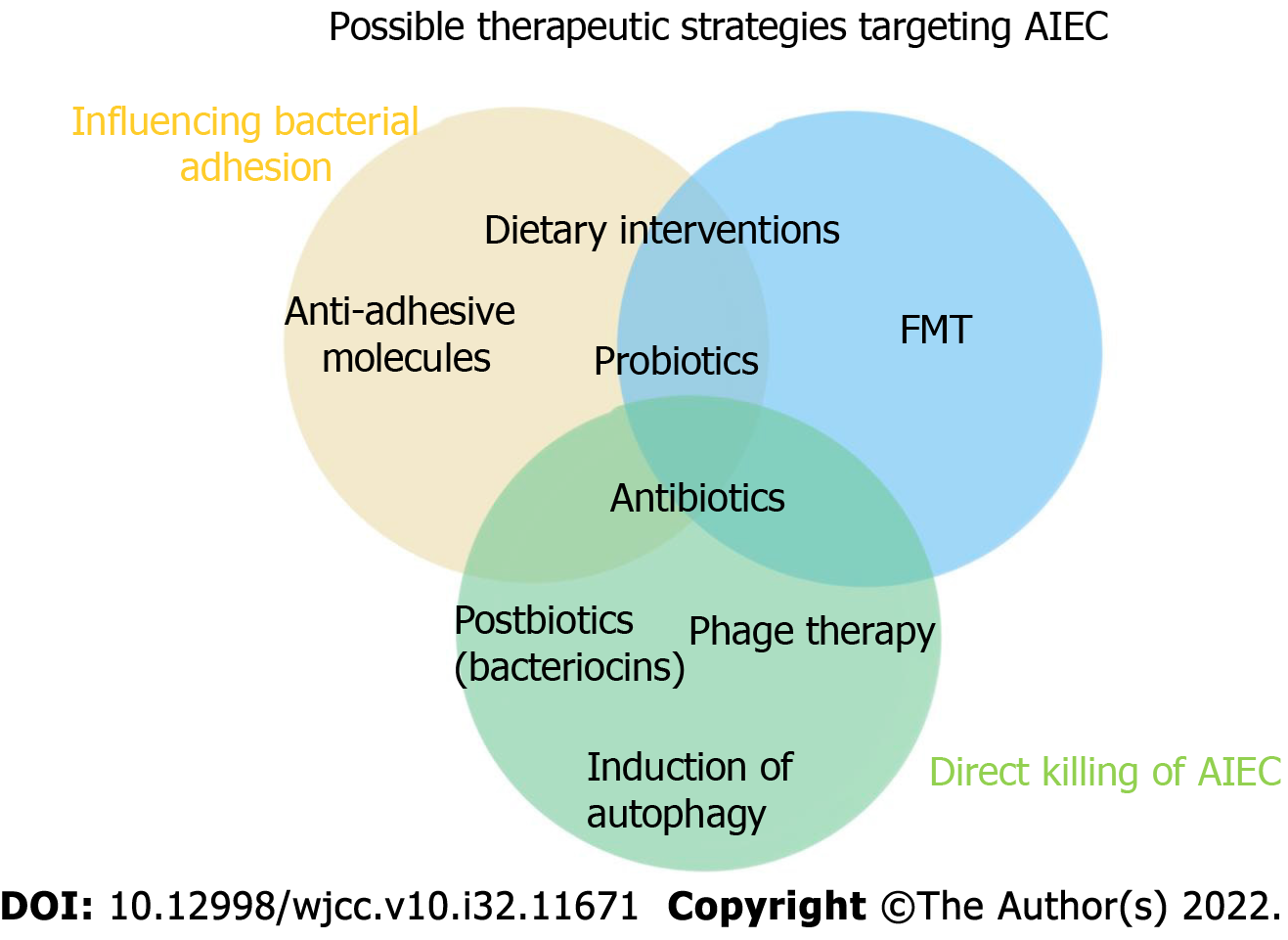
Dietary interventions: Among the factors that affect the composition and structure of the host intestinal flora, diet plays a crucial role. Dietary interventions are considered to be low-cost and easy ways to regulate the intestinal microbiota and treat disorders related to microbiota disorders[108]. In Europe, pure enteral nutrition has been listed as a first-line treatment for CD in children; children taking this treatment show higher disease remission rates than children taking corticosteroids, and the safety profile of the enteral diet is higher. A number of randomized controlled trials have shown that a low fermentable, oligo-, di-, monosaccharide and polyol diet intervention can fully alleviate the symptoms of irritable bowel syndrome (IBS) in patients, accompanied by increases in Bifidobacterium adolescentis, Bifidobacterium longum, and Faecalibacterium. The abundance of Prausnitzii and Bacteroides spp. decreased, and the level of fecal butyrate decreased[109]. In a crossover trial with patients in remission or with mild UC, researchers found significant improvements in the quality of life after 4 wk of targeted intake of either a low-fat or high-fiber diet. In addition, a low-fat diet significantly reduced patients' levels of expressed inflammatory markers and attenuated the dysregulation of their gut microbiota. Studies based on IBD animal models have shown that proteolytic diets induce remission associated with decreased abundance of pathogens such as E. coli and Clostridium perfringens, increased abundance of Clostridium hiranonis, a secondary bile acid producer, and elevated levels of chicholic acid and deoxycholic acid[110].
Among environmental factors, vitamin D plays a key role in maintaining the integrity of the intestinal mucosal barrier through tight junctions. Recent studies have shown that incubation of Caco-2BBe cells with 1,25(OH)2 vitamin D3 protected the cells against AICE-induced destruction of cross-epithelial resistance and tight junction proteins. Vitamin D3-deficient mice treated with DSS showed significant epithelial dysfunction and increased colitis susceptibility, aggravating the damage caused by AIEC colonization. Thus, vitamin D deficiency after AIEC infection may disrupt the mucosal barrier, leading to increased susceptibility to mucosal damage and an increased risk of IBD[111].
Some food ingredients, such as lactoferrin in milk, have antibacterial and/or anti-inflammatory properties that inhibit bacterial adhesion and invasion. Serum-derived bovine immunoglobulin/isolate has recently been extensively used to cause LF82-/DSS-induced attenuated colitis and inhibit the inflammatory cascade leading to IBD (Figure 1)[112].
Antibiotics: The efficacy and effects of antibiotics on IBD are not clear. In general, traditional antibiotics have shown poor efficacy against IBD, and they are currently used only to treat bacterial complications of CD, although antibiotics do provide relief, especially in CD patients with colitis. Antibiotic treatment, however, is no longer the main drug used to treat IBD, as the increase in multidrug-resistant bacteria promotes the growth of pathogenic bacteria[113]. In addition, long-term treatment of IBD with antibiotics, such as metronidazole, leads to profound adverse reactions and poor tolerability. Therefore, treatment with antibiotics may be effective only under certain conditions, such as for patients colonized with AIEC strains with high activity. Antibiotics such as ciprofloxacin and rifaximin induce fewer side effects than immunosuppressants and may be safer options for patients with CD infected with AIEC[114]. Further evaluation of antibiotics in IBD is needed, especially for specific cases, such as CD patients with AIEC colonization. In the near future, antibiotics may be used to specifically defend against invasive bacteria. Therefore, further reducing the side effects of antibiotics and selectively targeting specific bacteria are foci of future research (Figure 2)[112].
Probiotics, prebiotics, and postbiotics: Probiotics are living microbes that provide health benefits for the host when consumed in sufficient quantities. Prebiotics can be selectively utilized by bacteria in the host body and converted to benefit the health of the host. The American Gastroenterological Association recommends that probiotics be considered for the treatment of functional symptoms of IBD[115]. Anti-inflammatory probiotic administration has been validated in CD as a promising modulated microbiome composition. Studies have shown that the use of yeast probiotics can prevent colitis in mice[116]. Therefore, probiotics may be used in a novel approach for the treatment of CD patients with abnormal AIEC colonization in the ileum. In addition, prebiotics that are indigestible can stimulate the growth and metabolic activity of beneficial microbiota species. Recently, the safety of probiotics in CD patients has been questioned due to the breakdown of symbiotic flora tolerance in the context of acute inflammation[117]. In these cases, postbiotics (soluble factors produced by probiotics that trigger an immunomodulatory response) can act as therapeutic agents against CD because they can be administered in a purified intact form, ensuring therapeutic safety.
Immunoregulatory therapy: New approaches are emerging to suppress or manipulate abnormal immune responses in patients with IBD. For example, an anti-integrin α4β7 humanized monoclonal antibody (vedolizumab), which effectively prevents immune cell colonization in the intestinal mucosa, has shown promising results as a CD treatment[118]. In addition, a study has suggested that the injection of activated Tregs into CD patients holds potential therapeutic promise and may lead to the development of personalized immunotherapy. Notably, autologous hematopoietic stem cell transplantation has been reported to produce lasting remission in patients with CD that has been difficult to treat through conventional methods. Today, methods of manipulating a patient's microbiome through antibiotic therapy, fecal transplantation, nutrient interventions, or probiotics can be used alone or in combination with immunotherapy to alleviate symptoms of the disease or as a postoperative treatment to prevent recurrence[119].
Antiadhesion molecules: Antiadhesion molecules can specifically bind to cellular adhesion molecule-1, which is expressed by lymphoid cells, and block its interaction with SCRAMSCRAM1[120]. Then, lymphocytes are prevented from migrating and homing from the blood to inflammatory tissue, ultimately inhibiting intestinal inflammation. Vedolizumab, a representative anti-adhesion molecular agent, shows intestinal specificity and can specifically antagonize intestinal α4β7 integrin heterodimers, but its effect is relatively slow, with a clinical response time of at least 2 wk[121]. A study has shown that the clinical remission rate of patients with UC was 47.1% and that the mucosal healing rate was 40.9% after 6 wk of treatment with vedolizumab[122]. The clinical response rate of patients with CD was 14.5% 6 wk after treatment with vedolizumab. These results indicate that the efficacy of vedolizumab in the treatment of IBD is slow, which may be related to the vedolizumab mechanism of action. Although vedolizumab can specifically bind the α4β7 integrin heterodimer, its target is an upstream signaling pathway in the inflammatory response pathway, and it exerts no inhibitory effect on homing lymphocytes or their secreted cytokines; therefore, it cannot exert an anti-inflammatory effect in the short term[123]. However, these mechanisms of action contribute to the relatively long-lasing anti-inflammatory effect of vedolizumab. In addition, vedolizumab is the only novel biologic agent with high intestinal selectivity, which indicates a better clinical treatment effect for moderate to severe IBD patients with intestinal lesions, and the risk of vedolizumab inducing opportunistic infections is very low[124].
Fecal microbiota transplantation: Fecal microbiota transplantation (FMT) is the transplantation of fecal microbiota from a healthy donor into the gastrointestinal tract of a patient to correct bacterial dysregulation and reshape intestinal homeostasis. As an ancient intestinal microbiota intervention measure, FMT was first used according to The Elbow Reserve Urgent Prescription written by Ge Hong in the Eastern Jin Dynasty. Specifically, stool suspension was applied to treat food poisoning and severe diarrhea. Compared with oral probiotics, FMT has the advantages of a rich variety of transplanted bacteria, at high numbers, and with maximum retention of host functional bacteria. In contrast to applying specific bacteria to regulate the intestinal flora, the key to FMT effectiveness is the reliance on the whole flora to restore homeostasis in patients[125]. FMT is recognized as the most effective treatment for refractory Clostridium difficile infections (CDIs). FMT has been written into the US CDI treatment guidelines and has led to an 85%-90% response rate in one treatment and a 100% response rate in two treatments. In addition to CDI treatment, the greatest evidence showing the benefits of FMT has been obtained from studies of patients with IBD. Namely, a meta-analysis showed that the remission rate of patients with CD treated by FMT was 50.5% (42/83), which was significantly better than that of patients with UC [36% (201/555)][126]. Studies have shown that the choice of FMT transplantation has a direct impact on the efficacy of IBS treatment, and the transplantation of fresh or frozen feces administered by colonoscopy or nasal jejunal tube is superior to fecal bacteria capsules[127]. This approach can repair important components of the gut microbiome and reverse inappropriate immune stimulation in CD, making the gut ecosystem unsuitable for AIEC colonization. Changes in the intestinal flora are an important cause of IBD. Although the safety and efficacy of fecal transplantation are still being assessed, relevant studies have confirmed its efficacy.
Phage therapy: Bacteriophages are viruses that host bacteria; therefore, they are also called bacterial viruses. Phages can shape bacterial communities by inducing bacterial cell lysis and influencing bacterial phenotype acquisition. As an antibacterial therapy, phage therapy has the advantage of precise targeting compared with antibiotics and can be used to regulate intestinal flora and kill multiple types of drug-resistant bacteria. Phage therapy is used as an alternative or complementary therapy to antibiotics in parts of Eastern Europe[128]. Although phages have not been approved for the treatment of bacterial infections in Western countries, phage preparations are commercially available as prebiotics (e.g., prophage and prephage) for human use in the United States.
In gastroenterology, phage therapy research has focused on infectious diseases. Young rabbit models showed that a single phage, Phi_1, prophylactically and therapeutically controlled cholera without inducing detectable levels of resistance. AIEC is a common pathogenic bacterium in the ileal mucosa of CD patients, and it can bind to the CEACAM6 receptor expressed on the epithelial cell surface and induce colitis symptoms. Three phage mixtures targeting AIEC significantly reduced the colonization of the AIEC LF82 strain in the intestinal mucosa of CEABAC10 transgenic mice expressing the human CEACAM6 AIEC receptor, alleviating DSS-induced colitis symptoms and providing a potential new option for the treatment of CD by targeting the AIEC strain[129].
Extensive studies on the abundance and diversity of phages in the human gut have revealed some differences between the phages colonized in IBD patients and those colonized in healthy subjects, suggesting that phages may play a role in the development of IBD. Newly developed animal models have been used to investigate the dynamics of phage/bacterial populations in the gut, opening up a new field of research in this area[130].
Restoration/induction of autophagy: Autophagy is a conserved catabolic pathway that degrades cytoplasmic components and organelles in lysosomes. The basic function of autophagy is to maintain cell energy and survival by recycling amino acids and fatty acids. Autophagy protects cells by degrading damaged proteins and organelles, as well as pathogens within cells[131]. Therefore, autophagy dysfunction plays an important role in the pathogenesis of IBD, and the autophagy-related gene ATG16L1 and immune-related guanosine triphosphatase gene have been confirmed to be risk genes for CD[132].
