Published online Oct 10, 2014. doi: 10.5306/wjco.v5.i4.667
Revised: April 17, 2014
Accepted: June 10, 2014
Published online: October 10, 2014
Genome-wide association studies revealed that allelic variation in the α5-α3-β4 nicotine acetylcholine receptor (nAChR) cluster on chromosome 15q24-15q25.1 was associated with lung cancer risk. nAChRs are membrane ligand-gated cation channels whose activation is triggered by the binding of the endogenous neurotransmitter acetylcholine (ACh) or other biologic compounds including nicotine. nAChRs have been found on lung cancer cells, underscoring the idea that the non-neuronal nAChR pathway has important implications for lung cancer. Several studies focusing on the treatment with nAChR antagonists with improved selectivity might trigger novel strategies for the intervention and prevention of lung cancer. Here we review the genetic risk factors for lung cancer in the nAChR gene cluster, the roles of nicotine receptors, and the molecular mechanisms of acetylcholine receptor pathways to lead to more opportunities for intervention and prevention of lung cancer.
Core tip: Genome-wide association studies revealed that allelic variation in the α5-α3-β4 nicotine acetylcholine receptor (nAChR) cluster on chromosome 15q24-15q25.1 was associated with a higher risk for development of lung cancer. nAChRs are membrane ligand-gated cation channels whose activation is triggered by the binding of the endogenous neurotransmitter acetylcholine (ACh) or other biologic compounds including nicotine. nAChRs have been found on lung cancer cells, underscoring the idea that the non-neuronal nAChR signaling pathway has considerable implications for lung cancer. Several studies involving the design of nAChR antagonists with improved selectivity might identify novel strategies for the treatment of lung cancer.
- Citation: Niu XM, Lu S. Acetylcholine receptor pathway in lung cancer: New twists to an old story. World J Clin Oncol 2014; 5(4): 667-676
- URL: https://www.wjgnet.com/2218-4333/full/v5/i4/667.htm
- DOI: https://dx.doi.org/10.5306/wjco.v5.i4.667
Lung cancer is a multifactorial disease, resulting from a combination of genetic, environmental, and psychological factors. The evidence that genetic factors influence lung cancer risk has been demonstrated by many studies, tracing back to the landmark study in 1963[1]. This study demonstrated a 2.5-fold higher risk in smoking relatives of lung cancer cases compared with smoking relatives of controls[1], fully suggesting the importance of genetic factors in lung cancer risk.
More recently, some large genome-wide association (GWA) studies identified an association between single-nucleotide polymorphism (SNP) variation at 15q24-15q25.1 and susceptibility to lung cancer[2-11]. Genes mapping to this region of association include the α5-α3-β4 nicotine acetylcholine receptor (nAChR) cluster. nAChRs, encoded by the nicotinic acetylcholine receptor encoding (CHRN) genes[2-4,12], are members of the Cys-loop superfamily of ligand-gated ion channels[13], which are activated by endogenous agonist acetylcholine (ACh) and the exogenous compound nicotine[14] and nitrosamines, potential lung carcinogens in cigarette smoke and foods.
CHRN genes are expressed in both neuronal and non-neuronal tissues, suggesting that nAChRs may play an important role in processes other than synaptic transmission. Indeed, apart from the classic role at neuromuscular junctions, nAChRs have also been implicated in the regulation of cellular processes such as proliferation[15], cell-cell interaction, and cell death[16-18]. A decreased survival correlation with lung cancer may be explained by effects of the nAChRs pathway through CHRNA proteins on tumor cell proliferation[15], apoptosis, epithelial-mesenchymal transition, and proinvasive and angiogenic phenotypes[19]. This review will provide an overview for better understanding the genetic risk factors for lung cancer in the nAChR gene cluster, the roles of nicotine receptors, and the molecular mechanisms of the cholinergic pathways to lead to more opportunities for intervention and prevention of lung cancer.
Lung cancer is a heterogeneous and challenging disease to treat. With the arrival of genotyping and genomic profiling, management of non-small-cell lung cancer (NSCLC) has made several advances with the understanding of activating mutations in epidermal growth factor receptor (EGFR), fusion genes involving anaplastic lymphoma kinase (ALK), rearrangements in ROS-1. The next era of personalized treatment will involve a comprehensive genomic characterization of lung cancer. One important task in personalized medicine is to predict disease risk based on a person’s genome, e.g., on a large number of SNPs. GWA studies make SNP and phenotype data available to researchers.
Notably, the 15q24-15q25.1 region (CHRNA5/CHRNA3/CHRNB4) has been identified as a lung cancer risk spot by several GWA studies[2-11]. Of one GWA study in the International Agency for Research on Cancer (IARC), there is 14% increased lung cancer susceptibility associated with nAChR cluster variation irrespective of smoking status[3]. The same GWA result was identified by the other studies from the MD Anderson Cancer Center, United States[2], and deCODE Genetics, Iceland[4], respectively. Importantly, a non-synonymous variant rs16969968 of CHRNA5, inducing an amino acid substitution (D398N) in the second intracellular loop of the protein, can increase lung cancer risk by 30%[3]. The SNP rs8023462 in CHRNA3/CHRNB4 intergenic region, interfering CHRNA3/CHRNB4 gene expression by interacting with GATA transcription factors, was the first functional evidence for association with lung cancer risk[20].
To better understand the GWA studies for the nAChR gene cluster and lung cancer risk, we further investigate a catalog of all existing published GWA study data updated to December 20, 2013 from National Human Genome Research Institute (NHGRI) (http://www.genome.gov/), using the subject terms (CHRNA OR nAChR OR 15q25) AND (lung cancer OR lung adenocarcinoma OR lung adenocarcinoma (clinical stage) OR lung cancer (DNA repair capacity) OR lung cancer-asbestos exposure interaction). Twenty potentially relevant articles[2,3,6,8-11,21-33] were produced according to the first primary screening strategy, of which 7 GWA studies met the subject terms for further evaluating the relationship of the nAChR cluster with lung cancer susceptibility[2,3,6,8-11]. A flow diagram summarized the process of selecting and excluding articles with detailed reasons (Figure 1). All GWA studies for lung cancer at the 15q25.1 region are summarized in Table 1.
| Date | PubMedID | Ref. | Initialsample size | Replicationsample size | Chromosomeposition | Mappedgene | SNP | Context | P value | OR (95%CI) | Platform |
| 8/4/2009 | 19654303 | Broderick et al[9] | 1952 European ancestry cases, 1438 European ancestry controls | 5608 European ancestry cases, 6767 European ancestry controls | 78806023 | AGPHD 1 | rs8034191 | intron | 3.00E-26 | 1.29 (1.23-1.35) | Illumina |
| 11/2/2008 | 18978790 | McKay et al[6] | 2971 cases, 3746 controls | 2899 cases, 5573 controls | 78894339 | CHRNA 3 | rs1051730 | cds- synon | 1.00E-15 | 1.35 (1.25-1.45) | Illumina |
| 11/2/2008 | 18978787 | Wang et al[8] | 1952 cases, 1438 controls | 7579 cases, 8236 controls | 78908032 | CHRNA 3 | rs8042374 | intron | 8.00E-12 | NR | Illumina |
| 9/17/2008 | 18780872 | Liu et al[10] | 194 cases, 219 controls | 3878 cases, 4831 controls | 78806023 | AGPHD 1 | rs8034191 | intron | 1.00E-8 | 1.38 (1.17-1.64) | Affymetrix |
| 4/3/2008 | 18385676 | Amos et al[2] | 1154 cases, 1137 controls | 2724 cases, 3694 controls | 78806023 | AGPHD 1 | rs8034191 | intron | 3.00E-18 | 1.30 (1.15-1.47) | Illumina |
| 4/3/2008 | 18385738 | Hung et al[3] | 1926 cases, 2522 controls | 2513 cases, 4752 controls | 78806023 | AGPHD 1 | rs8034191 | intron | 5.00E-20 | 1.30 (1.23-1.37) | Illumina |
| 10/15/ 2009 | 19836008 | Landi et al[11] | 5739 European descent cases, 5848 European descent controls | 7561 European descent cases, 13818 European descent controls | 78894339 | CHRNA 3 | rs1051730 | cds- synon | 2.00E-51 | 1.31 (1.27-1.36) | Illumina |
Considering that the statistical power of individual GWA study has been limited by the modest effect sizes of genetic variants and the financial constraints on the numbers of variants, it is stringent to perform meta-analyses of existing genetic data on the nAChR gene cluster to better understand disease loci harboring common variants associated with lung cancer risk. A total of 12 qualified articles between 2008 and 2011 screened from 40 potentially relevant articles were selected, including 16 studies with 9 in Caucasians, 4 in East Asians, 2 in African-Americans, and 1 in mixed (Caucasian, African-American and Hispanic) populations[34]. CHRNA3 gene rs1051730-A allele, compared with G allele, is associated with a 36% higher risk for lung cancer (95%CI, 1.27-1.46; P < 0.0005)[34]. Surprisingly subgroup analyses suggested that rs1051730-A allele might be the factor for lung cancer susceptibility in Caucasians (OR = 1.32; 95%CI, 1.25-1.44; P < 0.0005), but not in East-Asians (OR = 1.51; 95%CI, 0.76-3.00; P = 0.237)[34]. Allele frequency of rs1051730 in Asians was lower than that in Caucasians. For rs1051730, the minor allele frequency (MAF) was reported to be 0.39, 0.15, and 0.04 in Whites, African Americans, and Chinese, respectively, based on the HapMap data (http://hapmap.ncbi.nlm.nih.gov/), which was reported in our previous study[35]. The Japanese finding[36] consistent with our reported Chinese study[35] also suggests that, like EGFR story, different genetic backgrounds may cause the discrepancy in lung cancer risk in different populations.
Owing to SNPs with weaker effects in influencing lung cancer risk could be missed given the stringent requirements for adjustment for multiple comparisons, nAChR pathway analysis has been proposed as a complementary approach to single SNP analyses in GWA studies. Using two lung cancer GWA studies data sets based on four studies: one a combined data set from Central Europe and Toronto (CETO)[3]; the other a combined data set from Germany (HGF study)[11,37] and MD Anderson Cancer Center (GRMD)[2], comparison of pathway analysis approaches demonstrated that the nAChR pathway was identified as associated with lung cancer in CETO and GRMD[38].
Although CHRNA5, CHRNA3 and CHRNB4 are clustered together at the same locus, they exhibit differential susceptibility to aberrant methylation. Treatment of cancer cells exhibiting CHRNA3 hypermethylation with DNA methylation inhibitors caused demethylation of the CHRNA3 promoter and gene reactivation, inducing apoptotic cell death[39]. Small hairpin RNA-mediated depletion of CHRNA3 in CHRNA3-expressing lung cancer cells led to resistance to apoptosis-inducing agents, underscoring the importance of epigenetic silencing of the CHRNA3 gene in human cancer[39]. Silencing of the CHRNA3-encoding gene may result in over-representation of other nAChR subunits, notably CHRNA7 and CHRNA5[39], which may stimulate cell survival and provide a proliferation advantage to tumor cells[40,41]. In contrast to CHRNA3 hypermethylation, elevated CHRNB4 methylation, insufficient to induce significant silencing of the gene, failed to inhibit gene expression. Conclusively, the 15q25.1 locus may be under epigenetic regulation and that its deregulation may lead to lung cancer.
Evidence that the nAChR cluster on 15q25 locus is associated with smoking status, nicotine dependence and the risk of lung cancer is inconsistent in different populations. The region of the nAChR cluster has been confirmed to be associated with a number of smoking-related traits, including nicotine dependence, cigarettes smoked per day, and heavy smoking, in some lung cancer GWA studies[4,42], and in some genome-wide meta-analyses in Caucasian populations[42-44]. For example, the Caucasian population with variant rs1051730 SNP in the nAChR cluster was related with lung cancer risk and nicotine dependence, approximately smoking one and two more cigarette per day than those without variant rs1051730 SNP[4]. Another study demonstrates further that association signals in the nAChR cluster affecting early smoking behaviors may have disparity with those contributing the mature nicotine dependence[45].
Surprisingly, a Japanese case-control study[36] reported associations between the selected SNPs in the nAChR cluster and risk of lung cancer and found that associations among never and ever smokers were similar. The association was consistent among non-smokers and smokers in our study[35]. These studies might argue for a role of the nAChR cluster in lung cancer that is independent of smoking behavior in Asians.
These findings in different populations suggest a role for racial differences in the association between smoking behaviors, nAChR cluster, and risk of lung cancer. Reasons underlying the racial difference in the genotype with smoking associations are unclear. This discrepancy may be due to differences in genetic and environmental backgrounds. Alternatively, other factors that have not been taken into account, such as food intake and passive smoking, differentiate the mode of contribution of the nAChR cluster in non-smokers.
The regulation of acetylcholine receptor pathways has been established from the primitive organisms, irrespective of neurons. There exist two types of AchRs, nAChRs and Muscarinic ACh receptors (mAChRs) of cholinergic signaling. nAChRs comprise five subunits, including ten α subunits (α1-α10), four β subunits (β1-β4), one δ, and one ε or γ subunit, which form hetero- or homo-pentamers enclosing a central ion channel. The nAChR subunit composition in turn further regulates the function and pharmacology of nAChRs[16,17].
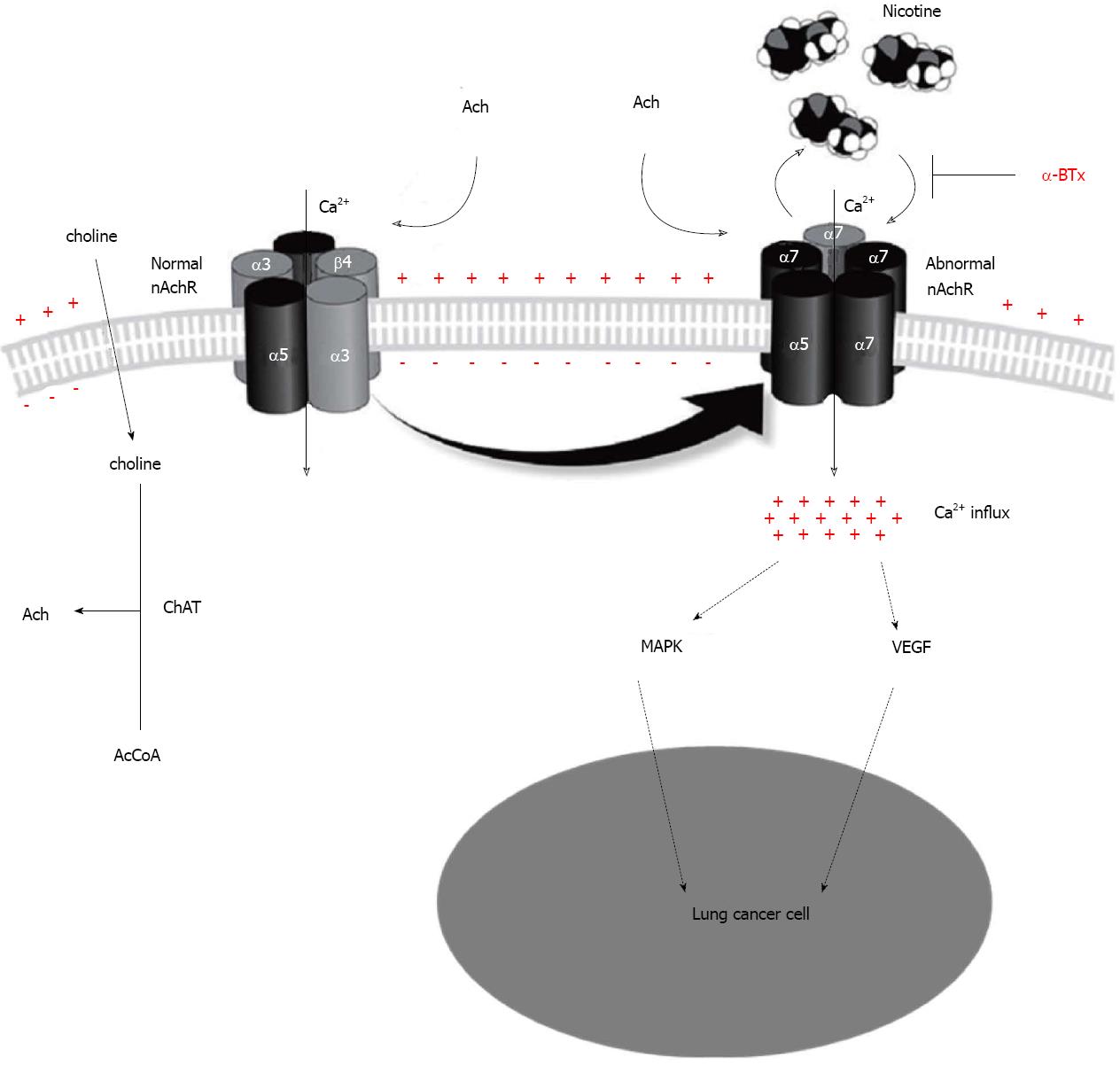
Acetylcholine receptor signaling in non-neuronal cells is comparable to acetylcholine receptor neurotransmission[46]. Both nAChR families are expressed in cancer cells[46], and both NSCLC and small-cell-lung cancer (SCLC) cell lines can synthesize and release Ach[47]. The widespread synthesis of ACh beyond the nervous system has changed the paradigm of ACh acting merely as a neurotransmitter, and it may also act as an autocrine or a paracrine messenger able to interact with nAChRs. For example, non-neuronal ACh is released from the living NSCLC or SCLC cells and binds to nAChRs of its source and neighbouring cells to mediate autocrine and paracrine regulatory loops, prolonging cell survival with subsequent cell proliferation through mitogen-activated protein kinase (MAPK) pathway[48-50] or with increase of vascular endothelial growth factor (VEGF) stimulating neoangiogenesis[19]. All these effects can be blocked at the level of the nAchRs[48,51,52]. This novel paradigm necessitates the opportunity of marker-guided lung cancer intervention and prevention strategies, making balance between nAChR-mediated stimulation and inhibition[16,53].
Ordinarily ACh is regarded as a classical neurotransmitter. ACh is synthesized intracellularly by the enzyme choline acetyltransferase (ChAT) from choline and acetyl-coenzyme A (AcCoA) before being released into the extracellular space to act on synaptic-adjacent cells. In contrast, the enzyme acetylcholinesterase (AChE) can rapidly clear the extracellular ACh pool into its inactive metabolites choline and acetate. This signaling system is targeted by various biological modulators which can inhibit ACh release or AChE activity[54]. ChAT is strongly up-regulated, whereas AChE is down-regulated in squamous cell carcinoma (SqCC)[54], which increases levels of Ach, providing endogenous proliferative stimuli to nAchRs.
All nAChRs allow the influx of different cations (Na+, K+, Ca2+), and the α7nAChR is selective for Ca2+. Binding of nicotine to α7nAChR can cause Ca2+ influx into lung cancer cells and the subsequent membrane depolarization activates voltage-gated Ca2+ channels, which activates the MAPK pathway[48]. Subsequently, MAPK activates complexes of the transcription factor NF-kB that induce entry into S phase to promote cancer cell proliferation[48]. Moreover, tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) has been found to promote Ca2+ influx thought binding to nAChRs, activating both protein kinase C (PKC) and the MAPK pathway to stimulate the proliferation of SCLC cells[49].
The role of nAChRs in the growth regulation of cancer was first identified in 1989[15], and reinforced further by the ability of generalized nAChR antagonists (e.g., hexamethonium and mecamylamine) to reverse the proliferative effects of nicotine. Nowadays, the mechanism of nicotine-induced tumor cell proliferation and associated angiogenesis is an active area of research. In particular, α7nAChR has been implicated in mediating the proliferative effects of nicotine, and α7nAChR antagonist a bungarotoxin (α-BTx) or methyllycaconitine can attenuate the proliferative effects of nicotine in NSCLC and SCLC cells[48,52]. These findings have been confirmed by the transfection of small interfering RNA targeting α7nAChR in lung cells[52]. This point implies that α7nAChR is being considered a target for cancer therapy[48], marking a new chapter in lung cancer research and the feasibility of using nAChRs as a viable molecular target for cancer therapy.
Beyond the above channel activity, nAChRs can be involved in other intracellular events involving various downstream nAChR-mediated signaling pathways, including Ca2+/calmodulin, PKC[49], MAPK[48-50], and VEGF[19]. It has been reported that EGFR was found to be on the list of high α6β3 tumors, and nAChR may trigger the MAPK pathway in which EGFR was involved, leading to promotion of cell growth and proliferation. Thus, cross-talk between signaling downstream of EGFR and nAChR activation via the MAPK pathway may together promote carcinogenesis[50]. The other findings suggest a strong connection between growth factor mediated angiogenic pathway and the cholinergic angiogenic pathway[19,55]. For instance, the endothelial cell (EC) migration induced by basic fibroblast growth factor (bFGF) and VEGF can be inhibited by blocking the nAChRs with α-BTx[19]. Figure 2 simply summarizes the above mentioned nAChRs pathway in lung cancer.
The carcinogenic nitrosamine N-nitrosodiethylamine (DeN) was identified recently to have the similar structure with ACh, acting as a high-affinity ligand for homo- and hetero-meric nAChRs in lung cancer cells[56]. Considering the fact that DeN is contained in numerous foods and drinks[57], such agents may lead to non-tobacco-related modulations of nAChRs. In addition, different nAChR subunit gene expression patterns were found between NSCLCs in smokers and non-smokers, and higher nAChR α6β3 expression was associated with NSCLC tumors from non-smokers, as compared with those from smokers[5].
Neurotransmitter γ-aminobutyric acid (GABA) is recognized as the most important inhibitory neurotransmitter in the brain, but it also acts as a tumor suppressor for pulmonary adenocarcinoma (PAC)[58] and carcinomas of the pancreas[59], breast[60] and colon[61]. As α4β2nAChR regulates the release of GABA, desensitization of this receptor may therefore lead to GABA deficiency[62]. Moreover, it has been shown that mRNA levels of the α4nAChRs were significantly lower in PAC tissues than in normal lung tissue or other types of NSCLCs[5]. Considering that oestrogen and phyto-oestrogens were reported to desensitize α4β2nAChR[63], the predominance of PAC in women may therefore at least in part be the result of impaired α4β2nAChR function. Recent reports of clinical trials of the nAChRα4β2 antagonist, which targets α4β2 receptors in the brain, have shown its clinical efficacy in smoking cessation[64]. It is possible that similar nAChR antagonists could block the effect on lung tumors, including lung cancer in women.
Pulmonary neuroendocrine cells (PNeCs) and SCLC can express high levels of α7nAChR[51]. α7nAChR is the central regulator of proliferation, apoptosis and migration in SCLC cells through stimulating the release of some autocrine growth factors, including serotonin and neuropeptides[51]. α7nAChR antagonist α-BTx can attenuate the proliferative effects of nicotine in SCLC cells[48,52].
nAChRs are associated with resistance to gemcitabine, cisplatin and paclitaxel in NSCLC cell lines. Our research is the first study to investigate whether or not a genetic variant in the 15q25 region has a prognostic effect on the survival outcome of patients with lung cancer[35]. The patients with CHRNA3 gene rs3743073G > T allele showed a higher risk of lung cancer and worst survival in Chinese Han patients with advanced stage NSCLC[35]. A Caucasian study showed that CHRNA3 (rs1051730) genotyping can improve customized chemotherapy based on tumor assessment of excision repair cross-complementing 1 (ERCC1) mRNA in stage IV NSCLC patients with a performance status of 0 (clinicaltrials.gov. identifier: NCT00174629)[65]. A further Korean survival study demonstrated that a functional SNP, rs6495309C > T, in the promoter of the CHRNA3 gene, was the prognostic factor for resected early stage NSCLC[66]. Compared with rs6495309 CC genotype, the patients in the studied Korean cohort with rs6495309 CT/TT genotype had a better 5-yr OS by 5% and better 5-yr DFS by 7%[66].
The use of nAChR antagonists that block the receptors still has some issues, because these receptors regulate many vital cell and organ functions, and deficiency or impairment of nAChR signaling will lead to overproduction of cytokines in some tissues and enhance tissue damage. Carefully designed animal studies are essential to investigate the potential side effects of nAChR antagonists on the brain, central nervous system, immune cells and muscle cells, which express high levels of nicotinic receptors.
In summary, strategies for marker-guided lung cancer intervention that targets nAChRs seem promising. nAChR antagonists could be potentially used in combination with established chemotherapeutic drugs to enhance the therapeutic response to chemotherapy. Future studies involving the design of nAChR antagonists with improved selectivity might trigger novel strategies for the intervention and prevention of lung cancer.
P- Reviewer: Sun ZF S- Editor: Ji FF L- Editor: Wang TQ E- Editor: Lu YJ
| 1. | TOKUHATA GK, LILIENFELD AM. Familial aggregation of lung cancer in humans. J Natl Cancer Inst. 1963;30:289-312. [PubMed] [Cited in This Article: ] |
| 2. | Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616-622. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633-637. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP, Manolescu A, Thorleifsson G, Stefansson H, Ingason A. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638-642. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Lam DC, Girard L, Ramirez R, Chau WS, Suen WS, Sheridan S, Tin VP, Chung LP, Wong MP, Shay JW. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67:4638-4647. [PubMed] [Cited in This Article: ] |
| 6. | McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J. Lung cancer susceptibility locus at 5p15.33. Nat Genet. 2008;40:1404-1406. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221-227. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407-1409. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, Amos CI, Houlston RS. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633-6641. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Liu P, Vikis HG, Wang D, Lu Y, Wang Y, Schwartz AG, Pinney SM, Yang P, de Andrade M, Petersen GM. Familial aggregation of common sequence variants on 15q24-25.1 in lung cancer. J Natl Cancer Inst. 2008;100:1326-1330. [PubMed] [DOI] [Cited in This Article: ] |
| 11. | Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679-691. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis. 2007;28:507-518. [PubMed] [Cited in This Article: ] |
| 13. | Lindstrom JM. Nicotinic acetylcholine receptors of muscles and nerves: comparison of their structures, functional roles, and vulnerability to pathology. Ann N Y Acad Sci. 2003;998:41-52. [PubMed] [Cited in This Article: ] |
| 14. | Arias HR, Bhumireddy P, Bouzat C. Molecular mechanisms and binding site locations for noncompetitive antagonists of nicotinic acetylcholine receptors. Int J Biochem Cell Biol. 2006;38:1254-1276. [PubMed] [Cited in This Article: ] |
| 15. | Schuller HM. Cell type specific, receptor-mediated modulation of growth kinetics in human lung cancer cell lines by nicotine and tobacco-related nitrosamines. Biochem Pharmacol. 1989;38:3439-3442. [PubMed] [Cited in This Article: ] |
| 16. | Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer. 2009;9:195-205. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73-120. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Catassi A, Servent D, Paleari L, Cesario A, Russo P. Multiple roles of nicotine on cell proliferation and inhibition of apoptosis: implications on lung carcinogenesis. Mutat Res. 2008;659:221-231. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Ng MK, Wu J, Chang E, Wang BY, Katzenberg-Clark R, Ishii-Watabe A, Cooke JP. A central role for nicotinic cholinergic regulation of growth factor-induced endothelial cell migration. Arterioscler Thromb Vasc Biol. 2007;27:106-112. [PubMed] [Cited in This Article: ] |
| 20. | Flora AV, Zambrano CA, Gallego X, Miyamoto JH, Johnson KA, Cowan KA, Stitzel JA, Ehringer MA. Functional characterization of SNPs in CHRNA3/B4 intergenic region associated with drug behaviors. Brain Res. 2013;1529:1-15. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Lan Q, Hsiung CA, Matsuo K, Hong YC, Seow A, Wang Z, Hosgood HD 3rd, Chen K, Wang JC, Chatterjee N. Genome-wide association analysis identifies new lung cancer susceptibility loci in never-smoking women in Asia. Nat Genet. 2012;44:1330-1335. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Timofeeva MN, Hung RJ, Rafnar T, Christiani DC, Field JK, Bickeböller H, Risch A, McKay JD, Wang Y, Dai J. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980-4995. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat Genet. 2011;43:792-796. [PubMed] [DOI] [Cited in This Article: ] |
| 24. | Li Y, Sheu CC, Ye Y, de Andrade M, Wang L, Chang SC, Aubry MC, Aakre JA, Allen MS, Chen F. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11:321-330. [PubMed] [DOI] [Cited in This Article: ] |
| 25. | Spinola M, Leoni VP, Galvan A, Korsching E, Conti B, Pastorino U, Ravagnani F, Columbano A, Skaug V, Haugen A. Genome-wide single nucleotide polymorphism analysis of lung cancer risk detects the KLF6 gene. Cancer Lett. 2007;251:311-316. [PubMed] [Cited in This Article: ] |
| 26. | Shiraishi K, Kunitoh H, Daigo Y, Takahashi A, Goto K, Sakamoto H, Ohnami S, Shimada Y, Ashikawa K, Saito A. A genome-wide association study identifies two new susceptibility loci for lung adenocarcinoma in the Japanese population. Nat Genet. 2012;44:900-903. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, Zo JI, Lee JS, Hosono N, Morizono T. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893-896. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W. The 5p15.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6:e1001051. [PubMed] [DOI] [Cited in This Article: ] |
| 29. | Galvan A, Falvella FS, Spinola M, Frullanti E, Leoni VP, Noci S, Alonso MR, Zolin A, Spada E, Milani S. A polygenic model with common variants may predict lung adenocarcinoma risk in humans. Int J Cancer. 2008;123:2327-2330. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Frullanti E, Galvan A, Falvella FS, Manenti G, Colombo F, Vannelli A, Incarbone M, Alloisio M, Nosotti M, Santambrogio L. Multiple genetic loci modulate lung adenocarcinoma clinical staging. Clin Cancer Res. 2011;17:2410-2416. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Wang LE, Gorlova OY, Ying J, Qiao Y, Weng SF, Lee AT, Gregersen PK, Spitz MR, Amos CI, Wei Q. Genome-wide association study reveals novel genetic determinants of DNA repair capacity in lung cancer. Cancer Res. 2013;73:256-264. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Wei S, Wang LE, McHugh MK, Han Y, Xiong M, Amos CI, Spitz MR, Wei QW. Genome-wide gene-environment interaction analysis for asbestos exposure in lung cancer susceptibility. Carcinogenesis. 2012;33:1531-1537. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Lee Y, Yoon KA, Joo J, Lee D, Bae K, Han JY, Lee JS. Prognostic implications of genetic variants in advanced non-small cell lung cancer: a genome-wide association study. Carcinogenesis. 2013;34:307-313. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Gu M, Dong X, Zhang X, Wang X, Qi Y, Yu J, Niu W. Strong association between two polymorphisms on 15q25.1 and lung cancer risk: a meta-analysis. PLoS One. 2012;7:e37970. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Niu X, Chen Z, Shen S, Liu Y, Zhou D, Zhang J, Li Z, Yu Y, Liao M, Lu S. Association of the CHRNA3 locus with lung cancer risk and prognosis in Chinese Han population. J Thorac Oncol. 2010;5:658-666. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Shiraishi K, Kohno T, Kunitoh H, Watanabe S, Goto K, Nishiwaki Y, Shimada Y, Hirose H, Saito I, Kuchiba A. Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis. 2009;30:65-70. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Sauter W, Rosenberger A, Beckmann L, Kropp S, Mittelstrass K, Timofeeva M, Wölke G, Steinwachs A, Scheiner D, Meese E. Matrix metalloproteinase 1 (MMP1) is associated with early-onset lung cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:1127-1135. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Fehringer G, Liu G, Briollais L, Brennan P, Amos CI, Spitz MR, Bickeböller H, Wichmann HE, Risch A, Hung RJ. Comparison of pathway analysis approaches using lung cancer GWAS data sets. PLoS One. 2012;7:e31816. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Paliwal A, Vaissière T, Krais A, Cuenin C, Cros MP, Zaridze D, Moukeria A, Boffetta P, Hainaut P, Brennan P. Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res. 2010;70:2779-2788. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | West KA, Brognard J, Clark AS, Linnoila IR, Yang X, Swain SM, Harris C, Belinsky S, Dennis PA. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111:81-90. [PubMed] [Cited in This Article: ] |
| 41. | Arias HR, Richards VE, Ng D, Ghafoori ME, Le V, Mousa SA. Role of non-neuronal nicotinic acetylcholine receptors in angiogenesis. Int J Biochem Cell Biol. 2009;41:1441-1451. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genet. 2010;6. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448-453. [PubMed] [DOI] [Cited in This Article: ] |
| 44. | Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, Vollenweider P, Preisig M, Wareham NJ, Zhao JH, Loos RJ, Barroso I, Khaw KT, Grundy S, Barter P, Mahley R, Kesaniemi A, McPherson R, Vincent JB, Strauss J, Kennedy JL, Farmer A, McGuffin P, Day R, Matthews K, Bakke P, Gulsvik A, Lucae S, Ising M, Brueckl T, Horstmann S, Wichmann HE, Rawal R, Dahmen N, Lamina C, Polasek O, Zgaga L, Huffman J, Campbell S, Kooner J, Chambers JC, Burnett MS, Devaney JM, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Epstein S, Wilson JF, Wild SH, Campbell H, Vitart V, Reilly MP, Li M, Qu L, Wilensky R, Matthai W, Hakonarson HH, Rader DJ, Franke A, Wittig M, Schäfer A, Uda M, Terracciano A, Xiao X, Busonero F, Scheet P, Schlessinger D, St Clair D, Rujescu D, Abecasis GR, Grabe HJ, Teumer A, Völzke H, Petersmann A, John U, Rudan I, Hayward C, Wright AF, Kolcic I, Wright BJ, Thompson JR, Balmforth AJ, Hall AS, Samani NJ, Anderson CA, Ahmad T, Mathew CG, Parkes M, Satsangi J, Caulfield M, Munroe PB, Farrall M, Dominiczak A, Worthington J, Thomson W, Eyre S, Barton A; Wellcome Trust Case Control Consortium, Mooser V, Francks C, Marchini J. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42:436-440. [PubMed] [DOI] [Cited in This Article: ] |
| 45. | Stephens SH, Hartz SM, Hoft NR, Saccone NL, Corley RC, Hewitt JK, Hopfer CJ, Breslau N, Coon H, Chen X. Distinct loci in the CHRNA5/CHRNA3/CHRNB4 gene cluster are associated with onset of regular smoking. Genet Epidemiol. 2013;37:846-859. [PubMed] [DOI] [Cited in This Article: ] |
| 46. | Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br J Pharmacol. 2008;154:1558-1571. [PubMed] [DOI] [Cited in This Article: ] |
| 47. | Song P, Sekhon HS, Lu A, Arredondo J, Sauer D, Gravett C, Mark GP, Grando SA, Spindel ER. M3 muscarinic receptor antagonists inhibit small cell lung carcinoma growth and mitogen-activated protein kinase phosphorylation induced by acetylcholine secretion. Cancer Res. 2007;67:3936-3944. [PubMed] [Cited in This Article: ] |
| 48. | Trombino S, Cesario A, Margaritora S, Granone P, Motta G, Falugi C, Russo P. Alpha7-nicotinic acetylcholine receptors affect growth regulation of human mesothelioma cells: role of mitogen-activated protein kinase pathway. Cancer Res. 2004;64:135-145. [PubMed] [Cited in This Article: ] |
| 49. | Arredondo J, Chernyavsky AI, Jolkovsky DL, Pinkerton KE, Grando SA. Receptor-mediated tobacco toxicity: cooperation of the Ras/Raf-1/MEK1/ERK and JAK-2/STAT-3 pathways downstream of alpha7 nicotinic receptor in oral keratinocytes. FASEB J. 2006;20:2093-2101. [PubMed] [Cited in This Article: ] |
| 50. | Nakayama H, Numakawa T, Ikeuchi T. Nicotine-induced phosphorylation of Akt through epidermal growth factor receptor and Src in PC12h cells. J Neurochem. 2002;83:1372-1379. [PubMed] [Cited in This Article: ] |
| 51. | Sartelet H, Maouche K, Totobenazara JL, Petit J, Burlet H, Monteau M, Tournier JM, Birembaut P. Expression of nicotinic receptors in normal and tumoral pulmonary neuroendocrine cells (PNEC). Pathol Res Pract. 2008;204:891-898. [PubMed] [DOI] [Cited in This Article: ] |
| 52. | Dasgupta P, Rastogi S, Pillai S, Ordonez-Ercan D, Morris M, Haura E, Chellappan S. Nicotine induces cell proliferation by beta-arrestin-mediated activation of Src and Rb-Raf-1 pathways. J Clin Invest. 2006;116:2208-2217. [PubMed] [Cited in This Article: ] |
| 53. | Benowitz NL. Nicotine addiction. N Engl J Med. 2010;362:2295-2303. [PubMed] [DOI] [Cited in This Article: ] |
| 54. | Song P, Sekhon HS, Fu XW, Maier M, Jia Y, Duan J, Proskosil BJ, Gravett C, Lindstrom J, Mark GP. Activated cholinergic signaling provides a target in squamous cell lung carcinoma. Cancer Res. 2008;68:4693-4700. [PubMed] [DOI] [Cited in This Article: ] |
| 55. | Cooke JP. Angiogenesis and the role of the endothelial nicotinic acetylcholine receptor. Life Sci. 2007;80:2347-2351. [PubMed] [Cited in This Article: ] |
| 56. | Schuller HM. Nitrosamines as nicotinic receptor ligands. Life Sci. 2007;80:2274-2280. [PubMed] [Cited in This Article: ] |
| 57. | Abnet CC. Carcinogenic food contaminants. Cancer Invest. 2007;25:189-196. [PubMed] [Cited in This Article: ] |
| 58. | Schuller HM, Al-Wadei HA, Majidi M. Gamma-aminobutyric acid, a potential tumor suppressor for small airway-derived lung adenocarcinoma. Carcinogenesis. 2008;29:1979-1985. [PubMed] [DOI] [Cited in This Article: ] |
| 59. | Schuller HM, Al-Wadei HA, Majidi M. GABA B receptor is a novel drug target for pancreatic cancer. Cancer. 2008;112:767-778. [PubMed] [Cited in This Article: ] |
| 60. | Drell TL, Joseph J, Lang K, Niggemann B, Zaenker KS, Entschladen F. Effects of neurotransmitters on the chemokinesis and chemotaxis of MDA-MB-468 human breast carcinoma cells. Breast Cancer Res Treat. 2003;80:63-70. [PubMed] [Cited in This Article: ] |
| 61. | Joseph J, Niggemann B, Zaenker KS, Entschladen F. The neurotransmitter gamma-aminobutyric acid is an inhibitory regulator for the migration of SW 480 colon carcinoma cells. Cancer Res. 2002;62:6467-6469. [PubMed] [Cited in This Article: ] |
| 62. | Kawai H, Berg DK. Nicotinic acetylcholine receptors containing alpha 7 subunits on rat cortical neurons do not undergo long-lasting inactivation even when up-regulated by chronic nicotine exposure. J Neurochem. 2001;78:1367-1378. [PubMed] [Cited in This Article: ] |
| 63. | Nakazawa K, Ohno Y. Block by phytoestrogens of recombinant human neuronal nicotinic receptors. J Pharmacol Sci. 2003;93:118-121. [PubMed] [Cited in This Article: ] |
| 64. | Tonstad S, Tønnesen P, Hajek P, Williams KE, Billing CB, Reeves KR. Effect of maintenance therapy with varenicline on smoking cessation: a randomized controlled trial. JAMA. 2006;296:64-71. [PubMed] [Cited in This Article: ] |
| 65. | Carcereny E, Ramirez JL, Sanchez-Ronco M, Isla D, Cobo M, Moran T, de Aguirre I, Okamoto T, Wei J, Provencio M. Blood-based CHRNA3 single nucleotide polymorphism and outcome in advanced non-small-cell lung cancer patients. Lung Cancer. 2010;68:491-497. [PubMed] [DOI] [Cited in This Article: ] |
| 66. | Jin G, Bae EY, Yang E, Lee EB, Lee WK, Choi JE, Jeon HS, Yoo SS, Lee SY, Lee J. A functional polymorphism on chromosome 15q25 associated with survival of early stage non-small-cell lung cancer. J Thorac Oncol. 2012;7:808-814. [PubMed] [DOI] [Cited in This Article: ] |
| 67. | Available from: http: //www.genome.gov/. [Cited in This Article: ] |