INTRODUCTION
Our mainstream picture of RNA virus evolution is determined by the quasispecies concept of Manfred Eigen. It was a very helpful theoretical model within the framework of physical chemistry to paint a picture of viruses with an evolution rate a multitude faster than evolution of cellular organisms. The core assumptions of quasispecies concept[1-3] are rather clear: (1) there is little knowledge about the origin of viruses or their role in the evolution of the biosphere. Viruses are selfish genetic elements that likely originated out of host organisms (escape theory); (2) viruses represent molecules that generate “information” through mutagenesis, i.e., replication-errors; (3) equations of mass action law govern chemical equilibrium in closed chemical systems. Percentage of components within this is determined by these equations; (4) in an environment of high mutation rate and thermodynamic conditions far from equilibrium, self-replicating entities reach maximum reproductive fitness; (5) the self replication entity is not a single molecule but a “cloud” that consists of variant reproductive molecules; (6) the distribution of these “clouds” in systems far from equilibrium depends on master fittest type and mutant spectra, i.e., dominant replicators, mutants closely related, and not closely related. Each of its sequence-syntax occupies a unique position in the sequence space; and (7) because the genetic code is a real language-not just a metaphor-information- and system-theoretical assumptions based on mathematical concepts of language therefore are the appropriate tools to explain quasispecies evolutionary dynamics.
If we look at the current empirical data, these assumptions do not explain many interactional behaviors and broad influences that have been observed in viral studies. In addition, the following issues are relevant: (1) viruses play large and essential roles in the biosphere evolution of host[4-7]. Phylogenetic analyses and comparative genomics suggest that viruses are older than cellular life[8-10]; (2) most viruses are symbiotic or even endosymbiotic inhabitants of cellular host organisms that can promote adaptability for the host[11-15]; (3) information representing natural codes is not the result of errors in replication processes. All empirical data indicate that natural codes depend on consortia of living agents that generate and use codes for information storage and interactional signalling. Natural codes are essential pre-requisites of everyday social interactions. If errors in natural code use occur, this does not enrich information but nearly all empirical data indicate information damage, deformation and decreased informational content. Today it seems rather curious to ground evolution of complexity on error and damage[16-19]; and (4) coherent with Ludwig Wittgensteins assumptions, the meaning of natural code sequences is its use (functional activity), i.e., depends on the context, not on its syntax[20]. Accordingly the meaning (functional activity) of syntactically identical viral nucleic acid sequences varies according to context such as epigenetic imprinting[21-23].
These observations have serious consequences for the “every variant” concept of Eigen’s quasi-species that include: (1) biological information of nucleic acid sequences does not occupy a unique position in sequence space but depends on contextual use; (2) because of its context-dependency biological information cannot be sufficiently described by information theory or similar mathematically based concepts of language; (3) (Evolutionary) algorithm-based machines cannot provide contextual real life simulations; (4) sequence space of real life nucleotide sequences is not the result of random assemblies; and (5) viral cloud building in natural habitats occurs different in comparison to abiotic molecule assembly.
Additionally quasispecies that cooperate can have various behavioural motifs not present in a pure physical chemistry. They can (1) compete with and exclude related populations; (2) have minority populations that are crucial for overall fitness[24,25]; (3) can display heterogeneity important for fitness that is not observed in the consensus type[26]; (4) can suppress their own replication through lethal defection[27]; and (5) can be composed of members that can complement and interfere with replication of the collective and many of these features can be observed in clinical infections such as humans with hepatitis C virus[28].
Therefore if quasispecies evolve with the above characteristics and are thus different from prior mathematic models, what then is an appropriate description that is also coherent with the abiotic/biotic split in animated nature? If species is the appropriate term in biology to describe essential common features of related groups of cellular organisms, then quasispecies remains an appropriate term to describe related groups of subcellular agents that play essential roles in evolution of the biosphere.
To denote the above crucial differences to the 20th century concept with its paradigmatic core of master copy (fittest type) and mutant spectra (variants) we propose the concept of quasispecies consortia (qs-c) in which behavioural motifs of cooperative RNA-agents are at the foundation of basic capabilities generating sequence space of de novo nucleotide sequences and for inserting, changing or deleting such sequence into host sequences.
FROM MOLECULAR ERRORS TO INTERACTIONAL MOTIFS: RNA PARASITES AS OPEN SPACE INVADERS
In most origin of life scenarios, RNA parasites are considered major barriers for the origin of code and life (systems) which compels these proposals to close off the action of parasites into self contained code systems. But RNA parasites can also provide new and highly dynamic code that is added to the system. Naturally evolved RNA sequences can never be completely specified (or closed), since they must interact with their environment, replicate and undergo adaptation while retaining code that can always be further parasitized. Open systems can thus embrace the capacity of parasites to add novelty. This contrasts sharply with closed systems which must limit all such parasites.
This open feature renders the ability to absolutely specify membership (absolute immunity) as basically indefinable. Any naturally evolved nucleotide sequence can never be fully secure from as yet undefined parasite agents. But a crucial inference out of this “insecurity” is that parasites provide the inherent capacity for novelty, i.e., the precondition for greater complexity. Since parasites are competent in code, they are not “mistakes”. This means that the accepted core requirement of biological innovation, variation, is not met by the explanation model of random mutation (error) and its selection. Instead, RNA parasites provide continued infection and colonization which result in added identity of new RNA to existing RNA groups and thus alter self identity (immunity). These RNAs are thus acting as competent agents-not mutations/errors-which seek to impose new and competent code (identity) onto the system. Such RNA’s, however, must interact to attain a stable colonization by inhibiting run-away parasite replication.
The core issue is thus to specify how RNA “agents” emerge from chemicals (ribozymes) to form the needed identity (such as for replicators) and also to form RNA groups that can support themselves and learn new membership.
For all extant life, these agents must have initially been RNA stem-loops. Single RNA stem-loop generation occurs by physical chemical properties solely as demonstrated by natural and randomized RNA experiments[29-31]. If numbers of stem-loops are able to build complex consortia with greater competence, they would then represent the initial cooperative interactions needed to develop living systems that are not present in a strictly chemical world. The resulting system must function to maintain itself. In consortia, the emergence of identity (ability to differentiate self vs non-self) is a crucial initial step. Thus we seek to understand how single RNA stem-loop RNAs can become competent RNA consortia. And in so doing, we invoke the central action of RNA parasites and follow how “parasite-derived” RNA stem-loops interact in social collectives promoting innovation, infection, immunity and complex multiple (group) identity[32].
CORRECTIVE AND COLLECTIVE POWER OF VIRUS VIA QS-C
The term quasispecies originated from models that described related viral RNA populations resulting from error based variation of the master fittest type[1-3]. It was not initially applied to consortia that showed cooperation. In the ensuing several decades, many laboratory observations were made that indicated more complex collective behaviours for viral quasispecies than were predicted by Eigen’s quasispecies equations. Two of the more active laboratories were those of John Holland and Esteban Domingo[33]. The most recent compilation of these studies outlines many of the collective behaviours that have been made with quasispecies[34]. The culmination study that most clearly reported that quasispecies have more complex collective behaviours seems to be the study from the Andino group of poliovirus pathogenesis in a mouse model in which diversity and cooperation were key to viral fitness[35,36]. Such studies led to the set of statements above on the cooperative nature of quasispecies. Thus quasispecies are collectives that have positive and negative interacting members that are bound together for a combined fitness that depends on diversity[36-38]. It is thus ironic that it is from the viruses, assumed for decades to be the most selfish of all genetic entities, we observe the characteristics of cooperative, collective behaviour. And it was the “fittest type” assumptions of Manfred Eigen[1] that generated quasispecies equations and theory which stimulated the development of this modern collective quasispecies view for over 40 years. But we are left with a conceptual contradiction. Modern quasispecies observations do not depend on the master (fittest type) and the consensus sequence. Consensus sequence may not predict the fitness of the diverse collective. In contrast to this diversity itself seems crucial.
QS-C PRODUCE HIGH RATES OF DIVERSITY: THIS IS NOT ERROR
With this clarification, it should become apparent that all RNA replicators (especially simple ones) must have high rates of diversity generation (not error). Novelty is then generated from new combinations of this diversity. Indeed, it sounds curious to use the term “error prone” for the high production rates of sequence novelties. An error is an inferior (“less better”) variant of an extant sequence. Such errors should only provide rare incremental improvement and be much less (if at all) able to generate de novo networks. With this error concept we also apply terms such as “damage”, “defect” and “incomplete” to variant information.
In contrast to this, cooperative RNA quasispecies produce and configure sequence novelties that are members of coherent populations and must generate an interacting diversity as prerequisite of innovation (variation), the driving forces of evolution. As an analogy, we might apply the limits of the “error” concept to innovative human endeavours. For example; poets produce novel poems by reconfiguring the commonly shared vocabulary. Must they also be error producers? Similarly, music composers and all artists in general produce novelty from common combinatorial rules and existing basic material tools. Do they also operate as error prone agents of fine arts? As we will see below, the qs-c concept requires interaction and diversity so it can even help us understand these processes of human innovation. But we must also apply the shared nature of “agents” that can create novelty. We will now consider how stem-loop RNAs can help us understand “agents” and their shared common use.
But in addition to stem-loop RNAs, all genetic entities that replicate via RNA will also be prone to qs-c (collective) behaviors. These behaviors will include cooperative and competitive interactions. RNA, however, is not simply providing syntax for genetic information. It is more than code. It can also provide: (1) structure (stem-loop); (2) identity (stem-loops, 5’, 3’ ends); and (3) functional (ribozyme, endonuclease, ligase) activity.
And it may be dynamic (e.g., pseudoknots). Because of this extended capacity relative to DNA, RNA can be considered as a more active entity, with behaviours that make it able to function as an “agent” to affect its own activity and survival[39,18]. At this point we adapt the framework from pure physics and chemistry to emergent biotic agent-based group building. In that light, DNA can be considered as a habitat for various RNA agents. It was from this perspective that we proposed that DNA should be considered as a habitat for these active RNA agents[18,32]. But this discussion of simple RNA replicators suggests that the concept of qs-c should also apply to the ideas and experiments concerning the “RNA world” hypothesis. Yet curiously, very little “RNA world” research has addressed any issues regarding quasispecies[40-42], let along the more modern qs-c idea. As many are starting to think that life originated in a cooperating situation[43], it is worth briefly considering if the qs-c concept will provide a different scenario for the origin of life.
RNA WORLD RECONSIDERED: INFECTIOUS STEM-LOOPS THAT OPERATE VIA QS-C
To evaluate the qs-c and infectious perspective on the RNA world hypothesis, we apply and explore the RNA-agent concept introduced above to the role of stem-loop ribozymes in the origin of life. The main objective is to incorporate the historically absent qs-c and parasitic perspective (with its inherent feature for group fitness) into the process that creates RNA societies. We will not explore early chemical evolution that might have led to the emergence of RNA molecules, but will instead assume RNA has come into existence and follow its features from this perspective.
One immediate consequence of this perspective is that we will be focused on collective features of RNA populations and will thus evaluate the chemical consequence of ribozyme qs societies, not individual replicators. This foundation immediately creates a situation in which “systems” of molecules with multiple behaviours will have the primary role in promoting the origin of life. It will also be important early on to consider how these systems maintain coherence (group identity, presented below), as this is an essential feature. Indeed, a basic and continuing theme will be that a core function of stem-loop RNAs is to provide molecular identity through all of evolution, including recent human evolution. This identity theme will persist throughout this chapter and will be frequently reintroduced.
The idea is then that individual members of stem-loop RNA societies were collectively able to invade (ligate into) each other to form a more stable and capable (ribozyme active) consortia with emergent, transformative and unpredictable abilities. These collectives would lead to the origin of various ribosome- and RNA cellular societies (still linked to its stem-loop tRNA origin). Such a scenario also introduces the basic role of cooperation in the origin of life. It does not, however, eliminate competition, preclusion or extinction which are also inherent features of qs-c behaviors. Furthermore, the identity and transmissive role for stem-loop RNAs set the early (precellular) foundation for the origin of viruses whose emergence will further drive host evolution via colonization.
The cooperative and parasitic features of qs-c will also promote the early participation of peptides in the identity and evolution of the RNPs. The maintenance of these RNPs as a coherent collective will generally be mediated by addiction modules, which underlie group identity and immunity in all living systems. Addiction modules are counterbalanced (former competing) genetic parasites which share a persistent life style in host genomes. Addiction modules are clearly the result of stable consortial interactions[4,44-46].
With this foundation, the emergence of genes, DNA, cells and individual fittest type selection can all be derived. But the emergence of DNA and cells and Darwinian evolution do not terminate the central role for transmissive RNA societies in the evolution of life. DNA becomes a habitat for these stem-loop “identity” RNAs and it is from this perspective that we can subsequently examine recent events in human evolution.
One issue should already be clear: This scenario posits that collective and cooperative behaviours were and remain essential for the emergence of living complexity. Qs-c then provides a conceptual foundation for the study of cooperating chemical biotic (in contrast to abiotic) networks in which mixtures of self-replicating RNA ribozymes can form highly cooperative and dynamic autocatalytic cycles[29,31]. Let us now put this into the perspective of virolution[11].
TO SURVIVE RNA STEM-LOOP REPLICATORS MUST FORM POPULATIONS THAT DYNAMICALLY GENERATE IDENTITY
In the origin of the RNA world, short RNA oligomers formed by chemical processes needed to become longer RNAs able to perform template based catalysis. It has been proposed that the initial chemical formation of hairpin-like RNAs (stem-loops) could provide ribozyme activity following a ligation based modular evolution that would yield ribozyme auto catalysis[47]. Indeed, below we present a series of studies that support this modular view.
But according to the parameters of qs-c evolution, for a consortium of RNA stem-loop replicators to survive, they must form a coherent population. They must share their identity and survival. The recognition of the stem-loop sequence itself by catalytic agents could provide such common identity. Alternatively, chemical markers or initiators of catalysis could also mark the common population for priming or replication. Thus it is very interesting that the smallest ribozyme so far reported consists of just 5 nucleotides able to catalyze aminoacylation of the 3’ end[48].
The addition of an amino acid to an RNA molecule has many interesting chemical implications. A ribozyme has rather limited chemical potential compared to proteins. This is mostly due to proton disassociation constant of various amino acid moieties which are not close to pH neutrality. Thus amino acids are much more capable as chemical catalyst for this reason. Without the participation of amino acids, ribozymes must attain complex folds, often with some dynamic character (pseudoknots) to be effective catalyst allowing them to cleave and ligate RNA. Given this chemical advantage, we might expect that RNA evolution was greatly facilitated (but not coded) by peptides that contribute catalytically. In addition, such a modified RNA would likely also provide a chemical marker that could distinguish this RNA population. Indeed this molecular identity idea is developed below as a way to better understand the origin of tRNA and its role in initiating replication of so many RNA viruses, as well as how this chemical marker could promote the symbolic genetic code.
BASIC GROUP-BUILDING OF RIBOZYMES THROUGH SELF-LIGATION OF RNA STEM-LOOP MODULES
A good starting point for the accumulation of complexity seems to be hairpin ribozymes whose activity can be controlled by external effectors[49]. Structural variation in these ribozymes allows progeny RNA to have different functions from their parental RNAs. The objective is to replicate RNA with RNA which hairpin ribozymes can perform as a sequence of ligation reactions that produce a longer ribozyme[50]. Along these lines, two short hairpin RNAs can catalyze their own ligation to form larger RNA constructs[30]. Thus we see interactions that promote more complex progeny. However, for a fully active ribozyme, complex RNA folding is needed. And such folding is cooperative[51].
Folded ribozymes can also interact with other small molecules promoting their function as riboswitches[52,53]. This includes amino acids which could promote either catalytic control or group identity marking. And the ribozyme folds can also be dynamic and context sensitive as seen in pseudoknots[54].
But ribozymes can also be invasive, including self invasive[55]. Thus stem-loop RNAs have many behaviors that would allow them to function as an identity group of agents involved in their own recognition and synthesis. Of particular interest is their ability to self-ligate[30] as this could promote the emergence of RNA societies with self-identity. We can also think of tRNA as stem-loop RNA with various functions and histories. Indeed, it appears that tRNAs evolved from two separate hairpins[56], in which each of the stem-loops interacts with a different ribosomal RNA subunit (presented below). This is a very interesting observation from an RNA society perspective. The invasive nature of intron ribozymes (endonuclease) also applies to tRNA from archaea, but here four distinct specificities are known[57]. This very much resembles an identity system in which introns are marking central cellular (self) agents (tRNAs) for group identity but should destroy similar tRNAs (viral, etc.) lacking the intron marking. It is thus also interesting that tRNA with various linked amino acids themselves have been proposed to have originated before the translation system as genomic 3’ tags needed for RNA ribozyme replication[58-60]. This early function can also be explained as having served as a tag for group identity and could better explain the polyphyletic nature of the origin of tRNA[61].
Interestingly viroids, the smallest virus-like agents, which infect plants, have striking similarities to Hepatitis Delta Virus[62]. This virus, the smallest genome of any animal virus, uses circular genome, secondary structure folding and replication by a rolling circle mechanism that is catalyzed by host enzymes and cis-acting ribozymes. Mobile genetic elements are also typical self-splicing ribozymes. They excise their own RNA, from precursors thus support their own identity. The module like structure is evident. The viroids rely only on RNA and its structural motif, so it is really the sequence itself and its secondary structure which represents the entire infectious agent[62].
THE ROLES OF RNA STEM-LOOP VARIATIONS IN QS-C BUILDING
This inherent capacity to form stem-loops with loops, bulges, junctions that are not immediately repaired or corrected opens the possibility to build abundance of varieties, which alter compositional patterns, identities, immunities and the whole row of progeny within a qs-c.
Let us consider a single stem-loop RNA that undergoes several rounds of replication. Potential identity effects of diversity may integrate the following: (1) altered self; (2) new interaction with other RNA; (3) complementing replication; (4) interfering with replication; (5) serving as primer for new replication pattern; (6) serving as target for ligase; (7) serving as target for cleavage; (8) serving as target for integration; (9) serving as initiation point of strand opening; and (10) serving as interaction point with peptide.
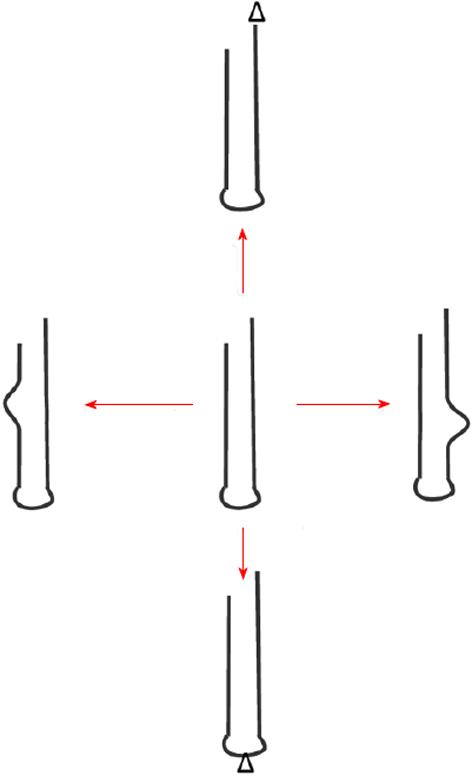
A variety of combinations of the above listed outcomes multiplies identity-generating and identity-shifting effects (Figure 1).
Figure 1 Self-generating RNA-stem-loop innovations: Generation of diversity (innovation) is shown.
This is different from errors (accident, damage). Red arrow denotes diverse products from central template by low fidelity replication. Bulges and triangles denote changes.
Each replication event-necessarily being low fidelity-produces its own peculiar version of diversified progeny, e.g., with a new “bulge” in the stem. This bulge then becomes available to provide a whole array of possible outcomes (including contradicting ones).
It might: (1) interact with the original template to either compliment or inhibit it; (2) provide an interaction point for other RNA progeny (including itself); (3) provide a target site for cleavage or ligation; (4) act in combination with other progeny to provide more complex catalytic (ribozyme) function; and (5) alter or provide a binding site to other participants, such as peptides (RNPs).
In other words, even a singly new altered RNA now has a whole array of possible and multiple usages (positive and negative). Whole actual use will depend on the circumstances and history of the population it is in, i.e., actual use depends on context.
If we add to this all the other diverse RNA progeny from these few rounds of replication-all in their own peculiar RNA region-all providing their own peculiar potential for use, we start to see the multiple potential for each individual RNA.
With such a scenario this combinations of possibilities very rapidly become too complex to follow the fate (fitness, usage) of any particular RNA. But this is the wrong, since it is an application of linear thinking. Instead, we need nonlinear thinking. Thus, if we think in sociological terms, then the RNA population (quasi species) can be considered as a “culture” that retains a common language which provides a level of group coherence (qs selection) on the basis of compatible cooperative organization. Each individual diverse RNA then becomes like a potentially new word for that language, i.e., new agent in the ensemble of interacting agents.
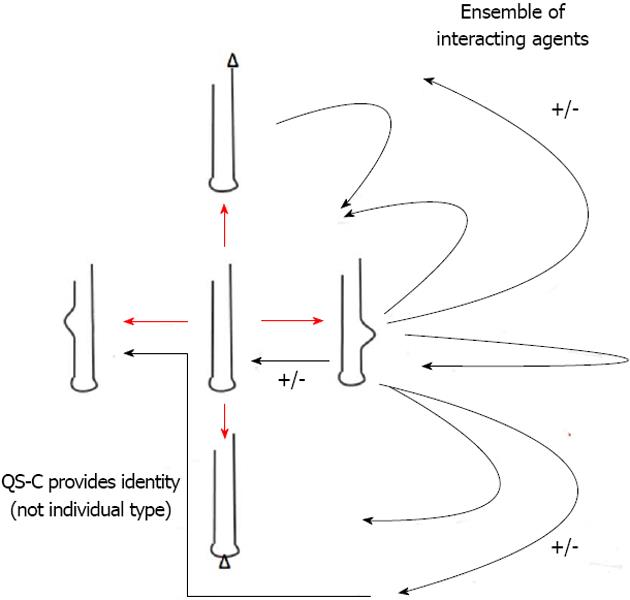
The “culture” is then free to use a variant RNA (possibly even with multiple “meaning”), however it can also reject it. In this case, the term qs-c can have yet another meaning, c for culture (Figure 2).
Figure 2 Crucial difference of quasispecies consortia with former quasispecies-concepts (fittest type-mutant spectra) is the basically consortial organisation of functional RNA ensembles.
Shown above are the possible consortial interactions (black arrows) of just one the diversified RNA-stem-loops. These multiple activities (shown as +/-) preclude individual fitness definitions but require emergence and adaptation of group membership-identities. Defectives with similar subviral RNA-(stem-loop) groups remain relevant in both evolutionary and developmental processes. As a result of this basic evolutionary process of RNA stem-loop consortia building we can look at the emergence of de novo identities. QS-C: Quasispecies consortia.
These RNA uses will also vary considerable with the history of prior RNAs as well as any possible interactions with outer RNA quasi species. And the use can also vary (and be lost) with time as the culture adopts new meanings.
RNA-STEM-LOOPS AS MODULE-LIKE POOL
RNA stem-loops thus serve as multiple use and re-useable tools in RNA secondary sequence structure. Once being invented/generated such a single stem-loop may become part of another stem-loop consortium. This leads us not solely to the abundance of small and long non-coding RNAs that serve as regulatory tools in all known relevant cellular processes, but also to recombination processes that drive evolution[63-68]. As a result we then can find a changed identity of the new consortium with altered features.
FROM RNAS TO RNAGENTS: FROM ABIOTIC CHEMICAL REACTIONS TO BIOTIC GROUP BEHAVIOUR
We have seen that from single stem-loop RNA populations of diverse stem-loops that acquire multifunctional “agent” behavior (RNAgents) can be generated. The emergence of RNAgents leads us to the emergence of life and life worlds. To understand the life-worlds of RNAgents means to understand a fascinating sphere of what was formerly not assumed to be possible: The mere sequences of molecules do not behave like physical/chemical entities in an abiotic world but behave as competent agents on genetic code syntax that cooperate and organize, constitute and innovate sequence structures[69,70]. Only groups of RNA stem-loops underly such selection. Single RNA stem-loops behave like a random assembly of nucleotides without selective forces[29,71,72]. Only if they are ligated to groups, i.e., consortia, are they competent. This means they share a culture of interactional patterns and a history of defined time scales[30,73]. Both culture and history are strongly influenced by ecosphere habitats that underlie selective forces and therefore are not reproducible features of abiotic matter.
The basic module of such RNAs are their complementary composition of base pairing stems and not-base pairing loops which results of an inherent property of RNA ensembles, the fold back to complementary polyRNAs, as demonstrated by the variety of ribozymes[74]. Also the genomes of plus strand RNA viruses are able to form secondary and higher-order RNA structures[75]. Interestingly, recent experiments demonstrated that single self-replicating RNAs are not as successful as cooperative ensembles and as a result cooperative RNA consortia outcompete selfish RNAgents[31]. Also, diversity in the backbone of RNA replicators seems essential to allow strand separation and replication[76]. Thus group cooperation is key at the very origins of RNA societies (this is coherent with empirical knowledge about the emergence of natural codes/languages: they are essentially social group features not solus ipse results; see Witzany 1995)[16].
Additionally, such RNA stem-loop consortia as found in long noncoding RNAs take multiple roles in RNA binding, DNA-binding, conformational switch and protein binding. Their modular nature results in functions such as control regimes, scaffolding to RNPs as well as guiding in target recognition within genomes[53]. It is very clear now that non-coding RNAs build a complex layer that determines the regulation of all steps and substeps of gene control in complex organisms[18].
SINGLE RNAGENTS JOIN GROUPS AND NETWORKS: THE FEATURES
The origin of spontaneously cooperating networks of stem-loop RNA replicators can be understood from the qs-c perspective in which cooperation is the essential behavioral motif that outcompetes selfish behavioral patterns[31]. Thus we see the emergence of networks at early stages in the evolution of life. In this review the term network will be used to include some distinct features, specifically network membership. Basically, for a network to be coherent and able to act collectively, it must limit membership to promote coordination. Otherwise it is simply a collection of uncoordinated agents and there will be no selection for maintaining the network coherence or existence. If we are examining a network composed of stem-loop RNAs, it will be necessary for the individual RNAs to have some feature or behaviors that maintains membership such as RNA replication and recognition. This requires interaction. If only one type of RNA is supported (e.g., high fidelity replication), there can be no complementation and complex function (i.e., ribozyme) for the collective. A diversity of behavior and type will be essential. Recall however, that these RNAs act as agents in which various (multiple) behaviors will be possible even for the same sequence. This means there is diversity of interaction as well as diversity of type is inherent to the network. Thus overall interaction of an RNA agent with the collective must promote coherence and continued existence. What then are the features that promote continued existence (selection) for a network?
This does not require that only positive (e.g., replication) interactions be supported. Negative interactions, including interference will also be needed. For example, highly efficient run-away replicons would overtake a qs-c and yield only one RNA type. Thus the qs would lose complementing functionality and would also consume all substrates if they were not regulated. This situation presents a problem in those habitats with limited substrates which likely is a very common state. Therefore, some level of self-regulation (negation) in the collective would promote the survival of the collective, especially if these RNAs could interact with the substrate in a regulatory, e.g., riboswitch-like manner.
That efficient replicators become susceptible to parasitic replicators would provide an inherently spontaneous process of self-regulation. Yet the collective will still need to promote replication when it is favored. Accordingly, it becomes important for members of the collective to be subjected to both positive and negative self-regulation via RNA-RNA interactions. However, here too there must be some limits to self-regulation as the collective cannot tolerate overly active self-regulating members that will extinguish the collective. Thus we see that being a successful member of a collective requires many (and multiple) behaviors associated with it.
On top of that, as a qs-c replicates, these features will drift with time in a dynamic manner. In this context we can see that a random RNA stem-loop or a stem-loop RNA from a different qs collective would likely not be coherent with the other members of a particular qs.
A qs society is generally rather specific for its members. Group selection has already occurred in generating the qs-c. Indeed, as many experiments with RNA viruses infecting humans and animals have shown, a particular qs will exclude other qs of the same virus, such self exclusion can provide an origin for immune functions[44,77]. Such society membership is also time dependent in that the serial passage of the same viral qs will usually result in subsequent qs that preclude prior individual members. This behavior has often been called a “Red Queen” behavior, but such a classical neo-Darwinian view does not incorporate or acknowledge the issue of group membership (qs-c coherence).
GROUP MEMBERSHIP IS THE PREREQUISITE FOR SELF/NON-SELF DIFFERENTIATION COMPETENCE
An important consequence out of this perspective is that the membership view allows us to understand the maintenance of minority types in the collective since these members can provide a needed but complementing catalytic control. Thus a qs society is a network that will naturally promote the emergence of membership, not the destruction of minority types. The important side effect is, that defective interfering agents can also contribute to membership control.
As previously proposed[45,46], group membership can also be promoted by the combined action of toxic agents linked to antitoxic agents. A common version of a toxic agent is an endonuclease that will cleave sequences that are recognized as foreign. The antitoxin in this case prevents the action of the endonuclease (e.g., via a bound protein or methylated base, dsRNA with another molecule, altered RNA fold, etc.). In this light, the endonuclease and ligation activities of stem-loop ribozymes are particularly interesting. A stem-loop ligase could provide a mechanism to recognize non-member stem-loop RNAs and destroy them by ligation.
Recall, however, that serial ligation can also be used to copy a stem-loop RNA. But such a situation has several very interesting implications. One of the problems with a society of stem-loop RNAs is that to attain their combined function, they need precise physical molecular placement relative to one another. This would normally require a high concentration dependence to counteract diffusion. By ligation, however, we could build a society of stem-loop RNAs that have covalently placed the various stem-loops in the correct functional (or dynamic/regulatory) context and have lost their concentration dependence. It seems likely that such a process would involve invasive self-colonizing stem-loop RNAs that results in one molecular entity with a common identity function. This would generate one entity that evolved from the ligation of a mixed set of stem-loop agents that now have a highly enhanced (collective) functional capacity. This collective would also have a highly enhanced capacity for persistence as it need not continually replicate individual stem-loop RNA agents to maintain its membership. The collective, however, would still need to oppose non-member or other parasite participation. Additionally, a collective might attain a conditional (regulated) replication capacity if it incorporates stem-loop RNA riboswitches. It is by such a process that we can now consider the origin of the ribosome.
BEFORE AND AFTER RIBOSOME EMERGENCE
A big problem with thinking that viruses are essential agents for the emergence of life, however, is the ribosome[4,9,78-81]. The ribosome really defines the cell from virus and seemed to preclude virus from early evolution of the cell[80]. It is now clear also that the ribosome acts as ribozyme[82]. Yet the ribosome itself is an ensemble of two rather complex stem-loop societies of riboagents[83], an ensemble that became “set” with the invention of DNA[84]. Therefore it makes sense now to re-evaluate the RNA virus first hypothesis[4] from the before ribosomal world (BR) and the after ribosomal world (AR). Of course, common themes of consortia, symbolic code, quasispecies, addiction modules, group identity, membership agents would provide the themes that could link the great BR-AR divide. Briones et al[47] developed an interesting four step model on this in scenarios of evolution on both mineral surfaces and inside vesicles, such as: (1) abiotic polymerization of RNA oligomers; (2) folding of the RNA oligomers and ubiquity of hairpin structures; (3) ligation based modular evolution of RNA and finally; and (4) template-dependent RNA polymerase. This unites the history and culture-dependent derived stem-loops integrated in the two ribosomal subunits[85].
MEMBERSHIP IS CRUCIAL FOR LIVING NETWORKS (SYSTEMS) TO EMERGE
In examining the literature relevant to qs, the RNA world and RNA network formation, we can indeed find some experimental evidence that supports qs and the spontaneous emergence of RNA networks. But almost completely lacking from such experiments is any evaluation of the membership issue. For example, quasispecies-like behavior has been observed with in vitro RNA replicator studies[86]. Non-enzymatic template (peptide) directed autocatalytic systems can show network behavior[87] and communities. RNA ribozyme replicator sets can also show lateral evolution[88]. Also rule-based computing simulations have been applied to similar systems in an effort to understand the emergence of parasites and antiparasites[6]. Along these lines, the hypercycle kinetic model was proposed to be a system of cross catalyzing RNA replicators which depend on cooperation for growth[3], but this is not a collective autocatalytic system as proposed above[89]. Hypercycles as proposed are not able to tolerate parasites, let alone depend on them for development. Yet the biggest problem of all such studies is that there is no assumption regarding the basic importance of network or group membership.
Without this network membership concept and its attending dynamic strategies, authentic collective action does not emerge, systems do not develop. The dynamic nature of network membership and collective action pose many unsolved problems for existing qs theory. For example, how is the multi-potential of an individual RNA to be evaluated within the qs-c if we cannot specify all the other interactions and how they change with time? We cannot apply our current ideas of fitness to this individual RNA as the historical and population context is key. Network membership needs to be prominently considered if we are to understand the origin of the ribosome and the genetic code.
Replicator identity marking via 3’ aminoacylated of stem-loop RNAs appears most able to explain the origin of a tRNA mediated genetic code. For in contrast to Darwinian evolution, network members will generally have multiple ancestral histories. These members will mostly originate from separate parasitic lineages that were able to penetrate defenses and join the network sometimes in mixtures. They don’t need to descend from one individual or even be from the same type of agent (virus, transposon, intron, intene, etc.).
From this perspective we can understand why the two halves of tRNA have distinct evolutionary histories, yet tRNA is a core agent for evolution of life. Thus neither the amino acid based (peptide) ancestors nor the RNA based ancestors need a common origin to participate in a symbiogenic network. Our qs-c concept supports such a network process and-additionally-network membership provides the basis for examining noncoding RNA based regulation needed for multicellular complexity[90].
VIRAL CORE COMPETENCIES: INNOVATION, INTEGRATION, REGULATION, EXAPTATION
Because infection derived domestications, such as the whole variety of retroposon derived non-coding RNA, are now known to be transcribed and to control the regulation of genes[91], these parasitic agents shape genome architecture and function-arrangements. We now realize life on planet Earth has always resided within a virosphere and that the evolution of species depended on the virosphere features of innovation and transfer[4,39]. Humans share similar gene number as C.elegans. And humans and mice share 98.5% coding DNA. Regulatory complexity thus seems to characterize the evolution of more complex eukaryotes and stems form parasitic elements. The remaining 1.5% protein coding information is differently regulated via these species specific non-coding elements in increasingly complex ways. These elements act together through a variety of combinations, situations and mechanisms to re-shape genome/gene functions through epigenetic imprinting and re-printing[23]. These regulatory RNAs mostly retain the stem-loop structure. Also interesting are reports that ingestion of small RNA gene regulators seems to be usual route of RNA transfer[92].
In this perspective we have applied qs-c concept to explain complex regulatory network formation in the origin of life and the cellular protein world of higher organisms. The real species that determine all these evolutionary patterns are viruses and virus-like (infectious) RNA qs-c[32,93,94].
Viral core competencies necessitates that they be competent in host genetic and epigenetic code. These competencies provide innovation, integration, regulation, exaptation of qs consortia to form networks in the host.
A very intriguing example of this is found in placentation of mammals[10,44]. From day 1-6 post fertilization, all vertebrate embryos are similar and divide to the morula stage. But on day 6 this morula hatches and becomes a trophoblast. From here on, embryo development differs significantly. The outer cell layer is the first committed tissue of the embryo and will become the placenta. Viruses have a real affinity (tissue specificity) for this layer. This is exactly where very high endogenous retrovirus (ERV) activity is found[95-97]. This tissue has been repurposed to invade the uterine wall, suppress mother’s (host) immune response, promote blood feeding (exchange), and alter mother’s behaviour and physiology and brain. About 1500 placental genes are thought to have been modified by altered (ERV mediated) network re-regulation[98]. In our own view, it was likely that a collection of ERVs were involved. Other viral agents associated with reproductive biology that we don’t yet understand, were also likely to have been involved.
Consider the example of human immunodeficiency virus 1 (HIV-1) as a virus that requires qs diversity and dynamics to solve very complex problems and dynamic situations. If we think of HIV-1 as a fittest type agent with only about 10000 bp of clonal RNA, such a “pure” virus could not defeat the complex human adaptive and innate immune systems. An HIV-1 limited to the “master copy” fittest type (consensus sequence from a successful human infection) would be unable to generate a qs-c, and would rapidly be eliminated by our immune system and pose no problem for vaccine development. Here, qs-c is providing fitness. It is this power of the consortia to defeat complex systems that when applied to stem-loop RNAgents can also provide the power for the generation of de novo diversity and cooperation needed to originate life.
CONCLUSION
In our expanding perspective from physics and chemistry to sociology, from elements/chemicals to emergence of agents, i.e., from RNAs to RNAgents, we found several indices that lead us to a new concept of RNA quasispecies. In contrast to former opinions and concepts that proposed single fittest type and its mutant spectra as the mechanisms of variation that drives evolution, qs-c depend on diversity, multiuse (counter) active agents and consortia membership. Thus, when we think of a commonly shared genetic code that is used and represented by consortia of RNA stem-loops, it is providing not only information storage but active group membership-identity. Therefore we term this membership-identity qs-c. In this perspective qs-RNA and virus evolution are inherently cooperative and modular. The essential players are not fittest types, but consortia of RNAgents that need diversity.
The basic rule here is that of innovation and group selection. The modular character of consortia building with its inherent self-ligation capability of basic stem-loop tools is based additionally on the participation and integration of defectives/mutants (errors/junk) as energy optimizing resource for module-like re-usage. In such dynamic RNA stem-loop populations several basic behavioral motifs are combined that are absent in pre-biotic chemistry; such as complementation, cooperation, competition (selection), preclusion and lethal defections. This broad range of behaviors require a diversity that also integrates the remaining “memory” (information-storing) parts of essential minorities. Under changing contexts minorities may have previously been or may yet become majorities. Thus memory of past experience is inherent in and used by qs-c.
In no other natural language are the agents that communicate (coordinate and organize) via repertoire of natural signs (language) also identical with the signs (words) themselves. This is precisely what we have proposed with stem-loop RNAs. This proposition defines a new phenomenon: at the beginning of life agents and “words” (information) are identical. What has been divided since invention of DNA and LUCAs (signs from agents), was formerly unified. The qs-c sociology thus describes this unified status and its interactions in their current DNA/protein habitats. Indeed it is difficult to formulate sentences about a status that never have been formulated before: Agents that represent sequences of signs are themselves subject of sequence generation (as described above). In the current RNA world (now residing in their DNA habitats) they are still alive. The contrast to the early RNA world, in that their available habitats (DNA/protein) have expanded indefinitely.
Now we can imagine the move from RNA physics/chemistry to RNA sociology. Although they consist of atoms that bind together by laws of physics and chemistry they don’t behave like abiotic ensembles but as semiotic subjects absent in abiotic world. “Words” are (stem-loop) agents and “sentences” are consortia of such agents. For example, some “sentences” result in tRNAs and the ribosomal subunits which assemble to ribosomes. Some result in RNA viruses, some in defectives RNAs that serve as effective non-coding RNAs in a regulation processes of host genomes. The high mutation rate is now recognized as freedom (from mechanistic determinism) to generate new sequence (consortia) space. “Error” now becomes innovation-competence for new generations of both RNAagents and biological information with unpredictable competence and membership.
ACKNOWLEDGMENTS
The authors are grateful to Professor Vera Kolb and to the reviewers for their constructive suggestions.