The IECs of patients with CD involving the ileum are often compromised by AIEC infection. Autophagy can promote adaptive immune clearance of infection, causing bacteria through the MHC-II presentation pathway, and can directly phagocytose and clear invasive pathogens[133]. Autophagy at the physiological level effectively inhibited the proliferation of AIEC, while IRGM- and ATG16L1-deficient cells showed a large amount of AIEC proliferation. Defects in autophagy led to decreased clearance of intracellular bacteria, resulting in the persistence of intracellular infection and recruitment of more inflammatory cells, as well as excessive secretion of cytokines and the formation of chronic granuloma. All these effects may be related to the pathogenesis of CD[134].
Other possible therapeutic interventions: Interference of Enterobacteriaceae adrenergic receptor QseC activity through the biochemical inhibitor LED209 alleviated experimental colitis[135]. In pathogenic IBD-associated E. coli strains, inactivation of QseC reduced in vitro virulence and persistence in a simple microbiome in vivo[136]. These results provide insights into the use of antiviral approaches to treat not only pathogens but also colitis-causing bacteria[137].
Gut microbiota imbalances play an important role in IBD, but no single pathogenic microorganism critical to IBD, which is specific to the IBD terminal ileum mucosa or can invade IECs, has been found. Invasive E. coli adhesion to macrophages is considered to be closely related to the pathogenesis of IBD. Further study of the specific biological characteristics of AIEC may contribute to a further under
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Nakaji K, Japan; Sahin Y, Turkey S-Editor: Wang LL L-Editor: Wang TQ P-Editor: Wang LL
| 1. | Rung GW, Williams D, Gelb DE, Grubb M. Isobaric spinal anesthesia for lumbar disk surgery. Anesth Analg. 1997;84:1165-1166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 2. | Mirsepasi-Lauridsen HC, Vallance BA, Krogfelt KA, Petersen AM. Escherichia coli Pathobionts Associated with Inflammatory Bowel Disease. Clin Microbiol Rev. 2019;32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 163] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 3. | Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16:38-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 364] [Article Influence: 91.0] [Reference Citation Analysis (0)] |
| 4. | Khan AA, Khan Z, Malik A, Kalam MA, Cash P, Ashraf MT, Alshamsan A. Colorectal cancer-inflammatory bowel disease nexus and felony of Escherichia coli. Life Sci. 2017;180:60-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Zhao Z, Xu S, Zhang W, Wu D, Yang G. Probiotic Escherichia coli NISSLE 1917 for inflammatory bowel disease applications. Food Funct. 2022;13:5914-5924. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 24] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 6. | Nadalian B, Yadegar A, Houri H, Olfatifar M, Shahrokh S, Asadzadeh Aghdaei H, Suzuki H, Zali MR. Prevalence of the pathobiont adherent-invasive Escherichia coli and inflammatory bowel disease: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2021;36:852-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 7. | Walczuk U, Sobieszczańska B, Turniak M, Rzeszutko M, Duda-Madej A, Iwańczak B. The prevalence of mucosa-associated diffusely adherent Escherichia coli in children with inflammatory bowel disease. Adv Clin Exp Med. 2019;28:899-905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Currò D, Ianiro G, Pecere S, Bibbò S, Cammarota G. Probiotics, fibre and herbal medicinal products for functional and inflammatory bowel disorders. Br J Pharmacol. 2017;174:1426-1449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 9. | Lee JG, Han DS, Jo SV, Lee AR, Park CH, Eun CS, Lee Y. Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: Potential impact on clinical outcomes. PLoS One. 2019;14:e0216165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 10. | Zheng L, Ji YY, Wen XL, Duan SL. Fecal microbiota transplantation in the metabolic diseases: Current status and perspectives. World J Gastroenterol. 2022;28:2546-2560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 6] [Cited by in F6Publishing: 14] [Article Influence: 7.0] [Reference Citation Analysis (4)] |
| 11. | Micenková L, Frankovičová L, Jaborníková I, Bosák J, Dítě P, Šmarda J, Vrba M, Ševčíková A, Kmeťová M, Šmajs D. Escherichia coli isolates from patients with inflammatory bowel disease: ExPEC virulence- and colicin-determinants are more frequent compared to healthy controls. Int J Med Microbiol. 2018;308:498-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Behrouzi A, Mazaheri H, Falsafi S, Tavassol ZH, Moshiri A, Siadat SD. Intestinal effect of the probiotic Escherichia coli strain Nissle 1917 and its OMV. J Diabetes Metab Disord. 2020;19:597-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Meheissen M, Header D, Abdelaty K. Phylogenetic and pathotype analysis of Escherichia coli stool isolates from Egyptian patients with inflammatory bowel disease. Germs. 2019;9:172-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Fang X, Monk JM, Mih N, Du B, Sastry AV, Kavvas E, Seif Y, Smarr L, Palsson BO. Escherichia coli B2 strains prevalent in inflammatory bowel disease patients have distinct metabolic capabilities that enable colonization of intestinal mucosa. BMC Syst Biol. 2018;12:66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 15. | Dubinsky V, Reshef L, Rabinowitz K, Wasserberg N, Dotan I, Gophna U. Escherichia coli strains from patients with inflammatory bowel diseases have disease-specific genomic adaptations. J Crohns Colitis. 2022;. [DOI] [Cited in This Article: ] |
| 16. | Barrios-Villa E, Martínez de la Peña CF, Lozano-Zaraín P, Cevallos MA, Torres C, Torres AG, Rocha-Gracia RDC. Comparative genomics of a subset of Adherent/Invasive Escherichia coli strains isolated from individuals without inflammatory bowel disease. Genomics. 2020;112:1813-1820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Renouf MJ, Cho YH, McPhee JB. Emergent Behavior of IBD-Associated Escherichia coli During Disease. Inflamm Bowel Dis. 2019;25:33-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Khan S, Imran A, Malik A, Chaudhary AA, Rub A, Jan AT, Syed JB, Rolfo C. Bacterial imbalance and gut pathologies: Association and contribution of E. coli in inflammatory bowel disease. Crit Rev Clin Lab Sci. 2019;56:1-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Naseer M, Poola S, Ali S, Samiullah S, Tahan V. Prebiotics and Probiotics in Inflammatory Bowel Disease: Where are we now and where are we going? Curr Clin Pharmacol. 2020;15:216-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 20. | Dogan B, Fu J, Zhang S, Scherl EJ, Simpson KW. Rifaximin decreases virulence of Crohn's disease-associated Escherichia coli and epithelial inflammatory responses. J Antibiot (Tokyo). 2018;71:485-494. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Kamali Dolatabadi R, Feizi A, Halaji M, Fazeli H, Adibi P. The Prevalence of Adherent-Invasive Escherichia coli and Its Association With Inflammatory Bowel Diseases: A Systematic Review and Meta-Analysis. Front Med (Lausanne). 2021;8:730243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 13] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Olivares-Morales MJ, De La Fuente MK, Dubois-Camacho K, Parada D, Diaz-Jiménez D, Torres-Riquelme A, Xu X, Chamorro-Veloso N, Naves R, Gonzalez MJ, Quera R, Figueroa C, Cidlowski JA, Vidal RM, Hermoso MA. Glucocorticoids Impair Phagocytosis and Inflammatory Response Against Crohn's Disease-Associated Adherent-Invasive Escherichia coli. Front Immunol. 2018;9:1026. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Dogan B, Belcher-Timme HF, Dogan EI, Jiang ZD, DuPont HL, Snyder N, Yang S, Chandler B, Scherl EJ, Simpson KW. Evaluation of Escherichia coli pathotypes associated with irritable bowel syndrome. FEMS Microbiol Lett. 2018;365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Ganji-Arjenaki M, Rafieian-Kopaei M. Probiotics are a good choice in remission of inflammatory bowel diseases: A meta analysis and systematic review. J Cell Physiol. 2018;233:2091-2103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 184] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 25. | Azimi T, Nasiri MJ, Chirani AS, Pouriran R, Dabiri H. The role of bacteria in the inflammatory bowel disease development: a narrative review. APMIS. 2018;126:275-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 26. | Viladomiu M, Metz ML, Lima SF, Jin WB, Chou L; JRI Live Cell Bank, Guo CJ, Diehl GE, Simpson KW, Scherl EJ, Longman RS. Adherent-invasive E. coli metabolism of propanediol in Crohn's disease regulates phagocytes to drive intestinal inflammation. Cell Host Microbe. 2021;29:607-619.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 54] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 27. | Derer S, Brethack AK, Pietsch C, Jendrek ST, Nitzsche T, Bokemeyer A, Hov JR, Schäffler H, Bettenworth D, Grassl GA, Sina C. Inflammatory Bowel Disease-associated GP2 Autoantibodies Inhibit Mucosal Immune Response to Adherent-invasive Bacteria. Inflamm Bowel Dis. 2020;26:1856-1868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 28. | Shaler CR, Elhenawy W, Coombes BK. The Unique Lifestyle of Crohn's Disease-Associated Adherent-Invasive Escherichia coli. J Mol Biol. 2019;431:2970-2981. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 29. | Cevallos SA, Lee JY, Velazquez EM, Foegeding NJ, Shelton CD, Tiffany CR, Parry BH, Stull-Lane AR, Olsan EE, Savage HP, Nguyen H, Ghanaat SS, Byndloss AJ, Agu IO, Tsolis RM, Byndloss MX, Bäumler AJ. 5-Aminosalicylic Acid Ameliorates Colitis and Checks Dysbiotic Escherichia coli Expansion by Activating PPAR-γ Signaling in the Intestinal Epithelium. mBio. 2021;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 43] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 30. | Luo X, Song H, Yang J, Han B, Feng Y, Leng Y, Chen Z. Encapsulation of Escherichia coli strain Nissle 1917 in a chitosan-alginate matrix by combining layer-by-layer assembly with CaCl2 cross-linking for an effective treatment of inflammatory bowel diseases. Colloids Surf B Biointerfaces. 2020;189:110818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Zheng L, Wen XL, Duan SL. Role of metabolites derived from gut microbiota in inflammatory bowel disease. World J Clin Cases. 2022;10:2660-2677. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 13] [Cited by in F6Publishing: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 32. | Bao CH, Wang CY, Li GN, Yan YL, Wang D, Jin XM, Wu LY, Liu HR, Wang XM, Shi Z, Wu HG. Effect of mild moxibustion on intestinal microbiota and NLRP6 inflammasome signaling in rats with post-inflammatory irritable bowel syndrome. World J Gastroenterol. 2019;25:4696-4714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 37] [Cited by in F6Publishing: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 33. | Costa RFA, Ferrari MLA, Bringer MA, Darfeuille-Michaud A, Martins FS, Barnich N. Characterization of mucosa-associated Escherichia coli strains isolated from Crohn's disease patients in Brazil. BMC Microbiol. 2020;20:178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Mirsepasi-Lauridsen HC, Struve C, Petersen AM, Krogfelt KA. Effect of α-Hemolysin Producing E. coli in Two Different Mouse Strains in a DSS Model of Inflammatory Bowel Disease. Microorganisms. 2020;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 35. | Metta V, Leta V, Mrudula KR, Prashanth LK, Goyal V, Borgohain R, Chung-Faye G, Chaudhuri KR. Gastrointestinal dysfunction in Parkinson's disease: molecular pathology and implications of gut microbiome, probiotics, and fecal microbiota transplantation. J Neurol. 2022;269:1154-1163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 50] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 36. | Elhenawy W, Tsai CN, Coombes BK. Host-Specific Adaptive Diversification of Crohn's Disease-Associated Adherent-Invasive Escherichia coli. Cell Host Microbe. 2019;25:301-312.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 37. | Dalmasso G, Nguyen HTT, Faïs T, Massier S, Barnich N, Delmas J, Bonnet R. Crohn's Disease-Associated Adherent-Invasive Escherichia coli Manipulate Host Autophagy by Impairing SUMOylation. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Siniagina M, Markelova M, Laikov A, Boulygina E, Khusnutdinova D, Kharchenko A, Misbakhova A, Grigoryeva T. Cultivated Escherichia coli diversity in intestinal microbiota of Crohn's disease patients and healthy individuals: Whole genome data. Data Brief. 2020;28:104948. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 39. | Ormsby MJ, Johnson SA, Carpena N, Meikle LM, Goldstone RJ, McIntosh A, Wessel HM, Hulme HE, McConnachie CC, Connolly JPR, Roe AJ, Hasson C, Boyd J, Fitzgerald E, Gerasimidis K, Morrison D, Hold GL, Hansen R, Walker D, Smith DGE, Wall DM. Propionic Acid Promotes the Virulent Phenotype of Crohn's Disease-Associated Adherent-Invasive Escherichia coli. Cell Rep. 2020;30:2297-2305.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 40. | Burstiner LS, Silver J, Burstiner LJ, Teymoorian A, Pallav K, Jones D, Owings A, Glover S. Escherichia coli O157: H7 sepsis following fecal microbiota transplant in an IgA-deficient inflammatory bowel disease patient. Gastroenterol Rep (Oxf). 2022;10:goab041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 41. | Xia Y, Wang J, Fang X, Dou T, Han L, Yang C. Combined analysis of metagenomic data revealed consistent changes of gut microbiome structure and function in inflammatory bowel disease. J Appl Microbiol. 2021;131:3018-3031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 42. | Cardoneanu A, Mihai C, Rezus E, Burlui A, Popa I, Cijevschi Prelipcean C. Gut microbiota changes in inflammatory bowel diseases and ankylosing spondilytis. J Gastrointestin Liver Dis. 2021;30:46-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 43. | Guo W, Li B, Zhou H, Zhang C, Wang X, Ni C. [Construction and characterization of a bio-detector for inflammatory bowel disease]. Sheng Wu Gong Cheng Xue Bao. 2018;34:1906-1914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 44. | Mazzarella G, Perna A, Marano A, Lucariello A, Rotondi Aufiero V, Sorrentino A, Melina R, Guerra G, Taccone FS, Iaquinto G, De Luca A. Pathogenic Role of Associated Adherent-Invasive Escherichia coli in Crohn's Disease. J Cell Physiol. 2017;232:2860-2868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 45. | Nagayama M, Yano T, Atarashi K, Tanoue T, Sekiya M, Kobayashi Y, Sakamoto H, Miura K, Sunada K, Kawaguchi T, Morita S, Sugita K, Narushima S, Barnich N, Isayama J, Kiridooshi Y, Shiota A, Suda W, Hattori M, Yamamoto H, Honda K. TH1 cell-inducing Escherichia coli strain identified from the small intestinal mucosa of patients with Crohn's disease. Gut Microbes. 2020;12:1788898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 46. | Dalmasso G, Nguyen HTT, Faïs T, Massier S, Chevarin C, Vazeille E, Barnich N, Delmas J, Bonnet R. Yersiniabactin Siderophore of Crohn's Disease-Associated Adherent-Invasive Escherichia coli Is Involved in Autophagy Activation in Host Cells. Int J Mol Sci. 2021;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 47. | Abdelhalim KA, Uzel A, Ünal NG. The role of major virulence factors and pathogenicity of adherent-invasive Escherichia coli in patients with Crohn's disease. Prz Gastroenterol. 2020;15:279-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 48. | Bukato O, Pobeguts O, Rakitina D, Baikova J, Butenko I, Silantyev A, Fisunov G, Govorun V. Proteomic dataset: Profiling of membrane fraction of Escherichia coli isolated from Crohn's disease patients after adhesion and invasion experiments. Data Brief. 2019;27:104417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 49. | Tyakht AV, Manolov AI, Kanygina AV, Ischenko DS, Kovarsky BA, Popenko AS, Pavlenko AV, Elizarova AV, Rakitina DV, Baikova JP, Ladygina VG, Kostryukova ES, Karpova IY, Semashko TA, Larin AK, Grigoryeva TV, Sinyagina MN, Malanin SY, Shcherbakov PL, Kharitonova AY, Khalif IL, Shapina MV, Maev IV, Andreev DN, Belousova EA, Buzunova YM, Alexeev DG, Govorun VM. Genetic diversity of Escherichia coli in gut microbiota of patients with Crohn's disease discovered using metagenomic and genomic analyses. BMC Genomics. 2018;19:968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 50. | Liu G, Gu K, Wang F, Jia G, Zhao H, Chen X, Wu C, Zhang R, Tian G, Cai J, Tang J, Wang J. Tryptophan Ameliorates Barrier Integrity and Alleviates the Inflammatory Response to Enterotoxigenic Escherichia coli K88 Through the CaSR/Rac1/PLC-γ1 Signaling Pathway in Porcine Intestinal Epithelial Cells. Front Immunol. 2021;12:748497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 51. | Fang X, Monk JM, Nurk S, Akseshina M, Zhu Q, Gemmell C, Gianetto-Hill C, Leung N, Szubin R, Sanders J, Beck PL, Li W, Sandborn WJ, Gray-Owen SD, Knight R, Allen-Vercoe E, Palsson BO, Smarr L. Metagenomics-Based, Strain-Level Analysis of Escherichia coli From a Time-Series of Microbiome Samples From a Crohn's Disease Patient. Front Microbiol. 2018;9:2559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 52. | Zhang S, Fu J, Dogan B, Scherl EJ, Simpson KW. 5-Aminosalicylic acid downregulates the growth and virulence of Escherichia coli associated with IBD and colorectal cancer, and upregulates host anti-inflammatory activity. J Antibiot (Tokyo). 2018;71:950-961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 53. | Ormsby MJ, Logan M, Johnson SA, McIntosh A, Fallata G, Papadopoulou R, Papachristou E, Hold GL, Hansen R, Ijaz UZ, Russell RK, Gerasimidis K, Wall DM. Inflammation associated ethanolamine facilitates infection by Crohn's disease-linked adherent-invasive Escherichia coli. EBioMedicine. 2019;43:325-332. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 54. | Elhenawy W, Hordienko S, Gould S, Oberc AM, Tsai CN, Hubbard TP, Waldor MK, Coombes BK. High-throughput fitness screening and transcriptomics identify a role for a type IV secretion system in the pathogenesis of Crohn's disease-associated Escherichia coli. Nat Commun. 2021;12:2032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 55. | Manchester AC, Dogan B, Guo Y, Simpson KW. Escherichia coli-associated granulomatous colitis in dogs treated according to antimicrobial susceptibility profiling. J Vet Intern Med. 2021;35:150-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 56. | Abdelhalim KA, Uzel A, Gülşen Ünal N. Virulence determinants and genetic diversity of adherent-invasive Escherichia coli (AIEC) strains isolated from patients with Crohn's disease. Microb Pathog. 2020;145:104233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Demarre G, Prudent V, Schenk H, Rousseau E, Bringer MA, Barnich N, Tran Van Nhieu G, Rimsky S, De Monte S, Espéli O. The Crohn's disease-associated Escherichia coli strain LF82 relies on SOS and stringent responses to survive, multiply and tolerate antibiotics within macrophages. PLoS Pathog. 2019;15:e1008123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Hajam IA, Ali F, Young J, Garcia MA, Cannavino C, Ramchandar N, Liu GY. Anti-Inflammatory Properties of Plasma from Children with Short Bowel Syndrome. Pathogens. 2021;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Zhilu X, Xiangqian D, Keli Y, Caroline C, Jingwan Z, Yu L, Tao Z, Cheung CL, Yang S, Fengrui Z, Kl CF, Jy SJ, Jun Y, Anthony B, Nicolas B, Jean-Frédéric C, Sunny Hei W, Yinglei M, Siew C N. Association of Adherent-invasive Escherichia coli with severe Gut Mucosal dysbiosis in Hong Kong Chinese population with Crohn's disease. Gut Microbes. 2021;13:1994833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Mancini NL, Rajeev S, Jayme TS, Wang A, Keita ÅV, Workentine ML, Hamed S, Söderholm JD, Lopes F, Shutt TE, Shearer J, McKay DM. Crohn's Disease Pathobiont Adherent-Invasive E coli Disrupts Epithelial Mitochondrial Networks With Implications for Gut Permeability. Cell Mol Gastroenterol Hepatol. 2021;11:551-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 61. | Simčič S, Berlec A, Stopinšek S, Štrukelj B, Orel R. Engineered and wild-type L. lactis promote anti-inflammatory cytokine signalling in inflammatory bowel disease patient's mucosa. World J Microbiol Biotechnol. 2019;35:45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 62. | Sarabi Asiabar A, Asadzadeh Aghdaei H, Sabokbar A, Zali MR, Feizabadi MM. Investigation of adherent-invasive E. coli in patients with Crohn's disease. Med J Islam Repub Iran. 2018;32:11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 63. | Zaidi D, Huynh HQ, Carroll MW, Mandal R, Wishart DS, Wine E. Gut Microenvironment and Bacterial Invasion in Paediatric Inflammatory Bowel Diseases. J Pediatr Gastroenterol Nutr. 2020;71:624-632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 64. | Marchukov D, Misselwitz B. [Insights into the Pathogenesis of Inflammatory Bowel Diseases: Genetics and Microbiota]. Ther Umsch. 2019;75:273-279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 65. | Axelrad JE, Joelson A, Nobel YR, Lawlor G, Green PHR, Lichtiger S, Lebwohl B. Enteric Infection in Relapse of Inflammatory Bowel Disease: The Utility of Stool Microbial PCR Testing. Inflamm Bowel Dis. 2017;23:1034-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 66. | Bekić D, Belošić Halle Ž. Infliximab rescue therapy in a patient with acute severe ulcerative colitis and coronavirus disease 2019 followed by Escherichia coli 0157:H7 infection: a case report. Croat Med J. 2021;62:634-637. [PubMed] [Cited in This Article: ] |
| 67. | Zhang B, Liu Y, Lan X, Xu X, Zhang X, Li X, Zhao Y, Li G, Du C, Lu S, Wang H. Oral Escherichia coli expressing IL-35 meliorates experimental colitis in mice. J Transl Med. 2018;16:71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 68. | Kim DH, Park J, Kim S, Yoon MY, Ma HW, Park IS, Son M, Kim JH, Kim TI, Kim WH, Yoon SS, Kim SW, Cheon JH. An Escherichia coli strain with extra catalase activity protects against murine colitis by scavenging hydrogen peroxide and regulating regulatory t cell/interleukin-17 pathways. Free Radic Biol Med. 2021;174:110-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 69. | Dadlani A, Bandikatla S, Koch JA. A Rare Case of Escherichia coli Chest Wall Abscess With Rib Osteomyelitis in a Patient With Crohn's Disease. Cureus. 2021;13:e13860. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (1)] |
| 70. | de Sousa Figueiredo MB, Pradel E, George F, Mahieux S, Houcke I, Pottier M, Fradin C, Neut C, Daniel C, Bongiovanni A, Foligné B, Titécat M. Adherent-Invasive and Non-Invasive Escherichia coli Isolates Differ in Their Effects on Caenorhabditis elegans' Lifespan. Microorganisms. 2021;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 71. | Rolhion N. A milestone in screening for adherent-invasive E. coli colonization in patients with Crohn's disease? United European Gastroenterol J. 2021;9:995-996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 72. | Shen Q, Huang Z, Yao J, Jin Y. Extracellular vesicles-mediated interaction within intestinal microenvironment in inflammatory bowel disease. J Adv Res. 2022;37:221-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 73. | Galtier M, De Sordi L, Sivignon A, de Vallée A, Maura D, Neut C, Rahmouni O, Wannerberger K, Darfeuille-Michaud A, Desreumaux P, Barnich N, Debarbieux L. Bacteriophages Targeting Adherent Invasive Escherichia coli Strains as a Promising New Treatment for Crohn's Disease. J Crohns Colitis. 2017;11:840-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 70] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 74. | McCoy CS, Mannion AJ, Feng Y, Madden CM, Artim SC, Au GG, Dolan M, Haupt JL, Burns MA, Sheh A, Fox JG. Cytotoxic Escherichia coli strains encoding colibactin, cytotoxic necrotizing factor, and cytolethal distending toxin colonize laboratory common marmosets (Callithrix jacchus). Sci Rep. 2021;11:2309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 75. | Aygun H, Karamese M, Ozic C, Uyar F. The effects of mucosal media on some pathogenic traits of Crohn's disease-associated Escherichia coli LF82. Future Microbiol. 2018;13:141-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 76. | Baldelli V, Scaldaferri F, Putignani L, Del Chierico F. The Role of Enterobacteriaceae in Gut Microbiota Dysbiosis in Inflammatory Bowel Diseases. Microorganisms. 2021;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 98] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 77. | Agus A, Richard D, Faïs T, Vazeille E, Chervy M, Bonnin V, Dalmasso G, Denizot J, Billard E, Bonnet R, Buisson A, Barnich N, Delmas J. Propionate catabolism by CD-associated adherent-invasive E. coli counteracts its anti-inflammatory effect. Gut Microbes. 2021;13:1-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 78. | Astley DJ, Masters N, Kuballa A, Katouli M. Commonality of adherent-invasive Escherichia coli isolated from patients with extraintestinal infections, healthy individuals and the environment. Eur J Clin Microbiol Infect Dis. 2021;40:181-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 79. | Leccese G, Bibi A, Mazza S, Facciotti F, Caprioli F, Landini P, Paroni M. Probiotic Lactobacillus and Bifidobacterium Strains Counteract Adherent-Invasive Escherichia coli (AIEC) Virulence and Hamper IL-23/Th17 Axis in Ulcerative Colitis, but Not in Crohn's Disease. Cells. 2020;9. [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 80. | Park HB, Wei Z, Oh J, Xu H, Kim CS, Wang R, Wyche TP, Piizzi G, Flavell RA, Crawford JM. Sulfamethoxazole drug stress upregulates antioxidant immunomodulatory metabolites in Escherichia coli. Nat Microbiol. 2020;5:1319-1329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 81. | Camprubí-Font C, Martinez-Medina M. Why the discovery of adherent-invasive Escherichia coli molecular markers is so challenging? World J Biol Chem. 2020;11:1-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 82. | Lin Q, Fu Q, Li X, Luo Y, Luo J, Chen D, Mao X, Yu B, Zheng P, Huang Z, Yu J, Yan H, He J. Human β-Defensin 118 Attenuates Escherichia coli K88-Induced Inflammation and Intestinal Injury in Mice. Probiotics Antimicrob Proteins. 2021;13:586-597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 83. | Perna A, Hay E, Contieri M, De Luca A, Guerra G, Lucariello A. Adherent-invasive Escherichia coli (AIEC): Cause or consequence of inflammation, dysbiosis, and rupture of cellular joints in patients with IBD? J Cell Physiol. 2020;235:5041-5049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 84. | Oberc AM, Fiebig-Comyn AA, Tsai CN, Elhenawy W, Coombes BK. Antibiotics Potentiate Adherent-Invasive E. coli Infection and Expansion. Inflamm Bowel Dis. 2019;25:711-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 85. | Lu J, Dong B, Chen A, He F, Peng B, Wu Z, Cao J, Li W. Escherichia coli promotes DSS induced murine colitis recovery through activation of the TLR4/NF κB signaling pathway. Mol Med Rep. 2019;19:2021-2028. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 86. | Lopez LR, Barlogio CJ, Broberg CA, Wang J, Arthur JC. A nadA Mutation Confers Nicotinic Acid Auxotrophy in Pro-carcinogenic Intestinal Escherichia coli NC101. Front Microbiol. 2021;12:670005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 87. | Chervy M, Barnich N, Denizot J. Adherent-Invasive E. coli: Update on the Lifestyle of a Troublemaker in Crohn's Disease. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 88. | López-Siles M, Camprubí-Font C, Gómez Del Pulgar EM, Sabat Mir M, Busquets D, Sanz Y, Martinez-Medina M. Prevalence, Abundance, and Virulence of Adherent-Invasive Escherichia coli in Ulcerative Colitis, Colorectal Cancer, and Coeliac Disease. Front Immunol. 2022;13:748839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 9] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 89. | Stange EF, von Bünau R, Erhardt A. Ulcerative colitis-Maintanance of Remission With the Probiotic Escherichia coli Strain Nissle 1917. Gastroenterology. 2021;160:2632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 90. | Rodríguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, Fernández-Caballero JA, García F, Rodríguez-Cabezas ME, Gálvez J. The Administration of Escherichia coli Nissle 1917 Ameliorates Development of DSS-Induced Colitis in Mice. Front Pharmacol. 2018;9:468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 91. | Elhenawy W, Oberc A, Coombes BK. A polymicrobial view of disease potential in Crohn's-associated adherent-invasive E. coli. Gut Microbes. 2018;9:166-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 92. | Rakitina DV, Manolov AI, Kanygina AV, Garushyants SK, Baikova JP, Alexeev DG, Ladygina VG, Kostryukova ES, Larin AK, Semashko TA, Karpova IY, Babenko VV, Ismagilova RK, Malanin SY, Gelfand MS, Ilina EN, Gorodnichev RB, Lisitsyna ES, Aleshkin GI, Scherbakov PL, Khalif IL, Shapina MV, Maev IV, Andreev DN, Govorun VM. Genome analysis of E. coli isolated from Crohn's disease patients. BMC Genomics. 2017;18:544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 93. | Dacquay LC, Tsang D, Chan D, Parkinson J, Philpott DJ, McMillen DR. E.coli Nissle increases transcription of flagella assembly and formate hydrogenlyase genes in response to colitis. Gut Microbes. 2021;13:1994832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 94. | Moshkovskaya M, Vakhrusheva T, Rakitina D, Baykova J, Panasenko O, Basyreva L, Gusev S, Gusev A, Mikhalchik E, Smolina N, Dobretsov G, Scherbakov P, Parfenov A, Fadeeva N, Pobeguts O, Govorun V. Neutrophil activation by Escherichia coli isolates from human intestine: effects of bacterial hydroperoxidase activity and surface hydrophobicity. FEBS Open Bio. 2020;10:414-426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 95. | Derke RM, Barron AJ, Billiot CE, Chaple IF, Lapi SE, Broderick NA, Gray MJ. The Cu(II) Reductase RclA Protects Escherichia coli against the Combination of Hypochlorous Acid and Intracellular Copper. mBio. 2020;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 96. | Zarei O, Arabestan MR, Majlesi A, Mohammadi Y, Alikhani MY. Determination of virulence determinants of Escherichia coli strains isolated from patients with colorectal cancer compared to the healthy subjects. Gastroenterol Hepatol Bed Bench. 2019;12:52-59. [PubMed] [Cited in This Article: ] |
| 97. | Yang Y, Liao Y, Ma Y, Gong W, Zhu G. The role of major virulence factors of AIEC involved in inflammatory bowl disease-a mini-review. Appl Microbiol Biotechnol. 2017;101:7781-7787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 98. | Sivignon A, Yu SY, Ballet N, Vandekerckove P, Barnich N, Guerardel Y. Heteropolysaccharides from S. cerevisiae show anti-adhesive properties against E. coli associated with Crohn's disease. Carbohydr Polym. 2021;271:118415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 99. | Lynch JP, Goers L, Lesser CF. Emerging strategies for engineering Escherichia coli Nissle 1917-based therapeutics. Trends Pharmacol Sci. 2022;43:772-786. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 28] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 100. | Steimle A, Menz S, Bender A, Ball B, Weber ANR, Hagemann T, Lange A, Maerz JK, Parusel R, Michaelis L, Schäfer A, Yao H, Löw HC, Beier S, Tesfazgi Mebrhatu M, Gronbach K, Wagner S, Voehringer D, Schaller M, Fehrenbacher B, Autenrieth IB, Oelschlaeger TA, Frick JS. Flagellin hypervariable region determines symbiotic properties of commensal Escherichia coli strains. PLoS Biol. 2019;17:e3000334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 101. | Kim D, Kim Y, Yoon SH. Development of a Genome-Scale Metabolic Model and Phenome Analysis of the Probiotic Escherichia coli Strain Nissle 1917. Int J Mol Sci. 2021;22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 102. | Chokr D, Cornu M, Neut C, Bortolus C, Charlet R, Desreumaux P, Speca S, Sendid B. Adherent invasive Escherichia coli (AIEC) strain LF82, but not Candida albicans, plays a profibrogenic role in the intestine. Gut Pathog. 2021;13:5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 103. | Veca R, O'Dea C, Burke J, Hatje E, Kuballa A, Katouli M. A Comparative Study of the Adherent-Invasive Escherichia coli Population and Gut Microbiota of Healthy Vegans vs Omnivores. Microorganisms. 2020;8. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 104. | Oh GM, Moon W, Seo KI, Jung K, Kim JH, Kim SE, Park MI, Park SJ. Therapeutic Potential of Escherichia coli Nissle 1917 in Clinically Remission-attained Ulcerative Colitis Patients: A Hospital-based Cohort Study. Korean J Gastroenterol. 2021;77:12-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 105. | He C, Wang H, Yu C, Peng C, Shu X, Liao W, Zhu Z. Alterations of Gut Microbiota in Patients With Intestinal Tuberculosis That Different From Crohn's Disease. Front Bioeng Biotechnol. 2021;9:673691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 106. | Buisson A, Vazeille E, Fumery M, Pariente B, Nancey S, Seksik P, Peyrin-Biroulet L, Allez M, Ballet N, Filippi J, Yzet C, Nachury M, Boschetti G, Billard E, Dubois A, Rodriguez S, Chevarin C, Goutte M, Bommelaer G, Pereira B, Hebuterne X, Barnich N; CEALIVE & REMIND study group. Faster and less invasive tools to identify patients with ileal colonization by adherent-invasive E. coli in Crohn's disease. United European Gastroenterol J. 2021;9:1007-1018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 107. | Mahmoudi H, Ghiasvand S, Zarei O, Hossainpour H, Alikhani MY. Identification of Quinolone and Colistin Resistance Genes in Escherichia Coli Strains Isolated from Mucosal Samples of Patients with Colorectal Cancer and Healthy Subjects. Recent Pat Antiinfect Drug Discov. 2020;15:30-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 108. | Delmas J, Gibold L, Faïs T, Batista S, Leremboure M, Sinel C, Vazeille E, Cattoir V, Buisson A, Barnich N, Dalmasso G, Bonnet R. Metabolic adaptation of adherent-invasive Escherichia coli to exposure to bile salts. Sci Rep. 2019;9:2175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 109. | Cochran L, Hill S, Lotti U, Allenspach K, Palma D, Forman M, Gary AT, Dogan B, McDonough SP, Simpson KW. Clinical characteristics and long-term outcome of E. coli-associated granulomatous ileocolitis in dogs: five cases (2010-2014). J Small Anim Pract. 2021;62:588-598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 110. | Ren J, Yan D, Wang Y, Zhang J, Li M, Xiong W, Jing X, Li P, Zhao W, Xiong X, Wu M, Zhong G. Inhibitor of Differentiation-2 Protein Ameliorates DSS-Induced Ulcerative Colitis by Inhibiting NF-κB Activation in Neutrophils. Front Immunol. 2021;12:760999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 2] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 111. | Alkaissi LY, Winberg ME, Heil SDS, Haapaniemi S, Myrelid P, Stange EF, Söderholm JD, Keita ÅV. Antagonism of Adherent Invasive E. coli LF82 With Human α-defensin 5 in the Follicle-associated Epithelium of Patients With Ileal Crohn's Disease. Inflamm Bowel Dis. 2021;27:1116-1127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 112. | Hughes ER, Winter MG, Alves da Silva L, Muramatsu MK, Jimenez AG, Gillis CC, Spiga L, Chanin RB, Santos RL, Zhu W, Winter SE. Reshaping of bacterial molecular hydrogen metabolism contributes to the outgrowth of commensal E. coli during gut inflammation. Elife. 2021;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 113. | Miki T, Hoshino Y, Sudo N, Ito M, Haneda T, Okada N. uvrY Deletion and Acetate Reduce Gut Colonization of Crohn's Disease-Associated Adherent-Invasive Escherichia coli by Decreasing Expression of Type 1 Fimbriae. Infect Immun. 2022;90:e0066221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 114. | Zhang S, Dogan B, Guo C, Herlekar D, Stewart K, Scherl EJ, Simpson KW. Short Chain Fatty Acids Modulate the Growth and Virulence of Pathosymbiont Escherichia coli and Host Response. Antibiotics (Basel). 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 115. | Touchon M, Perrin A, de Sousa JAM, Vangchhia B, Burn S, O'Brien CL, Denamur E, Gordon D, Rocha EP. Phylogenetic background and habitat drive the genetic diversification of Escherichia coli. PLoS Genet. 2020;16:e1008866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 116. | Zhang HJ, Xu B, Wang H, Wang GD, Jiang MZ, Lei C, Ding ML, Yu PF, Nie YZ, Wu KC, Sha SM, Li MB. IL-17 is a protection effector against the adherent-invasive Escherichia coli in murine colitis. Mol Immunol. 2018;93:166-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 117. | Mayorgas A, Dotti I, Martínez-Picola M, Esteller M, Bonet-Rossinyol Q, Ricart E, Salas A, Martínez-Medina M. A Novel Strategy to Study the Invasive Capability of Adherent-Invasive Escherichia coli by Using Human Primary Organoid-Derived Epithelial Monolayers. Front Immunol. 2021;12:646906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 118. | Gimier E, Chervy M, Agus A, Sivignon A, Billard E, Privat M, Viala S, Minet-Quinard R, Buisson A, Vazeille E, Barnich N, Denizot J. Methyl-donor supplementation prevents intestinal colonization by Adherent-Invasive E. coli in a mouse model of Crohn's disease. Sci Rep. 2020;10:12922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 119. | Souza ELS, Campos CLV, Reis DC, Cassali GD, Generoso SV, Cardoso VN, Azevedo V, Medeiros JD, Fernandes GR, Nicoli JR, Martins FS. Beneficial effects resulting from oral administration of Escherichia coli Nissle 1917 on a chronic colitis model. Benef Microbes. 2020;11:779-790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 120. | Ohno M, Hasegawa M, Hayashi A, Caballero-Flores G, Alteri CJ, Lawley TD, Kamada N, Núñez G, Inohara N. Lipopolysaccharide O structure of adherent and invasive Escherichia coli regulates intestinal inflammation via complement C3. PLoS Pathog. 2020;16:e1008928. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 121. | Vakhrusheva TV, Baikova YP, Balabushevich NG, Gusev SA, Lomakina GY, Sholina EA, Moshkovskaya MA, Shcherbakov PL, Pobeguts OV, Mikhal'chik EV. Binding of Mucin by E. coli from Human Gut. Bull Exp Biol Med. 2018;165:235-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 122. | Uchino M, Ikeuchi H, Hata K, Minagawa T, Horio Y, Kuwahara R, Nakamura S, Watanabe K, Saruta M, Fujii T, Kobayashi T, Sugimoto K, Hirai F, Esaki M, Hiraoka S, Matsuoka K, Shinzaki S, Matsuura M, Inoue N, Nakase H, Watanabe M. Intestinal cancer in patients with Crohn's disease: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2021;36:329-336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 123. | Yang H, Mirsepasi-Lauridsen HC, Struve C, Allaire JM, Sivignon A, Vogl W, Bosman ES, Ma C, Fotovati A, Reid GS, Li X, Petersen AM, Gouin SG, Barnich N, Jacobson K, Yu HB, Krogfelt KA, Vallance BA. Ulcerative Colitis-associated E. coli pathobionts potentiate colitis in susceptible hosts. Gut Microbes. 2020;12:1847976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 124. | Prudent V, Demarre G, Vazeille E, Wery M, Quenech'Du N, Ravet A, Dauverd-Girault J, van Dijk E, Bringer MA, Descrimes M, Barnich N, Rimsky S, Morillon A, Espéli O. The Crohn's disease-related bacterial strain LF82 assembles biofilm-like communities to protect itself from phagolysosomal attack. Commun Biol. 2021;4:627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 125. | Do J, Zafar H, Saier MH Jr. Comparative genomics of transport proteins in probiotic and pathogenic Escherichia coli and Salmonella enterica strains. Microb Pathog. 2017;107:106-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 126. | Sevrin G, Massier S, Chassaing B, Agus A, Delmas J, Denizot J, Billard E, Barnich N. Adaptation of adherent-invasive E. coli to gut environment: Impact on flagellum expression and bacterial colonization ability. Gut Microbes. 2020;11:364-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 127. | Kosari F, Taheri M, Moradi A, Hakimi Alni R, Alikhani MY. Evaluation of cinnamon extract effects on clbB gene expression and biofilm formation in Escherichia coli strains isolated from colon cancer patients. BMC Cancer. 2020;20:267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 128. | Zheng L, Wen XL, Dai YC. Mechanism of Jianpi Qingchang Huashi Recipe in treating ulcerative colitis: A study based on network pharmacology and molecular docking. World J Clin Cases. 2021;9:7653-7670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 10] [Cited by in F6Publishing: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 129. | Shawki A, Ramirez R, Spalinger MR, Ruegger PM, Sayoc-Becerra A, Santos AN, Chatterjee P, Canale V, Mitchell JD, Macbeth JC, Gries CM, Tremblay ML, Hsiao A, Borneman J, McCole DF. The autoimmune susceptibility gene, PTPN2, restricts expansion of a novel mouse adherent-invasive E. coli. Gut Microbes. 2020;11:1547-1566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 130. | Zhang YL, Cai LT, Qi JY, Lin YZ, Dai YC, Jiao N, Chen YL, Zheng L, Wang BB, Zhu LX, Tang ZP, Zhu RX. Gut microbiota contributes to the distinction between two traditional Chinese medicine syndromes of ulcerative colitis. World J Gastroenterol. 2019;25:3242-3255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 30] [Cited by in F6Publishing: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 131. | Lashermes A, Boudieu L, Barbier J, Sion B, Gelot A, Barnich N, Ardid D, Carvalho FA. Adherent-Invasive E. coli enhances colonic hypersensitivity and P2X receptors expression during post-infectious period. Gut Microbes. 2018;9:26-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 132. | Zheng L, Zhang YL, Dai YC, Chen X, Chen DL, Dai YT, Tang ZP. Jianpi Qingchang decoction alleviates ulcerative colitis by inhibiting nuclear factor-κB activation. World J Gastroenterol. 2017;23:1180-1188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 34] [Cited by in F6Publishing: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 133. | Chen DL, Dai YC, Zheng L, Chen YL, Zhang YL, Tang ZP. Features of the gut microbiota in ulcerative colitis patients with depression: A pilot study. Medicine (Baltimore). 2021;100:e24845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 134. | Zheng L, Zhang YL, Chen X, Chen DL, Dai YC, Tang ZP. Astragalus Polysaccharides Protects Thapsigargin-induced Endoplasmic Reticulum Stress in HT29 Cells. Open Life Sci. 2019;14:494-501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 135. | Larabi A, Dalmasso G, Delmas J, Barnich N, Nguyen HTT. Exosomes transfer miRNAs from cell-to-cell to inhibit autophagy during infection with Crohn's disease-associated adherent-invasive E. coli. Gut Microbes. 2020;11:1677-1694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 136. | Alvarez Dorta D, Chalopin T, Sivignon A, de Ruyck J, Dumych TI, Bilyy RO, Deniaud D, Barnich N, Bouckaert J, Gouin SG. Physiochemical Tuning of Potent Escherichia coli Anti-Adhesives by Microencapsulation and Methylene Homologation. ChemMedChem. 2017;12:986-998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 137. | Yu M, Kim J, Ahn JH, Moon Y. Nononcogenic restoration of the intestinal barrier by E. coli-delivered human EGF. JCI Insight. 2019;4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |