Published online Aug 10, 2013. doi: 10.5306/wjco.v4.i3.58
Revised: May 27, 2013
Accepted: June 5, 2013
Published online: August 10, 2013
Processing time: 214 Days and 20.1 Hours
E3 ubiquitin ligases are a large family of proteins that catalyze the ubiquitination of many protein substrates for targeted degradation by the 26S proteasome. Therefore, E3 ubiquitin ligases play an essential role in a variety of biological processes including cell cycle regulation, proliferation and apoptosis. E3 ubiquitin ligases are often found overexpressed in human cancers, including lung cancer, and their deregulation has been shown to contribute to cancer development. However, the lack of specific inhibitors in clinical trials is a major issue in targeting E3 ubiquitin ligases with currently only one E3 ubiquitin ligase inhibitor being tested in the clinical setting. In this review, we focus on E3 ubiquitin ligases that have been found deregulated in lung cancer. Furthermore, we discuss the processes in which they are involved and evaluate them as potential anti-cancer targets. By better understanding the mechanisms by which E3 ubiquitin ligases regulate biological processes and their exact role in carcinogenesis, we can improve the development of specific E3 ubiquitin ligase inhibitors and pave the way for novel treatment strategies for cancer patients.
Core tip: E3 ubiquitin ligases catalyze ubiquitination of proteins for degradation by the 26S proteasome. They are important for many biological processes including cell cycle regulation, proliferation and apoptosis. They are often overexpressed and deregulated in lung cancer, which contributes to cancer development. These processes underline their potential as anti-cancer targets. There is only one E3 ubiquitin ligase inhibitor in clinical trial. A better understanding of how E3 ubiquitin ligases regulate biological processes and of their exact role in carcinogenesis, will help to develop specific E3 ubiquitin ligase inhibitors to improve treatment strategies for cancer patients.
- Citation: Snoek BC, Wilt LH, Jansen G, Peters GJ. Role of E3 ubiquitin ligases in lung cancer. World J Clin Oncol 2013; 4(3): 58-69
- URL: https://www.wjgnet.com/2218-4333/full/v4/i3/58.htm
- DOI: https://dx.doi.org/10.5306/wjco.v4.i3.58
The ubiquitin-proteasome system (UPS) regulates multiple biological aspects of cell survival by mediating the degradation of targeted proteins and thereby maintaining cellular homeostasis[1]. In numerous cancer types, the deregulation of UPS components has been observed and their overexpression is often associated with chemoresistance and poor prognosis[2-5]. For example, the E3 ubiquitin ligase murine double minute 2 (MDM2), which is involved in the regulation of p53 levels, is frequently overexpressed in tumors and is predicted to be a negative prognostic marker for the development of several human cancers including breast carcinoma and prostate carcinoma[6-9].
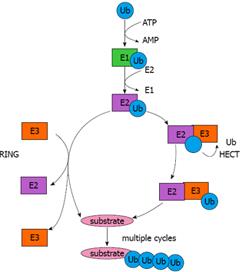
In the early 1980s, Ciechanover et al[10] and Hershko et al[11,12] obtained the initial understanding of ubiquitin-mediated protein degradation and identified several components of the ubiquitin system. A set of interconnected studies between 1984 and 1990 revealed the biological significance of protein degradation mediated by the ubiquitin system[13,14]. The mechanism by which ubiquitin molecules are covalently attached to targeted proteins can be delineated as a three-step enzymatic cascade (Figure 1). First, an ubiquitin-activating enzyme (E1) mediates the activation of the carboxyl-terminal glycine residue of ubiquitin in an ATP-dependent manner[10,15]. With the formation of a thiolester linkage, the activated ubiquitin is then transferred to E1 followed by the transfer of ubiquitin to a thiol site of an ubiquitin-conjugating enzyme (E2)[16]. Finally, an ubiquitin protein ligase (E3) confers substrate specificity by recognizing the target proteins and mediating the conjugation of (a) ubiquitin molecule(s) to a lysine residue on the targeted protein via an isopeptide bond[12]. Subsequently, the targeted protein is marked for degradation by the ATP-dependent 26S proteasome.
The addition of ubiquitin molecules onto targeted proteins is a modification that can be reversed. This reversal is called deubiquitination and is executed by proteases termed deubiquitinases (DUBs)[17]. DUBs specifically cleave ubiquitin after the terminal carboxyl group of ubiquitin and play a pivotal role in maintaining ubiquitin homeostasis[18,19]. Many DUBs have been shown to interact with E3 ligases, which suggests that a major function of DUBs is to control the stability of E3 ligases and subsequently destabilise the substrates of the cognate E3 ligase[20].
E3 ubiquitin ligases are often found overexpressed in a variety of human cancers, including lung cancer, and their deregulation has been shown to contribute to cancer development. As a result, increased attention is being paid to these E3 ubiquitin ligases and whether they can serve as potential anti-cancer targets. In targeted therapy, an ideal anti-cancer target should not only be overexpressed, but should meet additional criteria, such as its overexpression should be associated with poor prognosis, it plays a pivotal role in cancer genesis, inhibition induces apoptosis or growth reduction in the cancer cells, it is a “druggable” target (enzyme or cell surface molecule) that can be easily targeted, and finally, it is not expressed or is expressed at a very low level in normal cells.
E3 ubiquitin ligases can be divided in two major classes: the first class contains a C-terminal region Homologous to the E6-associated protein (E6-AP) carboxyl terminus (HECT), with an evolutionarily conserved cysteine residue required for the formation of a thiolester linkage with ubiquitin[21,22]. There are approximately 30 proteins containing the HECT domain. The second and largest class comprises E3 ubiquitin ligases that contain the really interesting new gene (RING) finger domain[23]. There are over 700 proteins containing the RING finger domain, but only a small part functions as an E3 ubiquitin ligase. Unlike RING proteins, most HECT proteins, if not all, are believed to function as E3 ubiquitin ligases. RING and HECT E3 ubiquitin ligases use different catalytic mechanisms to promote the transfer of ubiquitin to targeted substrates. RING E3 ubiquitin ligases can promote the direct transfer of ubiquitin from E2 to the targeted substrate, whereas HECT E3 ubiquitin ligases interact with the cognate E2, followed by the formation of a thiolester linkage with ubiquitin and subsequent transfer of ubiquitin to the targeted substrate (Figure 1).
The conjugation of one ubiquitin molecule to a protein is referred to as monoubiquitination, a process involved in protein trafficking, histone regulation, retrovirus budding and direct modulation of protein function[24]. As mentioned above, ubiquitin is attached to a lysine residue on the targeted substrate, however, ubiquitin itself also contains lysine residues that serve as self-conjugation sites. As a result, a chain of multiple ubiquitin molecules can be formed and appended to the targeted protein, which is referred to as polyubiquitination. Although monoubiquitination has been shown to be sufficient for the degradation of some proteins, polyubiquitination accelerates the degradation of most proteins[25-27]. Self-conjugation of ubiquitin can occur through different lysine residues and it has been shown that polyubiquitin chains resulting from different ubiquitin linkages have distinct functions. Linkage through lysine-48 is the primary target signal for proteasomal degradation[28], whereas ubiquitin chains linked through lysine-63 execute many functions including protein synthesis[29], kinase activation[30] and DNA repair[31]. In addition, linkage through other lysine residues has been suggested including the involvement of lysine-6 linkage in the regulation of DNA repair[32]. Furthermore, linkage through lysine-29 has been shown to be involved in protein degradation, however, its function is not similar to that of linkage via lysine-48[33].
E3 ubiquitin ligases can execute their function as a single peptide or they can act as multi-component complexes that function as RING-finger type E3 ubiquitin ligases. The distinct superfamily of E3 ubiquitin ligase complexes consists of the skp, cullin, F-box protein (SCF) family, the anaphase-promoting complex (APC) family, and the VHL-elongin C/elongin B (VCB) family[34]. The SCF family makes use of adaptor subunits called F-box proteins that control substrate recognition through distinct protein-protein interaction domains[35], whereas the APC family uses different adaptors and targets proteins that regulate mitosis[36]. The APC complex is composed of at least 10 subunits including yeast Apc2 and Apc11p[37], which are thought to show homology to subunits of the SCF complex[38]. The VCB-like complexes possess a similar architectural structure as the SCF and APC family and on that basis are referred to be E3 ubiquitin ligases.
Lung cancer is the most commonly diagnosed cancer as well as the most common cause of cancer related deaths worldwide, and can be divided in small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC)[39,40]. The major problem in the treatment of lung cancer is the emergence of intrinsic and acquired drug resistance[41]. Despite increased knowledge on the molecular mechanisms contributing to drug resistance and the development of novel agents, the overall 5-year survival of patients diagnosed with lung cancer is less than 15%. This highlights the relevance and necessity of novel agents that can be used in combinational therapies to circumvent drug resistance. One of the strategies that is currently being investigated is targeting components of the UPS system.
In lung cancer, the deregulation of various UPS components has been observed[2,42]. For instance, the mRNA expression of E1-like ubiquitin-activating enzyme (UBE1L) is often reduced in lung cancer cells[43,44]. UBE1L conjugates IFN-stimulated gene, 15-kDa protein (ISG15) and was shown to promote a complex between ISG15 and cyclin D1, which results in cyclin D1 inhibition and subsequent lung cancer growth suppression[45]. In addition, mRNA expression levels of E2 ligases UBE2C and UBE2T were found to be significantly upregulated in lung cancer tissues relative to normal lung tissues[46,47], whereas mRNA expression of E2 ligase Hrad6B was found to be significantly decreased[48]. These E2 ligases are involved in multiple biological processes: UBE2C (also known as UBCH10) initiates the degradation of APC/cyclosome (APC/C) substrates thereby regulating progression through mitosis[49], while UBE2T exerts its function in the fanconi anemia pathway by promoting monoubiquitination of the FANCD2 protein-a key step for efficient DNA repair[50]. In addition, Hrad6B is involved in UV mutagenesis, DNA repair[48,51] and was shown to be involved in histone methylation by promoting the ubiquitination of histone H2B[52].
In this review, we provide an overview of E3 ubiquitin ligases that have been found to be deregulated in lung cancer and a few of them meet some of the criteria of being an ideal anti-cancer target (Table 1). Furthermore, we will discuss the biological processes in which these E3 ubiquitin ligases are involved related to lung cancer as well as their potency to function as “druggable” targets for the treatment of lung cancer.
| Process | E3 ubiquitin ligase | Substrate | Ref. |
| Cell proliferation | c-Cbl | EGFR | 61, 62 |
| Nedd4 | PTEN | 65 | |
| Siah2 | HIPK2 | 56, 128 | |
| Cell cycle regulation | APC | OLC1 | 37, 68 |
| Cul3-based ligase | Rho GTPase; Rho BTB2 | 96 | |
| CCNB1IP1 | Cyclin B | 93 | |
| Parkin | Parkin, CDCrel-1 | 86, 88 | |
| SCF component: Fbxo7 | 79, 80 | ||
| SCF component: Skp2 | CdK: p27, p21, p53 | 72, 73, 74 | |
| Apoptosis | MDM2 | P53, pRb | 108, 117, 118, 161 |
| Parkin | 90, 91 | ||
| Pirh2 | P53 | 115, 162 | |
| SCF component: SAG | c-Jun | 132, 134, 136, 137 | |
| Siah2 | HIPK2 | 128, 129 | |
| Topors | P53 | 123 | |
| TRAF2 | RIP1 | 127, 163 | |
| XIAP | XIAP, AIF | 104 | |
| Gene regulation | Cul3-based ligase | IKBKb | 140 |
| DNA repair | FANCL | FANCD2 | 143-145 |
Tumorigenesis requires abnormal cellular proliferation. Consequently, signalling pathways controlling this complex process are the subjects of intensive research efforts. The RAS/MAPK pathway is one of the best-characterized signal transduction pathways involved in cellular proliferation. The GTPase RAS transmits extracellular signals from receptor tyrosine kinases, such as the epidermal growth factor receptor (EGFR), to downstream effector proteins (e.g., RAF, MEK and ERK) that besides cell proliferation also control differentiation, survival and apoptosis[53,54]. In drosophila, the RING E3 ubiquitin ligase SINA was shown to be a critical component in RAS signalling located most downstream in the pathway[55]. SINA has two human homologs: SIAH1 and SIAH2 that share 76% and 68% sequence similarity, respectively[56]. Interestingly, SIAH2 protein levels were increased in highly aggressive lung tumors compared to little or no expression in normal lung tissues[57]. Moreover, similar levels of SIAH2 mRNA transcripts were detected in multiple lung carcinoma cell lines. However, SIAH2 expression is not restricted to tumorigenic cells but is also expressed by cells lacking any tumorigenic potential suggesting SIAH2 to be involved in all human proliferative cells[58]. As expected, inhibition of SIAH2 suppressed proliferation and reduced the tumorigenesis of human lung cancer cells. Recently, researchers identified the SIAH2-specific substrate homeodomain-interacting protein kinase-2 (HIPK2) which is a serine/threonine kinase that promotes p53-regulated gene expression by phosphorylating p53 at serine 46[23]. They found that overexpression of HIPK2 in lung cancer cells reduced cellular proliferation. This is in line with the suggestion that HIPK2 is tightly regulated in a p53-dependent manner in order to prevent ERK-mediated cell proliferation in the presence of activated p53[59,60].
The RING E3 ubiquitin ligase c-Cbl is known for its function in cell signalling and protein ubiquitination of multiple substrates including EGFR[61]. By targeting EGFR for proteasomal degradation, c-Cbl negatively regulates EGF signalling and opposes cellular proliferation[62]. Recently, Tan et al[42] determined the genetic variation and functionality of c-Cbl in NSCLC. They found a significant loss of heterozygosity of the c-Cbl locus in tumor samples from lung cancer patients compared to normal lung tissues. In addition, they identified novel somatic missense mutations of c-Cbl in multiple regions of the protein including the catalytic RING finger domain and the N-terminal tyrosine kinase binding (TKB) domain, both of which are vital for its E3 ubiquitin ligase activity[63,64]. Furthermore, overexpression of these mutations in NSCLC cell lines resulted in enhanced proliferative potential and cell motility suggesting an essential role for c-Cbl in lung tumorigenesis and metastasis.
In NSCLC, loss of the PTEN tumor suppressor is frequently observed leading to constitutive activation of the AKT pathway which is involved in fundamental cellular processes including protein synthesis, cell proliferation and survival. Recently, the protein Neural precursor cell Expressed Developmentally Down-regulated 4-1 (Nedd4-1) was identified as the E3 ubiquitin ligase responsible for PTEN proteasomal degradation[65]. An additional study showed that Nedd4-1 is overexpressed in 80% of NSCLC tumors which correlates with the loss of PTEN protein[66]. Accordingly, knock-down of Nedd4-1 stabilized PTEN protein levels and, in addition, significantly reduced proliferation of NSCLC cells in vitro and tumor growth in vivo.
In order for a multicellular organism to develop normally, tight regulation of the cell cycle is required[67]. Key regulators of the cell cycle are cyclins, which bind and activate cyclin-dependent kinases (CDKs) resulting in cell cycle progression. The cell cycle consists of four distinct phases: G1, DNA synthesis (S phase), G2 and mitosis (M phase), with G1 and G2 functioning as “gap” phases separating S phase and M phase in time. Cells exit from mitosis upon degradation of mitotic cyclins, a process controlled by the RING E3 ubiquitin ligase anaphase-promoting complex (APC)[37]. As mentioned earlier, the APC complex consist of at least 10 subunits[37]. Among these subunits are the activator proteins cell-division cycle protein 20 (CDC20) and cadherin-1 (Cdh1), which regulate the activity and substrate specificity of APC. The E3 ubiquitin ligase APC, together with its regulatory subunits CDC20 and Cdh1, were found to be accountable for the degradation of the overexpressed in lung cancer 1 (OLC1) protein[68]. OLC1 is highly expressed in lung cancer tissues from patients with a history of cigarette smoking[68,69]. OLC1 degradation by the E3 ubiquitin ligase APC was compromised upon introducing cigarette smoke condensate (CSC). Several studies have revealed that OLC1 is involved in cytokinesis, a process following mitosis[70,71]. However, additional studies are required to clarify the exact role of OCL1 in lung tumorigenesis.
Another important regulator of cell cycle progression is the F-box protein Skp2. Skp2 is part of an SCF complex that targets cyclin-dependent kinase inhibitors p27, p21 and p57 for proteasomal degradation, thereby promoting G1 to S phase transition[72-74]. Overexpression of Skp2 is frequently observed in lung cancer tissues and is associated with the invasive and metastatic potential of NSCLC cells[75,76]. Accordingly, several studies have demonstrated that inhibition of Skp2 suppresses the growth of lung cancer cells[5,77,78].
The F-box protein Fbxo7 is also a component of an SCF complex and was found to selectively enhance CDK6 thereby regulating cell cycle progression[79,80]. Fbxo7 was reported to be upregulated in human lung cancers and to have transforming activity through cdk6[80]. Surprisingly, Fbxo7 does not seem to increase the degradation of the proteins with which it interacts but rather increases their assembly and activity. However, novel Fbxo7-interacting proteins have been identified and are currently being investigated as candidates for Fbxo7-mediated ubiquitination.
In an attempt to identify novel tumor suppressors, Cesari and colleagues identified increased mRNA levels of parkin in NSCLC cells[81]. Conversely, a different study revealed a loss of parkin transcripts in NSCLC tumor tissues and showed that parkin expression was able to inhibit tumorigenicity in mice[82]. The parkin gene is mainly studied due to its pivotal role in the onset of autosomal recessive juvenile parkinsonism (ARJP)[83]. The parkin protein contains a RING finger motif and an ubiquitin-like domain, and many alternatively spliced isoforms have been identified[84,85]. Interestingly, parkin has been shown to exhibit E3 ubiquitin ligase activity targeting itself[86] and several other substrates for proteasomal degradation thereby regulating apoptosis and cell cycle[87-91]. However, the exact role of parkin in lung tumorigenesis needs to be further elucidated.
In contrast with studies on Skp2 and Fbxo7, researchers found that low levels of the E3 ubiquitin ligase CCNB1IP1 in NSCLC correlates with a lower overall survival[92]. CCNB1IP1 contains a RING finger domain and regulates cell cycle by interacting with cyclin B and promoting its degradation[93]. The exact role of CCNB1IP1 in lung tumorigenesis is not known.
A major class of ubiquitin ligases are the Cullin-based E3 ubiquitin ligases, which are incorporated in the SCF and APC complexes[94]. In mammals, eight distinct cullin proteins have been identified; Cul1 to Cul7 and PARC[95]. The Cul1-based E3 ubiquitin ligases are the best characterized and have been shown to control the protein levels of tumor suppressors and oncogenes, and are involved in cell cycle regulation[94]. The Cul3-based E3 ubiquitin ligases have recently emerged as key regulators of mitosis[96]. The atypical Rho GTPase RhoBTB2 is one of the substrates of Cul3-based E3 ubiquitin ligase complexes and its gene expression is ablated in 50% of lung cancer cell lines[97]. It has been suggested that RhoBTB2 functions as a tumor suppressor by recruiting proteins to a Cul3 ubiquitin ligase complex for degradation. However, it is unknown whether the ablation of RhoBTB2 in lung cancer cells correlates with deregulated levels of the Cul3 ubiquitin ligase.
The ability of cells to undergo apoptosis is vital for tissue homeostasis and development[98]. An essential step in apoptosis is the activation of caspases, a family of cysteine proteases[99]. The inhibitors of apoptosis (IAP) proteins are a family that negatively regulate caspases, with the X-linked IAP (XIAP) protein as the best-studied member[100-103]. XIAP contains a RING finger domain and has been characterized as an E3 ubiquitin ligase[104]. Surprisingly, it was observed that high levels of XIAP correlate with a significant longer overall survival of NSCLC patients and is suggested to associate with less aggressive NSCLC[105]. These observations are conflicting with a study in leukemia patients where they demonstrate a correlation between XIAP expression and a decreased overall survival[106]. This implies alternate functions of XIAP in different types of cancer.
In response to physiological stress, the p53 protein is activated and promotes either apoptotic cell death or cell arrest[107]. The levels of p53 are tightly regulated by the E3 ubiquitin ligase MDM2 through an auto-regulatory negative feedback loop; a p53-regulated gene induces MDM2 expression while MDM2 targets p53 for degradation by the 26S proteasome thereby controlling p53-mediated biological responses[108]. MDM2 is often overexpressed in several human cancers including lung cancer[109]. It has been shown that protein expression levels of MDM2 are overexpressed in 70% of NSCLC tissues compared to adjacent normal lung tissues[110,111]. Recently, a single nucleotide polymorphism-SNP309-was identified in the promoter region of MDM2 and was shown to induce MDM2 overexpression thereby influencing p53 activity[112]. Interestingly, a subsequent study revealed an association between SNP309 and increased NSCLC risk, which was predominantly seen among woman[113].
The E3 ubiquitin ligase Pirh2 is another protein that promotes p53 degradation[114]. The Pirh2 gene is regulated by p53 and encodes a RING-finger containing protein that exerts the ubiquitination of p53 independently of MDM2. A study from Duan and colleagues showed that Pirh2 is overexpressed in the majority of lung cancer tissues when compared to normal lung tissues[115,116]. Furthermore, they found enhanced p53 ubiquitination which subsequently resulted in lower p53 expression in mouse lung tumors than in normal tissues. These results are consistent with their hypothesis that increased Pirh2 expression affects lung tumorigenesis by reducing p53 activity.
In addition to targeting p53 for proteasomal degradation, MDM2 has been shown to ubiquitinate the retinoblastoma protein (pRB) which plays a dual role in both apoptosis and cell proliferation[117,118]. Miwa and colleagues found that high expression levels of MDM2 correlated with low expression levels of pRB in a subset of NSCLC patients[117,119]. This correlation was mainly observed in NSCLC cells lacking wild-type p53. Miwa and co-workers suggest that MDM2-induced ubiquitination of pRB perturbs the pRB pathway and subsequently promotes carcinogenesis in a p53-independent manner.
Another protein that has been shown to interact with p53 is the RING finger protein topoisomerase I-binding protein (topors)[120-122]. This interaction results in p53 stabilization and consequent induction of either apoptosis or cell cycle arrest. Conversely, topors has been shown to possess E3 ubiquitin ligase activity targeting p53 for proteasomal degradation, although to a lesser extent than MDM2[123]. This insinuates that topors-induced p53 regulation does not only occur through ubiquitination but also by other mechanisms. Interestingly, preliminary studies have revealed an increase of the human topors gene (also known as LUN) in various lung cancer cell lines[122,124]. Furthermore, expression of LUN was slightly downregulated along with progression of primary NSCLC tumors and strongly downregulated in nodal metastasis[124]. It is suggested that LUN might play a role in inhibition of nodal metastasis as well as the oncogenesis of NSCLC.
Besides the well known caspase inhibitor XIAP and E3 ubiquitin ligases that interact with p53 there are more E3 ubiquitin ligases alternatively expressed in lung cancer that are involved in the apoptotic pathway. For example, the E3 ubiquitin ligase Tumor necrosis factor receptor-associated 2 (TRAF2) was identified as a candidate radiosensitizing target in lung cancer[125]. TRAF2 belongs to a family of seven TRAF members (TRAF1-7) that play a role in a variety of biological processes including immunity, inflammation and apoptosis[126,127]. The TRAF2 protein contains a RING finger domain and mediates several signalling pathways involved in apoptosis protection[127]. It was found that TRAF2 is overexpressed in both lung carcinoma tissues and lung cancer cell lines[125]. In addition, downregulation of TRAF2 in radioresistant lung cancer cells caused growth suppression and radiosensitization, suggesting that TRAF2 may be an attractive drug target for anticancer therapy and radiosensitization.
Moreover, the RING E3 ubiquitin ligase SIAH2 has been shown to play a role in apoptosis. Activity of the recently identified pro-apoptotic SIAH2-specific substrate HIPK2 is considered to play a role in restraining tumor development by targeting tumor cells toward apoptosis upon genotoxic stress[60]. Inhibition of HIPK2 in lung cancer cells resulted in protection against UV-induced apoptosis[128,129]. In accordance, overexpression of HIPK2 sensitized lung cancer cells to UV-induced apoptosis and reduced cellular proliferation.
Finally, it has been shown that the Sensitive to Apoptosis Gene (SAG) is significantly overexpressed in NSCLC tumor tissues compared to adjacent normal lung tissues[130,131]. The SAG protein contains a RING finger domain and has been characterized as a component of SCF E3 ubiquitin ligases targeting several substrates for proteasomal degradation[132,133]. Interestingly, high mRNA levels of SAG correlate with poor survival of NSCLC patients suggesting SAG as a potential prognostic marker in NSCLC. Furthermore, inhibition of SAG sensitizes radioresistant NSCLC cells to ionizing radiation[131]. Under stress conditions, SAG has been shown to function as a proliferating factor that inhibits apoptosis and promotes cell growth[134-137].
The nuclear factor-κB (NF-κB) is a key transcription factor that is thought to play a major role in carcinogenesis[138]. NF-κB controls genes that regulate a variety of biological processes including inflammation, innate and adaptive immunity, and stress responses. In lung cancer, NF-κB is frequently expressed and was found to be involved in the pathogenesis of lung cancer[139]. The inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKBKB) activates NF-κB and is a substrate of a Cul3 ubiquitin ligase complex. Recently, it was shown that genetic disruption of components of a Cul3 ubiquitin ligase complex results in elevated IKBKB levels and represents a mechanism of NF-κB activation in NSCLC[140]. Furthermore, inhibition of NF-κB was earlier shown to sensitize NSCLC cells to chemotherapy-induced apoptosis suggesting a possible role for the inactivation of NF-κB-induced pathways in the treatment of lung cancer[141].
Upon DNA lesions, DNA damage surveillance systems are triggered and subsequently promote the activation of a multitude of genome-protection pathways[142]. One of these pathways involves the fanconi anemia (FA) proteins that are found to form a multi-protein complex that functions as an E3 ubiquitin ligase[143]. This E3 ubiquitin ligase complex exerts its function by monoubiquitinating FANCD2 during DNA replication or following DNA damage, mainly triggered by DNA crosslinking agents such as mitomycin C or Cisplatin[143]. Monoubiquitinated FANCD2 can interact with FANCD1/BRCA2 and others to repair damaged DNA. It was found that the lung cancer cell line Calu-6 harbors an impaired FA-BRCA pathway resulting from alternatively expressed FANCL, a catalytic subunit of the E3 ubiquitin ligase complex[144,145]. The pathway integrity was re-established upon FANCL complementation and reduced the hypersensitivity of Calu-6 cells to mitomycin[145]. Based on these results, it is suggested that the status of the FA-BRCA pathway could play an important role in determining the sensitivity of cancer cells to DNA crosslinking agents.
Most of the E3 ubiquitin ligases described above are also involved in other biological processes. For example, besides its involvement in cell proliferation and apoptosis, Siah2 plays a role in the physiological responses to hypoxia and was shown to target a rate-limiting enzyme in the mitochondrial Krebs cycle[146,147]. In addition, c-Cbl is involved in cell proliferation but also plays a critical role in angiogenesis and is involved in immunity by targeting many protein substrates for proteasomal degradation[148-151]. However, at present the E3 ubiquitin ligases that have been found to be deregulated in lung cancer have not been described to be involved in biological processes other than the ones we have discussed in this review.
Currently, the major issue in targeting E3 ubiquitin ligases is the lack of specific inhibitors in clinical trials. Over the past years, many research efforts have focused on the development of proteasome inhibitors. At present, Bortezomib is the only selective and reversible proteasome inhibitor approved by the United States Food and Drug Administration (FDA) and the European Medicine Agency and it is being used for the treatment of relapsed/refractory multiple myeloma and mantle cell lymphoma[152,153]. However, its induced cytotoxicity is based on overall inhibition of proteolysis of many cellular proteins. By selectively inhibiting an E3 ubiquitin ligase the proteins are stabilized that are regulated by this E3 ubiquitin ligase and thereby circumvent undesired effects on other cellular proteins. The deregulation of E3 ubiquitin ligases has been shown to contribute to cancer development and they are often found overexpressed in lung cancer[4,5]. Altogether, targeting E3 ubiquitin ligases has gained increasing attention, which has led to the development of high-throughput screening assays to identify inhibitors of multiple E3 ubiquitin ligases[154,155]. For example, small molecule inhibitors of the E3 ubiquitin ligase MDM2 have been identified and developed such as cis-imidazolines, benzodiazepines and spiro-oxindoles[156,157]. These inhibitors selectively inhibit MDM2 E3 ubiquitin ligase driven polyubiquitination of p53 with barely any effect on other enzymes using ubiquitin. However, one major concern is the selectivity between normal and cancer cells. Although it is still unclear, the activation of p53 by these MDM2 inhibitors in normal cells induces growth arrest rather than apoptosis making it achievable to obtain a therapeutic window[158]. Excitingly, inhibitors targeting MDM2 are now in clinical trials and pave the way for novel treatment strategies for cancer patients including those diagnosed with lung cancer[157]. Eventually, these inhibitors can be utilized in combination with other therapies, e.g., chemotherapy in order to circumvent drug resistance which is an important problem in the treatment of patients with lung cancer.
In addition to MDM2, there are more E3 ubiquitin ligases that meet some of the criteria of being an ideal anti-cancer target. For example, the HECT E3 ubiquitin ligase Nedd4-1 is overexpressed in the majority of NSCLC tumors and its inhibition reduces the proliferation of NSCLC cells[66]. Furthermore, the SCF component SAG is frequently overexpressed in NSCLC tissues[131]. Importantly, high SAG expression is correlated with poor survival of NSCLC patients and could be a useful prognostic marker. In addition, other SCF components have been described in lung cancer. For example, the F-box protein Skp2 is often overexpressed in lung cancer tissues and is associated with the metastatic and invasive potential of NSCLC cells[75,76]. However, targeting SAG or Skp2 is challenging, since they are part of an SCF complex containing multiple components. Therefore, a general inhibitor against SAG or Skp2 may not have the desirable specificity against any SCF complex. More ideal anti-cancer targets would be the RING E3 ubiquitin ligase Pirh2 and TRAF2. Pirh2 is often overexpressed in many cancer tissues including lung cancer[159]. In addition, TRAF2 is overexpressed in the majority of lung cancer tissues and its downregulation suppresses cell growth and sensitizes otherwise radioresistant lung cancer cells. However, there are no inhibitors targeting these E3 ubiquitin ligases that are currently being tested in clinical trials.
Like E3 ligases, DUBs can be considered as potential anti-cancer targets. Although DUB inhibitors or activators have yet to successfully enter the clinic, multiple DUBs have been implicated in neoplastic disease such as ubiquitin-specific protease 4 (USP4), USP6, and USP8[160].
Although the biological functions of many E3 ubiquitin ligases are still not fully understood, it has become clear that some E3 ubiquitin ligases are promising anti-cancer targets. Despite the fact that we are still facing issues such as selectivity between normal and cancer cells and specificity between E3 ubiquitin ligase and protein substrates, the approval of Bortezomib and the recent entry of MDM2 inhibitors into clinical trials will further stimulate the development of specific E3 ubiquitin ligase inhibitors for the treatment of many cancers including lung cancer.
P- Reviewers Huang CS, Voortman J S- Editor Huang XZ L- Editor A E- Editor Lu YJ
| 1. | Ciechanover A, Orian A, Schwartz AL. The ubiquitin-mediated proteolytic pathway: mode of action and clinical implications. J Cell Biochem Suppl. 2000;34:40-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Christian PA, Fiandalo MV, Schwarze SR. Possible role of death receptor-mediated apoptosis by the E3 ubiquitin ligases Siah2 and POSH. Mol Cancer. 2011;10:57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438-444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 417] [Cited by in RCA: 478] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 4. | Mani A, Gelmann EP. The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol. 2005;23:4776-4789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 392] [Cited by in RCA: 412] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 5. | Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1051] [Cited by in RCA: 1135] [Article Influence: 59.7] [Reference Citation Analysis (0)] |
| 6. | Leite KR, Franco MF, Srougi M, Nesrallah LJ, Nesrallah A, Bevilacqua RG, Darini E, Carvalho CM, Meirelles MI, Santana I. Abnormal expression of MDM2 in prostate carcinoma. Mod Pathol. 2001;14:428-436. [PubMed] |
| 7. | Rayburn E, Zhang R, He J, Wang H. MDM2 and human malignancies: expression, clinical pathology, prognostic markers, and implications for chemotherapy. Curr Cancer Drug Targets. 2005;5:27-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 216] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 8. | Turbin DA, Cheang MC, Bajdik CD, Gelmon KA, Yorida E, De Luca A, Nielsen TO, Huntsman DG, Gilks CB. MDM2 protein expression is a negative prognostic marker in breast carcinoma. Mod Pathol. 2006;19:69-74. [PubMed] |
| 9. | Zhang H. MDM2 oncogene as a novel target for human cancer therapy. Curr Pharm Des. 2000;6:393-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 86] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc Natl Acad Sci USA. 1981;78:761-765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 162] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Hershko A, Ciechanover A, Rose IA. Resolution of the ATP-dependent proteolytic system from reticulocytes: a component that interacts with ATP. Proc Natl Acad Sci USA. 1979;76:3107-3110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 148] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 12. | Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206-8214. [PubMed] |
| 13. | Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 403] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 14. | Finley D, Ciechanover A, Varshavsky A. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell. 1984;37:43-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 395] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 15. | Hershko A, Ciechanover A, Rose IA. Identification of the active amino acid residue of the polypeptide of ATP-dependent protein breakdown. J Biol Chem. 1981;256:1525-1528. [PubMed] |
| 16. | Haas AL, Warms JV, Hershko A, Rose IA. Ubiquitin-activating enzyme. Mechanism and role in protein-ubiquitin conjugation. J Biol Chem. 1982;257:2543-2548. [PubMed] |
| 17. | Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. J Biochem. 2003;134:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 18. | Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189-207. [PubMed] |
| 19. | Clague MJ, Coulson JM, Urbé S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 20. | Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1748] [Cited by in RCA: 1628] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 21. | Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373-428. [PubMed] |
| 22. | Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 759] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 23. | Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6346] [Cited by in RCA: 6682] [Article Influence: 247.5] [Reference Citation Analysis (0)] |
| 24. | Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 906] [Cited by in RCA: 918] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 25. | Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 148] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 26. | Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1279] [Cited by in RCA: 1295] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 27. | Shaeffer JR, Kania MA. Degradation of monoubiquitinated alpha-globin by 26S proteasomes. Biochemistry. 1995;34:4015-4021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 41] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Miller J, Gordon C. The regulation of proteasome degradation by multi-ubiquitin chain binding proteins. FEBS Lett. 2005;579:3224-3230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102:67-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 298] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 30. | Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1450] [Cited by in RCA: 1459] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 31. | Hofmann RM, Pickart CM. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell. 1999;96:645-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 636] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 32. | Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:3916-3924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 181] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 33. | Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442-17456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 654] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 34. | Tyers M, Willems AR. One ring to rule a superfamily of E3 ubiquitin ligases. Science. 1999;284:601, 603-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 120] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 35. | Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1038] [Cited by in RCA: 1083] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 36. | Peters JM. SCF and APC: the Yin and Yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 199] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 37. | Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039-2058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 518] [Cited by in RCA: 536] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 38. | Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters JM. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci USA. 2000;97:8973-8978. [PubMed] |
| 39. | Beasley MB, Brambilla E, Travis WD. The 2004 World Health Organization classification of lung tumors. Semin Roentgenol. 2005;40:90-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 420] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 40. | Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer. 2010;46:765-781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1579] [Cited by in RCA: 1612] [Article Influence: 107.5] [Reference Citation Analysis (0)] |
| 41. | Nishio K, Nakamura T, Koh Y, Suzuki T, Fukumoto H, Saijo N. Drug resistance in lung cancer. Curr Opin Oncol. 1999;11:109-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 42. | Tan YH, Krishnaswamy S, Nandi S, Kanteti R, Vora S, Onel K, Hasina R, Lo FY, El-Hashani E, Cervantes G. CBL is frequently altered in lung cancers: its relationship to mutations in MET and EGFR tyrosine kinases. PLoS One. 2010;5:e8972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 43. | McLaughlin PM, Helfrich W, Kok K, Mulder M, Hu SW, Brinker MG, Ruiters MH, de Leij LF, Buys CH. The ubiquitin-activating enzyme E1-like protein in lung cancer cell lines. Int J Cancer. 2000;85:871-876. [PubMed] |
| 44. | Pitha-Rowe I, Petty WJ, Feng Q, Koza-Taylor PH, Dimattia DA, Pinder L, Dragnev KH, Memoli N, Memoli V, Turi T. Microarray analyses uncover UBE1L as a candidate target gene for lung cancer chemoprevention. Cancer Res. 2004;64:8109-8115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 45. | Feng Q, Sekula D, Guo Y, Liu X, Black CC, Galimberti F, Shah SJ, Sempere LF, Memoli V, Andersen JB. UBE1L causes lung cancer growth suppression by targeting cyclin D1. Mol Cancer Ther. 2008;7:3780-3788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 46. | Hao J, Xu A, Xie X, Hao J, Tian T, Gao S, Xiao X, He D. Elevated expression of UBE2T in lung cancer tumors and cell lines. Tumour Biol. 2008;29:195-203. [PubMed] |
| 47. | Kadara H, Lacroix L, Behrens C, Solis L, Gu X, Lee JJ, Tahara E, Lotan D, Hong WK, Wistuba II. Identification of gene signatures and molecular markers for human lung cancer prognosis using an in vitro lung carcinogenesis system. Cancer Prev Res (Phila). 2009;2:702-711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 48. | Sasaki H, Moriyama S, Nakashima Y, Yukiue H, Fukai I, Fujii Y. Decreased Hrad6B expression in lung cancer. Acta Oncol. 2004;43:585-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 49. | Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363-1369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 217] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 50. | Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D’Andrea AD, Dutta A. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 216] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 51. | Koken MH, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers JH. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc Natl Acad Sci USA. 1991;88:8865-8869. [PubMed] |
| 52. | Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459-471. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 451] [Cited by in RCA: 409] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 53. | Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1186] [Cited by in RCA: 1177] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 54. | Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3375] [Cited by in RCA: 3431] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 55. | Carthew RW, Rubin GM. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63:561-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 257] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 56. | Hu G, Chung YL, Glover T, Valentine V, Look AT, Fearon ER. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics. 1997;46:103-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 120] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 57. | Ahmed AU, Schmidt RL, Park CH, Reed NR, Hesse SE, Thomas CF, Molina JR, Deschamps C, Yang P, Aubry MC. Effect of disrupting seven-in-absentia homolog 2 function on lung cancer cell growth. J Natl Cancer Inst. 2008;100:1606-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 58. | Schmidt RL, Park CH, Ahmed AU, Gundelach JH, Reed NR, Cheng S, Knudsen BE, Tang AH. Inhibition of RAS-mediated transformation and tumorigenesis by targeting the downstream E3 ubiquitin ligase seven in absentia homologue. Cancer Res. 2007;67:11798-11810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 59. | Marchetti A, Cecchinelli B, D’Angelo M, D’Orazi G, Crescenzi M, Sacchi A, Soddu S. p53 can inhibit cell proliferation through caspase-mediated cleavage of ERK2/MAPK. Cell Death Differ. 2004;11:596-607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 60. | D’Orazi G, Rinaldo C, Soddu S. Updates on HIPK2: a resourceful oncosuppressor for clearing cancer. J Exp Clin Cancer Res. 2012;31:63. [PubMed] |
| 61. | de Melker AA, van der Horst G, Calafat J, Jansen H, Borst J. c-Cbl ubiquitinates the EGF receptor at the plasma membrane and remains receptor associated throughout the endocytic route. J Cell Sci. 2001;114:2167-2178. [PubMed] |
| 62. | Ettenberg SA, Keane MM, Nau MM, Frankel M, Wang LM, Pierce JH, Lipkowitz S. cbl-b inhibits epidermal growth factor receptor signaling. Oncogene. 1999;18:1855-1866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 104] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 63. | Waterman H, Levkowitz G, Alroy I, Yarden Y. The RING finger of c-Cbl mediates desensitization of the epidermal growth factor receptor. J Biol Chem. 1999;274:22151-22154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 249] [Cited by in RCA: 246] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 64. | Lill NL, Douillard P, Awwad RA, Ota S, Lupher ML, Miyake S, Meissner-Lula N, Hsu VW, Band H. The evolutionarily conserved N-terminal region of Cbl is sufficient to enhance down-regulation of the epidermal growth factor receptor. J Biol Chem. 2000;275:367-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 117] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 65. | Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 66. | Amodio N, Scrima M, Palaia L, Salman AN, Quintiero A, Franco R, Botti G, Pirozzi P, Rocco G, De Rosa N. Oncogenic role of the E3 ubiquitin ligase NEDD4-1, a PTEN negative regulator, in non-small-cell lung carcinomas. Am J Pathol. 2010;177:2622-2634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 124] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 67. | Tyson JJ, Csikasz-Nagy A, Novak B. The dynamics of cell cycle regulation. Bioessays. 2002;24:1095-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 175] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 68. | Zhang X, Xiao T, Cheng S, Tong T, Gao Y. Cigarette smoke suppresses the ubiquitin-dependent degradation of OLC1. Biochem Biophys Res Commun. 2011;407:753-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 69. | Yuan J, Ma J, Zheng H, Shi T, Sun W, Zhang Q, Lin D, Zhang K, He J, Mao Y. Overexpression of OLC1, cigarette smoke, and human lung tumorigenesis. J Natl Cancer Inst. 2008;100:1592-1605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 70. | Agromayor M, Carlton JG, Phelan JP, Matthews DR, Carlin LM, Ameer-Beg S, Bowers K, Martin-Serrano J. Essential role of hIST1 in cytokinesis. Mol Biol Cell. 2009;20:1374-1387. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 117] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 71. | Bajorek M, Morita E, Skalicky JJ, Morham SG, Babst M, Sundquist WI. Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell. 2009;20:1360-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 109] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 72. | Tsvetkov LM, Yeh KH, Lee SJ, Sun H, Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr Biol. 1999;9:661-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 616] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 73. | Bornstein G, Bloom J, Sitry-Shevah D, Nakayama K, Pagano M, Hershko A. Role of the SCFSkp2 ubiquitin ligase in the degradation of p21Cip1 in S phase. J Biol Chem. 2003;278:25752-25757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 382] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 74. | Kamura T, Hara T, Kotoshiba S, Yada M, Ishida N, Imaki H, Hatakeyama S, Nakayama K, Nakayama KI. Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc Natl Acad Sci USA. 2003;100:10231-10236. [PubMed] |
| 75. | Yokoi S, Yasui K, Saito-Ohara F, Koshikawa K, Iizasa T, Fujisawa T, Terasaki T, Horii A, Takahashi T, Hirohashi S. A novel target gene, SKP2, within the 5p13 amplicon that is frequently detected in small cell lung cancers. Am J Pathol. 2002;161:207-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 76. | Yokoi S, Yasui K, Mori M, Iizasa T, Fujisawa T, Inazawa J. Amplification and overexpression of SKP2 are associated with metastasis of non-small-cell lung cancers to lymph nodes. Am J Pathol. 2004;165:175-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 77. | Sumimoto H, Yamagata S, Shimizu A, Miyoshi H, Mizuguchi H, Hayakawa T, Miyagishi M, Taira K, Kawakami Y. Gene therapy for human small-cell lung carcinoma by inactivation of Skp-2 with virally mediated RNA interference. Gene Ther. 2005;12:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 59] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 78. | Jiang F, Caraway NP, Li R, Katz RL. RNA silencing of S-phase kinase-interacting protein 2 inhibits proliferation and centrosome amplification in lung cancer cells. Oncogene. 2005;24:3409-3418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 79. | Cenciarelli C, Chiaur DS, Guardavaccaro D, Parks W, Vidal M, Pagano M. Identification of a family of human F-box proteins. Curr Biol. 1999;9:1177-1179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 290] [Article Influence: 11.2] [Reference Citation Analysis (1)] |
| 80. | Laman H. Fbxo7 gets proactive with cyclin D/cdk6. Cell Cycle. 2006;5:279-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 81. | Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci USA. 2003;100:5956-5961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 260] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 82. | Picchio MC, Martin ES, Cesari R, Calin GA, Yendamuri S, Kuroki T, Pentimalli F, Sarti M, Yoder K, Kaiser LR. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin Cancer Res. 2004;10:2720-2724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 83. | Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605-608. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3646] [Cited by in RCA: 3757] [Article Influence: 139.1] [Reference Citation Analysis (0)] |
| 84. | Mizuno Y, Hattori N, Mori H, Suzuki T, Tanaka K. Parkin and Parkinson’s disease. Curr Opin Neurol. 2001;14:477-482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 81] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 85. | Dagata V, Cavallaro S. Parkin transcript variants in rat and human brain. Neurochem Res. 2004;29:1715-1724. [PubMed] [DOI] [Full Text] |
| 86. | Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275:35661-35664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 576] [Cited by in RCA: 583] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 87. | Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97:13354-13359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 722] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 88. | Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37:735-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 303] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 89. | Shimura H, Hattori N, Kubo Si, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1464] [Cited by in RCA: 1477] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 90. | Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105:891-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 811] [Cited by in RCA: 816] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 91. | Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, Troncoso J, Johnson B, Saffary R, Goh EL. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968-7978. [PubMed] |
| 92. | Confalonieri S, Quarto M, Goisis G, Nuciforo P, Donzelli M, Jodice G, Pelosi G, Viale G, Pece S, Di Fiore PP. Alterations of ubiquitin ligases in human cancer and their association with the natural history of the tumor. Oncogene. 2009;28:2959-2968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 93. | Toby GG, Gherraby W, Coleman TR, Golemis EA. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol Cell Biol. 2003;23:2109-2122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 69] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 94. | Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 955] [Cited by in RCA: 941] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 95. | Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 421] [Cited by in RCA: 408] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 96. | Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, Sumara I, Peter M. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791-800. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 97. | Wilkins A, Ping Q, Carpenter CL. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 2004;18:856-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 98. | Steller H. Mechanisms and genes of cellular suicide. Science. 1995;267:1445-1449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1812] [Cited by in RCA: 1753] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 99. | Fan TJ, Han LH, Cong RS, Liang J. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai). 2005;37:719-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 851] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 100. | Uren AG, Pakusch M, Hawkins CJ, Puls KL, Vaux DL. Cloning and expression of apoptosis inhibitory protein homologs that function to inhibit apoptosis and/or bind tumor necrosis factor receptor-associated factors. Proc Natl Acad Sci USA. 1996;93:4974-4978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 387] [Cited by in RCA: 387] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 101. | Yang YL, Li XM. The IAP family: endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000;10:169-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 217] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 102. | Scott FL, Denault JB, Riedl SJ, Shin H, Renatus M, Salvesen GS. XIAP inhibits caspase-3 and -7 using two binding sites: evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 310] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 103. | Shiozaki EN, Chai J, Rigotti DJ, Riedl SJ, Li P, Srinivasula SM, Alnemri ES, Fairman R, Shi Y. Mechanism of XIAP-mediated inhibition of caspase-9. Mol Cell. 2003;11:519-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 518] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 104. | Galbán S, Duckett CS. XIAP as a ubiquitin ligase in cellular signaling. Cell Death Differ. 2010;17:54-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 188] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 105. | Ferreira CG, van der Valk P, Span SW, Ludwig I, Smit EF, Kruyt FA, Pinedo HM, van Tinteren H, Giaccone G. Expression of X-linked inhibitor of apoptosis as a novel prognostic marker in radically resected non-small cell lung cancer patients. Clin Cancer Res. 2001;7:2468-2474. [PubMed] |
| 106. | Tamm I, Kornblau SM, Segall H, Krajewski S, Welsh K, Kitada S, Scudiero DA, Tudor G, Qui YH, Monks A. Expression and prognostic significance of IAP-family genes in human cancers and myeloid leukemias. Clin Cancer Res. 2000;6:1796-1803. [PubMed] |
| 107. | Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96-107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 447] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 108. | Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1:1001-1008. [PubMed] |
| 109. | Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 158] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 110. | Gorgoulis VG, Zacharatos P, Kotsinas A, Mariatos G, Liloglou T, Vogiatzi T, Foukas P, Rassidakis G, Garinis G, Ioannides T. Altered expression of the cell cycle regulatory molecules pRb, p53 and MDM2 exert a synergetic effect on tumor growth and chromosomal instability in non-small cell lung carcinomas (NSCLCs). Mol Med. 2000;6:208-237. [PubMed] |
| 111. | Gorgoulis VG, Zacharatos P, Kotsinas A, Liloglou T, Kyroudi A, Veslemes M, Rassidakis A, Halazonetis TD, Field JK, Kittas C. Alterations of the p16-pRb pathway and the chromosome locus 9p21-22 in non-small-cell lung carcinomas: relationship with p53 and MDM2 protein expression. Am J Pathol. 1998;153:1749-1765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 82] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 112. | Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 926] [Cited by in RCA: 980] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 113. | Lind H, Zienolddiny S, Ekstrøm PO, Skaug V, Haugen A. Association of a functional polymorphism in the promoter of the MDM2 gene with risk of nonsmall cell lung cancer. Int J Cancer. 2006;119:718-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 120] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 114. | Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 527] [Cited by in RCA: 556] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 115. | Duan W, Gao L, Druhan LJ, Zhu WG, Morrison C, Otterson GA, Villalona-Calero MA. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J Natl Cancer Inst. 2004;96:1718-1721. [PubMed] |
| 116. | Duan W, Gao L, Wu X, Zhang Y, Otterson GA, Villalona-Calero MA. Differential response between the p53 ubiquitin-protein ligases Pirh2 and MdM2 following DNA damage in human cancer cells. Exp Cell Res. 2006;312:3370-3378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 117. | Uchida C, Miwa S, Kitagawa K, Hattori T, Isobe T, Otani S, Oda T, Sugimura H, Kamijo T, Ookawa K. Enhanced Mdm2 activity inhibits pRB function via ubiquitin-dependent degradation. EMBO J. 2005;24:160-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 151] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 118. | Hickman ES, Moroni MC, Helin K. The role of p53 and pRB in apoptosis and cancer. Curr Opin Genet Dev. 2002;12:60-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 133] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 119. | Miwa S, Uchida C, Kitagawa K, Hattori T, Oda T, Sugimura H, Yasuda H, Nakamura H, Chida K, Kitagawa M. Mdm2-mediated pRB downregulation is involved in carcinogenesis in a p53-independent manner. Biochem Biophys Res Commun. 2006;340:54-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 120. | Zhou R, Wen H, Ao SZ. Identification of a novel gene encoding a p53-associated protein. Gene. 1999;235:93-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 121. | Weger S, Hammer E, Heilbronn R. Topors, a p53 and topoisomerase I binding protein, interacts with the adeno-associated virus (AAV-2) Rep78/68 proteins and enhances AAV-2 gene expression. J Gen Virol. 2002;83:511-516. [PubMed] |
| 122. | Lin L, Ozaki T, Takada Y, Kageyama H, Nakamura Y, Hata A, Zhang JH, Simonds WF, Nakagawara A, Koseki H. topors, a p53 and topoisomerase I-binding RING finger protein, is a coactivator of p53 in growth suppression induced by DNA damage. Oncogene. 2005;24:3385-3396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 123. | Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, Liu LF, Lutzker S, Saleem A, Rubin EH. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 2004;279:36440-36444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 153] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 124. | Oyanagi H, Takenaka K, Ishikawa S, Kawano Y, Adachi Y, Ueda K, Wada H, Tanaka F. Expression of LUN gene that encodes a novel RING finger protein is correlated with development and progression of non-small cell lung cancer. Lung Cancer. 2004;46:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 125. | Zheng M, Morgan-Lappe SE, Yang J, Bockbrader KM, Pamarthy D, Thomas D, Fesik SW, Sun Y. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570-7578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 126. | Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci. 2002;115:679-688. [PubMed] |
| 127. | Bradley JR, Pober JS. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene. 2001;20:6482-6491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 492] [Cited by in RCA: 515] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 128. | Hofmann TG, Möller A, Sirma H, Zentgraf H, Taya Y, Dröge W, Will H, Schmitz ML. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat Cell Biol. 2002;4:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 473] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 129. | D’Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 544] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 130. | Sasaki H, Yukiue H, Kobayashi Y, Moriyama S, Nakashima Y, Kaji M, Fukai I, Kiriyama M, Yamakawa Y, Fujii Y. Expression of the sensitive to apoptosis gene, SAG, as a prognostic marker in nonsmall cell lung cancer. Int J Cancer. 2001;95:375-377. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 131. | Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin Cancer Res. 2010;16:814-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 132. | Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 133. | Gu Q, Bowden GT, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage-dependent targeting of c-Jun/AP1 and IkappaB-alpha/NF-kappaB. J Cell Biol. 2007;178:1009-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 134. | Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145-3155. [PubMed] |
| 135. | Yang ES, Park JW. Regulation of nitric oxide-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Res. 2006;40:279-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 136. | Chanalaris A, Sun Y, Latchman DS, Stephanou A. SAG attenuates apoptotic cell death caused by simulated ischaemia/reoxygenation in rat cardiomyocytes. J Mol Cell Cardiol. 2003;35:257-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 137. | Lee SJ, Yang ES, Kim SY, Kim SY, Shin SW, Park JW. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Biol Med. 2008;45:167-176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 138. | Gilmore TD. Introduction to NF-kappaB: players, pathways, perspectives. Oncogene. 2006;25:6680-6684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 425] [Cited by in RCA: 465] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 139. | Tang X, Liu D, Shishodia S, Ozburn N, Behrens C, Lee JJ, Hong WK, Aggarwal BB, Wistuba II. Nuclear factor-kappaB (NF-kappaB) is frequently expressed in lung cancer and preneoplastic lesions. Cancer. 2006;107:2637-2646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 162] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 140. | Thu KL, Pikor LA, Chari R, Wilson IM, Macaulay CE, English JC, Tsao MS, Gazdar AF, Lam S, Lam WL. Genetic disruption of KEAP1/CUL3 E3 ubiquitin ligase complex components is a key mechanism of NF-kappaB pathway activation in lung cancer. J Thorac Oncol. 2011;6:1521-1529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 141. | Jones DR, Broad RM, Madrid LV, Baldwin AS, Mayo MW. Inhibition of NF-kappaB sensitizes non-small cell lung cancer cells to chemotherapy-induced apoptosis. Ann Thorac Surg. 2000;70:930-936; discussion 930-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 142. | Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH. DNA damage response and transcription. DNA Repair (Amst). 2011;10:743-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 127] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 143. | Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223-4233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 273] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 144. | Meetei AR, de Winter JP, Medhurst AL, Wallisch M, Waisfisz Q, van de Vrugt HJ, Oostra AB, Yan Z, Ling C, Bishop CE. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 432] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 145. | Zhang J, Wang X, Lin CJ, Couch FJ, Fei P. Altered expression of FANCL confers mitomycin C sensitivity in Calu-6 lung cancer cells. Cancer Biol Ther. 2006;5:1632-1636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 146. | Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941-952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 329] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 147. | Habelhah H, Laine A, Erdjument-Bromage H, Tempst P, Gershwin ME, Bowtell DD, Ronai Z. Regulation of 2-oxoglutarate (alpha-ketoglutarate) dehydrogenase stability by the RING finger ubiquitin ligase Siah. J Biol Chem. 2004;279:53782-53788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 148. | Singh AJ, Meyer RD, Navruzbekov G, Shelke R, Duan L, Band H, Leeman SE, Rahimi N. A critical role for the E3-ligase activity of c-Cbl in VEGFR-2-mediated PLCgamma1 activation and angiogenesis. Proc Natl Acad Sci USA. 2007;104:5413-5418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 149. | Chiou SH, Shahi P, Wagner RT, Hu H, Lapteva N, Seethammagari M, Sun SC, Levitt JM, Spencer DM. The E3 ligase c-Cbl regulates dendritic cell activation. EMBO Rep. 2011;12:971-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 150. | Marois L, Vaillancourt M, Marois S, Proulx S, Paré G, Rollet-Labelle E, Naccache PH. The ubiquitin ligase c-Cbl down-regulates FcgammaRIIa activation in human neutrophils. J Immunol. 2009;182:2374-2384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 151. | Miura-Shimura Y, Duan L, Rao NL, Reddi AL, Shimura H, Rottapel R, Druker BJ, Tsygankov A, Band V, Band H. Cbl-mediated ubiquitinylation and negative regulation of Vav. J Biol Chem. 2003;278:38495-38504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 152. | Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8:508-513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 493] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 153. | Orlowski RZ, Kuhn DJ. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res. 2008;14:1649-1657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 447] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 154. | Goldenberg SJ, Marblestone JG, Mattern MR, Nicholson B. Strategies for the identification of ubiquitin ligase inhibitors. Biochem Soc Trans. 2010;38:132-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 155. | Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Ther. 2003;2:623-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 156. | Lai Z, Yang T, Kim YB, Sielecki TM, Diamond MA, Strack P, Rolfe M, Caligiuri M, Benfield PA, Auger KR. Differentiation of Hdm2-mediated p53 ubiquitination and Hdm2 autoubiquitination activity by small molecular weight inhibitors. Proc Natl Acad Sci USA. 2002;99:14734-14739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 157. | Patel S, Player MR. Small-molecule inhibitors of the p53-HDM2 interaction for the treatment of cancer. Expert Opin Investig Drugs. 2008;17:1865-1882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 158. | Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y. Structure-based design of potent non-peptide MDM2 inhibitors. J Am Chem Soc. 2005;127:10130-10131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 529] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 159. | Jung YS, Qian Y, Chen X. Pirh2 RING-finger E3 ubiquitin ligase: its role in tumorigenesis and cancer therapy. FEBS Lett. 2012;586:1397-1402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 160. | Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 161. | Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844-848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3508] [Cited by in RCA: 3660] [Article Influence: 174.3] [Reference Citation Analysis (0)] |
| 162. | Hakem A, Bohgaki M, Lemmers B, Tai E, Salmena L, Matysiak-Zablocki E, Jung YS, Karaskova J, Kaustov L, Duan S. Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS Genet. 2011;7:e1002360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 163. | Lee TH, Shank J, usson N, Kelliher MA. The kinase activity of Rip1 is not required for tumor necrosis factor-alpha-induced IkappaB kinase or p38 MAP kinase activation or for the ubiquitination of Rip1 by Traf2. J Biol Chem. 2004;279:33185-33191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 208] [Article Influence: 9.9] [Reference Citation Analysis (0)] |