Published online Jun 24, 2020. doi: 10.5306/wjco.v11.i6.348
Peer-review started: January 31, 2020
First decision: April 18, 2020
Revised: May 18, 2020
Accepted: May 21, 2020
Article in press: May 21, 2020
Published online: June 24, 2020
MicroRNAs (miRNAs) are short noncoding RNAs that regulate the expression of genes by sequence-specific binding to mRNA to either promote or block its translation; they can also act as tumor suppressors (e.g., let-7b, miR-29a, miR-99, mir-100, miR-155, and miR-181) and/or oncogenes (e.g., miR-29a, miR-125b, miR-143-p3, mir-155, miR-181, miR-183, miR-196b, and miR-223) in childhood acute leukemia (AL). Differentially expressed miRNAs are important factors associated with the initiation and progression of AL. As shown in many studies, they can be used as noninvasive diagnostic and prognostic biomarkers, which are useful in monitoring early stages of AL development or during therapy (e.g., miR-125b, miR-146b, miR-181c, and miR-4786), accurate classification of different cellular or molecular AL subgroups (e.g., let-7b, miR-98, miR-100, miR-128b, and miR-223), and identification and development of new therapeutic agents (e.g., mir-10, miR-125b, miR-203, miR-210, miR-335). Specific miRNA patterns have also been described for commonly used AL therapy drugs (e.g., miR-125b and miR-223 for doxorubicin, miR-335 and miR-1208 for prednisolone, and miR-203 for imatinib), uncovering miRNAs that are associated with treatment response. In the current review, the role of miRNAs in the development, progression, and therapy monitoring of pediatric ALs will be presented and discussed.
Core tip: MicroRNAs (miRNAs) are small endogenous gene expression regulators. By performing the function of oncogenes and/or tumor suppressors, they have become interesting candidates for early diagnosis, accurate classification, and predictors of prognosis in the most common childhood cancers, i.e. acute leukemia. The possibility of using modulation of miRNA levels in targeted therapy is also important. Because they are noninvasive and relatively easy to determine in biological samples, miRNAs are gaining increasing attention from scientists and clinicians.
- Citation: Szczepanek J. Role of microRNA dysregulation in childhood acute leukemias: Diagnostics, monitoring and therapeutics: A comprehensive review. World J Clin Oncol 2020; 11(6): 348-369
- URL: https://www.wjgnet.com/2218-4333/full/v11/i6/348.htm
- DOI: https://dx.doi.org/10.5306/wjco.v11.i6.348
Childhood acute leukemias (ALs) are a group of diseases with varied immunophenotypes and specific genetic abnormalities[1]. Acute lymphoblastic leukemia (ALL), the most common type of childhood leukemia, develops from early forms of B- or T-cells at different stages of maturity[2]. Most of the remaining cases account for acute myeloid leukemia (AML), which develops from myeloid cells that form white blood cells[3] (other than lymphocytes), red blood cells, or platelets[1]. The 5-year survival rate for children with AL has greatly increased over time and is now more than 90% overall for ALL and in the range of 60% to 70% for AML[4,5]. Survival rates vary depending on the subtype of AL and other prognostic factors[1]. Relapse risk can be predicted by clinical and pharmacogenetic features, early treatment response to tailor chemotherapy intensity, and genetic characteristics of leukemic cells[6-8].
MicroRNAs (miRNAs, miRs) are a family of small (about 22-nucleotide), endogenous, noncoding RNAs that negatively regulate gene expression in a sequence-specific manner[9,10]. It has been predicted that miRNAs, which account for at least 1% of human protein-coding genes, can control more than a third of the protein-coding genes[11-13]. At the posttranscriptional level, miRNAs exhibit temporally and spatially regulated expression patterns[9,14]. Discovering miRNA molecules, identifying their targets, and predicting their functional and regulatory mechanisms are critical for understanding biological processes (e.g., diverse development, cell growth control proliferation, differentiation, and apoptosis) and their roles in the etiopathology of disease[15,16].
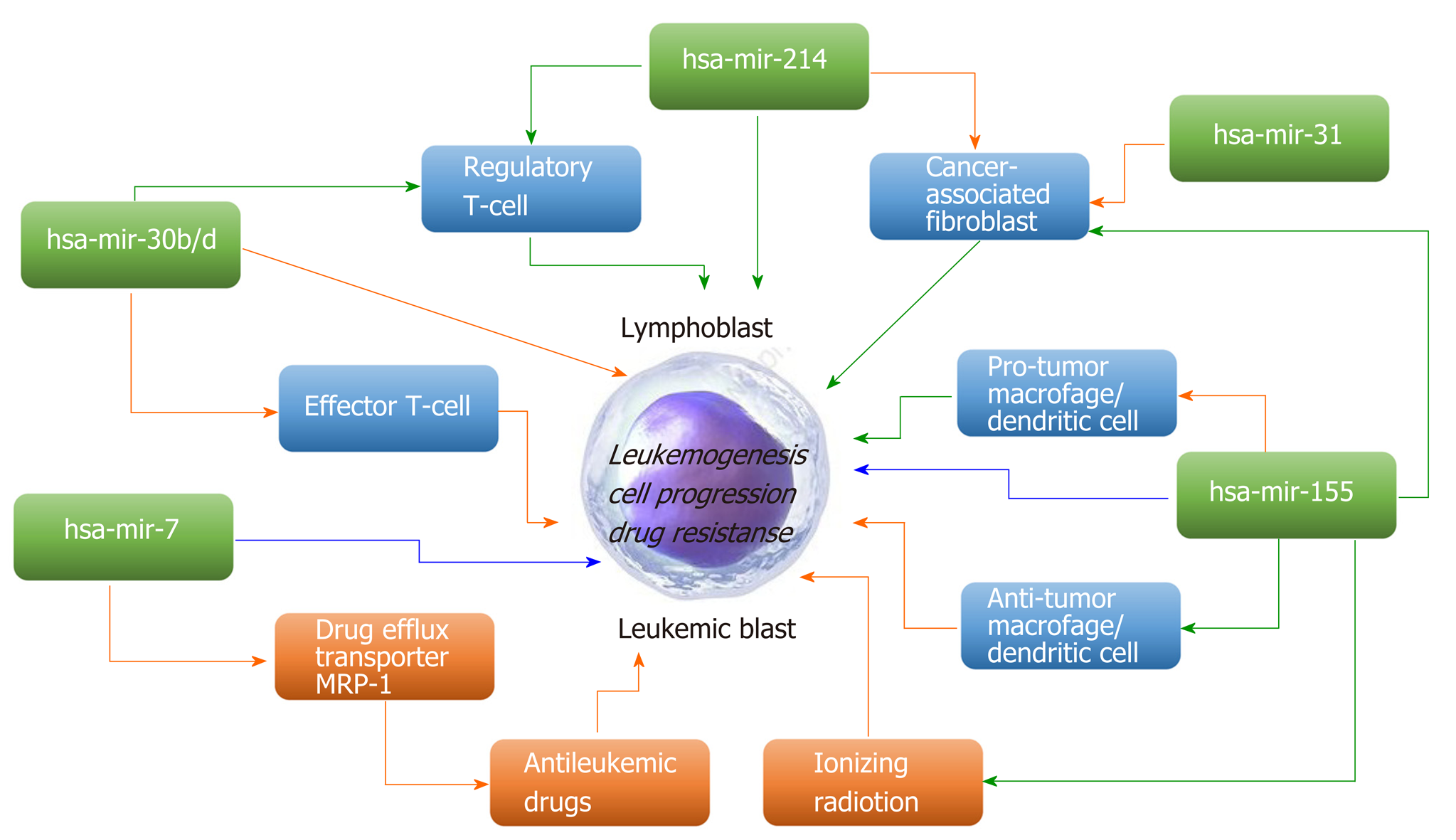
Great interest has emerged in modulating miRNA expression for therapeutic purposes. Many scientific studies have shown that these molecules play an important role as regulators of genes in many organisms, and miRNAs have already been implicated in a growing number of human diseases, including cancers[12,16,17]. miRNAs can act as either oncogenes and/or tumor suppressors, contributing to malignant transformation in solid and hematological tumors[18,19]. miRNAs that typically target a tumor suppressor are classified as oncomiRs and are generally upregulated in different types of cancer. MiRNAs, which downregulate oncogenes, are defined as tumor suppressor miRs[19]. Promoter methylation, mutation, or deletion or defective miRNA processing often leads to loss of suppressor miRs in cancer[20]. Some miRNAs may exert contrasting oncogenic/tumor-suppressive effects on cancer-modifying extrinsic factors and the leukemic cells themselves (Figure 1).
Deregulation of miRNA expression patterns is a hallmark of hematological malignancies and can be useful for the classification of AL genetic subtypes, which are important for differential diagnosis, prognosis, and treatment monitoring. These molecules control the levels of potentially large numbers of proteins, many of which might be important drug targets, and are useful in the development of new therapeutic regimens[21]. Cancer-specific miRNA signatures correlated with diagnosis, progression, prognosis, and response to treatment were determined for many cancers, including childhood leukemia. Although the importance of miRNA molecules in the genetic basis of leukemia is documented in an increasing number of publications, the role of miRNAs in pediatric AL still needs to be established. Profiling the expression levels of miRNA molecules at several levels with unprecedented resolution, depth, and speed is possible thanks to the development of high-throughput technologies, such as next-generation sequencing, microarrays, mass spectrometry, and new bioinformatics tools[15,22].
In the current review, the role of miRNAs in the development, progression, and therapy monitoring of pediatric ALs will be presented and discussed.
It is very important in the treatment of AL to treat patients in the right risk groups, which allows both maximizing the effectiveness of therapy and minimizing its toxicity. At present, the exact stratification of patients into relevant genetic subclasses, and subsequent risk groups, is still a laborious and expensive diagnostic process, including comprehensive morphology, immunophenotype, cytogenetics, response to induction therapy, and genetic testing[23-26].
Currently, several types of classification can be specified in AL. The main ones are: (A) French-American-British (FAB) morphological classification, distinguishing three ALL subtypes (L1-L3) and eight AML subtypes (M0-M7); (B) Cytochemical classification; (C) Immunological classification; (D) Cytogenetic classification; and (E) Molecular classification[27-29]. The World Health Organization system divides ALL into several groups, the most common of which are B-cell ALL (B-ALL) and T-cell ALL (T-ALL). B-ALL can be further subclassified according to distinct patterns of genomic alterations and gene expression signatures (most frequent abnormalities: ETV6–RUNX1 fusion, TFPT–PBX1 fusion, BCR–ABL1 fusion; and MLL fusions)[30,31]. In T-ALL, several subgroups have been recognized, e.g., immature/LYL1, TAL1, HOX11, HOX11L2, and HOX. The World Health Organization classification includes several AML subtypes with recurrent genetic abnormalities [e.g., AML with t(8;21)(q22;q22) RUNX1-RUNX1T1, AML with inv(16)(p13.1;q22) or t(16;16)(p13.1;q22) CBFB-MYH11, APL with t(15;17)(q22;q12) PML-RARA, and AML with t(9;11)(p22;q23) MLLT3-MLL][32,33]. In addition, molecular changes cannot be detected by classical karyotyping, such as WT1, NPM1, CEBPA, and DNMT3A mutations. Based on genetic classifications for subgroups of AML, it is supposed that two of the major subgroups of AML patients are most important: Those with disruptions of the CBF complex and those with disruptions of the FLT3 gene[34,35]. Classification systems are still unsatisfactory because most of the recurrences occur in the intermediate and standard risk groups[26].
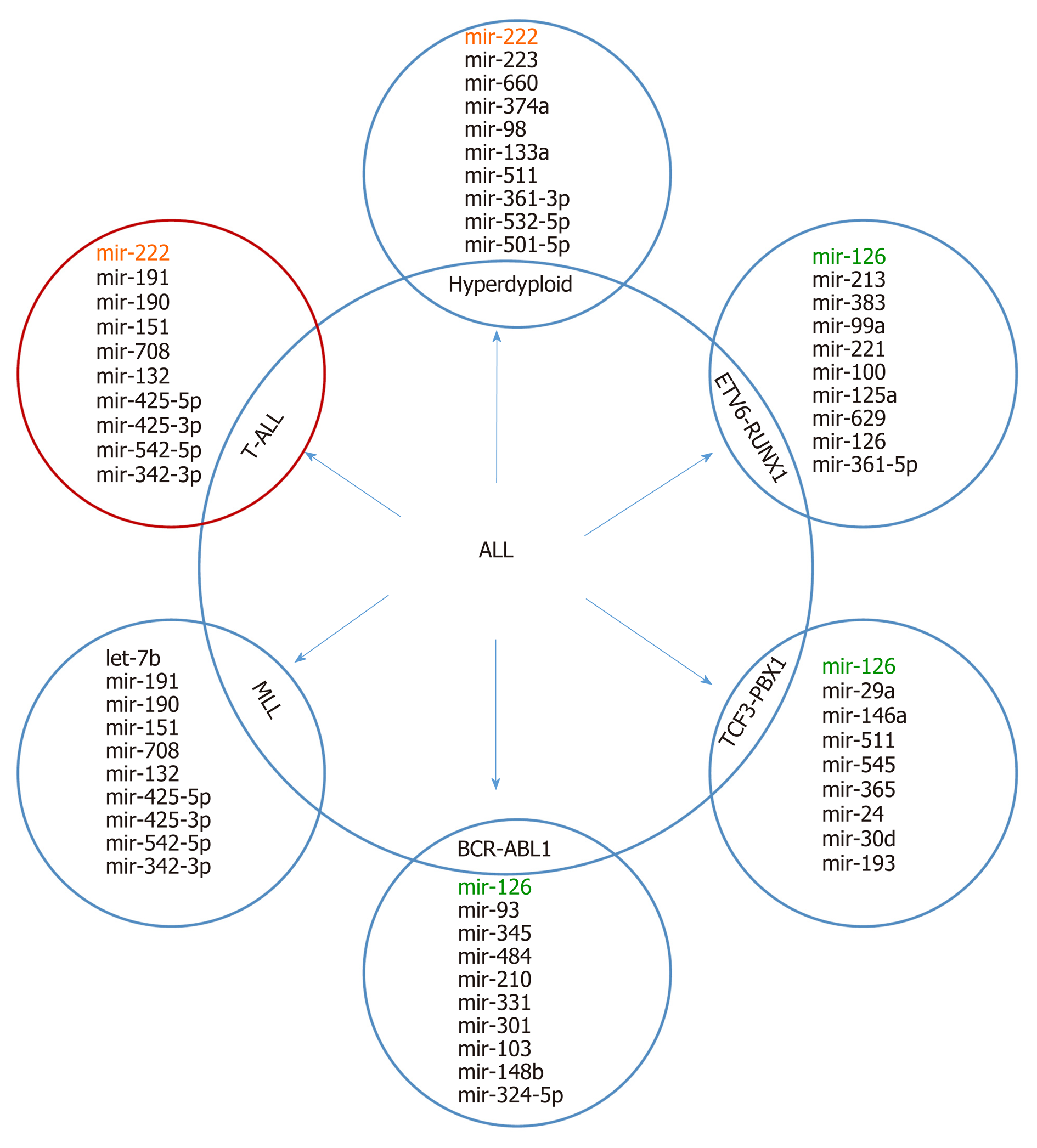
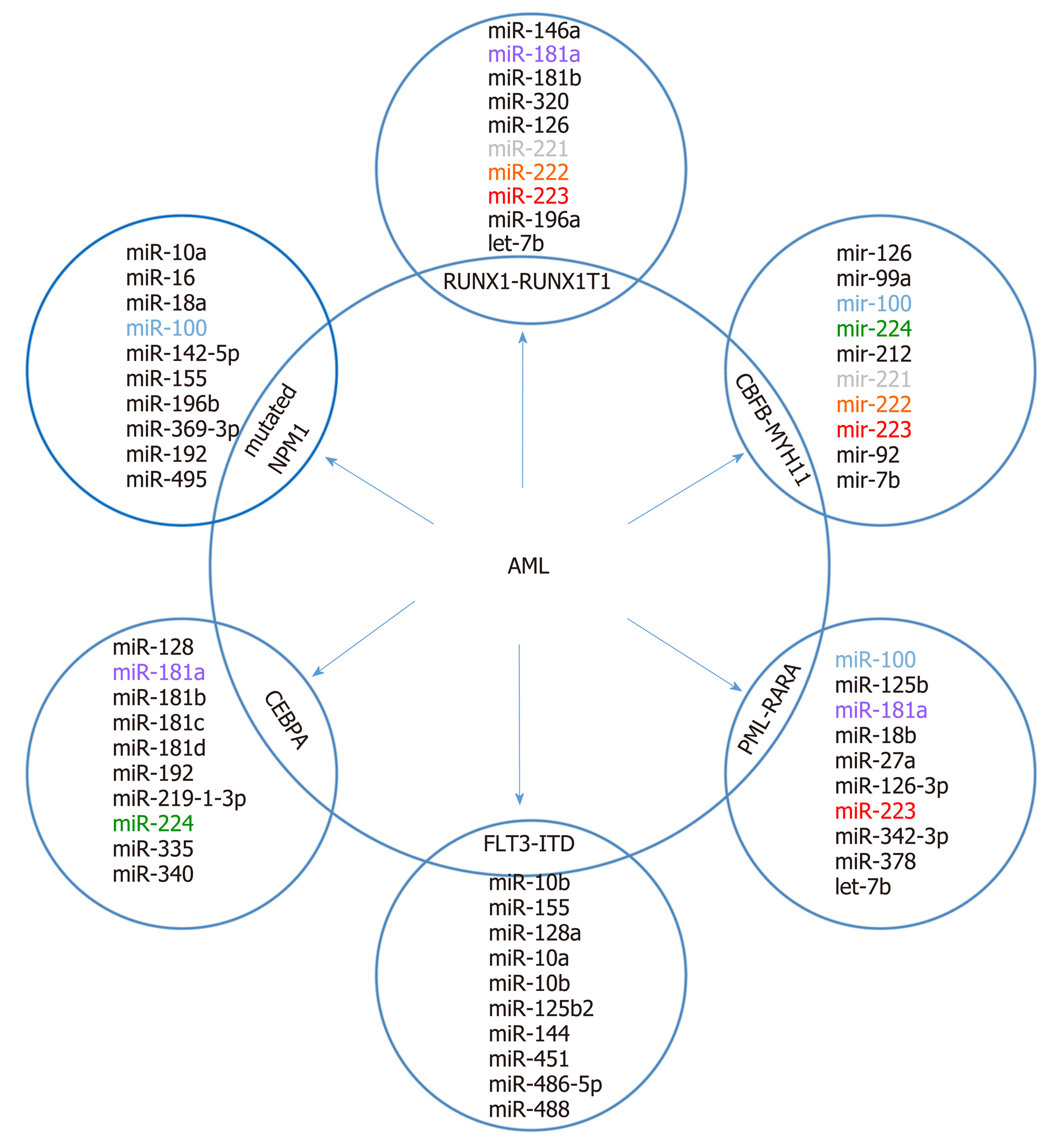
It is supposed that the cancer-specific miRNA patterns rather than mRNA signatures are a more accurate method of classifying cancer subtypes[36,37]. Molecular classification based on expression alterations (genes or miRNA) allows the determination of the genetic profile of myeloid and lymphoid lineage, distinguishing B-linear and T-linear ALL tumors, as well as listing subclasses within leukemia subtypes. Based on miRNA analyses, a correlation between immunophenotype and chromosome disorders and various miRNA expression patterns in all subtypes of AL was demonstrated. A top-ten discriminative miRNA set was proposed for AL types and their main subgroup, as shown in Figure 2 and 3.
The type and subtype of childhood leukemia plays a major role in both treatment options and patient prognosis. Determining the type (ALL or AML) and subtype of the leukemia is performed by testing samples of the blood, bone marrow, and sometimes lymph nodes or cerebrospinal fluid[23]. In recent years, there has been significant progress in the use of comprehensive transcriptomic and genomic methods (expression, single nucleotide polymorphism, and comparative genomic hybridization arrays; whole exome, whole transcriptome, and whole genome sequencing) to identify leukemia subtypes and inherited and somatic genomic changes, but nevertheless, much work is needed to define the intrinsic and extrinsic determinants of leukemia progression, prognosis, and drug response[8,38,39]. A summary of miRNA marker functions, based on the example of pediatric ALL, is presented in Table 1.
| miRNA name | Description | Ref. |
| hsa-let-7b | In patients with ALL, the expression of microRNA let-7b is regulated by methylation of CpG islands in the region of the genomic promoter. The microRNA let-7b may act as a tumor suppressor, whose low expression is involved in ALL development, indicating the microRNA let-7b may become a new therapeutic target for ALL | [154] |
| hsa-mir-17 | Differential expression of miR-17~92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia | [155] |
| hsa-mir-99a, hsa-mir-100 | miR-100 and miR-99a have critical roles in altering cellular processes by targeting both the FKBP51 and IGF1R/mTOR signaling pathways in vitro and might represent a potential novel strategy for ALL treatment | [43] |
| hsa-mir-101 | Hsp90 co-chaperone – p23, is regulated by hsa-miR-101, which is downregulated in childhood ALL cases | [156] |
| hsa-mir-124 | miR-124 contributes to glucocorticoid resistance in acute lymphoblastic leukemia by promoting proliferation, inhibiting apoptosis, and targeting the glucocorticoid receptor | [157] |
| hsa-mir-126 | miR-126 plays a critical but 2-faceted role in leukemia and thereby uncovers a new layer of miRNA regulation in cancer. miR-126 depletion can sensitize AML cells to standard chemotherapy, which suggests that miR-126 represents a promising therapeutic target | [158] |
| hsa-mir-142 | Upregulation of miR-142-3p decreased MLL-AF4 expression in the RS4;11 leukemic cell line. Ectopic expression of miR-142-3p remarkably suppressed cell proliferation and induced apoptosis, and exogenous expression of miR-142-3p strongly reduced the expression of MLL-AF4 target genes such as homeobox A HOXA9, HOXA7, and HOXA10 | [159] |
| hsa-mir-181a | Ectopic expression of miR-181a resulted in decreased CD10 hyperexpression in ETV6/RUNX1+ primary patient samples. miR-181a could target ETV6/RUNX1 and cause a reduction in the level of that oncoprotein, cell growth arrest, an increase in apoptosis, and induction of cell differentiation in ETV6/RUNX1+ cell line | [48] |
| hsa-mir-181a | miR-181a play role as negative regulator for the TGF-β1 signaling pathway | [47] |
| hsa-mir-196b | miR-196b becomes nonfunctional in T-cell ALL as a consequence of mutations in 3'-UTR of the c-myc gene in T-cell ALL cellular models | [160] |
| hsa-mir-196b, hsa-mir-1290 | miR196b and miR-1290 target the IGFBP3 3'-UTR and participate in the antitumor effect of resveratrol via regulation of IGFBP3 expression in acute lymphoblastic leukemia. miR-196b/miR-1290 are potential therapeutic targets for ALL | [161] |
| hsa-mir-221 hsa-mir-222 | Overexpression of miR-221 in ALL cells prompted cell-cycle progression and sensitization of ALL cells to cytotoxic agents. Niche-influenced miR-221/222 may define a novel therapeutic target in ALL | [162] |
| hsa-mir-520 h | POLD1 and MCM2 were found to be regulated by miR-520H via E2F1. High expression of POLD1, MCM2, and PLK4 might play positive roles in the recurrence of ALL | [163] |
| hsa-mir-595 | miR-595 suppresses the cellular uptake and cytotoxic effects of methotrexate by targeting SLC19A1 in CEM/C1 cells | [164] |
| hsa-mir-664 | miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukemia. miR-664 may represent a potential therapeutic target for T-ALL intervention | [115] |
| hsa-mir-708 | The expression level of miR-708 reflects differences among the clinical types of common-ALL, and CNTFR, NNAT, and GNG12 were identified as targets of miR-708 | [165] |
| hsa-mir-2909 | The miR-2909-KLF4 molecular axis is able to differentiate between the pathogeneses of pediatric B- and T-cell ALLs, which may represent a new diagnostic/prognostic marker | [166] |
Certain miRNA profiles have been identified to associate specifically with AL types. Zhang et al[40] showed a general expression pattern and assigned 21 upregulated and 11 downregulated miRNAs for primary ALL and 17 upregulated and 18 downregulated miRNAs for primary AML, but only 17 miRNAs showed convergent expression between two types of pediatric AL. By defining sets of characteristic genes for each leukemia subtype, including miR-34a, miR-128a, miR-128b, and miR-146a in ALL and miR-100, miR-125b, miR-335, miR-146a, and miR-99a in AML, they emphasized that these miRNA sets are significantly different from those selected for adult leukemias. This observation is consistent with reports of another group of researchers. Zhang et al[40] selected lineage-specific markers for the most common cytogenetic and FAB classification subtypes of AML. The overexpression of several miRs was significantly related to the degree of cell maturity and differentiation. High expression levels of miR-335 in M1, miR-126 in M2 (and AML1-ETO+), and miR-125b in M3 (and PML/RARA status) were observed[40]. In that study, the authors indicated that miR-125b and miR-126 may serve as favorable prognosticators for M3 and M2 patients, respectively. Mi et al[41] demonstrated, based on a genome-wide miRNA expression analysis on AL samples, that the expression signatures of only two miRNAs could accurately discriminate ALL from AML (accuracy rate > 95%). They proposed the possibility of using such lineage discriminatory miRNAs to develop a rapid and accurate diagnostic test of ALL vs AML in the future; however, the possibility of a more accurate diagnosis of childhood AL based on the miRNA signature remains to be realized. Mi et al[41] used a large-scale genome-wide miRNA expression assay to identify 27 differentially expressed miRNAs as markers for diagnosis and treatment. Of these, miR-128a and miR-128b were significantly overexpressed in ALL compared to AML, whereas let-7b and miR-223 were significantly downregulated[41].
Schotte et al[42] revealed distinct miRNA expression profiles for seven different subtypes of pediatric ALL. They showed that the precursor B-ALL has a specific expression pattern characterized by low expression levels of miR-127 and miR-143. This feature allowed distinguishing this cell line from control and CD34+ cells. In turn, T-ALL cells showed differential expression of 28 miRNAs. High expression of various miRNAs (e.g., miR-223, miR-222/222*, miR-98, and miR-511) was observed in ALL with hyperdiploidy. Interestingly, the miRNA signature of TEL-AML1-positive and hyper diploid cases partly overlapped, which may suggest a common underlying biology. Interestingly, specific classification profiles have been described for each of the pediatric AL subtypes, except for BCR-ABL1-positive and "B-other" ALL.
Almeida et al[21] used massive parallel sequencing to describe novel sets of 16 miRNAs correlated with childhood ALL subtypes. Among the subtype dis-criminators, ten (miR-708-5p, miR-497-5p, miR-151a-5p, miR-151b, miR-371b-5p, miR-455-5p, miR-195-5p, miR-1266-5p, miR-574-5p, and miR-425-5p) were downregulated and six (miR-450b-5p, miR-450a-5p, miR-542-5p, miR-424-5p, miR-629-5p, and miR-29c-5p) were upregulated in pediatric T-ALL. Researchers also assigned individual molecules to six functional categories, of which three were associated with induced miRNAs and three with repressed miRNAs in T-ALL (encompassed 45 genes that were shared by induced and repressed miRNAs; 210 genes were targeted by overexpressed miRNAs, and 143 genes were targeted by downregulated miRNAs). Differentially expressed genes and their targets represented relevant biological pathways, including viral carcinogenesis, cell cycle, and B-cell receptor signaling pathways for induced miRNAs and TGF-β signaling, apoptosis, NF-kappa B signaling, and cell differentiation and hematopoiesis processes[21]. Almeida et al[21] also identified miR-29c-5p as the best discriminator of the pediatric leukemia cell line.
Li et al[43] showed that the downregulation of miR-100 and miR-99a expression levels was a significant feature of leukemic blasts and was strongly correlated with the patient's 5-year survival. For these two marker molecules, differences in expression patterns between ALL and AML were noted, and white blood cell (WBC) count, ALL type (T-cell or B-cell), the MLL-rearranged gene, and the BCR-ABL fusion gene were correlated with changes in miR-100 and miR-99a levels. In addition, it has been shown in ex vivo experiments that upregulation of the expression of these molecules inhibited the expression of IGF1R and mTOR and their downstream oncogene MCL1[43]. de Oliveira et al[44] focused on assessing expression selected based on previous studies of miRNA markers[45], including miR-92a, miR-100, miR-125a-5p, miR-128a, miR-181b, miR-196b, and let-7e. As noted in leukemic blasts, miR-100, miR-196b, and let-7e showed lower expression levels, and miR-128 and miR-181 showed higher expression levels than normal bone marrow cells. Overexpression of miR-196 was observed for T-ALL. A high expression level was characteristic for patients who presented a WBC count < 50000/mm3 at diagnosis (P = 0.01) and was also associated with the presence of t(12;21) and the absence of a hyperdiploid karyotype. This suggests the possibility of a t(12;21)-specific regulation of miR-100. Differentiated expression of miR-181b and miR-128a was associated with the presence of t(4;11)[44].
Swellam et al[46] investigated the expression signature of miRNA-125b-1 and miRNA-203 among childhood ALL, and they proposed these miRNAs as useful molecular markers for the diagnosis of childhood ALL. They noticed that the expression level of miRNA-125b-1 was significantly higher in peripheral blood (PB) isolated from 43 newly diagnosed children with ALL, while the miRNA-203 level was significantly lower in childhood ALL compared to control samples. Moreover, miRNA-125-1 was increased in T-ALL compared to other ALL phenotypes, and the miRNA-203 expression level was high in T-ALL followed by pre-B-ALL[46].
Nabhan et al[47] focused on analyzing the involvement of just one marker molecule in the development of leukemia, i.e. miR-181a can both function as a tumor suppressor or an oncogene. The function of miR-181a is associated with cell metabolism and expression levels of target mRNAs. A decrease in the expression level of miR-181a was observed in the serum of children diagnosed with ALL. Investigators, as the first team, linked the miR-181a expression signature with Smad7 and TGF-β1 protein levels in the serum of childhood ALL. They found that miR-181a expression achieved a highly significant positive and a significant negative correlation with TGF-β1 and Smad7, respectively[47]. Furthermore, miR-181a was identified by Yang et al[48] as the most differentially downregulated miRNA (among the 17 identified miRNAs) in the PB of childhood ALL patients with the t(12;21) translocation. Nabhan et al[47] suggested that the diagnostic accuracy of pediatric ALL can be improved by using a small set of miRNA markers. Based on the data, they calculated that the combined use of miR-181a and Smad7 increased the sensitivity of diagnosis to 90%, whereas the combined use of miR-181a and TGF-β1 increased sensitivity to 100%.
It is supposed that differentiated miRNA expression profiles contribute to AML heterogeneity and have diagnostic and clinical significance. In numerous studies, comparisons of miRNA profiles between AML blasts and normal cells and among AML with recurrent genetic abnormalities were made[49-56]. In AML, miRNA signatures can also distinguish between cytogenetic subtypes[51-53,57] or molecular subtypes (e.g., NPM1, CEBPA, or FLT3 mutations)[52]. Most of the reported miRNAs in pediatric AML are presented in Figure 3. The prognostic significance of miRNA expression patterns in AML was confirmed[58-66].
Zhu et al[67] indicated that the 3-miRNA signature contributed to pediatric cytogenetically normal AML and can be a reliable prognostic biomarker. They found that high expression of miR-146b predicts poor prognosis and that miR-181c and miR-4786 are significantly related to favorable prognosis factors. The results indicate that the 3-miRNA-based signature is a reliable prognostic biomarker for pediatric AML. The upregulation of miR-10a, miR-10b, and miR-196b[52,68] and the downregulation of miR-192[51,68] can be biomarkers for NPM1 mutations. Patients with t(8;21) had overexpression of miR-126[53,56] and miR-146a[49,57]. High expression level of miR-155 was strongly associated with FLT3-ITD alteration[50-52,69].
The results of scientific research have led to the selection of biomarkers for diagnostic classification and prognostic evaluation, and have thus brought closer personalized medicine. Nevertheless, there is a constant need to search for sensitive and specific determinants for childhood AL progression[3,70].
One of the most commonly identified biomarkers associated with the development of AL is miR-125b, which is an important molecular regulator in normal cell homeostasis, cell metastasis, and disease pathogenesis and progression[71,72]. Playing either an oncogenic or a tumor-suppressive role through numerous target genes is crucial in abnormal proliferation, metastasis, and invasion of cells in hematological malignancies[71]. So et al[73] reported that miR-125b overexpression (through repressing IRF4) promotes myeloid and B-cell leukemia by inducing tumorigenesis, immortality, and self-renewal of progenitor cells. By targeting ARID3a in B-ALL cases with the t(11;14)(q24;q32) translocation, upregulation of miR-125b blocked differentiation and helped avoid apoptosis by blockade of caspase activation by a mechanism independent of p53 and BAK1. Moreover, high levels of miR-125b were linked to an increase in the expression of pluripotency-associated factors (OCT4, SOX2, KLF4, and NANOG)[74]. By suppressing TNFAIP3, the NF-κB-mediated increase in B-cell proliferation and dysregulation of glucose metabolism result in a reduction in apoptosis in T-ALL[75]. MiR-125b also blocks differentiation in myeloid progenitor cells with the t(2;11)(p21;q23) translocation[76]. In AML, the induction of leukemogenesis can be mediated by: (1) Pathways including CDX2, miR-125b, and CBFβ (high levels of CDX2 activate miR-125b transcription, which in turn inhibits CBFβ translation)[77]; (2) Targeting STAT3 transcription factors (also JUND and BAK1)[78]; and (3) Suppressing ABTB antiproliferative factors and deregulating genes involved in the p53 pathway, including BAK1 and TP53INP1[79].
One of the potential miRs related to important regulators in myeloid development is miR-223, a known regulator of myelopoiesis. Experimental works have shown that its expression increases with the degree of cell differentiation[80]. The high relative expression of miR-223 in AML1 samples was found by Ramsingh et al[81], but overexpression of miR-223 was not a common feature of leukemic cells. Danen-van Oorschot et al[82] observed levels of miR-29a, miR-155, miR-196a, and miR-196b in clinically relevant cytogenetic and molecular subgroups of 82 pediatric AML samples and observed higher expression of miR-196a/b in leukemic cells with MLL gene rearrangements, NPM1 mutations, and FLT3-ITD in cytogenetically normal AML. Downregulation of miR-196a/b expression was observed in CEBPA mutated cases. Differentiated expression of these miRs was linked to HOXA and HOXB cluster genes involved in myeloid transformation. In FLT3-ITD and NPM1-mutated cases, miR-155 was overexpressed, and lower expression of miR-29a in MLL-rearranged pediatric AML was found[82]. A broad miRNA profiling experiment of cytogenetically distinct childhood AML cases was conducted by Daschkey et al[49], where they selected miR-126, miR-146a, miR-181a/b, miR-100, and miR-125b as markers of the MLL-rearranged AML subtype. Emmrich et al[83] pointed out that miR-9 is an important regulator of t(8;21)-mediated leukemogenesis. Low levels of miR-9 were observed in AML patients with t(8;21), but high expression levels were noted in cases with MLL rearrangements. As a potential mechanism of regulation of leukemic cell proliferation and differentiation, repression of the oncogenic LIN28B/HMGA2 axis (including target genes CDH1, NFKB1, BACE1, and RES) was identified.
Children with AL are often put into risk groups (low, intermediate, or high risk), with more intensive treatment given to higher risk patients. Generally, children at low risk have a better outcome than those at very high risk. However, it is important to know that even children in higher risk groups can often still be cured. AL is a heterogeneous disease; hence, different treatment results are observed in patients with similar histopathological diagnoses, stages of development, and similar treatment protocols. The correct stratification of patients into high-risk groups with recurrence (more intensive treatment) and low-risk patients (avoidance of therapy toxicity) is of great importance[84,85]. The previous stratification does not meet the abovementioned expectations, which is why high hopes have been placed on the possibilities resulting from research using genomics and transcriptomics methods[84].
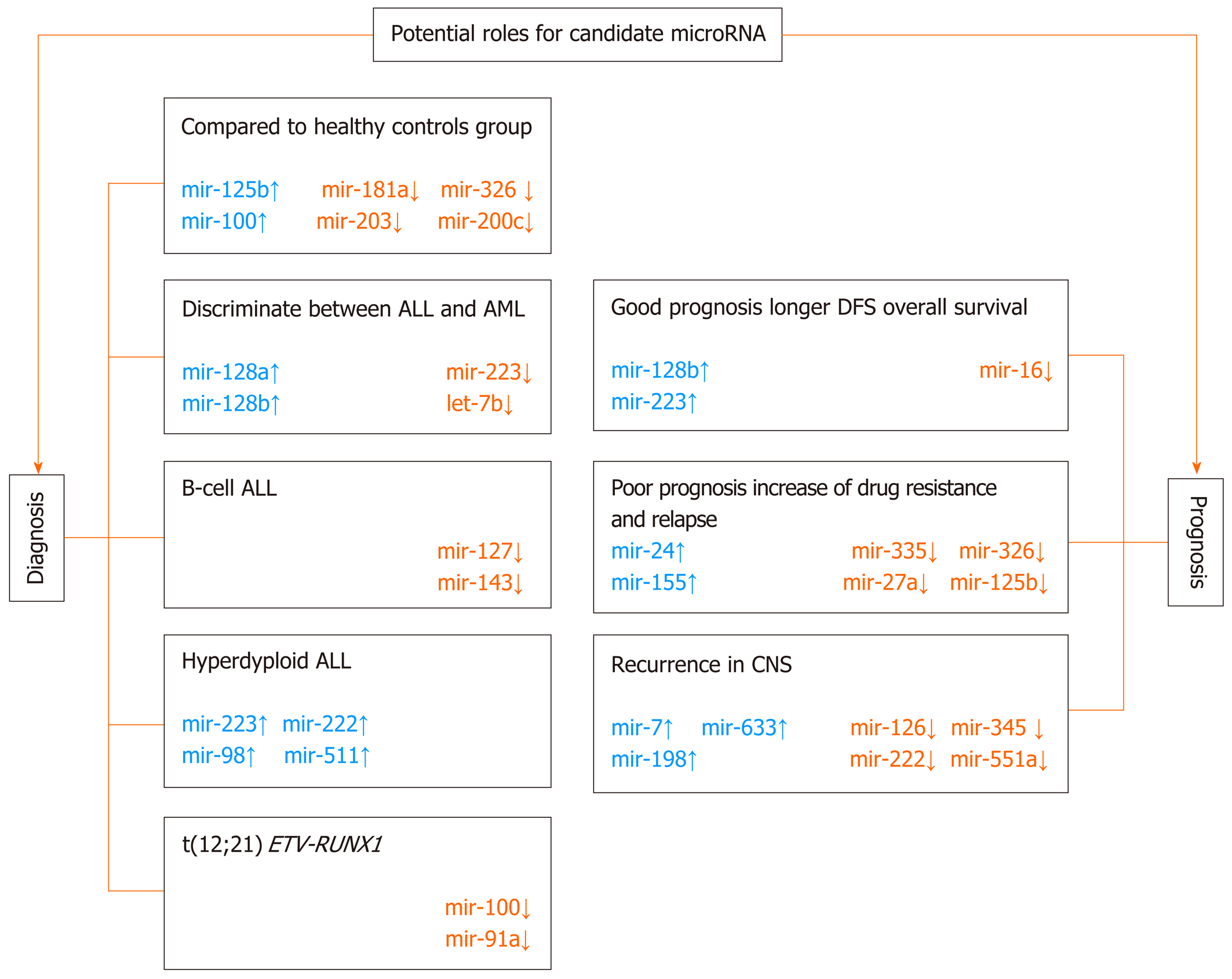
More and more often in pediatric AL, the expression signature of the set of miRNA molecules has been correlated with well-known prognostic factors, which include: WBC count, age at diagnosis, cytogenetic and molecular genetic analyses of blast cells, prednisolone response on day 8, and immunophenotype. The possible model of diagnostic and prognostic management in pediatric ALL based on the miRNA signature is presented in Figure 4.
In recent years, several studies have been conducted to identify miRNAs as predictors of the risk of leukemia recurrence. One of the first verified biomarkers was miR-16, which is implicated in apoptosis induction by targeting BCL-2 and cell cycle arrest[86-88]. Kaddar et al[89] obtained statistically significant relationships between low miR-16 levels and low WBC counts and good molecular markers. They found that, in the entire ALL group, miR-16 was significantly downregulated in the group of patients with leukocytes below 50 G/L and with hyperploidy or t(12;21). In T-ALL, overexpression of miR-16 expression was also related to corticosteroid resistance. After all, they could not assign a specific miR-16 expression profile to B-cell and T-ALL subgroups[89]. In the next study, Organista-Nava et al[90] conducted a multivariate analysis (including age at diagnosis, gender, and WBC of miR-24 expression, a well-known promotor of the survival of hematopoietic cells). Targets of miR-24 are pro-apoptotic (FAF-1, caspase 9, Bim, and Apaf-1) and cell cycle progression (enhanced MYC, E2F2, CCNB1, and CDC2 or inhibited p27Kip1 and VH) proteins[91-93]. In a previous study, it was reported that miR-214 expression is associated with cytogenetic and molecular subtypes of adult AML (e.g., AMLs with t(8;21), t(15;17), inv(16), NPM1, and CEBPA mutations)[52]. Organista-Nava et al[90] indicate miR-214 as an independent marker for predicting the clinical outcome in both AML and ALL patients. Upregulation of miR-24 was significantly associated with poor prognosis, shorter overall survival (OS), and a high risk of leukemia relapse.
One of the most interesting research results in this field were presented by the team of Nemes et al[94]. Researchers collected PB and bone marrow samples from 24 ALL patients from all phases of treatment [collected at diagnosis, at conventional response checkpoints (on days 15, 33), and before beginning protocol M]. Differentiated expression of set miRNAs (miRNA-16, miRNA-21, miRNA-24, miRNA-29b, miRNA-128b, miRNA-142-3p, miRNA-155, and miRNA-223) with potential roles in hematologic malignancies was analyzed. Based on the results obtained, it was concluded that miR-223 (involved in the regulation of the cell cycle or different signaling mechanisms, such as E2F1, CEBPα, and E2A) and miR-128b (play roles in the regulation of the PI3K-AKT-mTOR signaling pathway through downregulation of PTEN) expression signatures could be possible predictors of ALL relapse. Nemes et al[94] determined in general that high levels of miR-128b and low levels of miR-223 show a significant correlation with good prednisolone response and better prognosis in childhood ALL. In particular, they found an extreme high expression level of miR-128b at diagnosis, which significantly decreased as patients entered remission where normal levels of miR-128b expression for mononuclear cells were detected. Conversely, miR-223 expression can be undetectable at diagnosis, but during treatment and in remission, the level of miRNA is standard and then decreases again at relapse[94].
Among the newly typed prognostic and therapeutic markers for pediatric AL is miR-335, which acts as a tumor suppressor and targets genes that participate in most of the important biological processes associated with human cancer and is involved in pathways such as p53, mitogen activated protein kinase, TGF-β, Wnt, epidermal growth factor, mammalian target of rapamycin, Toll-like receptor, and focal adhesion[95]. Using genome-wide miRNA microarray analysis, Yan et al[96] found that low expression level of miR-335 was associated with unfavorable prognosis, poorer 5-year event-free survival, and glucocorticoid resistance in ALL. Moreover, the study highlighted the potential mechanism of silencing the expression of miR-335. According to previous observations of recurrent leukemia[97-99], epigenetic silencing through DNA methylation was suggested. Yan et al[96] used the results of in vitro experiments to show that exogenous expression of miR-335 in leukemic cells increases sensitization to prednisolone-mediated apoptosis, and they concluded that reintroducing miR-335 expression or overriding MAPK1 activity could become a promising therapeutic target for ALL treatment. The role of miR-335 in AML was analyzed by Zhou et al[100]. They found that: (1) Overexpression of miR-335 (independent of its methylation) was negatively correlated with decreased ID4 expression; (2) Aberrant miR-335 and ID4 (direct target) expression independently affected chemotherapy response and leukemia-free/OS in patients with AML; and (3) miR-335/ID4 dysregulation facilitated leukemogenesis through the activation of the PI3K/Akt signaling pathway. These results are similar to the Yan et al[96] study, which used ex vivo experiments to show that it was possible to reduce pro-proliferative and antiapoptotic effects and restore the physiological role of mir-335 (through restoration of ID4 expression)[100].
MiR-155 is another candidate predictive and prognostic marker of AL outcome. MiR-155 is evolutionarily conserved, and as an inhibitor of lineage differentiation, it is one of the most critical regulators of posttranscriptional gene expression in B cells[101,102]. It has also been confirmed to be associated with pathogenesis, aggressiveness and progression in CLL[103,104]; poor survival in adult and pediatric AML[105]; and poor clinical prognosis in Hodgkin's lymphomas[106] and B-cell-type Diffuse large B-cell lymphoma[107,108]. El-Khazragy et al[109] tried to connect the upregulation of miRNA-155a and miRNA-181a expression to ALL outcome. They found a significant correlation of high levels of minimal residual disease (MRD) and poor prognosis, but only overexpression of miRNA-155a was significantly related to high blast numbers (> 25%), unfavorable cytogenetic abnormality, total WBC, higher relapse rate, a higher MRD after 15 d, and poor prognosis[109]. Moreover, expression of both miR-155a and miR-181a was downregulated after chemotherapy, suggesting their potential use as biomarkers of therapeutic response in pediatric ALL.
A consistent and strong association of miR-155 overexpression with poor prognosis in pediatric AML was also confirmed[110-112]. An interesting relationship between the level of expression of miR-155, its biological function and prognosis in AML was noted by Narayan et al[105,111]. Researchers conducted experiments on cells obtained from patients diagnosed with AML and on a murine model, where they found that between 10- and 50-fold overexpression of miR-155 (plays the role of oncogene) is associated with poorer OS and increased tumor burden. However, when the miR-155 expression level is lower than 10-fold, outcomes may be favorable. When the level is higher than 50-fold (observed only in the murine model), miR-155 shows suppressor activity. Overexpression of miR-155 is associated with activation of B cells with an inflammatory stimulus of lipopolysaccharide and IL-4. The consequence of differentiated miR-155 expression is downregulation of the expression of target genes regulated by intermediate (CEBPB, SPI1, and TLE2) or high (MYB and KIT) miR-155 expression levels. Narayan et al[105,111] first described a novel dose-dependent function of miR-155 in the progression of AML and pointed out that it can have important therapeutic implications.
miRNAs can also act as independent prognostic factors to predict clinical outcomes for T-ALL patients. Miao et al[113] identified highly expressed miR-590 as a candidate oncogenic marker for both adult and pediatric T-ALL. Through regulation of the RB1 gene, miR-590 increased the proliferation and invasion of T-cells[113]. A common oncogenic marker in adult and pediatric T-ALL is miR-149. Overexpression of miR-149 inhibits apoptosis and enhances proliferation via the target gene JUNB[114]. Another example is miR-664, whose overexpression inhibits the PLP2 gene in pediatric T-ALL. Deregulation of PLP2 results in changes in adhesion and migration of leukemic cells[115].
MiR-181 is often mentioned among AML prognostic markers. In adults, its relationship with prognosis in AML is quite well described[60,116]; however, only one published paper describes the correlation between miR-181 expression level and prognosis in the pediatric population. Liu et al[117] analyzed the physiological function and mechanism of miR-181 and found that these molecules are responsible for the G1/S transition and cell proliferation via the tumor suppressor ATM.
Zhang et al[65] found that high expression of oncogenic miR-99 is significantly related to the promotion of proliferation and apoptosis inhibition. Overexpression of miR-99a was observed in pediatric AML (FAB subtypes M1-M5). Interestingly, during complete remission, silencing expression of miR-99 was noted. Regulation of cell growth and differentiation was associated with modulation of the expression of such miR target tumor suppressor genes as CTDSPL and TRIB2.
Evolutionarily conserved miR-125b, as an important regulator of hematopoietic stem/progenitor cell apoptosis, confers a proliferative advantage to leukemic cells in both ALL and AML[118]. In AML, the level of miR-125b could be up to 90-fold higher in comparison to normal cells[76]. In patients with B-ALL carrying the t(11;14)(q24;q32) translocation, expression of miR-125b is also 30- to 600-fold higher in comparison to cases without the translocation[72,119]. In AML, as shown by Ufkin et al[120], miR-125a expression was downregulated in favorable and intermediate prognoses and associated with decreased survival. In addition, in vitro experiments have identified a potential mechanism for regulating miR-125a expression, which is excessive methylation. They undertook effective attempts at global demethylation using decitabine, which resulted in an increase in miR-125a levels as well as, in consequence, inhibition of cell cycle proliferation and progression with increased apoptosis. In their study, they revealed that the ErbB pathway is directly regulated by miR-125a. The authors suggested that further research into ErbB inhibitors and miR-125a molecules may contribute to the development of targeted AML therapy[120].
Lin et al[61] indicated miR-370 as a potential noninvasive diagnostic and prognostic miRNA marker for childhood AML cases because its expression in bone marrow and serum samples at diagnosis was significantly decreased. The level of miR-370 was correlated with the FAB classification subtype M7 (P = 0.02) and unfavorable karyotype and with poor prognosis, unfavorable relapse-free survival (RFS), and shorter OS. Lin et al[61] also noticed that serum miR-370 levels were more obvious in the subgroup of patients with intermediate-risk cytogenetics. Second, Lin et al[62] described miR-335 as an independent prognostic marker of RFS and OS in pediatric AML. His high expression was related to the M7 subtype, unfavorable karyotype presence and shorter RFS and OS. The function of the biomarker unfavorable prognosis was confirmed for oncogene miR-183 by Wang et al[63]. High expression of miR-183 (especially in M7 AML) was associated with promotion of cell proliferation and G1/S transition and inhibition of apoptosis. Short RFS and OS were linked markedly with silencing of PDCD6 expression, a direct and functional target of miR-183. Wang et al[64] also proposed miR-375 as a prognostic factor for unfavorable cytogenetic risks in M7 ALL. Zhu et al[66] analyzed the clinical significance of miR-29a expression, a well-known gene that can act as either an oncogene or tumor suppressor. Low expression of this gene was characteristic of the M7 subtype of childhood AML and had shorter RFS and OS, while high expression was associated with good prognosis.
In summary, the most frequently identified prognostic miRNA markers of pediatric AL include the following: (1) miR-7, miR-16, miR-33, miR-100, miR-130b, miR-181, miR-215, miR-216, miR-369-5p, miR-496, miR-518d, miR-599, and miR-708 are unfavorable factors[40,42,89,121,122]; and (2) miR-10a, miR-23a, miR-27a, miR-128b, miR-134, miR-150, miR-191, miR-214, miR-223, miR-342, miR-484, miR-486, miR-487, miR-572 miR-580, miR-627, miR-624, let-7g, and let-7i are favorable factors[40,42,121,122].
Table 1 summarizes miRNAs involved in ALL prognosis, their impact in predicting relapse or progression, and their relationship with OS and treatment outcome.
The correct assignment of patients to risk groups presents another challenge, which therapy is the most appropriate to use. Anticipating the response to the administered drug is a challenge in current oncology[123-126]. AL is curable in children in 60%-90% of cases[127,128]. Several research groups have attempted to search for sets of miRNAs involved in therapy failure. Assuming that the phenomenon of drug resistance is a primary feature of leukemia blasts, it is possible to predict the ineffectiveness of treatment[128]. MiRNA profiling is increasingly used in research on multifactorial phenomena, such as drug resistance, due to its ability to simultaneously analyze the most important genes for this process[129]. The mechanism of drug resistance consists of many complex processes that overlap one another, including: (1) A change in the function of receptors; (2) Heterogeneity of cancer cells; (3) Tissue microenvironment; (4) Intercellular interactions; (5) Disorders in transport and signal transduction; and (6) A change in the expression of key genes[130]. However, the genetic basis of chemotherapy resistance is still not well understood. Based on the genetic profile, it is possible to predict the response to the treatment used by establishing a correlation between the expression of specific sets of miRNAs and the sensitivity/resistance to a given anticancer agent or the effectiveness of the treatment (Table 2)[25,84].
| Drug name | Most discriminating microRNAs |
| Prednisolone | miR-99a, miR-125a, mir-128b, miR-210, miR-221, miR-550, miR-595, miR-633, miR-638, miR-652 |
| L-asparaginase | miR-210, miR-454 |
| Vincristine | miR-9, miR-99a, miR-100, miR-124, miR-125b, miR-126, miR-128b, miR-141, miR-200c, miR-218, miR-625, miR-633, miR-629, miR-1206 |
| Prednisone | miR-15a, miR-18a, miR-16-1, miR-128b, miR-193a, miR-218, miR-223, miR-532, miR-550, miR-625, miR-633, miR-638 |
| Daunorubicin | miR-99a, miR-100, miR-125b, miR-126, miR-199b, miR-203, miR-210, miR-335, miR-383, let-7c |
| Doxorubicin | miR-125b |
| Methotrexate | miR-595, miR-1206, miR-6083 |
| Cytarabin | miR-562 |
| Dexamethasone | miR-210 |
| Imatinib | miR-203 |
| Staurosporine | miR-125b |
Multidrug resistance followed by relapse is regarded as one of the most important clinical problems for effective treatment in patients diagnosed with AL and is still the main cause of cancer death in children[131]. MiRNAs are considered a relevant regulator response to drug administration. Ghodousi et al[132] conducted an analysis of the expression of ABCA2 and ABCA3 transporters and their potential regulators, miR-326 and miR-200c. First, they confirmed that the expression levels of both miR-326 and miR-200c were significantly lower in patients with ALL diagnosis. They showed a significant downregulation of miR-326 in MRD+ and relapsed patients compared to the MRD- group, supporting the notion that decreased expression of miR-326 has an adverse impact on treatment response. A significant decrease in miR-200c expression level was observed between the MRD- patients and relapsed ALL patients. Ghodousi et al[132] pointed out that only the miR-326 level can be a negative prognostic biomarker that may discriminate between MRD+ and MRD- patients. The presented results (higher miR-128b expression correlated with good prednisolone response and better prognosis) suggest a correlation between miR-128b expression changes and steroid sensitivity of leukemic cells[94].
In several subsequent studies, miR-125b was selected as a potential biomarker for leukemia recurrence and was correlated with a high risk of therapy failure and poor survival prognosis[72,133-135]. Piatopoulou et al[135] evaluated the clinical significance of miR-125b for ALL prognosis and prediction of patients’ response to the Berlin-Frankfurt-Muenster (BFM) chemotherapy protocol. In their study, a low level of miR-125b was related to unfavorable prognosis, but after treatment with the BFM protocol, overexpression of this miR was detected. Higher expression on day 33/diagnosis was related to a higher risk for disease short-term relapse and worse survival, strongly suggesting that miR-125b could also be used as a clinically useful predictor of resistance to BFM chemotherapy[135]. Gefen et al[134] established upregulation of the miR-125b-2 cluster (miR-125b, miR-99a, and let-7c) in ETV6/RUNX1-positive childhood ALL. Overexpression of the cluster was an independent leukemia event. Increased miR-125b-2 levels protected cells from death. The antiapoptotic activity can be associated with a marked inhibition of caspase 3 activation and the cleavage of its substrate PARP. Differentiated expression of miR-125b-2 was also related to resistance to antileukemic agents staurosporine and doxorubicin.
In a study by Schotte et al[42], changes in the expression of miR-454 were only related to L-asparaginase resistance, but the strong overexpression of three miRs (miR-125b, miR-99a, and miR-100) was correlated with vincristine (VCR) and daunorubicin resistance. The strong association of miR-125b upregulation was especially evident in ETV6-RUNX1+ patients, who were VCR-resistant[42]. The overexpression of miR-125b was linked to inhibition of VCR-induced apoptosis and induction of the proliferation of CD34+ cells. Schotte et al[42] noticed that the interference of miR-125b function might provide a way to sensitize patients to VCR. Moreover, researchers identified 14 miR signatures (independent of ALL subtype) useful for prognosis and prediction in ALL. Among others, miR-10a, miR-134, and miR-214 were correlated with a favorable outcome, which confirmed earlier reports linking these miRs with caspase-dependent proapoptotic activity of miR-10a[136], inhibition of USF2-mediated cell proliferation by miR-10a and miR-214[137,138], and oncogene SOX2 downregulation by miR-134[139]. Differentiated expression of six miRNAs (miR-33, miR-215, miR-369-5p, miR-496, miR-518d, and miR-599) was related to an unfavorable long-term clinical outcome in ALL. Schotte et al[42] colleagues selected two miRs as potential targets for targeted therapy. They noted that it may be of clinical interest. The use of a demethylating agent increased the level of miR-10a in MLL-rearranged patients[42,99], and the application of antagomirs decreased the level of overexpressed miRNA in poor prognosis samples (e.g., miR-33)[42].
Akbari Moqadam et al[140] analyzed miR-125b, miR-99a, and miR-100 expression levels in correlation with VCR resistance in ETV6-RUNX1+ Reh cells. Only the combination set of miRs influenced cell sensitivity to drug administration. MiRNA overexpression resulted in lower expression of their directly regulated target genes (DNTT, NUCKS1, MALAT1, SNRPE, PNO1, SET, KIF5B, PRPS2, RPS11, RPL38, and RPL23A) in VCR-resistant ALL cells[140]. In vitro, the restoration of miR-100 and miR-99a in ALL cells suppressed cell proliferation and increased dexamethasone-induced cell apoptosis[43]. It has also been shown that the sensitivity of BCR-ABL1(Ph+) cells to treatment with tyrosine-kinase inhibitors (such as imatinib) can be increased in vitro by restoring miR-203[141]. Increasing the miR-203 expression level negatively regulates the expression level of target oncogenes (ABL1 and BCR-ABL1) and thus inhibits cell proliferation[142].
One of the major problems in childhood AL is the risk of relapse, and the molecular mechanism is still poorly understood. Intensive scientific work is being conducted on cognition factors related to therapy response and the biology of recurrence. Han et al[121] carried out a genome-wide miRNA array analysis to identify the miRNA expression patterns correlated with relapse or complete remission in childhood ALL. They identified a set of 70 differentially expressed miRNAs in samples at relapse or complete remission (CR) compared with the initial diagnosis of the same patients. The expression levels of miR-223, miR-23a, let-7g, miR-181, miR-708, and miR-130b were compared in samples at relapse vs diagnosis and miR-27a, miR-223, miR-23a, miR-181, and miR-128b levels were compared in CR samples and diagnostic samples. In the relapse samples, strong downregulation of miR-223, miR-23a, and let-7g and upregulation of the miR-181 family, miR-708, and miR-130b were confirmed. Han et al[121] found that miR-223 and miR-27a were overexpressed in patients during CR. Moreover, low levels of both miR-223 and miR-27a at the time of diagnosis were confirmed in patients who subsequently relapsed. Furthermore, Han et al[121] identified a high expression level of miR-708 in both standard risk and middle risk ALL and a low expression level in high-risk patients. The lowest level of miR-708 expression at initial diagnosis was confirmed among the four immunophenotypes, pro-B-ALL, pre-B-ALL, common ALL, and T-ALL[121]. These results suggest that miR-708, miR-27a, and miR-223 expression levels at initial diagnosis could be independent and reliable prediction factors of the OS rate in childhood ALL and could also be used to predict the risk of relapse before patients undergo therapy. These miRNAs and their targets (e.g., IKZF1, IL-15, and CASP8AP2) might be helpful in the optimization of therapeutic protocols and novel targets for the development of new antileukemic agents[121].
A multifaceted study of miRNome was conducted by Zhang et al[40], who described specific expression patterns for both pediatric ALL and AML, selected prognostic markers, and determined the relationship of miRs with the risk of recurrence in the central nervous system and a lack of sensitivity to prednisone. Authors developed the signature of the overexpressed miR-7, miR-198, and miR-633 and downregulated miR-126, miR-345, miR-222, and miR-551a, in which changes in expression level were related to central nervous system relapse in ALL. Zhang et al[40] identified a set of eight differentiated genes, including miR-18a, miR-532, miR-218, miR-625, miR-193a, miR-638, miR-550, and miR-633, that differentiated patients according to a good or poor prednisone response. Interestingly, researchers were unable to confirm the association with ALs for mir-15 and mir-16, frequently mentioned regulators of apoptosis in hematopoietic cells and prednisolone resistance modulators[86,87,89,110,112]. Xu et al[143] performed a study to determine an early marker of relapse in pediatric ALL and found miR-7, miR-216, and let-7i (high expression) and miR-486, miR-191, miR-150, miR-487, and miR-342 (low expression).
Pharmacogenomics of tumors (a field dealing with the assessment of innate genetic determinants of different drug effects) enables the discovery of new anticancer drugs and their genetic targets on the basis of a well-defined mechanism of oncogenesis[126]. The search for candidate molecules among miRNAs can also be used in the typing of a molecular target, drug design, determination of the in vitro and in vivo effects of the drug on the global level of gene expression, identification of mechanisms of action, detection of potential toxicity, and determination of the pharmacodynamic effect[144-146].
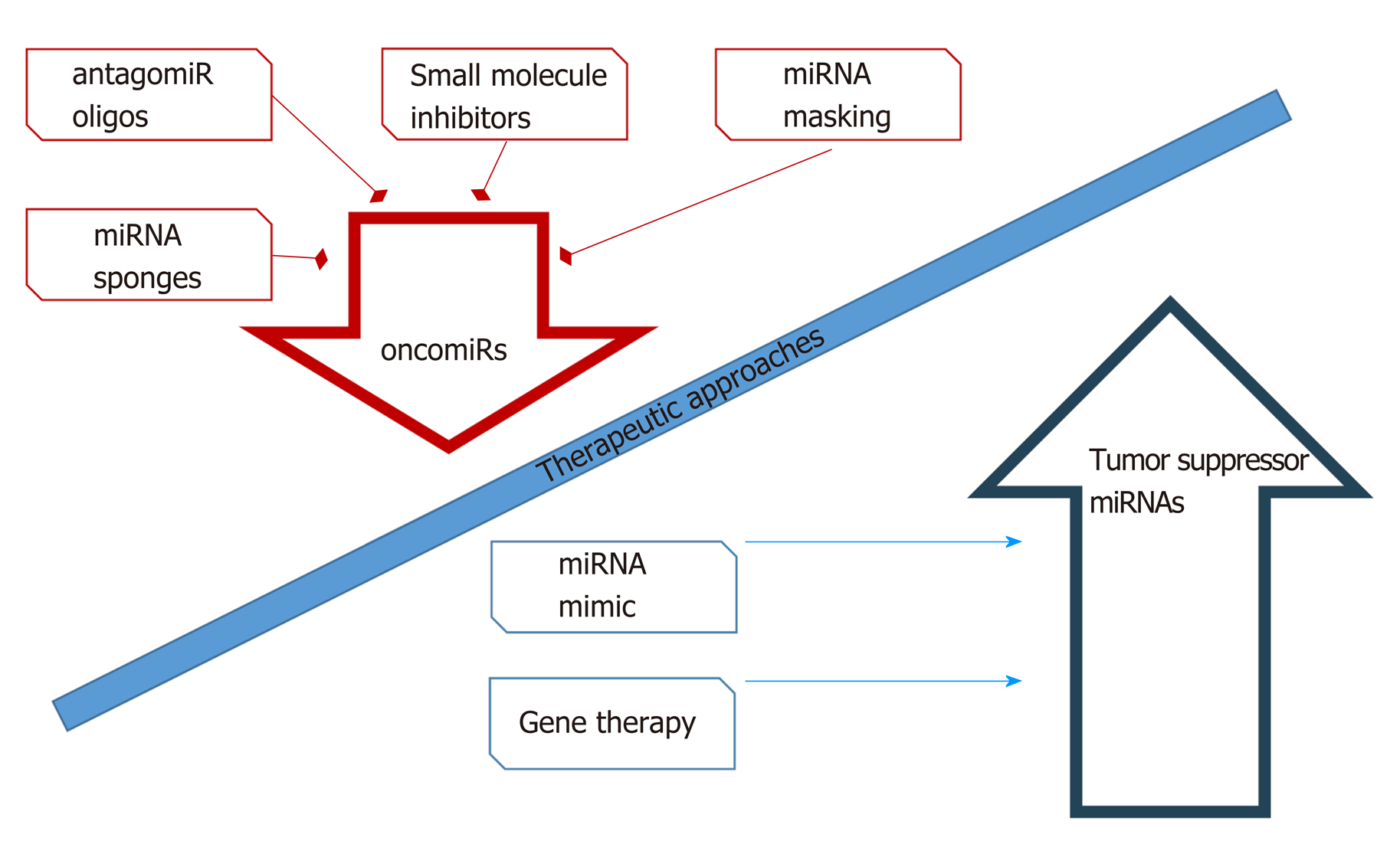
miRNAs are one of the most promising molecules in the development of new antileukemic agents and therapeutic protocols. miRNAs, as differentially expressed molecules, can be regulated by two anti-ancient mechanisms. The first involves inhibition of miRNA activity by: (1) The use of miRNA inhibitors and oligomers, including RNA, DNA, and DNA analogues (miRNA antisense therapy), small molecule inhibitors, and miRNA sponges; or (2) miRNA masking. The second mechanism includes enhancement of miRNA function (miRNA replacement therapy) by: (1) The use of modified miRNA mimetics, such as plasmids; or (2) Lentiviral vectors carrying miRNA sequences[147] (Figure 5).
Currently available therapeutic strategies based on the level of miRNA and its function in a cancer cell include transfection of mimic miRNAs or miRNA inhibitors in vitro, which have the effect of increasing or decreasing the expression level of the candidate miRNA, respectively. Therapy based on modulation of miRNA levels is at a promising early ex vivo stage. An effective method of delivering inhibitors and activators of miRNA expression to leukemia cells has not yet been developed. The safety and efficacy of such a therapy have not been studied, and it is difficult to predict the long-term effects of such a treatment.
Research using miRNA profiling in disease is currently in an intensive phase, and this technique shows important and promising directions for future research. As our knowledge of the mechanisms and course of childhood AL increases along with advances in genetics and molecular biology and the development of sensitive analytical methods, the accurate assessment of their real relationship with long oncological results in the near future may be possible.
Research on miRNA patterns at individual stages of leukemogenesis can lead to a better understanding of the disease process itself, as well as to the development of modern classification and more effective therapy. As shown, miRNAs are differentially expressed in distinct stages of lymphopoiesis and myelopoiesis and provide a new look at the molecular pathways leading to AL development and the molecular pathogenesis of pediatric ALs. The aberrant miRNA signatures observed in AL can be used to define biomarkers for diagnosis, classification, prognosis, and therapy monitoring of this disease. MiRNA expression level monitoring carried out in the last two decades has shown that hematological cancers can be precisely classified based on a genetic signature that is associated with morphological, immunophenotypic, cytogenetic, molecular, and other cellular traits. Circulating miRNAs can be detected with the use of noninvasive and easily applicable methods with high accuracy and sensitivity.
MiRNAs are becoming increasingly popular among scientists and oncohematologists due to their important role in the etiology of AL. However, the prospect of their routine use in diagnostics requires further research to identify a small group of sensitive and specific biomarkers of diagnostic and prognostic significance. Undoubtedly, the results of numerous studies indicate a few promising candidates, but nevertheless, there is still no one single miRNA or small set that has an accuracy close to 100% to diagnose AL or differentiate its subtypes, regardless of other diagnostic factors. A much more promising direction for further research on miRNAs is to utilize their potential for effective and personalized medicine. In the case of miRNA, the therapeutic challenge is also to improve targeting (one miRNA usually regulates the expression of many genes) as well as to increase the circulation time of the molecule. The therapeutic use of miRNA therefore requires a combination with a suitable nanoparticle that directs miR as well as protects against inactivation and degradation[148].
One of the trends in modern diagnostics and antileukemia therapy is the exact use of nanotechnology, which primarily offers the opportunity to improve sensitivity, selectivity, and bioavailability and thus effectiveness[149]. The advantage of nanoparticles is their size, which allows them to cross biological barriers more effectively. Anticancer drug components include new classes of therapeutic agents such as small interfering RNA, miRs, and single strand DNA[148]. An additional improvement is the possibility of functionalizing the surface of the nanoconstruct with specific ligands[150]. Nevertheless, nanomedicine is the domain of solid tumors[149,151]. In the case of leukemia, however, nanoconstructions are used as noninvasive methods of diagnosis and treatment. An example would be the construction of a modern nanoparticle containing antagomiR-126 [nanoconjugate: LNP@antagomiR126@Anti-CD45.2 (lipopolyplex NPs)] for adult AML therapy[152].
Nevertheless, the introduction of miRNA assays into diagnostics and therapy is a challenge and requires further refinement, above all, of analysis standards. In this field, convergent results of analyses of various research teams obtained in independent groups of patients with AL diagnosis are a great achievement. This is possible despite conducting the experiment in different conditions and with different parameters in independent diagnostic laboratories. Therefore, the possibility of using miRNA in the classification and assessment of risk groups in the case of childhood AL does exist. Significant technical progress does not go hand in hand with clinical trials. There is a long way to go in understanding miRNA regulatory mechanisms in childhood AL. We still need to acquire and integrate data in the field of miRNA-mRNA–protein interaction, phenotypic observation, posttranscriptional regulatory interactions, and functional analysis[15]. Much effort should also be made to standardize and validate laboratory procedures for determining miRNA levels.
Finally, although the survival rates for pediatric ALs has improved, there is still a need for identifying novel reliable, sensitive, and specific molecular markers, such as miRNAs, that can be used in a personalized approach to early diagnosis (perhaps even prevention), risk group stratification, and prediction of treatment response.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: Poland
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Farhat S, Moschovi MA, Yao DF S-Editor: Dou Y L-Editor: Filipodia E-Editor: Zhang YL
| 1. | Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29:551-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 572] [Cited by in F6Publishing: 592] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 2. | Wiemels J. Perspectives on the causes of childhood leukemia. Chem Biol Interact. 2012;196:59-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 158] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 3. | Grobbelaar C, Ford AM. The Role of MicroRNA in Paediatric Acute Lymphoblastic Leukaemia: Challenges for Diagnosis and Therapy. J Oncol. 2019;2019:8941471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 4. | Kantarjian H, O'Brien S, Cortes J, Wierda W, Faderl S, Garcia-Manero G, Issa JP, Estey E, Keating M, Freireich EJ. Therapeutic advances in leukemia and myelodysplastic syndrome over the past 40 years. Cancer. 2008;113:1933-1952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 70] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Rubnitz JE, Inaba H, Leung WH, Pounds S, Cao X, Campana D, Ribeiro RC, Pui CH. Definition of cure in childhood acute myeloid leukemia. Cancer. 2014;120:2490-2496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Bhojwani D, Howard SC, Pui CH. High-risk childhood acute lymphoblastic leukemia. Clin Lymphoma Myeloma. 2009;9 Suppl 3:S222-S230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 7. | Teachey DT, Hunger SP. Predicting relapse risk in childhood acute lymphoblastic leukaemia. Br J Haematol. 2013;162:606-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 8. | Szczepanek J, Styczyński J, Haus O, Tretyn A, Wysocki M. Relapse of acute lymphoblastic leukemia in children in the context of microarray analyses. Arch Immunol Ther Exp (Warsz). 2011;59:61-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26799] [Article Influence: 1340.0] [Reference Citation Analysis (0)] |
| 10. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4964] [Cited by in F6Publishing: 5118] [Article Influence: 255.9] [Reference Citation Analysis (0)] |
| 11. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8847] [Cited by in F6Publishing: 9072] [Article Influence: 477.5] [Reference Citation Analysis (0)] |
| 12. | John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 13. | Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 872] [Cited by in F6Publishing: 844] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 14. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14460] [Cited by in F6Publishing: 15337] [Article Influence: 1022.5] [Reference Citation Analysis (1)] |
| 15. | Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform. 2014;15:1-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 388] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 16. | John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2680] [Cited by in F6Publishing: 2794] [Article Influence: 139.7] [Reference Citation Analysis (0)] |
| 17. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5705] [Cited by in F6Publishing: 5849] [Article Influence: 324.9] [Reference Citation Analysis (0)] |
| 18. | Simeon V, Todoerti K, La Rocca F, Caivano A, Trino S, Lionetti M, Agnelli L, De Luca L, Laurenzana I, Neri A, Musto P. Molecular Classification and Pharmacogenetics of Primary Plasma Cell Leukemia: An Initial Approach toward Precision Medicine. Int J Mol Sci. 2015;16:17514-17534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 19. | Svoronos AA, Engelman DM, Slack FJ. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016;76:3666-3670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 417] [Cited by in F6Publishing: 529] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 20. | Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1765] [Cited by in F6Publishing: 1899] [Article Influence: 105.5] [Reference Citation Analysis (0)] |
| 21. | Almeida RS, Costa E Silva M, Coutinho LL, Garcia Gomes R, Pedrosa F, Massaro JD, Donadi EA, Lucena-Silva N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol Oncol. 2019;37:103-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 22. | Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38 Suppl:S2-S7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 344] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 23. | Haferlach T, Kern W, Schnittger S, Schoch C. Modern diagnostics in acute leukemias. Crit Rev Oncol Hematol. 2005;56:223-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 24. | Willenbrock H, Juncker AS, Schmiegelow K, Knudsen S, Ryder LP. Prediction of immunophenotype, treatment response, and relapse in childhood acute lymphoblastic leukemia using DNA microarrays. Leukemia. 2004;18:1270-1277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 25. | Carroll WL, Bhojwani D, Min DJ, Moskowitz N, Raetz EA. Childhood acute lymphoblastic leukemia in the age of genomics. Pediatr Blood Cancer. 2006;46:570-578. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Mullighan CG, Flotho C, Downing JR. Genomic assessment of pediatric acute leukemia. Cancer J. 2005;11:268-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 27. | Pinkel D. Genotypic classification of childhood acute lymphoid leukemia. Leukemia. 1999;13 Suppl 1:S90-S91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 28. | Starý J, Zuna J, Zaliova M. New biological and genetic classification and therapeutically relevant categories in childhood B-cell precursor acute lymphoblastic leukemia. F1000Res. 2018;7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Vrooman LM, Blonquist TM, Harris MH, Stevenson KE, Place AE, Hunt SK, O'Brien JE, Asselin BL, Athale UH, Clavell LA, Cole PD, Kelly KM, Laverdiere C, Leclerc JM, Michon B, Schorin MA, Sulis ML, Welch JJG, Neuberg DS, Sallan SE, Silverman LB. Refining risk classification in childhood B acute lymphoblastic leukemia: results of DFCI ALL Consortium Protocol 05-001. Blood Adv. 2018;2:1449-1458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 69] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 30. | Iacobucci I, Mullighan CG. Genetic Basis of Acute Lymphoblastic Leukemia. J Clin Oncol. 2017;35:975-983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 323] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 31. | Puumala SE, Ross JA, Aplenc R, Spector LG. Epidemiology of childhood acute myeloid leukemia. Pediatr Blood Cancer. 2013;60:728-733. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 32. | Foucar K, Anastasi J. Acute Myeloid Leukemia With Recurrent Cytogenetic Abnormalities. Am J Clin Pathol. 2015;144:6-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 33. | Yang JJ, Park TS, Wan TS. Recurrent Cytogenetic Abnormalities in Acute Myeloid Leukemia. Methods Mol Biol. 2017;1541:223-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 34. | Bullinger L, Rücker FG, Kurz S, Du J, Scholl C, Sander S, Corbacioglu A, Lottaz C, Krauter J, Fröhling S, Ganser A, Schlenk RF, Döhner K, Pollack JR, Döhner H. Gene-expression profiling identifies distinct subclasses of core binding factor acute myeloid leukemia. Blood. 2007;110:1291-1300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 35. | Mead AJ, Linch DC, Hills RK, Wheatley K, Burnett AK, Gale RE. FLT3 tyrosine kinase domain mutations are biologically distinct from and have a significantly more favorable prognosis than FLT3 internal tandem duplications in patients with acute myeloid leukemia. Blood. 2007;110:1262-1270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 221] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 36. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5384] [Cited by in F6Publishing: 5479] [Article Influence: 304.4] [Reference Citation Analysis (0)] |
| 37. | Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: the potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2:330-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 38. | Mullighan CG. How advanced are we in targeting novel subtypes of ALL? Best Pract Res Clin Haematol. 2019;32:101095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 39. | Szczepanek J, Jarzab M, Oczko-Wojciechowska M, Kowalska M, Tretyn A, Haus O, Pogorzala M, Wysocki M, Jarzab B, Styczynski J. Gene expression signatures and ex vivo drug sensitivity profiles in children with acute lymphoblastic leukemia. J Appl Genet. 2012;53:83-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 40. | Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 41. | Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104:19971-19976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 347] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 42. | Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R, Den Boer ML. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96:703-711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 43. | Li XJ, Luo XQ, Han BW, Duan FT, Wei PP, Chen YQ. MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. Br J Cancer. 2013;109:2189-2198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 92] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 44. | de Oliveira JC, Scrideli CA, Brassesco MS, Morales AG, Pezuk JA, Queiroz Rde P, Yunes JA, Brandalise SR, Tone LG. Differential miRNA expression in childhood acute lymphoblastic leukemia and association with clinical and biological features. Leuk Res. 2012;36:293-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 45. | de Oliveira JC, Brassesco MS, Scrideli CA, Tone LG, Narendran A. MicroRNA expression and activity in pediatric acute lymphoblastic leukemia (ALL). Pediatr Blood Cancer. 2012;59:599-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 46. | Swellam M, Hashim M, Mahmoud MS, Ramadan A, Hassan NM. Aberrant Expression of Some Circulating miRNAs in Childhood Acute Lymphoblastic Leukemia. Biochem Genet. 2018;56:283-294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Nabhan M, Louka ML, Khairy E, Tash F, Ali-Labib R, El-Habashy S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene. 2017;628:253-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 48. | Yang YL, Yen CT, Pai CH, Chen HY, Yu SL, Lin CY, Hu CY, Jou ST, Lin DT, Lin SR, Lin SW. A Double Negative Loop Comprising ETV6/RUNX1 and MIR181A1 Contributes to Differentiation Block in t(12;21)-Positive Acute Lymphoblastic Leukemia. PLoS One. 2015;10:e0142863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Daschkey S, Röttgers S, Giri A, Bradtke J, Teigler-Schlegel A, Meister G, Borkhardt A, Landgraf P. MicroRNAs distinguish cytogenetic subgroups in pediatric AML and contribute to complex regulatory networks in AML-relevant pathways. PLoS One. 2013;8:e56334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 50. | Faraoni I, Laterza S, Ardiri D, Ciardi C, Fazi F, Lo-Coco F. MiR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: correlation with NPM1 and FLT3 mutation status. J Hematol Oncol. 2012;5:26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 51. | Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183-3189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 478] [Cited by in F6Publishing: 475] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 52. | Jongen-Lavrencic M, Sun SM, Dijkstra MK, Valk PJ, Löwenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078-5085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 313] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 53. | Li Z, Lu J, Sun M, Mi S, Zhang H, Luo RT, Chen P, Wang Y, Yan M, Qian Z, Neilly MB, Jin J, Zhang Y, Bohlander SK, Zhang DE, Larson RA, Le Beau MM, Thirman MJ, Golub TR, Rowley JD, Chen J. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci USA. 2008;105:15535-15540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 352] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 54. | Mendler JH, Maharry K, Radmacher MD, Mrózek K, Becker H, Metzeler KH, Schwind S, Whitman SP, Khalife J, Kohlschmidt J, Nicolet D, Powell BL, Carter TH, Wetzler M, Moore JO, Kolitz JE, Baer MR, Carroll AJ, Larson RA, Caligiuri MA, Marcucci G, Bloomfield CD. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol. 2012;30:3109-3118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 203] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 55. | Sun SM, Rockova V, Bullinger L, Dijkstra MK, Döhner H, Löwenberg B, Jongen-Lavrencic M. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia. 2013;27:100-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 56. | Cammarata G, Augugliaro L, Salemi D, Agueli C, La Rosa M, Dagnino L, Civiletto G, Messana F, Marfia A, Bica MG, Cascio L, Floridia PM, Mineo AM, Russo M, Fabbiano F, Santoro A. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 57. | Dixon-McIver A, East P, Mein CA, Cazier JB, Molloy G, Chaplin T, Andrew Lister T, Young BD, Debernardi S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008;3:e2141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 207] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 58. | Marcucci G, Maharry K, Radmacher MD, Mrózek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA, Bloomfield CD. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. J Clin Oncol. 2008;26:5078-5087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 59. | Marcucci G, Radmacher MD, Maharry K, Mrózek K, Ruppert AS, Paschka P, Vukosavljevic T, Whitman SP, Baldus CD, Langer C, Liu CG, Carroll AJ, Powell BL, Garzon R, Croce CM, Kolitz JE, Caligiuri MA, Larson RA, Bloomfield CD. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1919-1928. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 334] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 60. | Schwind S, Maharry K, Radmacher MD, Mrózek K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler KH, Paschka P, Baldus CD, Liu S, Garzon R, Powell BL, Kolitz JE, Carroll AJ, Caligiuri MA, Larson RA, Marcucci G, Bloomfield CD. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28:5257-5264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 61. | Lin X, Wang Z, Wang Y, Feng W. Serum MicroRNA-370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int J Clin Exp Pathol. 2015;8:14658-14666. [PubMed] [Cited in This Article: ] |
| 62. | Lin X, Wang Z, Zhang R, Feng W. High serum microRNA-335 level predicts aggressive tumor progression and unfavorable prognosis in pediatric acute myeloid leukemia. Clin Transl Oncol. 2015;17:358-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 63. | Wang X, Zuo D, Yuan Y, Yang X, Hong Z, Zhang R. MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J Cancer Res Clin Oncol. 2017;143:169-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 64. | Wang Z, Hong Z, Gao F, Feng W. Upregulation of microRNA-375 is associated with poor prognosis in pediatric acute myeloid leukemia. Mol Cell Biochem. 2013;383:59-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 65. | Zhang L, Li X, Ke Z, Huang L, Liang Y, Wu J, Zhang X, Chen Y, Zhang H, Luo X. MiR-99a may serve as a potential oncogene in pediatric myeloid leukemia. Cancer Cell Int. 2013;13:110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 66. | Zhu C, Wang Y, Kuai W, Sun X, Chen H, Hong Z. Prognostic value of miR-29a expression in pediatric acute myeloid leukemia. Clin Biochem. 2013;46:49-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 67. | Zhu R, Zhao W, Fan F, Tang L, Liu J, Luo T, Deng J, Hu Y. A 3-miRNA signature predicts prognosis of pediatric and adolescent cytogenetically normal acute myeloid leukemia. Oncotarget. 2017;8:38902-38913. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 68. | Russ AC, Sander S, Lück SC, Lang KM, Bauer M, Rücker FG, Kestler HA, Schlenk RF, Döhner H, Holzmann K, Döhner K, Bullinger L. Integrative nucleophosmin mutation-associated microRNA and gene expression pattern analysis identifies novel microRNA - target gene interactions in acute myeloid leukemia. Haematologica. 2011;96:1783-1791. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 69. | Trino S, Lamorte D, Caivano A, Laurenzana I, Tagliaferri D, Falco G, Del Vecchio L, Musto P, De Luca L. MicroRNAs as New Biomarkers for Diagnosis and Prognosis, and as Potential Therapeutic Targets in Acute Myeloid Leukemia. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 57] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 70. | Xu LH, Guo Y, Zhang XL, Chen JJ, Hu SY. Blood-Based Circulating MicroRNAs are Potential Diagnostic Biomarkers for Leukemia: Result from a Meta-Analysis. Cell Physiol Biochem. 2016;38:939-949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 71. | Sun YM, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 263] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 72. | Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci USA. 2010;107:21558-21563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 222] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 73. | So AY, Sookram R, Chaudhuri AA, Minisandram A, Cheng D, Xie C, Lim EL, Flores YG, Jiang S, Kim JT, Keown C, Ramakrishnan P, Baltimore D. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 2014;124:1502-1512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 74. | Puissegur MP, Eichner R, Quelen C, Coyaud E, Mari B, Lebrigand K, Broccardo C, Nguyen-Khac F, Bousquet M, Brousset P. B-cell regulator of immunoglobulin heavy-chain transcription (Bright)/ARID3a is a direct target of the oncomir microRNA-125b in progenitor B-cells. Leukemia. 2012;26:2224-2232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 75. | Liu Z, Smith KR, Khong HT, Huang J, Ahn EE, Zhou M, Tan M. miR-125b regulates differentiation and metabolic reprogramming of T cell acute lymphoblastic leukemia by directly targeting A20. Oncotarget. 2016;7:78667-78679. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 76. | Bousquet M, Quelen C, Rosati R, Mansat-De Mas V, La Starza R, Bastard C, Lippert E, Talmant P, Lafage-Pochitaloff M, Leroux D, Gervais C, Viguié F, Lai JL, Terre C, Beverlo B, Sambani C, Hagemeijer A, Marynen P, Delsol G, Dastugue N, Mecucci C, Brousset P. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J Exp Med. 2008;205:2499-2506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 209] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 77. | Lin KY, Zhang XJ, Feng DD, Zhang H, Zeng CW, Han BW, Zhou AD, Qu LH, Xu L, Chen YQ. miR-125b, a target of CDX2, regulates cell differentiation through repression of the core binding factor in hematopoietic malignancies. J Biol Chem. 2011;286:38253-38263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 78. | Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, Scherr M, Eder M. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood. 2011;117:4338-4348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 79. | Bousquet M, Nguyen D, Chen C, Shields L, Lodish HF. MicroRNA-125b transforms myeloid cell lines by repressing multiple mRNA. Haematologica. 2012;97:1713-1721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 80. | Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125-1129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 896] [Cited by in F6Publishing: 934] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 81. | Ramsingh G, Koboldt DC, Trissal M, Chiappinelli KB, Wylie T, Koul S, Chang LW, Nagarajan R, Fehniger TA, Goodfellow P, Magrini V, Wilson RK, Ding L, Ley TJ, Mardis ER, Link DC. Complete characterization of the microRNAome in a patient with acute myeloid leukemia. Blood. 2010;116:5316-5326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 82. | Danen-van Oorschot AA, Kuipers JE, Arentsen-Peters S, Schotte D, de Haas V, Trka J, Baruchel A, Reinhardt D, Pieters R, Zwaan CM, van den Heuvel-Eibrink MM. Differentially expressed miRNAs in cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Pediatr Blood Cancer. 2012;58:715-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 83. | Emmrich S, Katsman-Kuipers JE, Henke K, Khatib ME, Jammal R, Engeland F, Dasci F, Zwaan CM, den Boer ML, Verboon L, Stary J, Baruchel A, de Haas V, Danen-van Oorschot AA, Fornerod M, Pieters R, Reinhardt D, Klusmann JH, van den Heuvel-Eibrink MM. miR-9 is a tumor suppressor in pediatric AML with t(8;21). Leukemia. 2014;28:1022-1032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 84. | Chung CH, Levy S, Chaurand P, Carbone DP. Genomics and proteomics: emerging technologies in clinical cancer research. Crit Rev Oncol Hematol. 2007;61:1-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 85. | Stegmaier K. Genomic approaches in acute leukemia. Best Pract Res Clin Haematol. 2006;19:263-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 86. | Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166-5171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 571] [Cited by in F6Publishing: 567] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 87. | Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944-13949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2701] [Cited by in F6Publishing: 2623] [Article Influence: 138.1] [Reference Citation Analysis (0)] |
| 88. | Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240-2252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 414] [Cited by in F6Publishing: 434] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 89. | Kaddar T, Chien WW, Bertrand Y, Pages MP, Rouault JP, Salles G, Ffrench M, Magaud JP. Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leuk Res. 2009;33:1217-1223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 90. | Organista-Nava J, Gómez-Gómez Y, Illades-Aguiar B, Del Carmen Alarcón-Romero L, Saavedra-Herrera MV, Rivera-Ramírez AB, Garzón-Barrientos VH, Leyva-Vázquez MA. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol Rep. 2015;33:1639-1649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 91. | Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290-1297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1089] [Cited by in F6Publishing: 1129] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 92. | Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, Becker KG, Gorospe M, Hide W, Lieberman J. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35:610-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 449] [Cited by in F6Publishing: 480] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 93. | Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 94. | Nemes K, Csóka M, Nagy N, Márk Á, Váradi Z, Dankó T, Kovács G, Kopper L, Sebestyén A. Expression of certain leukemia/lymphoma related microRNAs and its correlation with prognosis in childhood acute lymphoblastic leukemia. Pathol Oncol Res. 2015;21:597-604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 95. | Yan Z, Xiong Y, Xu W, Gao J, Cheng Y, Wang Z, Chen F, Zheng G. Identification of hsa-miR-335 as a prognostic signature in gastric cancer. PLoS One. 2012;7:e40037. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 96. | Yan J, Jiang N, Huang G, Tay JL, Lin B, Bi C, Koh GS, Li Z, Tan J, Chung TH, Lu Y, Ariffin H, Kham SK, Yeoh AE, Chng WJ. Deregulated MIR335 that targets MAPK1 is implicated in poor outcome of paediatric acute lymphoblastic leukaemia. Br J Haematol. 2013;163:93-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 97. | Agirre X, Martínez-Climent JÁ, Odero MD, Prósper F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia. 2012;26:395-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 98. | Roman-Gomez J, Agirre X, Jiménez-Velasco A, Arqueros V, Vilas-Zornoza A, Rodriguez-Otero P, Martin-Subero I, Garate L, Cordeu L, San José-Eneriz E, Martin V, Castillejo JA, Bandrés E, Calasanz MJ, Siebert R, Heiniger A, Torres A, Prosper F. Epigenetic regulation of microRNAs in acute lymphoblastic leukemia. J Clin Oncol. 2009;27:1316-1322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 99. | Stumpel DJ, Schotte D, Lange-Turenhout EA, Schneider P, Seslija L, de Menezes RX, Marquez VE, Pieters R, den Boer ML, Stam RW. Hypermethylation of specific microRNA genes in MLL-rearranged infant acute lymphoblastic leukemia: major matters at a micro scale. Leukemia. 2011;25:429-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 100. | Zhou JD, Li XX, Zhang TJ, Xu ZJ, Zhang ZH, Gu Y, Wen XM, Zhang W, Ji RB, Deng ZQ, Lin J, Qian J. MicroRNA-335/ID4 dysregulation predicts clinical outcome and facilitates leukemogenesis by activating PI3K/Akt signaling pathway in acute myeloid leukemia. Aging (Albany NY). 2019;11:3376-3391. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 101. | Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847-859. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 604] [Cited by in F6Publishing: 620] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 102. | Georgantas RW, Hildreth R, Morisot S, Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM, Civin CI. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750-2755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 372] [Cited by in F6Publishing: 399] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 103. | Vargova K, Pesta M, Obrtlikova P, Dusilkova N, Minarik L, Vargova J, Berkova A, Zemanova Z, Michalova K, Spacek M, Trneny M, Stopka T. MiR-155/miR-150 network regulates progression through the disease phases of chronic lymphocytic leukemia. Blood Cancer J. 2017;7:e585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 104. | Cui B, Chen L, Zhang S, Mraz M, Fecteau JF, Yu J, Ghia EM, Zhang L, Bao L, Rassenti LZ, Messer K, Calin GA, Croce CM, Kipps TJ. MicroRNA-155 influences B-cell receptor signaling and associates with aggressive disease in chronic lymphocytic leukemia. Blood. 2014;124:546-554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 105. | Narayan N, Bracken CP, Ekert PG. MicroRNA-155 expression and function in AML: An evolving paradigm. Exp Hematol. 2018;62:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 106. | Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 512] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 107. | Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627-3632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1026] [Cited by in F6Publishing: 1022] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 108. | Hezaveh K, Kloetgen A, Bernhart SH, Mahapatra KD, Lenze D, Richter J, Haake A, Bergmann AK, Brors B, Burkhardt B, Claviez A, Drexler HG, Eils R, Haas S, Hoffmann S, Karsch D, Klapper W, Kleinheinz K, Korbel J, Kretzmer H, Kreuz M, Küppers R, Lawerenz C, Leich E, Loeffler M, Mantovani-Loeffler L, López C, McHardy AC, Möller P, Rohde M, Rosenstiel P, Rosenwald A, Schilhabel M, Schlesner M, Scholz I, Stadler PF, Stilgenbauer S, Sungalee S, Szczepanowski M, Trümper L, Weniger MA, Siebert R, Borkhardt A, Hummel M, Hoell JI; ICGC MMML-Seq Project. Alterations of microRNA and microRNA-regulated messenger RNA expression in germinal center B-cell lymphomas determined by integrative sequencing analysis. Haematologica. 2016;101:1380-1389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 109. | El-Khazragy N, Noshi MA, Abdel-Malak C, Zahran RF, Swellam M. miRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J Cell Biochem. 2019;120:6315-6321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 110. | Ramamurthy R, Hughes M, Morris V, Bolouri H, Gerbing RB, Wang YC, Loken MR, Raimondi SC, Hirsch BA, Gamis AS, Oehler VG, Alonzo TA, Meshinchi S. miR-155 expression and correlation with clinical outcome in pediatric AML: A report from Children's Oncology Group. Pediatr Blood Cancer. 2016;63:2096-2103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 111. | Narayan N, Morenos L, Phipson B, Willis SN, Brumatti G, Eggers S, Lalaoui N, Brown LM, Kosasih HJ, Bartolo RC, Zhou L, Catchpoole D, Saffery R, Oshlack A, Goodall GJ, Ekert PG. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017;31:808-820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 112. | Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrózek K, Nicolet D, Kohlschmidt J, Whitman SP, Mendler JH, Schwind S, Becker H, Eisfeld AK, Carroll AJ, Powell BL, Kolitz JE, Garzon R, Caligiuri MA, Stone RM, Bloomfield CD. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013;31:2086-2093. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 143] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 113. | Miao MH, Ji XQ, Zhang H, Xu J, Zhu H, Shao XJ. miR-590 promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia by inhibiting RB1. Oncotarget. 2016;7:39527-39534. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 114. | Fan SJ, Li HB, Cui G, Kong XL, Sun LL, Zhao YQ, Li YH, Zhou J. miRNA-149* promotes cell proliferation and suppresses apoptosis by mediating JunB in T-cell acute lymphoblastic leukemia. Leuk Res. 2016;41:62-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 115. | Zhu H, Miao MH, Ji XQ, Xue J, Shao XJ. miR-664 negatively regulates PLP2 and promotes cell proliferation and invasion in T-cell acute lymphoblastic leukaemia. Biochem Biophys Res Commun. 2015;459:340-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 116. | Guo Q, Luan J, Li N, Zhang Z, Zhu X, Zhao L, Wei R, Sun L, Shi Y, Yin X, Ding N, Jiang G, Li X. MicroRNA-181 as a prognostic biomarker for survival in acute myeloid leukemia: a meta-analysis. Oncotarget. 2017;8:89130-89141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 117. | Liu X, Liao W, Peng H, Luo X, Luo Z, Jiang H, Xu L. miR-181a promotes G1/S transition and cell proliferation in pediatric acute myeloid leukemia by targeting ATM. J Cancer Res Clin Oncol. 2016;142:77-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 118. | Guo S, Lu J, Schlanger R, Zhang H, Wang JY, Fox MC, Purton LE, Fleming HH, Cobb B, Merkenschlager M, Golub TR, Scadden DT. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci USA. 2010;107:14229-14234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 261] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 119. | Chapiro E, Russell LJ, Struski S, Cavé H, Radford-Weiss I, Valle VD, Lachenaud J, Brousset P, Bernard OA, Harrison CJ, Nguyen-Khac F. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362-1364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 120. | Ufkin ML, Peterson S, Yang X, Driscoll H, Duarte C, Sathyanarayana P. miR-125a regulates cell cycle, proliferation, and apoptosis by targeting the ErbB pathway in acute myeloid leukemia. Leuk Res. 2014;38:402-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 121. | Han BW, Feng DD, Li ZG, Luo XQ, Zhang H, Li XJ, Zhang XJ, Zheng LL, Zeng CW, Lin KY, Zhang P, Xu L, Chen YQ. A set of miRNAs that involve in the pathways of drug resistance and leukemic stem-cell differentiation is associated with the risk of relapse and glucocorticoid response in childhood ALL. Hum Mol Genet. 2011;20:4903-4915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 98] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 122. | Schotte D, Akbari Moqadam F, Lange-Turenhout EA, Chen C, van Ijcken WF, Pieters R, den Boer ML. Discovery of new microRNAs by small RNAome deep sequencing in childhood acute lymphoblastic leukemia. Leukemia. 2011;25:1389-1399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 123. | Bank PC, Swen JJ, Guchelaar HJ. Pharmacogenetic biomarkers for predicting drug response. Expert Rev Mol Diagn. 2014;14:723-735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 124. | Ma Y, Ding Z, Qian Y, Shi X, Castranova V, Harner EJ, Guo L. Predicting cancer drug response by proteomic profiling. Clin Cancer Res. 2006;12:4583-4589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 70] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 125. | Robert J, Morvan VL, Smith D, Pourquier P, Bonnet J. Predicting drug response and toxicity based on gene polymorphisms. Crit Rev Oncol Hematol. 2005;54:171-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 84] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 126. | Robert J, Vekris A, Pourquier P, Bonnet J. Predicting drug response based on gene expression. Crit Rev Oncol Hematol. 2004;51:205-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 127. | Greaves M. Childhood leukaemia. BMJ. 2002;324:283-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 177] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 128. | Pui CH. Recent research advances in childhood acute lymphoblastic leukemia. J Formos Med Assoc. 2010;109:777-787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 129. | Zhu L, Liu J, Liang F, Rayner S, Xiong J. Predicting response to preoperative chemotherapy agents by identifying drug action on modeled microRNA regulation networks. PLoS One. 2014;9:e98140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 130. | Alaoui-Jamali MA, Dupré I, Qiang H. Prediction of drug sensitivity and drug resistance in cancer by transcriptional and proteomic profiling. Drug Resist Updat. 2004;7:245-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 131. | Bhatla T, Jones CL, Meyer JA, Vitanza NA, Raetz EA, Carroll WL. The biology of relapsed acute lymphoblastic leukemia: opportunities for therapeutic interventions. J Pediatr Hematol Oncol. 2014;36:413-418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 132. | Ghodousi ES, Rahgozar S. MicroRNA-326 and microRNA-200c: Two novel biomarkers for diagnosis and prognosis of pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;119:6024-6032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 133. | El-Khazragy N, Elshimy AA, Hassan SS, Matbouly S, Safwat G, Zannoun M, Riad RA. Dysregulation of miR-125b predicts poor response to therapy in pediatric acute lymphoblastic leukemia. J Cell Biochem. 2018;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 134. | Gefen N, Binder V, Zaliova M, Linka Y, Morrow M, Novosel A, Edry L, Hertzberg L, Shomron N, Williams O, Trka J, Borkhardt A, Izraeli S. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia. 2010;24:89-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 135. | Piatopoulou D, Avgeris M, Marmarinos A, Xagorari M, Baka M, Doganis D, Kossiva L, Scorilas A, Gourgiotis D. miR-125b predicts childhood acute lymphoblastic leukaemia poor response to BFM chemotherapy treatment. Br J Cancer. 2017;117:801-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 136. | Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782-10788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 171] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 137. | Agirre X, Jiménez-Velasco A, San José-Enériz E, Garate L, Bandrés E, Cordeu L, Aparicio O, Saez B, Navarro G, Vilas-Zornoza A, Pérez-Roger I, García-Foncillas J, Torres A, Heiniger A, Calasanz MJ, Fortes P, Román-Gómez J, Prósper F. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830-1840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 165] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 138. | Yang Z, Chen S, Luan X, Li Y, Liu M, Li X, Liu T, Tang H. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life. 2009;61:1075-1082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 139. | Zhang L, Zheng Y, Sun Y, Zhang Y, Yan J, Chen Z, Jiang H. MiR-134-Mbd3 axis regulates the induction of pluripotency. J Cell Mol Med. 2016;20:1150-1158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 140. | Akbari Moqadam F, Lange-Turenhout EA, Ariës IM, Pieters R, den Boer ML. MiR-125b, miR-100 and miR-99a co-regulate vincristine resistance in childhood acute lymphoblastic leukemia. Leuk Res. 2013;37:1315-1321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 141. | Li Y, Yuan Y, Tao K, Wang X, Xiao Q, Huang Z, Zhong L, Cao W, Wen J, Feng W. Inhibition of BCR/ABL protein expression by miR-203 sensitizes for imatinib mesylate. PLoS One. 2013;8:e61858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 142. | Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496-506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 388] [Cited by in F6Publishing: 372] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 143. | Xu L, Liang YN, Luo XQ, Liu XD, Guo HX. [Association of miRNAs expression profiles with prognosis and relapse in childhood acute lymphoblastic leukemia]. Zhonghua Xue Ye Xue Za Zhi. 2011;32:178-181. [PubMed] [Cited in This Article: ] |
| 144. | Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306-6315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 272] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 145. | Harrison CJ, Foroni L. Cytogenetics and molecular genetics of acute lymphoblastic leukemia. Rev Clin Exp Hematol. 2002;6:91-113; discussion 200-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 80] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 146. | Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122:3407-3415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 177] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 147. | Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology (Review). Int J Oncol. 2016;49:5-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 159] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 148. | Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Kamm RD, Mattu C, Chiono V. MicroRNA delivery through nanoparticles. J Control Release. 2019;313:80-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 202] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 149. | Vinhas R, Mendes R, Fernandes AR, Baptista PV. Nanoparticles-Emerging Potential for Managing Leukemia and Lymphoma. Front Bioeng Biotechnol. 2017;5:79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 150. | Pérez-Herrero E, Fernández-Medarde A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur J Pharm Biopharm. 2015;93:52-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 864] [Cited by in F6Publishing: 966] [Article Influence: 107.3] [Reference Citation Analysis (0)] |
| 151. | Ganju A, Khan S, Hafeez BB, Behrman SW, Yallapu MM, Chauhan SC, Jaggi M. miRNA nanotherapeutics for cancer. Drug Discov Today. 2017;22:424-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 204] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 152. | Dorrance AM, Neviani P, Ferenchak GJ, Huang X, Nicolet D, Maharry KS, Ozer HG, Hoellarbauer P, Khalife J, Hill EB, Yadav M, Bolon BN, Lee RJ, Lee LJ, Croce CM, Garzon R, Caligiuri MA, Bloomfield CD, Marcucci G. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29:2143-2153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 89] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 153. | Ghelani HS, Rachchh MA, Gokani RH. MicroRNAs as newer therapeutic targets: A big hope from a tiny player. J Pharmacol Pharmacother. 2012;3:217-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 154. | Xu WQ, Huang YM, Xiao HF. [Expression Analysis and Epigenetics of MicroRNA let-7b in Acute Lymphoblastic Leukemia]. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2015;23:1535-1541. [PubMed] [Cited in This Article: ] |
| 155. | Scherr M, Elder A, Battmer K, Barzan D, Bomken S, Ricke-Hoch M, Schröder A, Venturini L, Blair HJ, Vormoor J, Ottmann O, Ganser A, Pich A, Hilfiker-Kleiner D, Heidenreich O, Eder M. Differential expression of miR-17~92 identifies BCL2 as a therapeutic target in BCR-ABL-positive B-lineage acute lymphoblastic leukemia. Leukemia. 2014;28:554-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 156. | Liu X, Zou L, Zhu L, Zhang H, Du C, Li Z, Gao C, Zhao X, Bao S, Zheng H. miRNA mediated up-regulation of cochaperone p23 acts as an anti-apoptotic factor in childhood acute lymphoblastic leukemia. Leuk Res. 2012;36:1098-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 157. | Liang YN, Tang YL, Ke ZY, Chen YQ, Luo XQ, Zhang H, Huang LB. MiR-124 contributes to glucocorticoid resistance in acute lymphoblastic leukemia by promoting proliferation, inhibiting apoptosis and targeting the glucocorticoid receptor. J Steroid Biochem Mol Biol. 2017;172:62-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 158. | Li Z, Chen P, Su R, Li Y, Hu C, Wang Y, Arnovitz S, He M, Gurbuxani S, Zuo Z, Elkahloun AG, Li S, Weng H, Huang H, Neilly MB, Wang S, Olson EN, Larson RA, Le Beau MM, Zhang J, Jiang X, Wei M, Jin J, Liu PP, Chen J. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood. 2015;126:2005-2015. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 59] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 159. | Dou L, Li J, Zheng D, Li Y, Gao X, Xu C, Gao L, Wang L, Yu L. MicroRNA-142-3p inhibits cell proliferation in human acute lymphoblastic leukemia by targeting the MLL-AF4 oncogene. Mol Biol Rep. 2013;40:6811-6819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 160. | Bhatia S, Kaul D, Varma N. Functional genomics of tumor suppressor miR-196b in T-cell acute lymphoblastic leukemia. Mol Cell Biochem. 2011;346:103-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 161. | Zhou W, Wang S, Ying Y, Zhou R, Mao P. miR-196b/miR-1290 participate in the antitumor effect of resveratrol via regulation of IGFBP3 expression in acute lymphoblastic leukemia. Oncol Rep. 2017;37:1075-1083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 162. | Moses BS, Evans R, Slone WL, Piktel D, Martinez I, Craig MD, Gibson LF. Bone Marrow Microenvironment Niche Regulates miR-221/222 in Acute Lymphoblastic Leukemia. Mol Cancer Res. 2016;14:909-919. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 163. | Li S, Wang C, Wang W, Liu W, Zhang G. Abnormally high expression of POLD1, MCM2, and PLK4 promotes relapse of acute lymphoblastic leukemia. Medicine (Baltimore). 2018;97:e10734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 164. | Wang SM, Sun LL, Wu WS, Yan D. MiR-595 Suppresses the Cellular Uptake and Cytotoxic Effects of Methotrexate by Targeting SLC19A1 in CEM/C1 Cells. Basic Clin Pharmacol Toxicol. 2018;123:8-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 165. | Li X, Li D, Zhuang Y, Shi Q, Wei W, Ju X. Overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr Blood Cancer. 2013;60:2060-2067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 166. | Malik D, Kaul D, Chauhan N, Marwaha RK. miR-2909-mediated regulation of KLF4: a novel molecular mechanism for differentiating between B-cell and T-cell pediatric acute lymphoblastic leukemias. Mol Cancer. 2014;13:175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |