Published online Apr 28, 2021. doi: 10.37126/aige.v2.i2.12
Peer-review started: February 15, 2021
First decision: March 16, 2021
Revised: March 30, 2021
Accepted: April 20, 2021
Article in press: April 20, 2021
Published online: April 28, 2021
In recent years, artificial intelligence has been extensively applied in the diagnosis of gastric cancer based on medical imaging. In particular, using deep learning as one of the mainstream approaches in image processing has made remarkable progress. In this paper, we also provide a comprehensive literature survey using four electronic databases, PubMed, EMBASE, Web of Science, and Cochrane. The literature search is performed until November 2020. This article provides a summary of the existing algorithm of image recognition, reviews the available datasets used in gastric cancer diagnosis and the current trends in applications of deep learning theory in image recognition of gastric cancer. covers the theory of deep learning on endoscopic image recognition. We further evaluate the advantages and disadvantages of the current algorithms and summarize the characteristics of the existing image datasets, then combined with the latest progress in deep learning theory, and propose suggestions on the applications of optimization algorithms. Based on the existing research and application, the label, quantity, size, resolutions, and other aspects of the image dataset are also discussed. The future developments of this field are analyzed from two perspectives including algorithm optimization and data support, aiming to improve the diagnosis accuracy and reduce the risk of misdiagnosis.
Core Tip: Gastric cancer is a life-threatening disease with a high mortality rate. With the development of deep learning in the image processing of gastrointestinal endoscope, the efficiency and accuracy of gastric cancer diagnosis through imaging technology have been greatly improved. At present, there is no comprehensive summary on the graphic recognition method for gastric cancer based on deep learning. In this review, some gastric cancer image databases and mainstream gastric cancer recognition models were summarized to make a prospect for the application of deep learning in this field.
- Citation: Li Y, Zhou D, Liu TT, Shen XZ. Application of deep learning in image recognition and diagnosis of gastric cancer. Artif Intell Gastrointest Endosc 2021; 2(2): 12-24
- URL: https://www.wjgnet.com/2689-7164/full/v2/i2/12.htm
- DOI: https://dx.doi.org/10.37126/aige.v2.i2.12
Gastric cancer is a life-threatening disease with a high mortality rate[1]. Globally, more than 900000 individuals develop gastric cancer each year out of which more than 700000 Lose their lives. Gastric cancer is second only to lung cancer in terms of mortality[2]. Unlike the developing countries, the number of diagnosed cases and the mortality rate of this cancer are declining in the developed countries such as those in the EU and North America[3,4].
Around 50% of the world's gastric cancer cases are diagnosed in Southeast Asia[5]. In China, gastric cancer is also second to lung cancer in terms of the number of annual cases, for instance, 424000 new patients are annually diagnosed with gastric cancer, accounting for more than 40% of the global total, out of which 392000 Lose their lives ranking the fifth and the sixth worldwide in annual morbidity and mortality, respectively[6].
The diagnosis of gastric cancer mainly relies on clinical manifestation, pathological images and medical imaging[7]. Compared with other methods such as pathological diagnosis, medical imaging provides a simple non-invasive and reliable method for the diagnosis of gastric cancer which is more accessible and efficient, easier to operate and has almost no side effects for the patients[8].
Doctors make a judgment based on medical imaging which mainly depend on their experience from similar cases, hence, occasional misdiagnosis is inevitable[9,10]. With the rapid development of computer technology and artificial intelligence, deep learning techniques are extremely effective in various branches of image processing and have been used in medical imaging to improve cancer diagnosis[11-13]. Danaee et al[14] established a deep learning model for colorectal cancer image recognition, the results showed that the deep learning method can achieve more effective information and is far more efficient than the way of manual extraction. Burke et al[15] found that deep learning could classify and predict mutations of NSCLC based on histopathological images, and the recognition efficiency of deep learning was much higher than that of manual recognition. Muhammad Owais et al[16] proposed a deep learning model to classify a variety of gastrointestinal diseases by recognizing endoscopic videos. This model can simultaneously extract spatiotemporal features to achieve better classification performance. Experimental results of the proposed models showed superior performance to the latest technology and indicated its potential in clinical application[16].
Endoscopic images are mostly used in gastric cancer diagnosis[17]. Endoscopic images contain a lot of useful structural information which can be used for deep learning algorithm, the algorithm can carry out purposeful image recognition[18]. Most of the image recognition based on gastric cancer diagnosis methods adopt supervised deep learning algorithms, mainly because the monitored network in supervised learning makes full use of the labeled sample data in the training and can obtain more accurate segmentation results[19].
In fact, the purpose of medical image recognition is to identify the tumor and we call this process image segmentation[20-22]. Accurate segmentation of tumor images is an important step in diagnosis, surgical planning and postoperative evaluation[23,24]. Endoscopic images segmentation can provide more comprehensive information for the diagnosis and treatment of gastric cancer, alleviate the doctor's heavy work for reading film and improve the accuracy of diagnosis[25]. However, due to the variety and complexity of gastric tumor types, segmentation has become an important and difficult problem in computer-aided diagnosis. Compared with the traditional segmentation methods, the deep learning segmentation method of gastric tumor image has achieved obvious improved performance and rapid development[26,27].
As mentioned above, the deep learning method based on supervised learning can fully mine the effective information of existing data. However, when the amount of existing data cannot meet the requirements of model training, it is necessary to find ways to increase the data scale[28]. The deep learning based on unsupervised learning can generate samples, which are similar to the existing samples in dimension and structure, but not identical. At present, relevant research results have been obtained[29]. Researchers use semi-supervised and unsupervised image recognition algorithms to generate samples like training samples, to improve the accuracy of gastric cancer tumor recognition and enhance the robustness of the model[30].
In this paper, deep learning-based diagnosis of gastric cancer based on endoscope images is summarized and analyzed. The adopted segmentation networks in the previous works can be divided into three categories: the supervised network, semi-supervised network, and unsupervised network. The basic idea of the recognition method, the basic structure of the network, the experimental results, as well as their advantages and disadvantages are summarized. The performance of typical methods above-mentioned in recognition is compared. Finally, we hope to provide insights and concluding remarks on the development of deep-learning-based diagnosis of gastric cancer.
To promote the progress of image recognition and make an objective comparison of available image recognition methods for gastric cancer diagnosis, we investigate the commonly used datasets including the GR-AIDS provided by Medical Image Computing and Computer Assisted Intervention Society as well as those internal datasets.
The GR-AIDS dataset established by Sun Yat-Sen University Cancer Center consists of 1036496 endoscopic images from 84424 individuals. This dataset is used according to the 8:1:1 pattern, the data is randomly selected for training and internal validation datasets for GR-AIDS development as well as for evaluating GR-AIDS performance[31].
Using clinical data collected from Gil Hospital, Jang Hyung Lee et al[32] also established a data set containing 200 normal cases, 367 cancer cases, and 220 ulcer cases. The data was divided into training sets of 180, 200, 337 images and test sets of 20, 30, 20 images. To improve the local contrast of the image and enhance the edge definition in each area of the image, histogram equalization was adopted to further enhance the image, the images’ size was adjusted to 224 × 224 pixels [32].
Hirasawa et al[32] collected 13,584 endoscopic images of gastric cancer to build an image database. To evaluate the diagnostic accuracy, an independent test set of 2296 gastric images was collected from 69 patients with continuous gastric cancer lesions constructed as convolutional neural network (CNN). The image has an in-plane resolution of 512 × 512[33].
Cho et al[34] collected 5017 images from 1269 patients, of which 812 images from 212 patients were used as the test data set. An additional 200 images from 200 patients were collected and used for prospective validation. The resolution of the images is 512 × 512. The information for all major databases is shown in Table 1[34].
| Database | Time collected | Number of samples | Resolution | Training set | Test set |
| GR-AIDS[31] | 2019 | 1036496 | 512 × 512 | 829197 | 103650 |
| Jang Hyung Lee[32] | 2019 | 787 | 224 × 224 | 717 | 70 |
| Toshiaki Hirasawa[33] | 2018 | 13584 | 512 × 512 | 13584 | 2496 |
| Bum-Joo Cho[34] | 2019 | 5017 | 512 × 512 | 4205 | 812 |
| Hiroya Ueyama[35] | 2020 | 7874 | 512 × 512 | 5574 | 2300 |
| Lan Li[36] | 2020 | 2088 | 512 × 512 | 1747 | 341 |
| Mads Sylvest Bergholt[37] | 2011 | 1063 | 512 × 512 | 850 | 213 |
To evaluate the effectiveness of each model in diagnosing gastric cancer, the following evaluation indicators are commonly used in the related literature (Table 2): DICE Similarity Coefficient (DICE, 1945), Jaccard Coefficient (Jaccard, 1912), Volumetric Over-lap Error (VOE), and Relative Volume Difference (RVD).
| Index | Description | Usage | Unit |
| DICE | Repeat rate between the segmentation results and markers | Commonly | % |
| RMSD | The root mean square of the symmetrical position surface distance between the segmentation results and the markers | Commonly | mm |
| VOE | The degree of overlap between the segmentation results and the actual segmentation results represents the error rate | Commonly | % |
| RVD | The difference in volume between the segmentation results and the markers | Rarely | % |
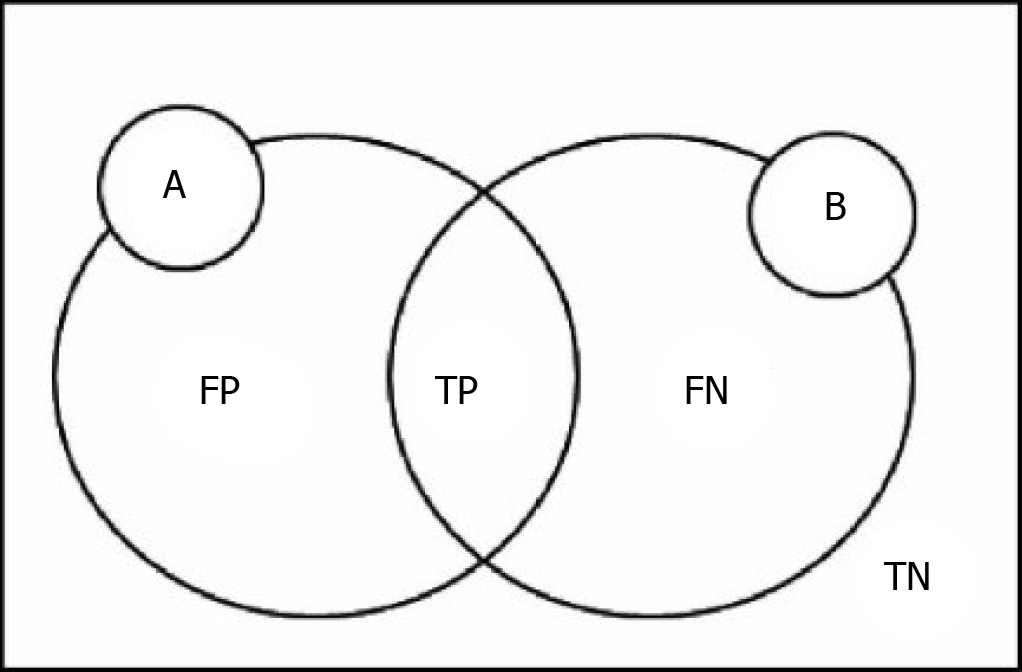
Here we define the following variables: P and N are used for judgment of the model results, T and F evaluation model of the judgment is correct, FP is on behalf of the false-positive cases, FN represents false-negative cases, TP is on behalf of the real example, TN represents true negative cases[38]. A represents the theory of segmentation, results for comparison with the resulting image. B represents the segmentation results[39]. The relationship among them is shown in Figure 1.
DICE coefficient: DICE coefficient also known as the overlap index, is one of the most commonly used indexes for verification of image segmentation. The DICE coefficient represents the repetition rate between the segmentation results and the markers. The value range of DICE is 0-1, where 0 indicates that the experimental segmentation result significantly deviates from the labeled result, and 1 indicates that the experimental segmentation result completely coincides with the labeled result[40]. DICE coefficient is defined as the following:
DICE = (2|A ∩ B|)/(|A| + |B|) = (2TP)/(2TP + FP + FN)
Jaccard coefficient: Jaccard coefficient represents the similarity and difference between the segmentation result and the standard. The larger the coefficient, the higher the sample similarity. Besides, the Jaccard coefficient and DICE coefficient are correlated[41]. Jaccard coefficient is defined as the following:
JAC = (|A ∩ B|)/(|A| U |B|) = TP/(TP + FP + FN) = DICE/(2 - DICE)
VOE: VOE stands for error rate, derived from Jaccard. VOE is represented as %, where 0% indicates complete segmentation. If there is no overlap between the segmentation result and the markers, the VOE is 100%[42]. VOE is defined as the following:
VOE = 1 - (|A ∩ B|)/(|A| U |B|) = 1 - TP/(TP + FP + FN)
RVD: RVD represents the noise difference between the segmentation result and the markers. RVD is presented as %, where 0% denotes the same volume between the segmentation result and the markers[42]. The formula is:
RVD = (|B| - |A|)/|A| = FP/(TP + FN)
The specific concepts of all indicators are shown in Table 2.
Deep neural networks are often trained based on deep learning algorithms using large labeled datasets (i.e., images in this case)[43]. The network is therefore able to learn how features are related to the target[44]. Since the data is already labeled, this learning method is referred to as supervised learning. Most of the existing studies on diagnosing gastric cancer are based on supervised learning in image recognition tasks[45-47]. This is because the network makes full use of the labeled dataset in the training, hence can obtain more accurate segmentation results.
Recent research works showed that CNN achieves outstanding performance in various image recognition tasks[48,49]. Toshiaki Hirasawa built a CNN-based diagnostic system based on a single-shot Multi-Box detector, Adejub, with a total sensitivity of 92.2% and trained their CNN using 13584 endoscopic images of gastric cancer. The trained CNN correctly called 71 out of 77 cases of gastric cancer, i.e., a total sensitivity of 92.2%, also detected 161 non-cancerous samples as gastric cancer, i.e., a positive predictive value of 30.6%. The CNN also correctly detected 70 of 71 cases of gastric cancer (98.6%) with a diameter of 6 mm or larger, as well as all invasive cancers[33]. Ueyama et al[35] also constructed an AI-assisted CNN based computer-aided diagnosis system with narrow band imaging-magnifying endoscopy images.
The above studies show that the CNN-based approach is far more accurate than human in recognition of cancer. This makes us believe that the method based on deep CNN can effectively solve the identification problem of gastric cancer.
However, the issue with the CNN is that only partial features could be extracted[50]. Due to the imbalanced information of gastric cancer image data, extracting the local features does not reflect all the information and might harm the efficiency of the image recognition. To address the problem, Shelhamer et al[51] proposed full convolutional neural network (FCN) for image segmentation. This network attempts to recover the category of each pixel from the abstract feature, in other words, instead of image-level classification, the network uses pixel-level classification[51]. This addresses the semantic level image segmentation problem and is the core component of many advanced semantic segmentation models[52,53].
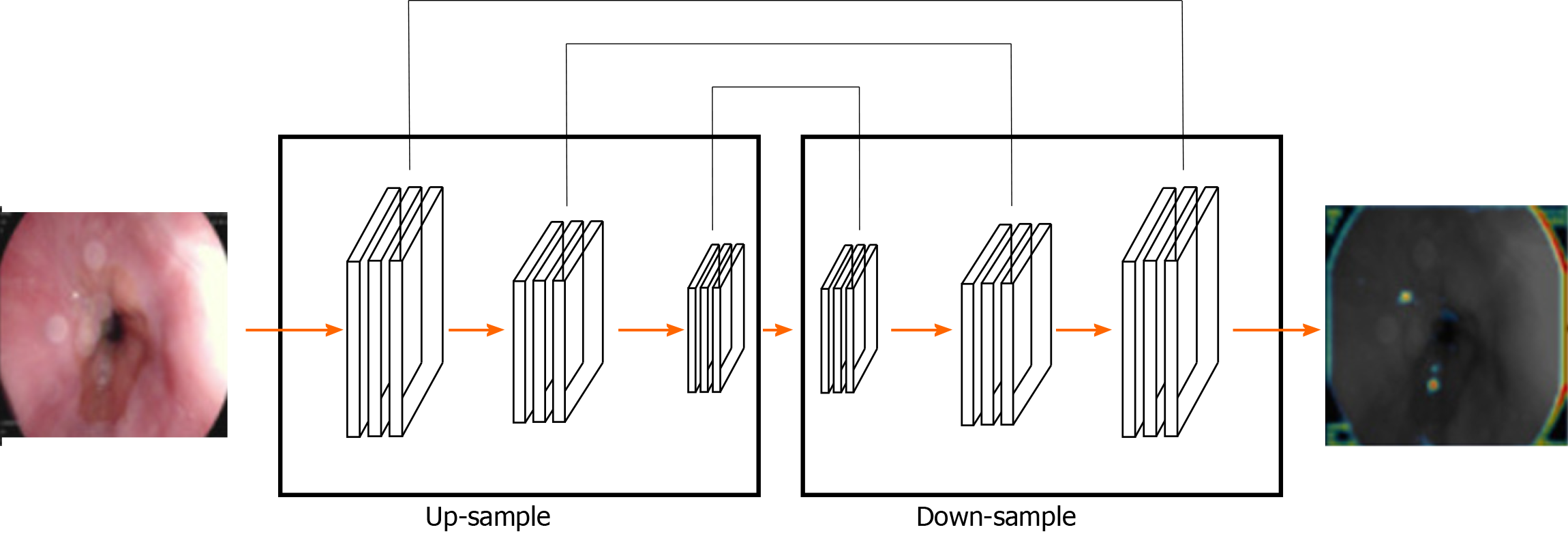
The segmentation method of gastric cancer images based on the FCN network is mainly based on the idea of code-decoding design[54]. In practice, the image is classified at the pixel level and the network is pre-trained with supervision. In this method, the input image can have any arbitrary size and the output of the same size can be generated through effective reasoning and learning[55]. Typical FCN network-based image segmentation architecture for gastric cancer is shown in Figure 2.
The FCN is improved based on the CNN by transforming the last three full connections into three convolutional layers. The success of FCN network is largely attributed to the excellent ability of CNN network to extract hierarchical representation. In the concrete implementation process, the network realizes the segmentation of gastric tumor by down-sampling and up-sampling through convolution-deconvolution operation. The down-sampling path consists of convolution layer and maximum or average pooling layer, which can extract high-level semantic information, but its spatial resolution is often low. The up-sample path consists of convolution and a deconvolution layer (also known as transpose convolution) and uses the output of the down-sample path to predicting the fraction of each class at the pixel level[56,57]. However, the output image of deconvolution operations might be very rough and lost a lot of detail. The skip structure of the FCN network presented in the classified forecast comes from the deep layer (thick) semantic information and information from the appearance of the shallow layer (fine), thus, achieving a more accurate and robust segmentation result. As a deep neural network, FCN has shown good performance in many challenging medical image segmentation tasks, including liver tumor segmentation[58,59].
One of the most important features of the FCN is the use of skip structure. It is used to fuse the feature information of both the high and low layers. Through the cross-layer connection structure, the texture information of the shallow layer and the semantic information of the deep layer of the network are then combined to achieve the precise segmentation task[60,61]. Jang Hyung Lee improved the original FCN framework by applying the pre-trained Inception, Res-Net, and VGG-Net models on ImageNet. The areas under the operating characteristic curves of each receiver are 0.95, 0.97, and 0.85, respectively, hence, Res-Net shows the highest level of performance. Under normal conditions, the classification between normal and ulcer or cancer, is more than 90 percent accurate[32].
The deep network structure leads to the problem of decreased training accuracy[62]. In Sun et al[63] the basic form of convolution is replaced with the deformable convolution and Atrous convolution in a specific layer to adapt to the non-rigid characteristics and large receiving fields. The Atrous space pyramid pooling module and the semantic-level embedded network based on encoder/decoder are used for multi-scale segmentation. Besides, they proposed a lightweight decoder to fuse the context information and further used dense up-sampled convolution for the boundary optimization at the end of the decoder. The model achieves 91.60% pixel-level accuracy and 82.65% average degree of the intersection[63].
Cho et al[34] established the Inception-ResNET-V2 model, which is an FCN model. In this model, they divided the images into five categories: advanced gastric cancer, early gastric cancer, high atypical hyperplasia, low atypical hyperplasia and non-neoplastic. For the above five categories, the Inception- ResNet-v2 model has a weighted average accuracy of 84.6%. The mean area under the curve of the model for differentiating gastric cancer and neoplasm was 0.877 and 0.927, respectively[34].
The above works show that FCN addresses the issue with the CNN hence can extract the local features. This is why the FCN is considered as the mainstream in gastric cancer image classification methods.
In addition to the application of FCN to address the shortcomings of CNN, researchers also tried other approaches such as fusion of multiple CNN methods to obtain an Ensemble of CNN algorithm to get more accurate classification results. Nguyen et al[64] trained three different CNN model architectures, including VGG-based, Inception-based Network and Dense-Net. In their study, the VGG-based network was used as a conventional deep CNN for classification problems, which consists of a linear stack of the convolutional layer. The network-based on Dense-net can be used as a very deep CNN with a short path, which is also helpful to train the network and extract more abstract and effective image features easily. The three models were trained separately, the AVERAGE combination rule is then used to combine the classification results of the three CNN-based Models. The final result was 70.369% of overall classification accuracy, 68.452% of sensitivity and 72.571% of specificity. The overall classification accuracy is higher than that generated by the listed model based on a single CNN[64].
Both the use of a fully convolutional network and the fusion of several CNN algorithms are significantly effective in improving the accuracy of gastric cancer image recognition. They are also effective in addressing the issues with the quality of images in the database. Table 3 shows the performance comparison of gastric cancer image recognition by using CNN, FCN, and Ensemble CNN.
| Methods | DICE/% | VOE/% | RMSD/mm |
| Toshiaki Hirasawa (CNN) | 0.5738 | 0.5977 | 6.491 |
| Hiroya Ueyama (CNN) | 0.6327 | 0.5373 | 7.257 |
| Jang Hyung Lee (FCN) | 0.8102 | 0.319 | 2.468 |
| Bum-Joo Cho (FCN) | 0.9350 | 0.1221 | - |
| Dat Tien Nguyen (ECNN) | 0.8947 | 0.113 | - |
Most gastric cancer image recognition methods adopt supervised learning algorithms because the monitored network makes full use of the labeled sample data in the training and can obtain more accurate segmentation results. Nevertheless, there are very few accurately labeled image datasets, hence researchers have carried out studies based on semi-supervised and unsupervised image recognition algorithms for gastric cancer. In such studies, they trained a small number of samples through generative models to generate similar samples to improve the accuracy and robustness of gastric cancer tumor recognition[65].
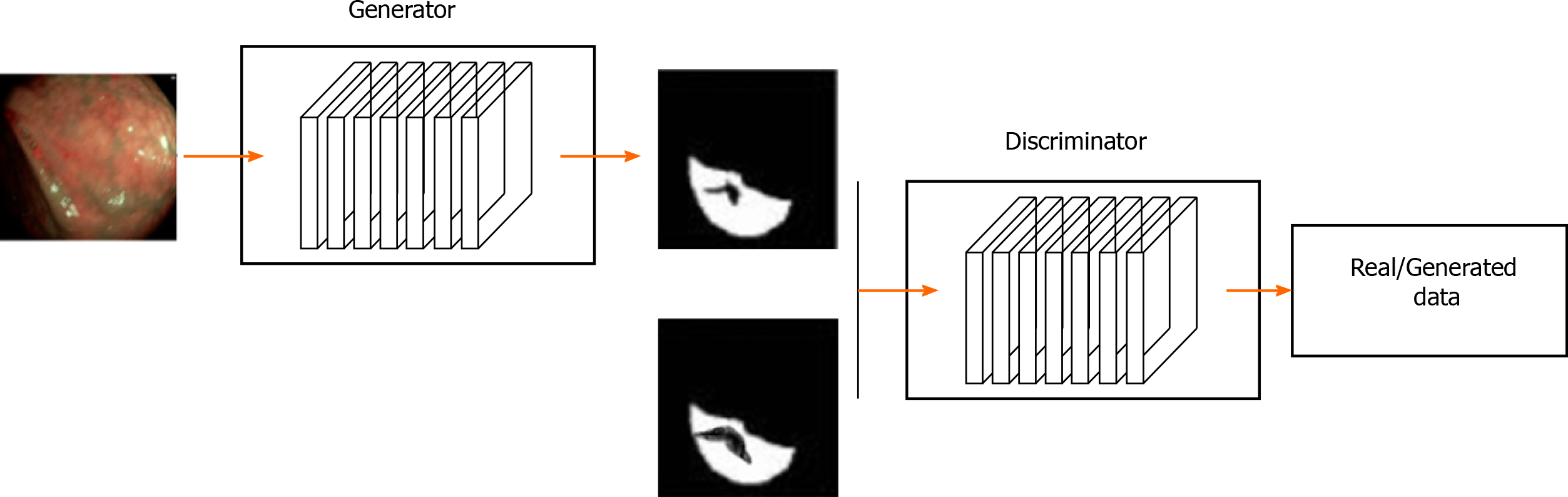
Generative adversarial network (GAN) is a generative model proposed by Goodfellow et al[66] It uses an unsupervised training method that is trained by adversarial learning. The objective is to estimate the potential distribution of data samples and generate new data samples. GAN is composed of a generation model (Goodfellow et al[66], 2014) and a discrimination model (Denton et al, 2015). The generation model learns the distribution of a given noise (generally refers to uniform distribution or normal distribution) and synthesizes it, whereas the discrimination model distinguishes the real data from generated data. In theory, the former is trying to produce data that is closed to the real data. The latter is also constantly strengthening the "counterfeit detection" ability[67]. The success of GAN lies in its ability to capture high-level semantic information using adversarial learning techniques. Luc et al[68] first applied GAN to image segmentation. However, GAN has several drawbacks: (1) Crash problem: when the generation model crashes, all different inputs are mapped to the same data[69]; and (2) Instability: It causes the same input to produce different outputs. The main reason is due to gradient vanishing problem during the optimization process[66,70].
Although batch normalization is often used to solve the instability of GAN, it is often not enough to achieve optimal stability of GAN performance. Therefore, many GAN derived models have emerged to solve these gaps, e.g., conditional GAN, deep convolutional GAN, information maxi-mizing GAN, Wassertein GAN, etc[71]. In the GAN-based image recognition for gastric cancer, the generator is used to perform the segmentation task. The discriminator is then used to train the refining generator. A typical gastric cancer image recognition architecture based on the generative adversarial network is illustrated in Figure 3.
Since its proposal generative adversarial network has been widely considered and rapidly developed in different application areas. In medical image processing, it is very challenging to construct a large enough dataset due to the difficulty of data acquisition and annotation[72]. To overcome this problem, traditional image enhancement technology such as geometric transformation is often used to generate new data. This technique cannot learn biological changes in medical data and can produce images that are not credible[73]. Although GAN is unable to know in advance hypothesis distribution due to the limitation of segmentation performance improvement. it can automatically infer real data sets, further expand the scale and diversity of data, and provide a new method for data expansion, thus improving the efficiency of model training[74,75].
Almalioglu et al[76] showed that the poor resolution of the capsule endoscope is a limiting factor in the accuracy of diagnosis. They designed an image synthesis technology based on GAN to enrich the training data. First, the standard data expansion method was used to enlarge the dataset. Then the dataset was used to train GAN and the proposed Endol2h method was used to synthesize gastric cancer images with higher resolution[76]. Wang proposed an unsupervised image classification method for tumors based on prototype migration generated against the network (Prototype Transfer Generative Adversarial Network). Using different data acquisition devices and parameter settings caused differences in the stye of tumor image and data distribution. These differences can be reduced by designing the target domain to generate network, training process through the domain discriminant and performing generator reconstruction between source domain and target domain. The method achieved an average accuracy of 87.6% for unsupervised breast tumor image dichotomy under different magnifications and shows good scalability[77].
In conclusion, the GAN-based image segmentation method for gastric cancer can generate realistic gastric cancer images through the GAN network in the training stage, thus avoid the imbalance of the training samples. Moreover, due to the amplification of limited labeled sample data, the deep network is well-trained and achieves a high segmentation efficiency. However, there are still many problems in GAN, such as the instability of training and the breakdown of the training network. Therefore, researchers have optimized the original GAN network to reduce data noise or deal with class imbalance and other problems. In order to solve the problem that medical images are often polluted by different amounts and types of praise, T.Y Zhang et al. propose a novel Noise Adaptation Generative Adversarial Network (NAGAN), which contains a generator and two discriminators. The generator aims to map the data from source domain to target domain. Among the two discriminators, one discriminator enforces the generated images to have the same noise patterns as those from the target domain, and the second discriminator enforces the content to be preserved in the generated images. They apply the proposed NAGAN on both optical coherence tomography images and ultrasound images. Results show that the method is able to translate the noise style[74]. In the traditional GAN network training, the small number of samples of the minority classes in the training data makes the learning of optimal classification challenging, while the more frequently occurring samples of the majority class hamper the generalization of the classification boundary between infrequently occurring target objects and classes. Mina Rezaei et al. developed a novel generative multi-adversarial network, called Ensemble-GAN, for mitigating this class imbalance problem in the semantic segmentation of abdominal images. The Ensemble-GAN framework is composed of a single-generator and a multi-discriminator variant for handling the class imbalance problem to provide a better generalization than existing approaches[73]. In addition, there are other studies on the optimization of GAN network in medical image segmentation. Klages et al[78] proposed the patch-based generative adversarial neural network models, this model can significantly reduce errors in data generation. Nuo Tong et al[79] proposed the self-paced Dense-Net with boundary constraint for automated multi-organ segmentation on abdominal CT images. Specifically, a learning-based attention mechanism and dense connection block are seamlessly integrated into the proposed self-paced Dense-Net to improve the learning capability and efficiency of the backbone network. In a word, in the process of optimizing GAN network, whether it is optimizing generator or discriminant, the purpose of optimization is to generate new data which is as equal to the real data as possible. Therefore, more studies will be devoted to the optimization of GAN network to provide strong support for improving the image recognition of gastric cancer.
Table 4 shows comparison results of the three current mainstream methods for image recognition of gastric cancer.
| Model features | Contributions | Advantages | Disadvantages | Scope of application |
| CNN | The topology can be extracted from a two-dimensional image, and the backpropagation algorithm is used to optimize the network structure and solve the unknown parameters in the network | Shared convolution kernel, processing high-dimensional data without pressure; Feature extraction can be done automatically | When the network layer is too deep, the parameters near the input layer will be changed slowly by using BP propagation to modify parameters. A gradient descent algorithm is used to make the training results converge to the local minimum rather than the global minimum. The pooling layer will lose a lot of valuable information | Suitable for data scenarios with similar network structures |
| FCN | The end-to-end convolutional network is extended to semantic segmentation. The deconvolution layer is used for up-sampling; A skip connection is proposed to improve the roughness of the upper sampling | Can accept any size; Input image; Jump junction; The structure combines fine layers and coarse; Rough layers, generating precise segmentation | The receptive field is too small to obtain the global information;Small storage overhead | Applicable to large sample data |
| GAN | With adversarial learning criteria, there are two No's: The same network, not a single network | Can produce a clearer, more realistic sample; any generated network can be trained | Training is unstable and difficult to train; GAN is not suitable for processing data in discrete form | Suitable for data generation (e.g., there are not many data sets with labels), image style transfer; Image denoising and restoration; Used to counter attacks |
At the present, the development direction of deep learning in image recognition of gastric cancer mainly focuses on the following aspects: (1) Training of deep learning algorithms relies on the availability of large datasets, because medical images are often difficult to obtain, medical professionals need to spend a lot of time on data collection and annotation which is time-consuming and costly. Besides, medical workers need not only to provide a large amount of data support but also to make use of all the effective information in the data as much as possible. Deep neural networks enable full mining of the information content of the data. Using deep networks seems to be the dominant future research direction in this field; (2) Multimodal gastric image segmentation combined with several different deep neural networks are used to extract the deeper information of the image and improve the accuracy of tumor segmentation and recognition. This is a promising major research direction in this field; and (3) Currently, most of the medical image segmentation techniques use supervised deep learning algorithms. However, for some of the rare diseases lacking a large number of data samples, supervised deep learning algorithms cannot reach their full efficiency. To overcome the issue with the lack of large datasets, some researchers utilize semi-supervised or unsupervised techniques such as GAN and combine the generated adversarial network with other higher performance networks. This might be another emerging research trend in this area.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and Hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Nayyar A, Taira K, Tanabe S S-Editor: Wang JL L-Editor: A P-Editor: Wang LL
| 1. | Higgins AJ, Lees P. Arachidonic acid metabolites in carrageenin-induced equine inflammatory exudate. J Vet Pharmacol Ther. 1984;7:65-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 64] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 2. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18694] [Cited by in F6Publishing: 20838] [Article Influence: 2315.3] [Reference Citation Analysis (2)] |
| 3. | Ferlay J, Colombet M, Soerjomataram I, Dyba T, Randi G, Bettio M, Gavin A, Visser O, Bray F. Cancer incidence and mortality patterns in Europe: Estimates for 40 countries and 25 major cancers in 2018. Eur J Cancer. 2018;103:356-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1625] [Cited by in F6Publishing: 1466] [Article Influence: 244.3] [Reference Citation Analysis (0)] |
| 4. | Howlader N NA, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER Cancer Statistics Review, 1975-2017. Bethesda: National Cancer Institute, 2019. Available from: https://seer.cancer.gov/csr/1975_2017/. [Cited in This Article: ] |
| 5. | Rahman R, Asombang AW, Ibdah JA. Characteristics of gastric cancer in Asia. World J Gastroenterol. 2014;20:4483-4490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 265] [Cited by in F6Publishing: 290] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 6. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11444] [Cited by in F6Publishing: 12500] [Article Influence: 1562.5] [Reference Citation Analysis (0)] |
| 7. | Homeida AM, Cooke RG. Pharmacological aspects of metaldehyde poisoning in mice. J Vet Pharmacol Ther. 1982;5:77-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Luo X, Mori K, Peters TM. Advanced Endoscopic Navigation: Surgical Big Data, Methodology, and Applications. Annu Rev Biomed Eng. 2018;20:221-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, Pech O, Meining A, Neuhaus H, Bisschops R, Schoon EJ, de With PH, Bergman JJ. The Argos project: The development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 75] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 10. | Qu JY, Li Z, Su JR, Ma MJ, Xu CQ, Zhang AJ, Liu CX, Yuan HP, Chu YL, Lang CC, Huang LY, Lu L, Li YQ, Zuo XL. Development and Validation of an Automatic Image-Recognition Endoscopic Report Generation System: A Multicenter Study. Clin Transl Gastroenterol. 2020;12:e00282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Gan T, Liu S, Yang J, Zeng B, Yang L. A pilot trial of Convolution Neural Network for automatic retention-monitoring of capsule endoscopes in the stomach and duodenal bulb. Sci Rep. 2020;10:4103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging. 2016;35:1285-1298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3594] [Cited by in F6Publishing: 1748] [Article Influence: 218.5] [Reference Citation Analysis (0)] |
| 13. | Bernal J, Kushibar K, Asfaw DS, Valverde S, Oliver A, Martí R, Lladó X. Deep convolutional neural networks for brain image analysis on magnetic resonance imaging: a review. Artif Intell Med. 2019;95:64-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 119] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 14. | Danaee P, Ghaeini R, Hendrix DA. A Deep Learning Approach for Cancer Detection and Relevant Gene Identification. Pac Symp Biocomput. 2017;22:219-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 111] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 15. | Burke HB, Goodman PH, Rosen DB, Henson DE, Weinstein JN, Harrell FE Jr, Marks JR, Winchester DP, Bostwick DG. Artificial neural networks improve the accuracy of cancer survival prediction. Cancer. 1997;79:857-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 16. | Owais M, Arsalan M, Choi J, Mahmood T, Park KR. Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis. J Clin Med. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 17. | Lu S, Cottone CM, Yoon R, Jefferson FA, Sung JM, Okhunov Z, Tapiero S, Patel RM, Landman J, Clayman RV. Endockscope: A Disruptive Endoscopic Technology. J Endourol. 2019;33:960-965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Wang KW, Dong M. Potential applications of artificial intelligence in colorectal polyps and cancer: Recent advances and prospects. World J Gastroenterol. 2020;26:5090-5100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 22] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Yasuda Y, Tokunaga K, Koga T, Sakamoto C, Goldberg IG, Saitoh N, Nakao M. Computational analysis of morphological and molecular features in gastric cancer tissues. Cancer Med. 2020;9:2223-2234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Yao Y, Gou S, Tian R, Zhang X, He S. Automated Classification and Segmentation in Colorectal Images Based on Self-Paced Transfer Network. Biomed Res Int. 2021;2021:6683931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Baig R, Bibi M, Hamid A, Kausar S, Khalid S. Deep Learning Approaches Towards Skin Lesion Segmentation and Classification from Dermoscopic Images - A Review. Curr Med Imaging. 2020;16:513-533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 22. | Zhang K, Liu X, Liu F, He L, Zhang L, Yang Y, Li W, Wang S, Liu L, Liu Z, Wu X, Lin H. An Interpretable and Expandable Deep Learning Diagnostic System for Multiple Ocular Diseases: Qualitative Study. J Med Internet Res. 2018;20:e11144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 23. | Weng S, Xu X, Li J, Wong STC. Combining deep learning and coherent anti-Stokes Raman scattering imaging for automated differential diagnosis of lung cancer. J Biomed Opt. 2017;22:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 24. | Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, Chang EI. Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics. 2017;18:281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 164] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 25. | Yonekura A, Kawanaka H, Prasath VBS, Aronow BJ, Takase H. Automatic disease stage classification of glioblastoma multiforme histopathological images using deep convolutional neural network. Biomed Eng Lett. 2018;8:321-327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Horiuchi Y, Aoyama K, Tokai Y, Hirasawa T, Yoshimizu S, Ishiyama A, Yoshio T, Tsuchida T, Fujisaki J, Tada T. Convolutional Neural Network for Differentiating Gastric Cancer from Gastritis Using Magnified Endoscopy with Narrow Band Imaging. Dig Dis Sci. 2020;65:1355-1363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 78] [Article Influence: 19.5] [Reference Citation Analysis (1)] |
| 27. | Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M. Deep-learning based detection of gastric precancerous conditions. Gut. 2020;69:4-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 63] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 28. | Wu T, Tegmark M. Toward an artificial intelligence physicist for unsupervised learning. Phys Rev E. 2019;100:033311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 29. | Zhang R, Zhang Y, Li X. Unsupervised Feature Selection via Adaptive Graph Learning and Constraint. IEEE Trans Neural Netw Learn Syst. 2020;PP. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 30. | Wang Z, Li M, Xu Z, Jiang Y, Gu H, Yu Y, Zhu H, Zhang H, Lu P, Xin J, Xu H, Liu C. Improvements to the gastric cancer tumor-node-metastasis staging system based on computer-aided unsupervised clustering. BMC Cancer. 2018;18:706. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 31. | Luo H, Xu G, Li C, He L, Luo L, Wang Z, Jing B, Deng Y, Jin Y, Li Y, Li B, Tan W, He C, Seeruttun SR, Wu Q, Huang J, Huang DW, Chen B, Lin SB, Chen QM, Yuan CM, Chen HX, Pu HY, Zhou F, He Y, Xu RH. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol. 2019;20:1645-1654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 209] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 32. | Lee JH, Kim YJ, Kim YW, Park S, Choi YI, Park DK, Kim KG, Chung JW. Spotting malignancies from gastric endoscopic images using deep learning. Surg Endosc. 2019;33:3790-3797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 33. | Hirasawa T, Aoyama K, Tanimoto T, Ishihara S, Shichijo S, Ozawa T, Ohnishi T, Fujishiro M, Matsuo K, Fujisaki J, Tada T. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer. 2018;21:653-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 373] [Article Influence: 62.2] [Reference Citation Analysis (0)] |
| 34. | Cho BJ, Bang CS, Park SW, Yang YJ, Seo SI, Lim H, Shin WG, Hong JT, Yoo YT, Hong SH, Choi JH, Lee JJ, Baik GH. Automated classification of gastric neoplasms in endoscopic images using a convolutional neural network. Endoscopy. 2019;51:1121-1129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 82] [Article Influence: 16.4] [Reference Citation Analysis (1)] |
| 35. | Ueyama H, Kato Y, Akazawa Y, Yatagai N, Komori H, Takeda T, Matsumoto K, Ueda K, Hojo M, Yao T, Nagahara A, Tada T. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J Gastroenterol Hepatol. 2021;36:482-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 63] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 36. | Li L, Chen Y, Shen Z, Zhang X, Sang J, Ding Y, Yang X, Li J, Chen M, Jin C, Chen C, Yu C. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer. 2020;23:126-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 112] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 37. | Bergholt MS, Zheng W, Lin K, Ho KY, Teh M, Yeoh KG, Yan So JB, Huang Z. In vivo diagnosis of gastric cancer using Raman endoscopy and ant colony optimization techniques. Int J Cancer. 2011;128:2673-2680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 38. | Liang Q, Nan Y, Coppola G, Zou K, Sun W, Zhang D, Wang Y, Yu G. Weakly Supervised Biomedical Image Segmentation by Reiterative Learning. IEEE J Biomed Health Inform. 2019;23:1205-1214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 39. | El-Khatib H, Popescu D, Ichim L. Deep Learning-Based Methods for Automatic Diagnosis of Skin Lesions. Sensors (Basel). 2020;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 40. | Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans Med Imaging. 2016;35:1240-1251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1538] [Cited by in F6Publishing: 814] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 41. | Polanski WH, Zolal A, Sitoci-Ficici KH, Hiepe P, Schackert G, Sobottka SB. Comparison of Automatic Segmentation Algorithms for the Subthalamic Nucleus. Stereotact Funct Neurosurg. 2020;98:256-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 42. | Liu T, Liu J, Ma Y, He J, Han J, Ding X, Chen CT. Spatial feature fusion convolutional network for liver and liver tumor segmentation from CT images. Med Phys. 2021;48:264-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 43. | Cuocolo R, Caruso M, Perillo T, Ugga L, Petretta M. Machine Learning in oncology: A clinical appraisal. Cancer Lett. 2020;481:55-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 80] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 44. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36149] [Cited by in F6Publishing: 17268] [Article Influence: 1918.7] [Reference Citation Analysis (0)] |
| 45. | Zhou T, Han G, Li BN, Lin Z, Ciaccio EJ, Green PH, Qin J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med. 2017;85:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 46. | Tan JW, Wang L, Chen Y, Xi W, Ji J, Xu X, Zou LK, Feng JX, Zhang J, Zhang H. Predicting Chemotherapeutic Response for Far-advanced Gastric Cancer by Radiomics with Deep Learning Semi-automatic Segmentation. J Cancer. 2020;11:7224-7236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 47. | Cho BJ, Bang CS, Lee JJ, Seo CW, Kim JH. Prediction of Submucosal Invasion for Gastric Neoplasms in Endoscopic Images Using Deep-Learning. J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 48. | Krizhevsky A, Sutskever I, Hinton G. ImageNet Classification with Deep Convolutional Neural Networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in Neural Information Processing Systems 25 (NIPS 2012). Red Hook: Curran Associates, 2012. [Cited in This Article: ] |
| 49. | Karpathy A, Toderici G, Shetty S, Leung T, Sukthankar R, Fei-Fei L. Large-Scale Video Classification with Convolutional Neural Networks. In: 2014 IEEE Conference on Computer Vision and Pattern Recognition; 2014 June 23-28; Columbus, USA. IEEE, 2014. [DOI] [Cited in This Article: ] |
| 50. | Xiang Y, Lin Z, Meng J. Automatic QRS complex detection using two-level convolutional neural network. Biomed Eng Online. 2018;17:13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 51. | Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell. 2016;39:1. [DOI] [Cited in This Article: ] [Cited by in Crossref: 4004] [Cited by in F6Publishing: 1588] [Article Influence: 226.9] [Reference Citation Analysis (0)] |
| 52. | Kim HK, Yoo KY, Park JH, Jung HY. Asymmetric Encoder-Decoder Structured FCN Based LiDAR to Color Image Generation. Sensors (Basel). 2019;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 53. | Zhu H, Adeli E, Shi F, Shen D; Alzheimer’s Disease Neuroimaging Initiative. FCN Based Label Correction for Multi-Atlas Guided Organ Segmentation. Neuroinformatics. 2020;18:319-331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 54. | Guo X, Nie R, Cao J, Zhou D, Qian W. Fully Convolutional Network-Based Multifocus Image Fusion. Neural Comput. 2018;30:1775-1800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 55. | Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys. 2017;44:5221-5233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 56. | Wang R, Cao S, Ma K, Zheng Y, Meng D. Pairwise learning for medical image segmentation. Med Image Anal. 2021;67:101876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 57. | Xue J, He K, Nie D, Adeli E, Shi Z, Lee SW, Zheng Y, Liu X, Li D, Shen D. Cascaded MultiTask 3-D Fully Convolutional Networks for Pancreas Segmentation. IEEE Trans Cybern. 2021;51:2153-2165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 58. | Wu W, Wu S, Zhou Z, Zhang R, Zhang Y. 3D Liver Tumor Segmentation in CT Images Using Improved Fuzzy C-Means and Graph Cuts. Biomed Res Int. 2017;2017:5207685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 59. | Baazaoui A, Barhoumi W, Zagrouba E. Semi-Automated Segmentation of Single and Multiple Tumors in Liver CT Images Using Entropy-Based Fuzzy Region Growing. IRBM. 2017;38:98-108. [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 60. | Öztürk Ş, Özkaya U. Skin Lesion Segmentation with Improved Convolutional Neural Network. J Digit Imaging. 2020;33:958-970. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 29] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 61. | Oda M, Tanaka K, Takabatake H, Mori M, Natori H, Mori K. Realistic endoscopic image generation method using virtual-to-real image-domain translation. Healthc Technol Lett. 2019;6:214-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 62. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology 2020; 158: 915-929. e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 174] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 63. | Sun M, Zhang G, Dang H, Qi X, Zhou X, Chang Q. Accurate Gastric Cancer Segmentation in Digital Pathology Images Using Deformable Convolution and Multi-Scale Embedding Networks. IEEE Access. 2019;7:75530-75541. [DOI] [Cited in This Article: ] |
| 64. | Nguyen DT, Lee MB, Pham TD, Batchuluun G, Arsalan M, Park KR. Enhanced Image-Based Endoscopic Pathological Site Classification Using an Ensemble of Deep Learning Models. Sensors (Basel). 2020;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 65. | Teramoto A, Tsukamoto T, Yamada A, Kiriyama Y, Imaizumi K, Saito K, Fujita H. Deep learning approach to classification of lung cytological images: Two-step training using actual and synthesized images by progressive growing of generative adversarial networks. PLoS One. 2020;15:e0229951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 66. | Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. In: Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2. Montreal: MIT Press, 2014: 2672-2680. [Cited in This Article: ] |
| 67. | Enokiya Y, Iwamoto Y, Chen YW, Han XH. Automatic Liver Segmentation Using U-Net with Wasserstein GANs. Int J Image Graph. 2019;7:94-101. [DOI] [Cited in This Article: ] |
| 68. | Luc P, Couprie C, Chintala S, Verbeek J. Semantic segmentation using adversarial networks. 2016 Preprint. Available from: arXiv:1611.08408. [Cited in This Article: ] |
| 69. | Borji A. Pros and cons of GAN evaluation measures. CVIU. 2019;179:41-65. [DOI] [Cited in This Article: ] |
| 70. | Metz L, Poole B, Pfau D, Sohl-Dickstein J. Unrolled generative adversarial networks. 2016 Preprint. Available from: arXiv:1611.02163. [Cited in This Article: ] |
| 71. | Poorneshwaran JM, Santhosh Kumar S, Ram K, Joseph J, Sivaprakasam M. Polyp Segmentation using Generative Adversarial Network. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019:7201-7204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 72. | Tang C, Zhang W, Wang L, Cai A, Liang N, Li L, Yan B. Generative adversarial network-based sinogram super-resolution for computed tomography imaging. Phys Med Biol. 2020;65:235006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 73. | Rezaei M, Näppi JJ, Lippert C, Meinel C, Yoshida H. Generative multi-adversarial network for striking the right balance in abdominal image segmentation. Int J Comput Assist Radiol Surg. 2020;15:1847-1858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 74. | Zhang T, Cheng J, Fu H, Gu Z, Xiao Y, Zhou K, Gao S, Zheng R, Liu J. Noise Adaptation Generative Adversarial Network for Medical Image Analysis. IEEE Trans Med Imaging. 2020;39:1149-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 75. | Han L, Huang Y, Dou H, Wang S, Ahamad S, Luo H, Liu Q, Fan J, Zhang J. Semi-supervised segmentation of lesion from breast ultrasound images with attentional generative adversarial network. Comput Methods Programs Biomed. 2020;189:105275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 76. | Almalioglu Y, Bengisu Ozyoruk K, Gokce A, Incetan K, Irem Gokceler G, Ali Simsek M, Ararat K, Chen RJ, Durr NJ, Mahmood F, Turan M. EndoL2H: Deep Super-Resolution for Capsule Endoscopy. IEEE Trans Med Imaging. 2020;39:4297-4309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 77. | Wang D. Research on Key Technologies of Medical Image Classification Based on Unsupervised and Semi-supervised Framework. M.D. Thesis, Jilin University. 2020 Available from: http://cdmd.cnki.com.cn/Article/CDMD-10183-1020754186.htm. [Cited in This Article: ] |
| 78. | Klages P, Benslimane I, Riyahi S, Jiang J, Hunt M, Deasy JO, Veeraraghavan H, Tyagi N. Patch-based generative adversarial neural network models for head and neck MR-only planning. Med Phys. 2020;47:626-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 79. | Tong N, Gou S, Niu T, Yang S, Sheng K. Self-paced DenseNet with boundary constraint for automated multi-organ segmentation on abdominal CT images. Phys Med Biol. 2020;65:135011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |