Published online Feb 26, 2014. doi: 10.4331/wjbc.v5.i1.40
Revised: November 27, 2013
Accepted: December 17, 2013
Published online: February 26, 2014
Processing time: 142 Days and 1 Hours
The messenger RNA 3’-untranslated region (3’UTR) plays an important role in regulation of gene expression on the posttranscriptional level. The 3’UTR controls gene expression via orchestrated interaction between the structural components of mRNAs (cis-element) and the specific trans-acting factors (RNA binding proteins and non-coding RNAs). The crosstalk of these factors is based on the binding sequences and/or direct protein-protein interaction, or just functional interaction. Much new evidence that has accumulated supports the idea that several RNA binding factors can bind to common mRNA targets: to the non-overlapping binding sites or to common sites in a competitive fashion. Various factors capable of binding to the same RNA can cooperate or be antagonistic in their actions. The outcome of the collective function of all factors bound to the same mRNA 3’UTR depends on many circumstances, such as their expression levels, affinity to the binding sites, and localization in the cell, which can be controlled by various physiological conditions. Moreover, the functional and/or physical interactions of the factors binding to 3’UTR can change the character of their actions. These interactions vary during the cell cycle and in response to changing physiological conditions. Abnormal functioning of the factors can lead to disease. In this review we will discuss how alterations of these factors or their interaction can affect cancer development and promote or enhance the malignant phenotype of cancer cells. Understanding these alterations and their impact on 3’UTR-directed posttranscriptional gene regulation will uncover promising new targets for therapeutic intervention and diagnostics. We will also discuss emerging new tools in cancer diagnostics and therapy based on 3’UTR binding factors and approaches to improve them.
Core tip: The messenger RNA 3’-untranslated region (3’UTR) plays an important role in regulation of gene expression on the posttranscriptional level. 3’UTR controls gene expression via orchestrated interaction between structural components mRNAs (cis-element) and specific trans-acting factors (RNA binding proteins and non-coding RNAs). Alteration of any of these components can lead to various pathologies. In this review we will discuss how alteration of these factors or a change in the crosstalk between them can affect cancer development and promote or enhance the malignant phenotype of cancer cells. Understanding these regulatory mechanisms and their impact on 3’UTR-directed posttranscriptional gene regulation may uncover promising new targets for therapeutic intervention and diagnostics.
- Citation: Vislovukh A, Vargas TR, Polesskaya A, Groisman I. Role of 3’-untranslated region translational control in cancer development, diagnostics and treatment. World J Biol Chem 2014; 5(1): 40-57
- URL: https://www.wjgnet.com/1949-8454/full/v5/i1/40.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v5.i1.40
During tumor growth, characteristic alterations in gene expression result in modification of the quantity of the corresponding proteins. The alterations have been extensively documented at the mRNA transcription and protein degradation levels; both have a strong impact on the accumulation of critical proteins involved in tumorigenesis. While translational control is a key mechanism involved in the regulation of the gene expression[1], the impact of the misregulation of gene expression during carcinogenesis at the translational level has long been widely underestimated.
Translation of mRNA into proteins can be specifically regulated by a combination of RNA-binding factors (proteins and antisense RNA) that act positively or negatively on translation initiation and elongation, mRNA stability and mRNA localization. This regulation is mostly controlled by sequence elements in 3’-untranslated region (3’UTR) of the transcripts, located downstream from the open reading frame. The importance of the 3’UTR was not fully appreciated until the discovery of small non-coding regulatory RNAs (microRNAs or miRNAs). MiRNAs interact with a protein complex called RNA-induced silencing complex (RISC), which controls gene expression by binding to miRNA target sites in mRNA 3’UTRs. MiRNAs have proven to be not only important markers but also key players in the control of gene expression during cancer development. Multiple 3’UTR regulatory elements are usually involved in the regulation of translation. One of the best characterized of them is the cytoplasmic polyadenylation element (CPE) which, upon binding by the CPE-binding protein (CPEB), regulates specific target mRNAs. CPEB1 directly controls the mammalian cell cycle, particularly during senescence, suggesting a role in cancer and aging. According to the literature and to our unpublished data, members of CPEB family are misregulated in many cancers and can play important role in carcinogenesis[2,3].
The insulin-like growth factor (IGF)-2 mRNA-binding proteins 1, 2 and 3 [IGF2BP1-3/Insulin-like growth factor 2 mRNA-binding protein 1 (IMP1-3)] belong to another well-known family of proteins that bind to 3’UTR and control the expression of proteins important in the normal cell cycle and in cancerous transformation[4,5]. IGF2BP1-3 and IMPs are highly over-expressed in a number of cancers[6].
The aim of this review is to show that regulatory factors controlling gene expression via binding to 3’UTR do not act separately but in cooperation. Crosstalk of these factors is based on the binding sequences and direct protein-protein interaction. The functional and physical interactions of factors binding to 3’UTR can change the character of their action, according to physiological conditions[7]. Disruption of the coordinated action of these factors can have a big impact on the expression of proteins involved in cancer induction and development. A detailed understanding of these mechanisms can help in development of new tools for cancer diagnostics and treatment.
One of the main breakthroughs in cellular and molecular biology in the last decade was the discovery of gene expression regulation by non-coding RNAs. The number of classes of non-coding RNAs continues to grow rapidly. Major among them are miRNAs, piRNAs, endo-siRNAs, exo-siRNAs, rasiRNAs, scnRNAs, tasiRNAs, natsiRNAs, 21U-RNA, lncRNAs and tRNA-derived RNA fragments[8]. We will focus this review on miRNAs, which is the most widely studied group of non-coding regulatory RNAs. MiRNAs are small (21-23 nt) RNAs. MiRNAs originate from Pol II-transcribed precursors (pri-miRNAs). Then the Drosha enzyme recognizes a 70 nt stem-loop structure and produces pre-miRNA, which is transported from the nucleus by Еxportin 5. In the cytoplasm, Dicer enzyme forms a double-stranded 22 nt RNA from pre-miRNA. One of the RNA strands is degraded, whereas the other one inserts into the RISC complex, binds to the target sequence in 3’UTR, and carries out its regulatory function[9]. These tiny molecules are involved in the regulation of almost all cellular processes[10-12]. Since single miRNA can potentially have hundreds of targets, alteration of its expression can easily influence cellular homeostasis, which in the most extreme case may result in cell death or in malignant transformation of the cell. Indeed, the first evidence of involvement of miRNAs in tumorigenesis was shown in 2002 by Calin et al[13]. These authors found that in 68% of chronic lymphocytic leukemia (CLL) cases, deletions and down-regulation of miRNA genes miR-15 and miR-16 at 13q14 locus were observed. Since then, thousands of publications have been devoted to miRNAs involvement in various types of cancer.
Involvement of miRNA in cancer has been proven by genome-wide expression studies using microarray technology and techniques based on quantitative polymerase chain reaction (qPCR), which have helped to establish the miRNA profiles of normal and neoplastic tissues[14,15]. These studies revealed a global decrease in miRNA expression in many tumors. Various tumors also correlate with changes in specific miRNA expression. The above studies were supported by a number of investigations of individual types of neoplasms[16-29] (and many others). About 200 miRNAs have at least once been reported as being up- or down-regulated in tumors. Overall, these studies prove that each neoplasm could exhibit a distinct miRNA expression profile that differs from one of the other neoplasms and its normal tissue counterpart. However, a group of miRNAs was shown to have a similar expression profile in multiple cancers, suggesting that their involvement in tumorigenesis is common for many cancer types. At the same time, there are many miRNAs that are differentially misregulated in different cancers[30]. The reason for this is not yet clear, but it is likely that the function of a miRNA may vary because of tissue-specific expression of their targets. On the other hand, specific miRNAs can have different cofactors and build different networking in different cancers. Thus, it becomes possible for a given miRNA to act either as an oncogene or as a tumor suppressor, according to the context.
One of the best examples for tissue-specific target regulation is the let-7 family of miRNAs, which according to many reports acts as tumor suppressors[31-34]. It has been shown that let-7 is frequently down-regulated in many cancers, leading to up-regulation of the proto-oncogenes RAS[35], High Mobility Group A2 (HMGA2)[36-38], Myc[39], integrin beta 3[40], the oncofetal gene IMP-1[41] and the miRNAs maturation enzyme Dicer[42]. Let-7b was shown to down-regulate the expression of cyclin D1, D3, A and cyclin-dependent kinase (Cdk 4) in melanoma cells[43].
A similar effect was observed for the miR-34 family, another potential tumor-suppressor in a variety of cancers. Localized to chromosomes 1 (34a) and 11 (34c and b), this family is frequently deregulated in various cancers, including lung, ovarian, CLL and colorectal[44-47]. In addition, miR-34b/c polymorphism has been linked to risk of developing hepatocellular carcinoma[48]. The miR-34 family appears to be the direct transcriptional target of p53[49,50] and has few validated targets, SNAIL (zing finger protein SNAL1, epithelial-mesenchymal transition), Wnt, SIRT1 (silent mating type information regulation homolog), cyclin-dependent kinase 6 (CDK6) and others[51-54].
The miR-29 family (a, b and c) also has often been found to be decreased in tumors, such as CLL, hepatocellular carcinoma and breast cancer[55-57], and has been validated to target key components of cellular survival as MCL-1 (induced myeloid leukemia cell differentiation protein), cell cycle CDK6 and dedifferentiation Krüppel-like factor 4[26,55,58]. The most interesting observation concerning miR-29 is that it can globally alter methylation status through targeting of DNA methyltransferases 3A and B (DNA methyltransferases 3A and B) and lead to the derepression of critical tumor suppressors[59].
The miR-17-92 cluster acts as a group of oncogenes when over-expressed. This group includes 14 homologous miRNAs that are encoded by three gene clusters on chromosomes 7, 13 and X[25,60]. The cluster on chromosome 13 is amplified in human B cell lymphomas[61], which leads to increased expression of various miRNA members. Forced expression of the miR17-92 cluster along with myc proto-oncogene (MYC) accelerates tumor development in mouse B cell lymphoma[62], thus acts as an oncogene. Up-regulation of members of this large miRNA group protects cells from apoptosis by inhibiting the expression of E2F, p21 and Bim[63,64].
Among oncogenic miRNAs families, the most therapeutic and diagnostic potential is the miR-21 family, located on chromosome 17. It is over-expressed in several cancers, including breast, colorectal and lung[65-67], and has few validated targets: TPMI (tropomyosin), PDCD4 (program cell death protein 4) and PTEN (phosphatase and tensin homolog)[68-70].
These and other observations found in the literature prove that miRNAs play very important roles in cancer, although their mode of action can differ according to the composition of the targets and a combination of other factors. Knowledge of the mechanisms of miRNA action in particular cancers, especially understanding of their collaborators or inhibitors, will help to develop proper tools for miRNA-based therapy and diagnostics.
Epithelial-mesenchymal transition: To date, it is believed that one of the causes of failure in the treatment of cancer is the existence of cancer stem cells[71]. In cancer, epithelial-mesenchymal transition (EMT) is a process by which epithelial cells are reprogrammed to lose their differentiation and become undifferentiated stem cells with mesenchymal properties. Despite the fact that genes responsible for EMT are well known[72], data devoted to the involvement of miRNAs in this process are still accumulating. Thus, Nairismägi et al[73] showed that miR-580 and CPEB1/2 down-regulate TWIST1 expression, one of the main inductors of EMT in a cooperative way. Another miRNA that suppresses EMT belongs to the miR-200 family. These miRNAs increase E-cadherin expression by targeting the mRNA of the E-cadherin transcriptional repressors zinc finger E-box-binding homeobox 1 (ZEB1) and ZEB2[74,75]. It was later shown that the miR-200 family is downregulated in the initial stages of stromal invasion, but restored at metastatic sites[76]. In cases of hepatocellular carcinoma, miR-612 was found to have an inhibitory effect on EMT targeting of the AKT (also known as protein kinase B) signal cascade[77]. On the other hand, a set of miRNAs is correlated with EMT progression. MiR-21 is thus over-expressed during EMT, whereas blockage of miR-21 inhibits metastasis development[78,79]. During EMT, the Twist transcription factor induces expression of miR-10b. In turn, over-expression of miR-10b in non-metastatic breast tumors initiates intense invasion and metastasis[80]. Furthermore, in hepatocellular carcinoma, miR-106b promotes cell migration and metastasis by activating the EMT process[81].
Angiogenesis and proliferation: The tumor growth rate is one of the most critical characteristics that define the level of cell malignancy. However, while growing, a tumor must supply itself with nutrients, which are provided by active angiogenesis. Deregulation of miRNA expression is also involved in these processes. For instance, upregulation of the miR-17-92 cluster in adenocarcinoma leads to downregulation of its predicted targets: anti-angiogenic thrombospondin-1 and connective tissue growth factor, resulting in enhanced neovascularization[82]. Lee et al[83] showed that miR-378 increases cell survival, tumor growth and angiogenesis. Detailed analyses revealed that the main targets of the miR-378 were SuFu (inhibitor of Hedgehog signal cascade) and tumor suppressor Fus-1. Regulation of proliferation is mainly carried out by forced entry and progression of the cell cycle. Cyclins and their CDKs regulators are key players in the above-mentioned process. Thus, miRNAs can potentially inhibit key components, resulting in the inhibition of proliferation or in decreased expression of cyclin inhibitors. Indeed, Linsley et al[84] showed that the miR-16 family regulates cell cycle progression. Furthermore, the miR-497-195 cluster has been shown to target multiple cell cycle regulators, including CDKs, but it is transcriptionally silenced in hepatocellular carcinoma[85]. In the case of breast cancer, the same miRNA was able to decrease the cyclin E1 level[86]. Wee-1, a well-known cell cycle regulator, is also a target of miR-497[87]. There are other cases in which miRNAs inhibit cell cycle inhibitors. Thus, analyses of miRnome from a broad set of different cancer samples demonstrated that a number of miRNAs were over-expressed in all cases. Interestingly, one of the main targets of these miRNAs was RB1, a well-known negative regulator of the cell cycle[15].
Genetic mechanisms: It is well known that genome instability characterizes malignant cells. The discovery of DNA alteration involvement in trespassing of miRNA gene expression came from the observation that 30%-50% of miRNA genes are located in fragile sites[88,89]. Fragile DNA sites are regions that possess high levels of instability and are susceptible to such processes as genomic rearrangement, which include multiplication and deletion of loci, translocation, high rates of mutation etc[90]. Such a process is deletion of oncosuppressive miR-15a/miR-16a miRNAs that target the anti-apoptotic B-cell lymphoma 2 (BCL-2) protein[91], which was found in the majority of CLL cases[13]. Another rearrangement, translocation, was shown to alter the 17-92 cluster that contains a set of miRNAs among which is leukemogenic miR-19[92]. Translocation can also alter miRNA targets, which results in the disruption of miRNA-mediated proto-oncogenes repression. For instance, Mayr showed that translocation of High Mobility Group A2 (HMGA2) 3’UTR disrupts its repression by let-7 miRNA[37]. During amplification, the number of pro-oncogenic miRNA genes is often increased. Thus, miR-26a, a direct regulator of PTEN, is frequently amplified at the DNA level in human glioma[93]. Amplification of growth-promoting miR-23a is widely observed in gastric cancer[94]. While there is little data concerning the role of mutations in miRNA-mediated control in cancer, the number of publications dedicated to the role of single-nucleotide polymorphism (SNP) on miRNA action is growing fast. SNPs are single-nucleotide variations that naturally occur in the genome. They can potentially alter miRNA seed sequence, which results in alterations in miRNA target sites and deprivation of proto-oncogene expression control. It may also influence miRNA secondary structure and cause disruption of pri-miRNA recognition by miRNA processing enzymes. So far, numerous genomic studies have shown that SNPs in the miRNA seed sequence or target site may be associated with the risk for different types of cancer and in the prognosis of cancer treatment[95-99].
Transcriptional mechanisms: MiRNAs can be processed from RNA intron (mirtron) or transcribed as independent transcripts. In the latter case, an miRNA gene has its own promoter and is transcribed by Pol II[9]. Since tissue transcription factors in cancer are often misregulated, it is logical to assume that this also influences miRNA expression. Indeed, regulation of miRNA expression by such well-known cancer-related transcription factors such as E2F, RAS, MYC and P53 has been shown[100,101]. Moreover, miRNAs and their transcription factors often work in feedback loops. Thus, E2F is responsible for up-regulation of the above-mentioned 17-92 cluster of miRNA in gliomas. E2F1 acts as a transcriptional activator of the miR-17-92 cluster and binds directly to the miR-17-92 promoter[102]. However, the set of miRNAs produced from this cluster directly inhibits E2F1. This is an example of a negative feedback loop[102,103]. Since E2F1 activates its own transcription by a positive feedback loop, miRNAs in this case act as a fuse for E2F1 over-saturation. MiRNAs miR-449a and miR-449b are other targets of E2F1. In this case, both miRNAs form a negative feedback loop indirectly by targeting the pRb-E2F1 pathway through cell cycle arrest[104]. High expression of miR-375 and estrogen receptor α (ERα) in breast cancer cells is an example of a positive feedback circuit. MiR-375 targets dexamethasone-induced ras-related protein 1 mRNA, an ERα inhibitor, whereas ERα increases miR-375 expression[105].
Epigenetic mechanisms: Methylation of DNA, especially gene promoter regions of the genes, causes alteration in gene expression[106]. During cancer progression, two cases could potentially be realized: hypermethylation of oncosuppressors and hypomethylation of oncogenes. The fact that most miRNAs are associated with CpG islands[107] allows us to assume that miRNA genes are potential targets of DNA methylation machinery. Indeed, treatment of cells with inhibitors of DNA methylation (5-aza-2’deoxycytidine) led to upregulation of the subset of oncosuppressor miRNAs in human cancer cells[108]. Another example is the oncosuppressor miR-663 gene, which targets well-known proto-oncogenes such as EEF1A2, TGFβ, JunB and JunD[109-111]. It was found to be downregulated via methylation in samples of human acute myeloid leukemia, hepatocellular carcinoma and breast cancer, as well as in the K-562 leukemia cell line[112-115]. Similar processes occur with miR-129-2, a tumor-suppressive miRNA that is frequently methylated in lymphoid but not myeloid malignancies[116]. The process of hypomethylation can also be influenced in cancer-related alterations of miRNA expression. Thus, Li et al[117] observed hypomethylation of miR-200a/200b promoters with subsequent overexpression of these miRNAs. MiR-200a and miR-200b target SIP1, a protein product that suppresses E-cadherin expression and contributes to epithelial mesenchymal transition[74,117]. In renal cell carcinoma, the promoter of the well-known oncogene miR-21 was found to be hypomethylated, which correlates with upregulated miRNA expression level[118].
The stoichiometry of miRNAs and their targets: Each miRNA potentially targets hundreds of transcripts. Depending on the strength of the miRNA binding site, the target can be more or less inhibited. Thus, constant levels of miRNAs and mRNAs expression are in equilibrium, which provides cell homeostasis. However, several mechanisms that might decrease the miRNA level by using “miRNA sponges” have been discovered. The most well-known example is regulation of PTEN oncosuppressor expression by its pseudogene PTENP1, which harbors the same conserved miRNA binding site as PTEN mRNA[119]. In samples of colon cancer, a decrease in the PTENP1 pseudogene copy number was found, which potentially increases the miRNA pool that targets PTEN. A pseudogene sequestering the miRNA pool was also shown in the case of KRAS1P pseudogene that possess binding sites for miR-143 and let-7 family[120]. Another example of a “miRNA sponge” is circular RNAs (circRNAs). These non-coding RNAs are processed from introns during splicing and carry multiple miRNA binding sites. Hansen et al[121] has shown that ciRS-7 (circular RNA sponge for miR-7) contains more than 70 selectively conserved miRNA target sites and strongly inhibits miR-7 oncosuppressor activity[122,123].
Modulation of the protein expression on the posttranscriptional level during oncogenic transformation often depends on 3’UTR and takes place by changing cis-elements or trans-binding factors that dictate stability and translation efficiency of cancer-related protein mRNAs.
There are few well-characterized cis-elements present in the 3’UTR region. One of them is the CPE, which has a consensus sequence of U4-8A1-2U and is located in relatively close proximity to the ubiquitous nuclear polyadenylation hexanucleotide AAUAAA[124-126].
CPE binds CPEB, one member of a family of four conserved sequence-specific RNA-binding proteins that contain a zinc finger and two RNA recognition motifs[127]. During Xenopus oocyte maturation, CPEB controls meiosis progression from prophase I to metaphase II[127]. Translational control by CPEBs was later also shown to be involved in learning and memory[128,129] and in the regulation of the mammalian cell cycle[130]. CPEB is also implicated in senescence in mammals[131,132] and in controlling the translation of proteins involved in cell cycle checkpoints[133]. Xenopus studies have shown that CPEB can both promote and inhibit RNA translation by respectively elongating and shortening the poly(A) tail. The balance between the two CPEB-associated activities is altered during progression of the cell cycle, depending on post-transcriptional modifications as well as on the number and location of CPEs to which CPEB binds and recruits associated adenylating and de-adenylating protein complexes. The CPEB-containing complex in Xenopus include: symplekin, which may be a platform protein upon which multi-component complexes are assembled, poly(A) ribonuclease, a de-adenylating enzyme and germ-line-development factor 2 (Gld2), an atypical poly(A) polymerase[134,135]. The induction of cytoplasmic polyadenylation is mediated by activation of the serine/threonine kinase Aurora A/Eg2, possibly through repression of glycogen synthase kinase 3[136,137]. When phosphorylated on S174 or T171 (species-dependent), CPEB promotes polyadenylation by stimulating the activity of Gld-2[138]. The newly elongated tail bound by the poly(A)-binding protein promotes translation by augmenting the assembly of the eIF4F initiation complex[139].
CPEB family members were found to be misregulated in various cancers[3]. One of them, CPEB4, was recently shown to be not only over-expressed in pancreatic cancer and glioblastoma in comparison with healthy pancreatic and brain tissues, but also plays a role as a key regulator of cancer transformation and controls translation of hundreds of mRNAs. SiRNA down-regulation of CPEB4 expression in RWP-1 (human pancreatic cancer lines) and Capan pancreatic cancer cells reduce their ability to form tumors after injection into nude mice[2]. This group found that one of the most enriched CPEB4-associated mRNAs, tissue type plasminogen activators (tPA), which is known to be over-expressed in pancreatic tumors, has a short poly(A) tail in normal tissue, whereas in ductal tumors and pancreatic ductal adenocarcinoma cell lines, the tPA poly(A) tail is elongated. This observation supports the idea that misregulation of protein expression during cancer transformation can be controlled by the length of the poly(A) tail, which depends on the presence of CPEB proteins[2].
Insulin-like growth factor-2 mRNA-binding proteins (IGF2BPs or IMPs) are oncofetal proteins that were first discovered in human embryonic Rhabdomyosarcoma and are highly expressed in a number of human cancers[6]. IMPs belong to a conserved family of RNA-binding proteins implicated in the post-transcriptional regulation of multiple mRNAs, IGF2, MYC, CD44, PTEN, G1/S-specific cyclin-D1 (CCND1), CCND3, G1/S-specific cyclin-G1 (CCNG1) and others[4,5,140,141]. All these IMP targets are implicated in the proliferation and invasion of human cancer cells. Moreover, several studies have shown that IMPs participate in essential cell functions alienated during cancer transformation, such as cell polarization, migration, morphology, metabolism, proliferation and differentiation[142].
IMPs are mainly expressed in the embryo and are important during development. However, because of their abnormal re-expression in several types of cancer, IMPs are considered as oncofetal proteins. Typically, IMP1 and IMP3 have been implicated in colon, liver, kidney, pancreas and female reproductive tissue cancers. IMP3 is reported in over 50 publications as being over-expressed in multiple cancer types. IMP3 expression actually correlates with tumor aggressiveness. Concerning IMP2, a few studies have linked its expression to liposarcoma, liver cancer and endometrial adenocarcinomas[142].
IMPs are generally observed in the cytoplasm, where they associate with target mRNAs in cytoplasmic ribonucleoprotein complexes (mRNPs). Actually, in complex with a wide range of other RNA binding proteins (RBPs), IMPs are able to control mRNA turnover, transport, localization and translation.
Other studies provide evidence suggesting an important role for IMPs in cell migration. For instance, IMP2 binds and controls the expression of PINCH-2 (particularly interesting new cysteine-histidine-rich protein) and MURF-3 (muscle specific RING finger protein2) mRNAs to modulate cell motility[143].
Despite controversial observations regarding a potential nuclear role of IMPs, increasing evidence suggests that IMPs can recruit their target mRNAs in the nucleus during their transcription[144-146]. Moreover, a recent study actually shows that in contrast with IMP1 and IMP2, IMP3 has nuclear localization in a large number of human cancer cell lines. For example, IMP3 is almost 100% nuclear in hepatocellular carcinoma, breast and ovarian cancer cells[4].
Among other well known proteins that bind mostly to the AU-rich sequences in 3’UTR and are involved in cancer transformations are Hu/elav proteins, known to bind AU-rich sequences in the 3’UTR and enhance mRNA translation or increase its stability[147,148]. HuR is ubiquitously expressed and HuB, -C and -D are primarily neuronal. HuR is also known as embryonic lethal, abnormal vision, Drosophila-like 1. A link between HuR and malignant transformation has been suggested in cancers such as breast, colon, lung and ovary[149]. Their targets are involved in several processes, such as cell growth and survival, proliferation, stress response, senescence and cancer[150,151].
AU-binding factor 1 (AUF1), also known as heterogeneous nuclear ribonucleoprotein D, belongs to a big family of hnRNPs that includes hnRNNP A, B, C, D, E, F, H, I, K, L, M, Q and R. AUF1 binds to the AU-rich sequence in the 3’UTR of target mRNAs and promotes degradation of the target transcript, most probably by recruiting them to exosomes for degradation[152,153]. However, AUF1 was found to enhance stability and translation of some mRNAs[154,155]. AUF1 was also shown to be involved in several processes: cell cycle, stress response, apoptosis and carcinogenesis.
T-cell intracellular antigen 1 (TIA-1) TIA-1-related (TIAR) binds to AU/U-rich sequences in the 3’UTR of the target transcript and suppresses mRNA translation[156]. Under stress conditions, these proteins are thought to halt mRNA-to-protein aggregations known as stress granules[157].
Nuclear factors 90 interacts with AU rich sequences and suppresses translation of mRNAs involved in the cell cycle[158].
Tristetraprolin (TTP), zinc finger protein, binds AU-rich sequences in mRNAs to promote their decay. It is involved in the cell cycle, inflammation and carcinogenesis[159,160].
KH-type splices regulatory protein (KSRP). RBP binds to AU-rich sequences of target transcripts, promoting mRNA decay. Its targets encoded cytokines, transcription factors, proto-oncogenes and cell cycle regulators[161].
Nucleolin interacts with mRNAs bearing AU-rich or G-rich sequences and regulates mRNA stability and translation. Its targets are involved in the cell cycle, cell morphology, development, cell proliferation and cancer genesis[148].
Obviously, two or more RBP may functionally interplay among themselves and with microRNAs through binding to the same mRNA 3’UTR.
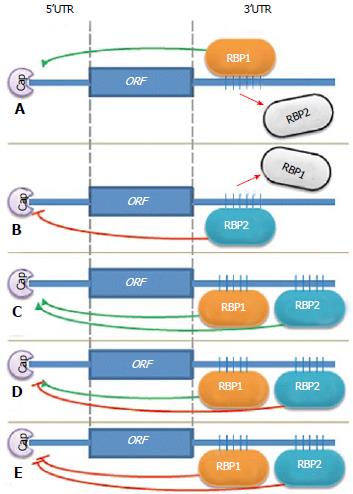
Significant evidence has accumulated to support the idea that several RNA binding proteins can bind the same mRNA target on either the non-overlapping binding sites or on common sites in a competitive fashion. Different RBPs that are capable of binding to the same RNA can cooperate or compete in their actions (Figure 1). The outcome of the combined action of all factors bound to the same mRNA 3’UTR depends on many circumstances, such as expression of different RBPs, their affinity for the binding sites, and their localization in the cells. This can be controlled by different physiological conditions.
For instance, interleukin (IL)-8 plays an integral role in promoting a malignant phenotype in breast cancer and its production is directly influenced by inflammatory cytokines in the tumor microenvironment. Subsequently, activation of the IL-1 receptor on malignant breast cancer cells strongly induces IL-8 mRNA expression. HuR, KSRP and TIAR were found to bind one or more locations within the IL-8 3’UTR, although affinity of the stabilizing factor HuR was 20-fold stronger than that of the KSRP destabilizing factor[162]. HuR, AUF1 and nucleolin bind to BCL-2 mRNAs. HuR and nucleolin both stabilized the BCL-2 transcript, while AUF1 enhanced degradation[163-166]. Thus HuR and nucleolin can have a cooperative effect that is antagonized by AUF1. Another example is related to regulation of GADD45A mRNA stability and translation efficiency. Nucleolin stabilized GADD45A mRNA and was antagonized by AUF1, which promotes decay of this mRNA, and by TIAR, which suppresses translation[167].
In addition, HuR and AUF1 formed a stable ribonucleoprotein complex in the nucleus, whereas in the cytoplasm, HuR and AUF1 bound to target mRNAs individually. HuR colocalizes with the translational apparatus and AUF1 with exosomes[168].
The nuclear localization of IMP3 depends on its protein partner, HNRNPM. Nuclear IMP3 is important for the efficient synthesis of CCND1, D3 and G1 proteins and for the proliferation of human cancer cells. Curiously, IMP3 can be differentially localized in normal versus cancerous adult cells, which in turn will determine the efficiency of protein synthesis of CCND1, D3 and G1 in these cells and have an impact on their proliferation[4]. These studies suggested that IMPs are controlling the transcript destiny of targeted mRNAs in the nucleus and subsequently influence their stability and translation in the cytoplasm. IMP is also found in complex with other RNA-binding proteins, such as HNRNP A2/B1, HNRNP A1, HNRNP A3, Polypyrimidine tract-binding protein 1, interleukin enhancer-binding factor 3, an RNA helicase DHX9 and a mRNA-stabilizing protein HuR[4]. Some of these IMP3 partners, such as HuR, are already known to positively regulate CCND1 mRNA stability and translation[168].
Members of the CPEB and PUF (drosophila pumilio (Pum) protein is a founder member of a novel family of RNA-binding proteins, known as the PUF family.) (Pomelia/Fem-3 mRNA-binding factor 1) families collaborate to regulate mRNA expression throughout eukaryotes. PUF was shown to directly interact with CPEB in C. elegans and humans (CPEB3) and to inhibit translation of common targets[169].
3’UTR binding factors can control translation efficiency via interaction with translation, initiation and elongation factors. An example of the interaction with initiation factors has been described for CPEB1 in a previous chapter. Recently, the eukaryotic translation elongation factor 1A1 (eEF1A1) was shown to be involved in EMT regulation. The main function of eEF1A1 is delivery of aminoacyl tRNA to the A-site of the ribosome[170-172]. However, Hussey et al[173] discovered a new mechanism of EMT control when eEF1A1 in complex with hnRNP E1 binds to the BAT element in the 3’UTR of the EMT, inducing Dab2 and ILEI transcripts. This results in the inhibition of eEF1A1 release from the ribosomal A site, which causes a stall in translational elongation of the above-mentioned transcripts[173].
Moreover, PUF and Ago can interact with eEF1A proteins to repress translation elongation in both C. elegans and mammalians. This repression occurred after translation initiation and led to ribosome accumulation within the open reading frame, roughly at the site where the nascent polypeptide emerged from the ribosomal exit tunnel. Together, these data suggest that a conserved PUF-Ago-eEF1A complex attenuates translation elongation[174].
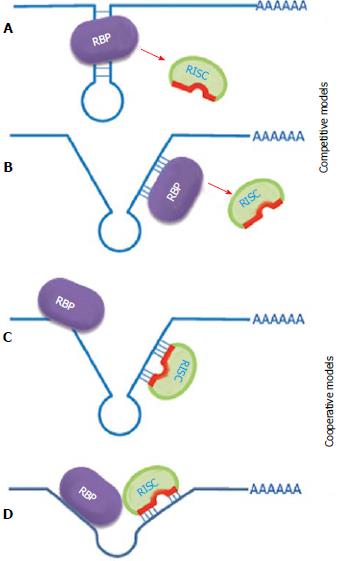
Proteins that bind to the same mRNAs 3’UTR can modulate the function of miRNAs. They either enhance the inhibitory function of miRNA or prevent it. On the other hand, miRNA also can assist the function of RNA binding protein or inhibit it. This can happen simply through binding site competition or collaboration (via RNA remodeling), direct protein-protein interaction of 3’UTR-binding complexes, or just functional interplay when a few factors act separately but their actions augment or negate each other (Figure 2).
Interestingly there are few cases described in the literature in which miRNA in collaboration with RNA binding proteins can change their mode of action during the cell cycle or under physiological conditions such as oxidative stress and others.
It has been shown that upon cell cycle arrest, the ARE (AU-rich element) in tumor necrosis factor-α mRNA acts as a translation activation signal, recruiting AGO (argonaute RISC catalytic component) and fragile X mental retardation-related protein 1 factors associated with miRNPs. Human miRNA mir-369-3 directs the association of these proteins with the AREs, leading to the activation of translation[7]. Moreover, two well-studied miRNAs, let-7 and the synthetic miRNA miRcxcr4, also induce translational up-regulation of target mRNA upon cell cycle arrest. However, they repress translation in proliferating cells. It has been proposed that translation regulation by miRNPs oscillates between repression and activation during the cell cycle[7].
Another example of inactivation, storage and reactivation is calcium transport protein (CAT)-1 mRNA targeting by mir-122 under stress conditions. The derepression of CAT-1 mRNA is accompanied by its release from cytoplasmic P-bodies and its recruitment to polysomes. Derepression requires binding of HuR, an AU-rich-element-binding protein, to the 3’UTR of CAT-1 mRNA[175]. Thus, interaction with RNA binding proteins can change the miRNA mode of action directed to the same target, according to conditions.
Some difficulty in understanding the affiliation of certain RBPs for oncogenes or tumor suppressors came from the observation that the same RBP interacting with different miRNAs in the regulation of different targets could lead to enhanced or suppressed cancer transformation, according to the nature of the target. The stimulation effect of Pomelia on miRNA function, most probably through mRNA remodeling, is directed towards the targets acting in opposite ways, as oncogene or tumor suppressor. It was shown by Kedde and coworkers that Pomelia RBP pumilio RNA-binding family member 1 (PUM1) and PUM2 promote the regulation of miR-221/222 on the p27kip1 check-point protein and tumor suppressor mRNAs by opening of the secondary structure of the p27 3’UTR and exposing the binding sequence to miR-221/222. This causes down-regulation of p27kip1 accumulation and stimulates cell proliferation and breast cancer development[176]. On the other hand, Pomelia collaborates with some miRNAs to repress E2F3, transcription factor and strong oncogene. This prevents cell proliferation and down-regulates bladder cancer development[177].
Another example of miRNA and RBP collaboration was shown by Nairismägi et al[73] who showed that miR-580 and CPEB1/2 down-regulate TWIST1 expression, one of the main inductors of EMT in a cooperative way. On the other hand, Dnd1 is an example of RBP that prevents binding of miRNA to their target sequences in a few genes, such as p27kip1 and LATS2, and suppresses formation of germ cell tumor[178]. It also prevents miR-21 function on its MutS protein homolog 2 target, which suppresses tumorigenesis in skin[179]. Thus by preventing miRNA down-regulation of tumor suppressors, Dnd1 inhibits the development of certain tumors.
The same RNA binding protein can cooperate or antagonize miRNA functions, according to the mRNA-target. One of the most investigated examples is HuR[147], which was found to recruit let-7 to suppress c-MYC mRNA translation[8] but competes with miR-494 and miR-548-3p for the regulation of nucleolin and TOP2A mRNA, respectively[180,181].
Some RBPs not working alone but in complex with other RNA binding proteins can prevent miRNA actions. IMP1 in complex with heterogeneous nuclear ribonucleoprotein U, synaptotagmin binding, cytoplasmic RNA interacting protein, YXB1 (transcriptional regulator ) and DHX9 [DEAH (Asp-Glu-Ala-His) box helicase 9] is able to stabilize the mRNA of MYC, possibly by inhibiting its translation-coupled degradation[182]. However, some studies showed that MYC is repressed by members of the let-7 microRNA family, suggesting a possible function of IMP1 in protecting MYC mRNAs from microRNA silencing. This was previously proposed as a mechanism for the stabilization of the BTRC (beta-transducin repeat containing E3 ubiquitin protein ligase) mRNA by IMP1[183,184].
Not only RNA-binding protein can influence miRNA function, but reciprocal action has already also been documented in the literature. Some miRNAs can affect the function of RNA binding protein. For example, interaction of mir-16 (a member of the mir-15/16 family of miRNPs) and an ARE-binding protein TTP (tristetraprolin) has been shown to occur through association with AGO/eiF2C family members. Mir-16 assists TTP in targeting ARE, which appears to be an essential step in ARE-mediated mRNA degradation[185].
From all of these examples, one can see that interaction among factors binding to 3’UTR brings a new level of complexity to the mechanisms of action of these factors and their influences on cancer transformation. It is becoming clear that to understand the true picture of the post-transcriptional control of certain genes via 3’UTR, especially that involved in cancer transformation, one needs to take into account all proteins and miRNAs binding to their 3’UTRs.
From the very early investigations that suggested miRNA involvement in cancer, scientists began to think about using it as a tool for cancer diagnosis and therapy. A number of studies have been initiated utilizing miRNA expression profiling to determine markers for diseases. An early study comparing a limited number of available miRNAs in cancer and normal tissues drove the conclusions that miRNAs expression signatures are able to classify tumors based on the development lineage and the differentiation state, suggesting miRNAs as a potential biomarker[14]. Following works used the miRNA expression profile to define a number of normal and cancerous tissues from thyroid, kidney, bladder, liver etc[186-193]. Furthermore, miRNA profiling has also been used to classify tumor subtypes in breast cancer in development[194,195]. Mir-342 is differentially expressed in breast cancer subtypes with high expression in Luminal B-type tumors and decreased expression in therapeutically difficult estrogen receptor/human epidermal growth factor receptor 2-negative tumors[196]. This observation suggested that select miRNAs expression could differentiate tumor subtypes that can be more sensitive or resistant to particular treatments.
Radiation therapy (RT) is one of the most often used procedures in cancer treatment; however, not all patients respond well to it. So, it is very important to develop markers that can predict a patient’s response to RT. MiRNA profiling has a big potential for this type of diagnosis.
One of the first reports identifies the let-7 family for its role in modulating sensitivity for RT in lung cancer[197]. It has been demonstrated that over-expression of let-7 promotes radio-sensitivity while knockdown increases resistance both in vitro and in vivo. Mir-181a has been identified as an important miRNA for radio-sensitivity in glioma cells. Transient over-expression of miR-181a prevented radio-sensitivity that correlated with decreased quantities of Bcl-2, an anti-apoptotic protein[198]. Similarly, over-expression of mir-451 in colorectal cancer cell lines decreases proliferation and increases RT sensitivity of colorectal cancer cells[199].
Chemotherapy is another widely used treatment in cancer therapy. The miRNA profile also has a big potential as a marker for chemo-sensitivity. Inhibition or introduction of some miRNAs to certain cancers can improve their chemo-sensitivity. Inhibition of mir-21 sensitizes U251 glioma cells to etoposide and glioma in mice to tumor necrosis factor-related apoptosis, inducing the ligand S-TRAIL (TNF-related apoptosis-inducing ligand)[200-202]. Mir-451 is downregulated in the glioblastoma stem cell population. Reintroduction of mir-451 in combination with the frequently used glioblastoma treatment imatinib inhibits the growth of glioblastoma stem cells and the formation of neurospheres[203].
Mir-122 was shown to be downregulated in hepatocellular carcinoma (HCC) cells, which promotes RT resistance as well as growth, proliferation and metastasis[204]. Insulin growth factor 1 tyrosine kinase receptor is targeted and suppressed by miR-122 in normal liver cells. However, depletion of mir-122 in HCC increases the IgfIR level. Reintroduction of mir-122 in HCC promotes sensitivity to the tyrosine kinase inhibitor sorafenib[204].
In colorectal cancer, a number of miRNAs have been associated with predicting the response to nucleoside analogs. Mir-143 is downregulated in colon cancer. It targets NF-κB, Bxl-2 and ERK5 and has been shown to increase sensitivity to fluorouracil in HCT-166 colon cancer cell lines[205]. In rectal cancer, mir-125b and mir-137 were associated with poor response to capecitabine, a pro-drug that is enzymatically converted to fluorouracil[206]. In colon cancer, mir-519c targets and suppresses ATP-binding cassette sub-family G member 2 (ABCG2) in cell lines that are sensitive to mitoxantrone, whereas mir-519c inhibition increases the ABCG2 level and chemoresistance. In the ABCG2 resistant cell line, mRNA possess a shortened 3’UTR, which results in the loss of a mir-519c target site and a high-level of ABCG2 protein[207].
All these examples clearly show that miRNA profiling of each cancer could provide useful information for choosing the right treatment strategy. Few bio-pharmaceutical companies are working on developing miRNA-profiling platforms for more detailed identification of cancer subtypes that could improve recommendation of treatment. There are more than 100 ongoing trials incorporating miRNA as biomarkers underway in various bio-pharmacological companies.
Direct miRNA therapeutics, the fundamental principle of miRNA therapy, involves either directed silencing or reduction in tumor-promoting miRNAs versus enrichment of tumor suppressive miRNAs. In vivo, these approaches include genetically engineered animals and different ways of delivery, such as viral vectors, nanoparticle-based delivery, mimics and antimiRs. Targeting miRNA for suppression through the use of antimiRs is perhaps the most promising model. Through complementary binding to the target miRNA (working strand), these molecules can repress the action of select miRNAs.
To improve stability and target specificity, investigators have developed various modifications. Three types of modification currently give the most promising results: replacement of 2-OH residues by 2’-O-methyl modified oligonucleotides, 2’-O-methoxyethyl and locked nucleic acid. In addition, conjugation of cholesterol may be used to improve target specificity[208].
Sponge is another tool for RNA-silencing. By having multiple target binding sites, sponges essentially compete with target mRNA for miRNA occupancy, thus decreasing binding miRNA to its real target[209].
To target a few miRNAs involved in the same cancer formation, investigators started using tiny 8-mer locked nucleic acids with a phosphorothioate backbone to enhance the stability level[210]. They were shown to inhibit families of miR-221/222 and let-7 with high specificity.
Viral vector-based delivery systems, including adenoviral, retroviral and lentiviral systems provide some advantages. For example, lentiviral let-7 delivery has been successfully used in murine models of lung cancer[32]. Several nanoparticles with lipid-based formulations were perhaps the most effective in delivery while minimizing toxicity. Lipid emulsions have been used to deliver miRNAs in lung cancer and lymphoma[211-213].
In spite of big efforts, only mir-122 has successfully reached the clinical trial in targeted therapy[214,215]. The systematic delivery of antimiR-122 could reduce the hepatitis C virus (HCV) viral load chimpanzee model of chronic HCV infection with minimal toxicity[216]. Santaris Pharma conducted a human phase IIa trial safety antiviral function using miravirsen (a locked nucleic acid-modified miR-122 antagonist).
RNA binding proteins similarly can be used as markers for proper cancer diagnostics, leading to better treatment selection. For example, IMP3 over-expression has been associated with distinct cancer types. Several studies have suggested IMP3 as an important marker for poor prognosis in cancer[217,218]. Moreover, it was demonstrated that IMP3 promotes cell growth, proliferation and resistance to ionic irradiation in an IGF2-dependent manner[219,220]. Since CPEB4 was found to be a key protein for pancreatic cancer and glioblastoma development, one can try to apply siRNA-dependent direct down-regulation of CPEB4 protein in this type of tumor using delivery methods that are discussed in this chapter.
In conclusion, 3’UTRs of human mRNAs contained many cis-elements that bind trans factors and are important for the development of various diseases, including cancer. Additional work is required to identify the complete set of 3’UTR cis-elements and the trans-regulatory factors that interact with them and to determine functional consequences of these interactions and their role in cancer transformation. Powerful transcriptome-wide computational and experimental methods are now being used to address these questions. Along with lower-throughput reductionist approaches, they should move us closer to a system biology understanding of how 3’UTRs contribute to gene regulation during cancer transformation. This will allow developing new, more powerful drugs in cancer therapy.
We are thankful to Vladimir Zakon, Noah Hardy and Boris Negrutskii for technical assistance and providing important information.
P- Reviewers: Chattopadhyay S, Yoshida K S- Editor: Cui XM L- Editor: Roemmele A E- Editor: Wu HL
| 1. | Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46:674-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 941] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 2. | Ortiz-Zapater E, Pineda D, Martínez-Bosch N, Fernández-Miranda G, Iglesias M, Alameda F, Moreno M, Eliscovich C, Eyras E, Real FX. Key contribution of CPEB4-mediated translational control to cancer progression. Nat Med. 2012;18:83-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 3. | D’Ambrogio A, Nagaoka K, Richter JD. Translational control of cell growth and malignancy by the CPEBs. Nat Rev Cancer. 2013;13:283-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 93] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 4. | Rivera Vargas T, Boudoukha S, Simon A, Souidi M, Cuvellier S, Pinna G, Polesskaya A. Post-transcriptional regulation of cyclins D1, D3 and G1 and proliferation of human cancer cells depend on IMP-3 nuclear localization. Oncogene. 2013;Jul; Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 5. | Vikesaa J, Hansen TV, Jønson L, Borup R, Wewer UM, Christiansen J, Nielsen FC. RNA-binding IMPs promote cell adhesion and invadopodia formation. EMBO J. 2006;25:1456-1468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 273] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 6. | Yisraeli JK. RNA localization and cell polarity: a cellular ‘pas de deux’. Biol Cell. 2005;97:3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545-1549. [PubMed] |
| 8. | Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2362] [Cited by in RCA: 2456] [Article Influence: 153.5] [Reference Citation Analysis (0)] |
| 9. | Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2112] [Cited by in RCA: 2436] [Article Influence: 162.4] [Reference Citation Analysis (0)] |
| 10. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2105] [Cited by in RCA: 2353] [Article Influence: 130.7] [Reference Citation Analysis (0)] |
| 11. | Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1960] [Cited by in RCA: 1737] [Article Influence: 108.6] [Reference Citation Analysis (0)] |
| 12. | Lee HJ. Exceptional stories of microRNAs. Exp Biol Med (Maywood). 2013;238:339-343. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524-15529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3675] [Cited by in RCA: 3787] [Article Influence: 164.7] [Reference Citation Analysis (0)] |
| 14. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7723] [Cited by in RCA: 7369] [Article Influence: 368.5] [Reference Citation Analysis (0)] |
| 15. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4471] [Cited by in RCA: 4518] [Article Influence: 237.8] [Reference Citation Analysis (0)] |
| 16. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6021] [Article Influence: 316.9] [Reference Citation Analysis (0)] |
| 17. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5601] [Article Influence: 294.8] [Reference Citation Analysis (0)] |
| 18. | Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553-5559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 19. | Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851-4860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 20. | Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, Liedberg F, Chebil G, Gudjonsson S, Borg A. MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236-2242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 182] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 21. | Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 954] [Cited by in RCA: 952] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 22. | Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875-886. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1370] [Cited by in RCA: 1307] [Article Influence: 76.9] [Reference Citation Analysis (1)] |
| 23. | Tetzlaff MT, Liu A, Xu X, Master SR, Baldwin DA, Tobias JW, Livolsi VA, Baloch ZW. Differential expression of miRNAs in papillary thyroid carcinoma compared to multinodular goiter using formalin fixed paraffin embedded tissues. Endocr Pathol. 2007;18:163-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 160] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 24. | Magrelli A, Azzalin G, Salvatore M, Viganotti M, Tosto F, Colombo T, Devito R, Di Masi A, Antoccia A, Lorenzetti S. Altered microRNA Expression Patterns in Hepatoblastoma Patients. Transl Oncol. 2009;2:157-163. [PubMed] |
| 25. | Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217-222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 883] [Cited by in RCA: 864] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 26. | Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133-6140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 665] [Cited by in RCA: 677] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 27. | Henson BJ, Bhattacharjee S, O’Dee DM, Feingold E, Gollin SM. Decreased expression of miR-125b and miR-100 in oral cancer cells contributes to malignancy. Genes Chromosomes Cancer. 2009;48:569-582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 172] [Cited by in RCA: 186] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 28. | Lerner M, Harada M, Lovén J, Castro J, Davis Z, Oscier D, Henriksson M, Sangfelt O, Grandér D, Corcoran MM. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16-1. Exp Cell Res. 2009;315:2941-2952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 29. | Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma -A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 209] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 30. | Ferdin J, Kunej T, Calin GA. Non-coding RNAs: identification of cancer-associated microRNAs by gene profiling. Technol Cancer Res Treat. 2010;9:123-138. [PubMed] |
| 31. | Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759-764. [PubMed] |
| 32. | Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105:3903-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 670] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 33. | Legesse-Miller A, Elemento O, Pfau SJ, Forman JJ, Tavazoie S, Coller HA. let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J Biol Chem. 2009;284:6605-6609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 34. | Slack F. let-7 microRNA reduces tumor growth. Cell Cycle. 2009;8:1823. [PubMed] |
| 35. | Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2664] [Cited by in RCA: 2707] [Article Influence: 135.4] [Reference Citation Analysis (0)] |
| 36. | Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025-1030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 950] [Cited by in RCA: 954] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 37. | Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576-1579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 919] [Cited by in RCA: 898] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 38. | Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585-2590. [PubMed] |
| 39. | Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762-9770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 590] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 40. | Müller DW, Bosserhoff AK. Integrin beta 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27:6698-6706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 41. | Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587-2591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 175] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 42. | Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 43. | Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 343] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 44. | Tanaka N, Toyooka S, Soh J, Kubo T, Yamamoto H, Maki Y, Muraoka T, Shien K, Furukawa M, Ueno T. Frequent methylation and oncogenic role of microRNA-34b/c in small-cell lung cancer. Lung Cancer. 2012;76:32-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 45. | Vogt M, Munding J, Grüner M, Liffers ST, Verdoodt B, Hauk J, Steinstraesser L, Tannapfel A, Hermeking H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 258] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 46. | Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, Kamat AA, Sood AK, Ellenson LH, Hermeking H. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119-1128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 263] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 47. | Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S, Barbarotto E, Cimmino A, Adair B, Wojcik SE. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305:59-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 216] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 48. | Xu Y, Liu L, Liu J, Zhang Y, Zhu J, Chen J, Liu S, Liu Z, Shi H, Shen H. A potentially functional polymorphism in the promoter region of miR-34b/c is associated with an increased risk for primary hepatocellular carcinoma. Int J Cancer. 2011;128:412-417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 156] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 49. | He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819-822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 439] [Cited by in RCA: 425] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 50. | Rokhlin OW, Scheinker VS, Taghiyev AF, Bumcrot D, Glover RA, Cohen MB. MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther. 2008;7:1288-1296. [PubMed] |
| 51. | Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, Hermeking H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256-4271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 478] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 52. | Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol. 2011;195:417-433. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 356] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 53. | Kim NH, Kim HS, Kim NG, Lee I, Choi HS, Li XY, Kang SE, Cha SY, Ryu JK, Na JM. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci Signal. 2011;4:ra71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 246] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 54. | Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421-13426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 950] [Cited by in RCA: 1077] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 55. | Cittelly DM, Finlay-Schultz J, Howe EN, Spoelstra NS, Axlund SD, Hendricks P, Jacobsen BM, Sartorius CA, Richer JK. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2013;32:2555-2564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 56. | Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590-11593. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 461] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 57. | Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 296] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 58. | Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630-2639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 275] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 59. | Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci USA. 2007;104:15805-15810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1300] [Cited by in RCA: 1278] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 60. | Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 448] [Cited by in RCA: 468] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 61. | Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087-3095. [PubMed] |
| 62. | He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2729] [Cited by in RCA: 2822] [Article Influence: 141.1] [Reference Citation Analysis (0)] |
| 63. | Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, Agarwal R, Paun BC, Jin Z, Olaru AV. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689-1700. [PubMed] |
| 64. | Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68:8191-8194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 314] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 65. | Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699-8707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1167] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 66. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1037] [Cited by in RCA: 1195] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 67. | Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12085-12090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 411] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 68. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1350] [Cited by in RCA: 1457] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 69. | Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282:14328-14336. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 808] [Cited by in RCA: 817] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 70. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2191] [Cited by in RCA: 2183] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 71. | Koch U, Krause M, Baumann M. Cancer stem cells at the crossroads of current cancer therapy failures--radiation oncology perspective. Semin Cancer Biol. 2010;20:116-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 72. | Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6575] [Cited by in RCA: 7880] [Article Influence: 492.5] [Reference Citation Analysis (0)] |
| 73. | Nairismägi ML, Vislovukh A, Meng Q, Kratassiouk G, Beldiman C, Petretich M, Groisman R, Füchtbauer EM, Harel-Bellan A, Groisman I. Translational control of TWIST1 expression in MCF-10A cell lines recapitulating breast cancer progression. Oncogene. 2012;31:4960-4966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 74. | Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2843] [Cited by in RCA: 3078] [Article Influence: 181.1] [Reference Citation Analysis (0)] |
| 75. | Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1717] [Cited by in RCA: 1845] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 76. | Paterson EL, Kazenwadel J, Bert AG, Khew-Goodall Y, Ruszkiewicz A, Goodall GJ. Down-regulation of the miRNA-200 family at the invasive front of colorectal cancers with degraded basement membrane indicates EMT is involved in cancer progression. Neoplasia. 2013;15:180-191. [PubMed] |
| 77. | Tao ZH, Wan JL, Zeng LY, Xie L, Sun HC, Qin LX, Wang L, Zhou J, Ren ZG, Li YX. miR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 78. | Bornachea O, Santos M, Martínez-Cruz AB, García-Escudero R, Dueñas M, Costa C, Segrelles C, Lorz C, Buitrago A, Saiz-Ladera C. EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Sci Rep. 2012;2:434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 79. | Zavadil J, Narasimhan M, Blumenberg M, Schneider RJ. Transforming growth factor-beta and microRNA: mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs. 2007;185:157-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 118] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 80. | Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1936] [Cited by in RCA: 1996] [Article Influence: 110.9] [Reference Citation Analysis (0)] |
| 81. | Yau WL, Lam CS, Ng L, Chow AK, Chan ST, Chan JY, Wo JY, Ng KT, Man K, Poon RT. Over-expression of miR-106b promotes cell migration and metastasis in hepatocellular carcinoma by activating epithelial-mesenchymal transition process. PLoS One. 2013;8:e57882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 82. | Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060-1065. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 838] [Cited by in RCA: 820] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 83. | Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104:20350-20355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 423] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 84. | Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240-2252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 444] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 85. | Furuta M, Kozaki K, Tanimoto K, Tanaka S, Arii S, Shimamura T, Niida A, Miyano S, Inazawa J. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 107] [Cited by in RCA: 122] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 86. | Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, Fang L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 87. | Creevey L, Ryan J, Harvey H, Bray IM, Meehan M, Khan AR, Stallings RL. MicroRNA-497 increases apoptosis in MYCN amplified neuroblastoma cells by targeting the key cell cycle regulator WEE1. Mol Cancer. 2013;12:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 88. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 89. | Mathelier A, Carbone A. Large scale chromosomal mapping of human microRNA structural clusters. Nucleic Acids Res. 2013;41:4392-4408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 90. | Bartova E, Galiova G, Legartova S, Stixova L, Jugova A, Kozubek S. Genome instability in the context of chromatin structure and fragile sites. Crit Rev Eukaryot Gene Expr. 2010;20:181-194. [PubMed] |
| 91. | Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944-13949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2701] [Cited by in RCA: 2707] [Article Influence: 135.4] [Reference Citation Analysis (0)] |
| 92. | Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat Cell Biol. 2010;12:372-379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 280] [Cited by in RCA: 270] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 93. | Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327-1337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 418] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 94. | An J, Pan Y, Yan Z, Li W, Cui J, Yuan J, Tian L, Xing R, Lu Y. MiR-23a in amplified 19p13.13 loci targets metallothionein 2A and promotes growth in gastric cancer cells. J Cell Biochem. 2013;114:2160-2169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 95. | Linhares JJ, Azevedo M, Siufi AA, de Carvalho CV, Wolgien Mdel C, Noronha EC, Bonetti TC, da Silva ID. Evaluation of single nucleotide polymorphisms in microRNAs (hsa-miR-196a2 rs11614913 C/T) from Brazilian women with breast cancer. BMC Med Genet. 2012;13:119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 96. | Srivastava K, Srivastava A. Comprehensive review of genetic association studies and meta-analyses on miRNA polymorphisms and cancer risk. PLoS One. 2012;7:e50966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 104] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 97. | Wynendaele J, Böhnke A, Leucci E, Nielsen SJ, Lambertz I, Hammer S, Sbrzesny N, Kubitza D, Wolf A, Gradhand E. An illegitimate microRNA target site within the 3’ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010;70:9641-9649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 98. | Mishra PJ, Mishra PJ, Banerjee D, Bertino JR. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: Introducing microRNA pharmacogenomics. Cell Cycle. 2008;7:853-858. [PubMed] |
| 99. | Preskill C, Weidhaas JB. SNPs in microRNA binding sites as prognostic and predictive cancer biomarkers. Crit Rev Oncog. 2013;18:327-340. [PubMed] |
| 100. | Emmrich S, Pützer BM. Checks and balances: E2F-microRNA crosstalk in cancer control. Cell Cycle. 2010;9:2555-2567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 101. | Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3165] [Cited by in RCA: 3664] [Article Influence: 244.3] [Reference Citation Analysis (0)] |
| 102. | Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130-2134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 364] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 103. | Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135-2143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 445] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 104. | Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, Yu Q. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388-2393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 196] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 105. | de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175-9184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 106. | Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu G, Ye CJ. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol. 2009;220:538-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 107. | Weber B, Stresemann C, Brueckner B, Lyko F. Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle. 2007;6:1001-1005. [PubMed] |
| 108. | Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220-2222. [PubMed] |
| 109. | Liu ZY, Zhang GL, Wang MM, Xiong YN, Cui HQ. MicroRNA-663 targets TGFB1 and regulates lung cancer proliferation. Asian Pac J Cancer Prev. 2011;12:2819-2823. [PubMed] |
| 110. | Tili E, Michaille JJ, Adair B, Alder H, Limagne E, Taccioli C, Ferracin M, Delmas D, Latruffe N, Croce CM. Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD. Carcinogenesis. 2010;31:1561-1566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 199] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 111. | Vislovukh A, Kratassiouk G, Porto E, Gralievska N, Beldiman C, Pinna G, El’skaya A, Harel-Bellan A, Negrutskii B, Groisman I. Proto-oncogenic isoform A2 of eukaryotic translation elongation factor eEF1 is a target of miR-663 and miR-744. Br J Cancer. 2013;108:2304-2311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 112. | Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, Länger F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 343] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 113. | Potapova A, Albat C, Hasemeier B, Haeussler K, Lamprecht S, Suerbaum S, Kreipe H, Lehmann U. Systematic cross-validation of 454 sequencing and pyrosequencing for the exact quantification of DNA methylation patterns with single CpG resolution. BMC Biotechnol. 2011;11:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 114. | Yan-Fang T, Jian N, Jun L, Na W, Pei-Fang X, Wen-Li Z, Dong W, Li P, Jian W, Xing F. The promoter of miR-663 is hypermethylated in Chinese pediatric acute myeloid leukemia (AML). BMC Med Genet. 2013;14:74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 115. | Yang Y, Wang LL, Li YH, Gao XN, Liu Y, Yu L. Effect of CpG island methylation on microRNA expression in the k-562 cell line. Biochem Genet. 2012;50:122-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 116. | Wong KY, Yim RL, Kwong YL, Leung CY, Hui PK, Cheung F, Liang R, Jin DY, Chim CS. Epigenetic inactivation of the MIR129-2 in hematological malignancies. J Hematol Oncol. 2013;6:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 117. | Li Z, Qi CF, Shin DM, Zingone A, Newbery HJ, Kovalchuk AL, Abbott CM, Morse HC. Eef1a2 promotes cell growth, inhibits apoptosis and activates JAK/STAT and AKT signaling in mouse plasmacytomas. PLoS One. 2010;5:e10755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 54] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 118. | The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2717] [Cited by in RCA: 2694] [Article Influence: 224.5] [Reference Citation Analysis (0)] |
| 119. | Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033-1038. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1939] [Cited by in RCA: 1867] [Article Influence: 124.5] [Reference Citation Analysis (0)] |
| 120. | Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4127] [Cited by in RCA: 5538] [Article Influence: 395.6] [Reference Citation Analysis (0)] |
| 121. | Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4631] [Cited by in RCA: 6039] [Article Influence: 503.3] [Reference Citation Analysis (0)] |
| 122. | Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y. miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 232] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 123. | Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G, Luo J, Ren T. MicroRNA-7-regulated TLR9 signaling-enhanced growth and metastatic potential of human lung cancer cells by altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt pathway. Mol Biol Cell. 2013;24:42-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 124. | McGrew LL, Dworkin-Rastl E, Dworkin MB, Richter JD. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803-815. [PubMed] |
| 125. | Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990;10:5634-5645. [PubMed] |
| 126. | Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582-2592. [PubMed] |
| 127. | Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617-627. [PubMed] |
| 128. | Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 134] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 129. | Costa-Mattioli M, Sonenberg N, Richter JD. Translational regulatory mechanisms in synaptic plasticity and memory storage. Prog Mol Biol Transl Sci. 2009;90:293-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 130. | Groisman I, Huang YS, Mendez R, Cao Q, Richter JD. Translational control of embryonic cell division by CPEB and maskin. Cold Spring Harb Symp Quant Biol. 2001;66:345-351. [PubMed] |
| 131. | Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes Dev. 2006;20:2701-2712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 132. | Burns DM, Richter JD. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 2008;22:3449-3460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 114] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 133. | Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447-456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 134. | Rouhana L, Wang L, Buter N, Kwak JE, Schiltz CA, Gonzalez T, Kelley AE, Landry CF, Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 85] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 135. | Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641-651. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 265] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 136. | Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000;404:302-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 286] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 137. | Sarkissian M, Mendez R, Richter JD. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev. 2004;18:48-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 138. | Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA. 2004;101:4407-4412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 117] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 139. | Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 381] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 140. | Stöhr N, Köhn M, Lederer M, Glass M, Reinke C, Singer RH, Hüttelmaier S. IGF2BP1 promotes cell migration by regulating MK5 and PTEN signaling. Genes Dev. 2012;26:176-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 141. | Liao B, Hu Y, Herrick DJ, Brewer G. The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem. 2005;280:18517-18524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 170] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 142. | Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657-2675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 576] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 143. | Boudoukha S, Cuvellier S, Polesskaya A. Role of the RNA-binding protein IMP-2 in muscle cell motility. Mol Cell Biol. 2010;30:5710-5725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 144. | Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199-207. [PubMed] |
| 145. | Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 465] [Cited by in RCA: 503] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 146. | Pan F, Hüttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to beta-actin mRNA during transcription. Mol Cell Biol. 2007;27:8340-8351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 147. | Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci. 2012;13:372-379. [PubMed] |
| 148. | Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1:214-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 338] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 149. | López de Silanes I, Lal A, Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2005;2:11-13. [PubMed] |
| 150. | Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc Natl Acad Sci USA. 2008;105:20297-20302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 183] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 151. | Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168-3181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 482] [Cited by in RCA: 482] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 152. | Gratacós FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA. 2010;1:457-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 155] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 153. | Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451-464. [PubMed] |
| 154. | Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279-1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 155. | Vázquez-Chantada M, Fernández-Ramos D, Embade N, Martínez-Lopez N, Varela-Rey M, Woodhoo A, Luka Z, Wagner C, Anglim PP, Finnell RH. HuR/methyl-HuR and AUF1 regulate the MAT expressed during liver proliferation, differentiation, and carcinogenesis. Gastroenterology. 2010;138:1943-1953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 106] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 156. | Kim HS, Headey SJ, Yoga YM, Scanlon MJ, Gorospe M, Wilce MC, Wilce JA. Distinct binding properties of TIAR RRMs and linker region. RNA Biol. 2013;10:579-589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 157. | Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383-5398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 809] [Cited by in RCA: 797] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 158. | Kuwano Y, Pullmann R, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225-238. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 94] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 159. | Ogilvie RL, Abelson M, Hau HH, Vlasova I, Blackshear PJ, Bohjanen PR. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J Immunol. 2005;174:953-961. [PubMed] |
| 160. | Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689-11699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 206] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 161. | Briata P, Chen CY, Giovarelli M, Pasero M, Trabucchi M, Ramos A, Gherzi R. KSRP, many functions for a single protein. Front Biosci (Landmark Ed). 2011;16:1787-1796. [PubMed] |
| 162. | Suswam EA, Nabors LB, Huang Y, Yang X, King PH. IL-1beta induces stabilization of IL-8 mRNA in malignant breast cancer cells via the 3’ untranslated region: Involvement of divergent RNA-binding factors HuR, KSRP and TIAR. Int J Cancer. 2005;113:911-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 83] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 163. | Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1). J Biol Chem. 2010;285:27182-27191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 164. | Lee JH, Jeon MH, Seo YJ, Lee YJ, Ko JH, Tsujimoto Y, Lee JH. CA repeats in the 3’-untranslated region of bcl-2 mRNA mediate constitutive decay of bcl-2 mRNA. J Biol Chem. 2004;279:42758-42764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 165. | Lossi L, Gambino G, Ferrini F, Alasia S, Merighi A. Posttranslational regulation of BCL2 levels in cerebellar granule cells: A mechanism of neuronal survival. Dev Neurobiol. 2009;69:855-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 166. | Ishimaru D, Ramalingam S, Sengupta TK, Bandyopadhyay S, Dellis S, Tholanikunnel BG, Fernandes DJ, Spicer EK. Regulation of Bcl-2 expression by HuR in HL60 leukemia cells and A431 carcinoma cells. Mol Cancer Res. 2009;7:1354-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 167. | Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45alpha by genotoxic stress. Mol Cell. 2006;22:117-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 168. | Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092-3102. [PubMed] |
| 169. | Campbell ZT, Menichelli E, Friend K, Wu J, Kimble J, Williamson JR, Wickens M. Identification of a conserved interface between PUF and CPEB proteins. J Biol Chem. 2012;287:18854-18862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 170. | Budkevich TV, Timchenko AA, Tiktopulo EI, Negrutskii BS, Shalak VF, Petrushenko ZM, Aksenov VL, Willumeit R, Kohlbrecher J, Serdyuk IN. Extended conformation of mammalian translation elongation factor 1A in solution. Biochemistry. 2002;41:15342-15349. [PubMed] |
| 171. | Kinzy TG, Merrick WC. Characterization of a limited trypsin digestion form of eukaryotic elongation factor 1 alpha. J Biol Chem. 1991;266:4099-4105. [PubMed] |
| 172. | Petrushenko ZM, Negrutskii BS, Ladokhin AS, Budkevich TV, Shalak VF, El’skaya AV. Evidence for the formation of an unusual ternary complex of rabbit liver EF-1alpha with GDP and deacylated tRNA. FEBS Lett. 1997;407:13-17. [PubMed] |
| 173. | Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419-431. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 174. | Friend K, Campbell ZT, Cooke A, Kroll-Conner P, Wickens MP, Kimble J. A conserved PUF-Ago-eEF1A complex attenuates translation elongation. Nat Struct Mol Biol. 2012;19:176-183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 175. | Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 1006] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 176. | Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami R. A Pumilio-induced RNA structure switch in p27-3’ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12:1014-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 322] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 177. | Miles WO, Tschöp K, Herr A, Ji JY, Dyson NJ. Pumilio facilitates miRNA regulation of the E2F3 oncogene. Genes Dev. 2012;26:356-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 178. | Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Ørom UA. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 557] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 179. | Bhandari A, Gordon W, Dizon D, Hopkin AS, Gordon E, Yu Z, Andersen B. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32:1497-1507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 180. | Tominaga K, Srikantan S, Lee EK, Subaran SS, Martindale JL, Abdelmohsen K, Gorospe M. Competitive regulation of nucleolin expression by HuR and miR-494. Mol Cell Biol. 2011;31:4219-4231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 181. | Srikantan S, Abdelmohsen K, Lee EK, Tominaga K, Subaran SS, Kuwano Y, Kulshrestha R, Panchakshari R, Kim HH, Yang X. Translational control of TOP2A influences doxorubicin efficacy. Mol Cell Biol. 2011;31:3790-3801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 182. | Köbel M, Weidensdorfer D, Reinke C, Lederer M, Schmitt WD, Zeng K, Thomssen C, Hauptmann S, Hüttelmaier S. Expression of the RNA-binding protein IMP1 correlates with poor prognosis in ovarian carcinoma. Oncogene. 2007;26:7584-7589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 183. | Elcheva I, Goswami S, Noubissi FK, Spiegelman VS. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35:240-246. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 184. | Mongroo PS, Noubissi FK, Cuatrecasas M, Kalabis J, King CE, Johnstone CN, Bowser MJ, Castells A, Spiegelman VS, Rustgi AK. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172-2182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 185. | Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 667] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 186. | Cohn DE, Fabbri M, Valeri N, Alder H, Ivanov I, Liu CG, Croce CM, Resnick KE. Comprehensive miRNA profiling of surgically staged endometrial cancer. Am J Obstet Gynecol. 2010;202:656.e1-656.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 187. | Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol. 2009;83:2357-2367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 188. | Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res. 2010;70:36-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 194] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 189. | Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416-6424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 388] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 190. | Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, Ogawa R, Harata K, Fujii Y. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34:537-542. [PubMed] |
| 191. | Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 347] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 192. | Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348-2360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 825] [Cited by in RCA: 899] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 193. | Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, Kim RY, Saria MG, Pastorino S, Kesari S. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689-700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 232] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 194. | Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199-227. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1217] [Cited by in RCA: 1212] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 195. | Yang R, Schlehe B, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, Volkmann J, Varon R, Weber BH, Niederacher D. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121:693-702. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 196. | Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11:R27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 341] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 197. | Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111-11116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 297] [Cited by in RCA: 314] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 198. | Chen G, Zhu W, Shi D, Lv L, Zhang C, Liu P, Hu W. MicroRNA-181a sensitizes human malignant glioma U87MG cells to radiation by targeting Bcl-2. Oncol Rep. 2010;23:997-1003. [PubMed] |
| 199. | Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X, Zarate R, Diaz-Gonzalez JA, Ramirez N, Sola JJ. microRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res. 2009;15:2281-2290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 284] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 200. | Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994-9000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 296] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 201. | Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 158] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 202. | Ren Y, Zhou X, Mei M, Yuan XB, Han L, Wang GX, Jia ZF, Xu P, Pu PY, Kang CS. MicroRNA-21 inhibitor sensitizes human glioblastoma cells U251 (PTEN-mutant) and LN229 (PTEN-wild type) to taxol. BMC Cancer. 2010;10:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 167] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 203. | Arnetoli M, Vooijs R, ten Bookum W, Galardi F, Gonnelli C, Gabbrielli R, Schat H, Verkleij JA. Arsenate tolerance in Silene paradoxa does not rely on phytochelatin-dependent sequestration. Environ Pollut. 2008;152:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 204. | Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015-32027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 412] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 205. | Borralho PM, Kren BT, Castro RE, da Silva IB, Steer CJ, Rodrigues CM. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689-6700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 206. | Svoboda M, Izakovicova Holla L, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541-547. [PubMed] |
| 207. | To KK, Robey RW, Knutsen T, Zhan Z, Ried T, Bates SE. Escape from hsa-miR-519c enables drug-resistant cells to maintain high expression of ABCG2. Mol Cancer Ther. 2009;8:2959-2968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 208. | Bijsterbosch MK, Rump ET, De Vrueh RL, Dorland R, van Veghel R, Tivel KL, Biessen EA, van Berkel TJ, Manoharan M. Modulation of plasma protein binding and in vivo liver cell uptake of phosphorothioate oligodeoxynucleotides by cholesterol conjugation. Nucleic Acids Res. 2000;28:2717-2725. [PubMed] |
| 209. | Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 217] [Cited by in RCA: 234] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 210. | Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet. 2011;43:371-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 499] [Cited by in RCA: 507] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 211. | Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, Weidhaas JB, Bader AG, Slack FJ. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116-1122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 553] [Cited by in RCA: 518] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 212. | Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, Slack FJ. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695-E1704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 393] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 213. | Wu DW, Cheng YW, Wang J, Chen CY, Lee H. Paxillin predicts survival and relapse in non-small cell lung cancer by microRNA-218 targeting. Cancer Res. 2010;70:10392-10401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 214. | Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896-899. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1268] [Cited by in RCA: 1279] [Article Influence: 75.2] [Reference Citation Analysis (0)] |
| 215. | Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153-1162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 520] [Cited by in RCA: 521] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 216. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1290] [Article Influence: 86.0] [Reference Citation Analysis (0)] |
| 217. | Kapoor S. IMP3: a new and important biomarker of systemic malignancies. Clin Cancer Res. 2008;14:5640; author reply 5640-5641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 218. | Findeis-Hosey JJ, Xu H. The use of insulin like-growth factor II messenger RNA binding protein-3 in diagnostic pathology. Hum Pathol. 2011;42:303-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 219. | Suvasini R, Shruti B, Thota B, Shinde SV, Friedmann-Morvinski D, Nawaz Z, Prasanna KV, Thennarasu K, Hegde AS, Arivazhagan A. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J Biol Chem. 2011;286:25882-25890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 220. | Liao B, Hu Y, Brewer G. RNA-binding protein insulin-like growth factor mRNA-binding protein 3 (IMP-3) promotes cell survival via insulin-like growth factor II signaling after ionizing radiation. J Biol Chem. 2011;286:31145-31152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |