Published online Jul 28, 2022. doi: 10.35713/aic.v3.i3.42
Peer-review started: February 14, 2022
First decision: March 12, 2022
Revised: April 9, 2022
Accepted: July 13, 2022
Article in press: June 13, 2022
Published online: July 28, 2022
The use of machine learning and deep learning has enabled many applications, previously thought of as being impossible. Among all medical fields, cancer care is arguably the most significantly impacted, with precision medicine now truly being a possibility. The effect of these technologies, loosely known as artificial intelligence, is particularly striking in fields involving images (such as radiology and pathology) and fields involving large amounts of data (such as genomics). Practicing oncologists are often confronted with new technologies claiming to predict response to therapy or predict the genomic make-up of patients. Underst-anding these new claims and technologies requires a deep understanding of the field. In this review, we provide an overview of the basis of deep learning. We describe various common tasks and their data requirements so that oncologists could be equipped to start such projects, as well as evaluate algorithms presented to them.
Core Tip: Designing projects and evaluating algorithms require a basic understanding of principles of machine learning. In addition, specific tasks have specific data requirements, annotation requirements, and applications. In this review, we describe the basic principles of machine learning, as well as explain various common tasks and their data requirements and applications in order to enable practicing oncologists to plan their own projects.
- Citation: Ramachandran A, Bhalla D, Rangarajan K, Pramanik R, Banerjee S, Arora C. Building and evaluating an artificial intelligence algorithm: A practical guide for practicing oncologists. Artif Intell Cancer 2022; 3(3): 42-53
- URL: https://www.wjgnet.com/2644-3228/full/v3/i3/42.htm
- DOI: https://dx.doi.org/10.35713/aic.v3.i3.42
Artificial intelligence (AI) has touched many areas of our everyday life. In medical practice also, it has shown great potential in several studies[1]. The implications of use of AI in oncology are profound, with applications ranging from assisting early screening of cancer to personalization of cancer therapy. As we enter this exciting transformation, practicing oncologists in any sub-field of oncology are oftentimes faced with various studies and products claiming to achieve certain results. Verifying these claims and implementing these in clinical practice remain an uphill task.
This is an educational review, through which we will attempt to familiarize the reader with AI technology in current use. We first explain some basic concepts, in order to understand the meaning of techniques labelled as AI, and then move into explaining the various tasks that can be performed by AI. In each we provide information on what kind of data would be required, what kind of effort would be required to annotate these images, as well as how to assess networks based on these tasks, for the benefit of those oncologists wishing to foray into the field for research, or for those wishing to implement these algorithms in their clinical practice.
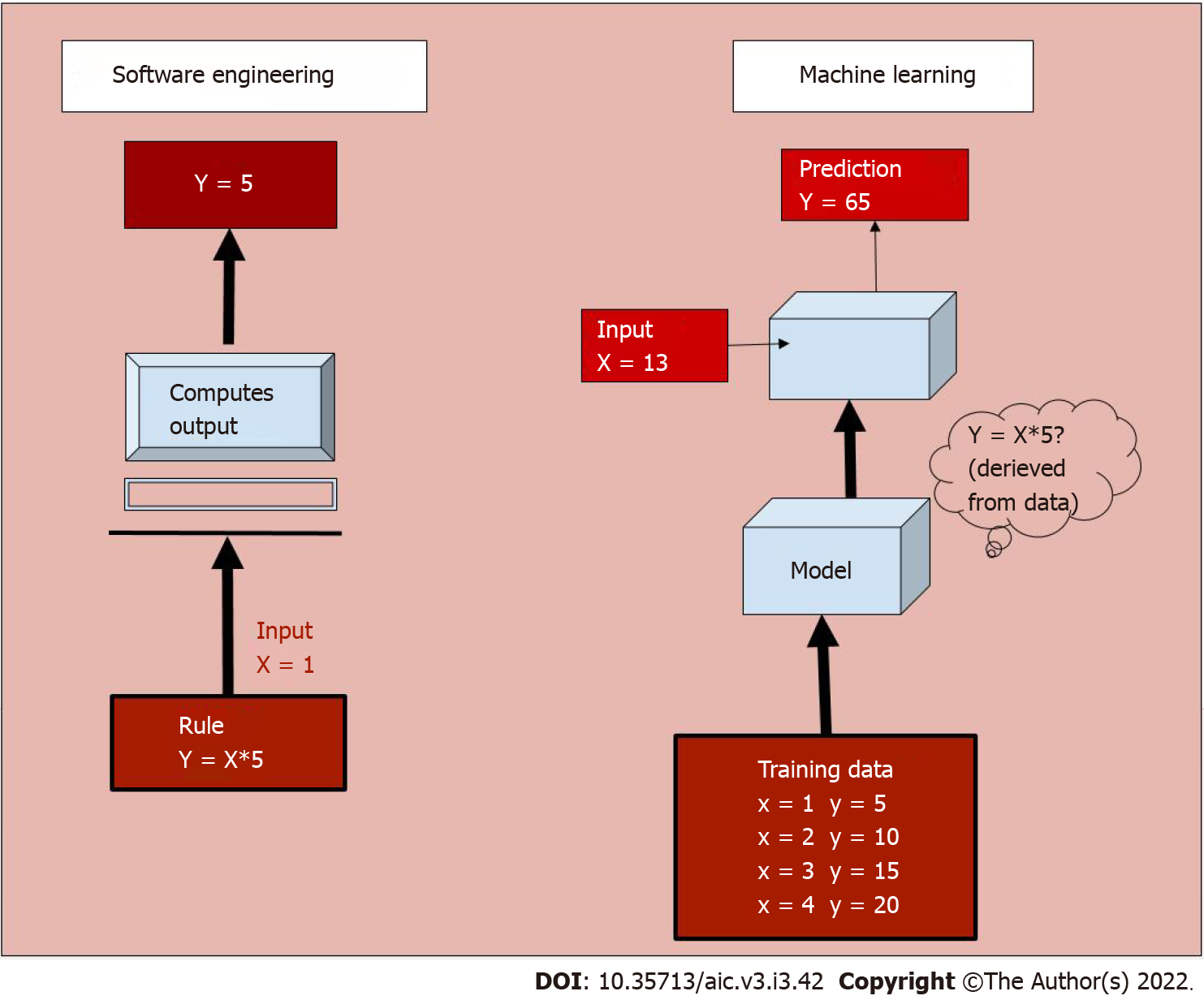
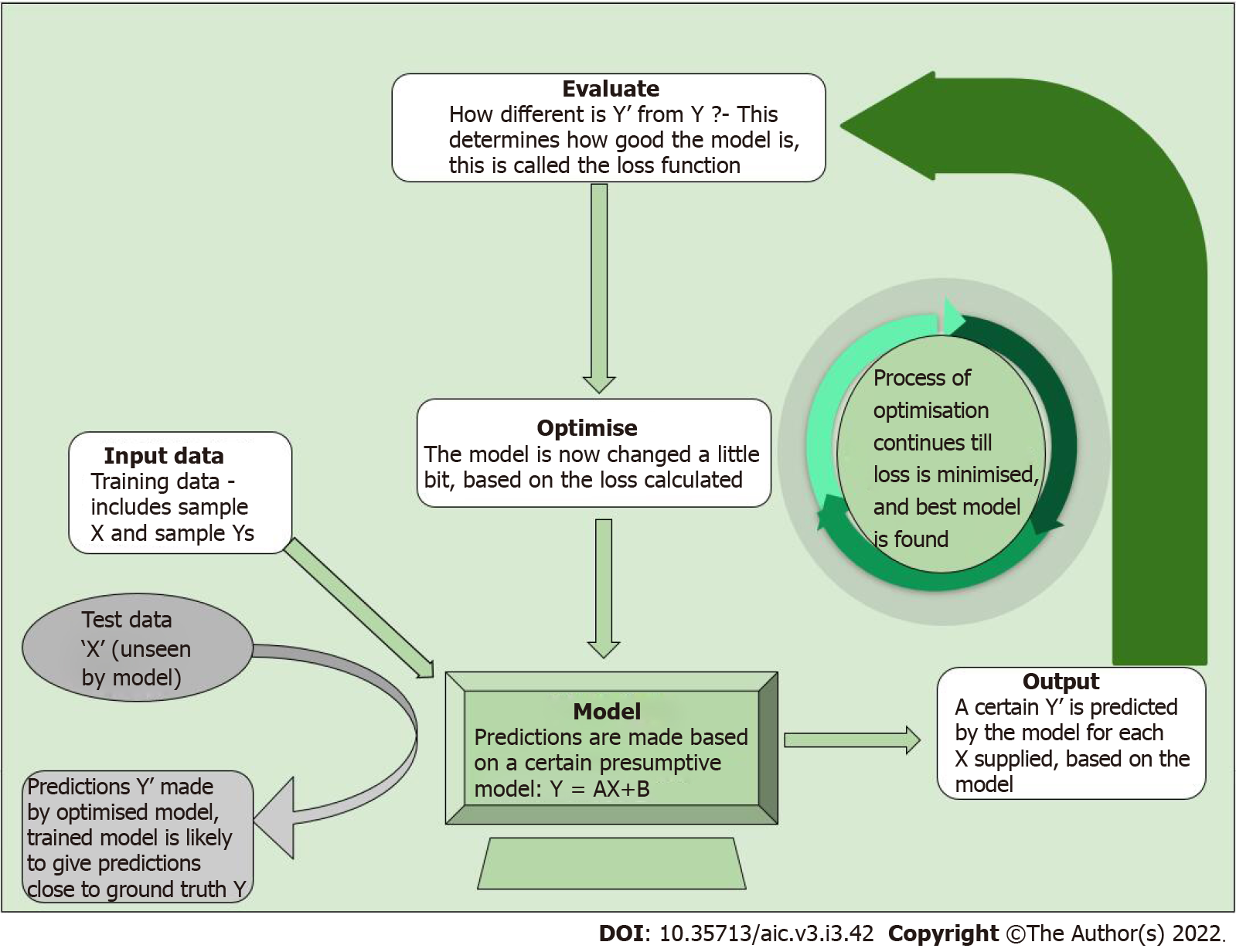
AI is a broad, non-specific term referring only to the “intelligence” in a specific task performed, irrespective of the method used. Machine learning (ML) is a subgroup of AI, and deep learning (DL) is a further subgroup of ML, which are data-driven approaches. Unlike traditional software engineering where a set of rules is defined upon which the computer’s outputs are based, ML involves learning of rules by “experience” without “explicit programming”[2,3]. What this means is that given large amounts of data which includes a set of inputs and the ideal outputs (training data), the task of ML is to understand a pattern within this set of inputs which results in outputs closest to the ideal output (Figure 1). The process of training the model is explained in Figure 2.
To understand this in medical terms, say the task of an AI system is to predict the survival of patients, given the stage of a particular tumour. If we were to use traditional software engineering, we would have to feed the median survival for each stage into the model, and teach the model to output the number corresponding to a particular stage. Whereas in case of a ML model, we would simply give as input, the stage and survival information of a few thousand patients. The model would learn the rules involved in making this prediction. While in the former case, we defined the rules (that is if stage= X then survival= Y), in the latter we only provided data, and the ML model deciphered the rules. While the former is rigid, that is, if a new therapy alters the survival, we would have to change the rules to accommodate the change, the latter learns with experience. As new data emerges, the ML model would learn to update the rules so that it can dynamically make accurate predictions. In addition, the ML model can take multiple inputs, say level of tumour markers, age, general condition, blood parameters into account, in addition to the stage of the patient, and personalise the survival prediction of a particular patient.
The above example also illustrates why ML models are data intensive. A good model needs to see a large amount of data, with adequate variability in parameters to make accurate predictions. For the same reason, AI has bloomed in disciplines which have large amount of digital data available, and these include ophthalmology, dermatology, pathology, radiology, and genomics. However, as curated digital data emerges in all fields, it is likely to touch and transform all fields of medical practice.
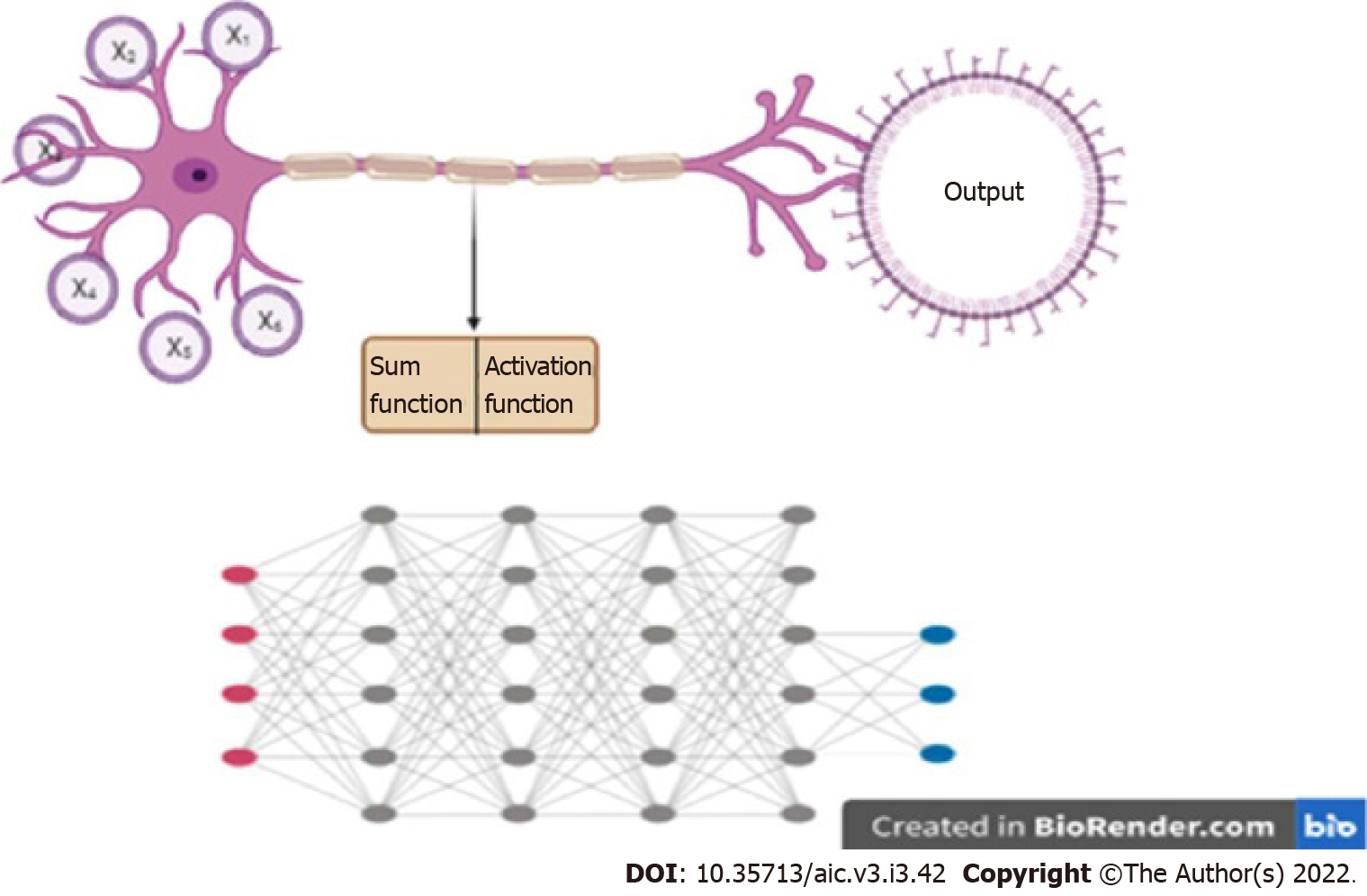
A particular kind of ML algorithm, called neural network, has been particularly effective in performing complex tasks. A neural network takes inspiration from a biological neuron, where it receives several inputs, performs a certain calculation, and goes through an activation function, where similar to a biological neuron, a decision on whether it should fire or not is made. When there are a number of layers of these mathematical functions, the network is known as a “deep neural network” (DNN), and the process is called “Deep Learning” (Figure 3). A DNN is capable of handling a large amount of data, and defining complex functions, which explains its ability to perform complex classification tasks and predictions.
A specific kind of DL, called convolutional neural network (CNN), has performed particularly well in image-related tasks. CNNs use “filters” which are applied to images, similar to the traditional image processing techniques. “Convolving” with a filter (a mathematical operation) results in highlighting certain features of an image. Given an input set of images, a CNN basically learns what set of filters highlights features of a particular image, most relevant to a given task. In other words, a CNN is learning the features of an image that may be crucial to making a decision. For example, in case of mammography, a CNN is trying to answer what features of a mammogram are most predictive of the presence of a cancer within.
Images contain information far beyond what meets the eye. While radiologists can interpret some of these features with the naked eye (such as margins, heterogeneity and density), pixel-by-pixel analysis of these images can yield significant amounts of hidden information. Studies have shown that these may be successfully correlated to outcomes such as patient survival and genomic mutations[4]. More specifically, it was shown by Choudhery et al[5] that in addition to differentiating among the molecular subtypes of breast cancer, texture features including entropy were significantly different among HER2 positive tumors showing complete response to chemotherapy. Other parameters such as standard deviation of signal intensity were found predictive in triple negative cancers. A similar study by Chen et al[6] in patients with non-small cell lung cancer treated with chemoradiotherapy showed that the ‘radiomics signature’ could predict failure of therapy. Therefore, using non-invasive imaging, it is thus possible to predict the mutations, response to specific drugs and best site of biopsy. Thus potentially, the therapy of the patient can be guided by markers mined from non-invasive imaging, making precision medicine a true possibility.
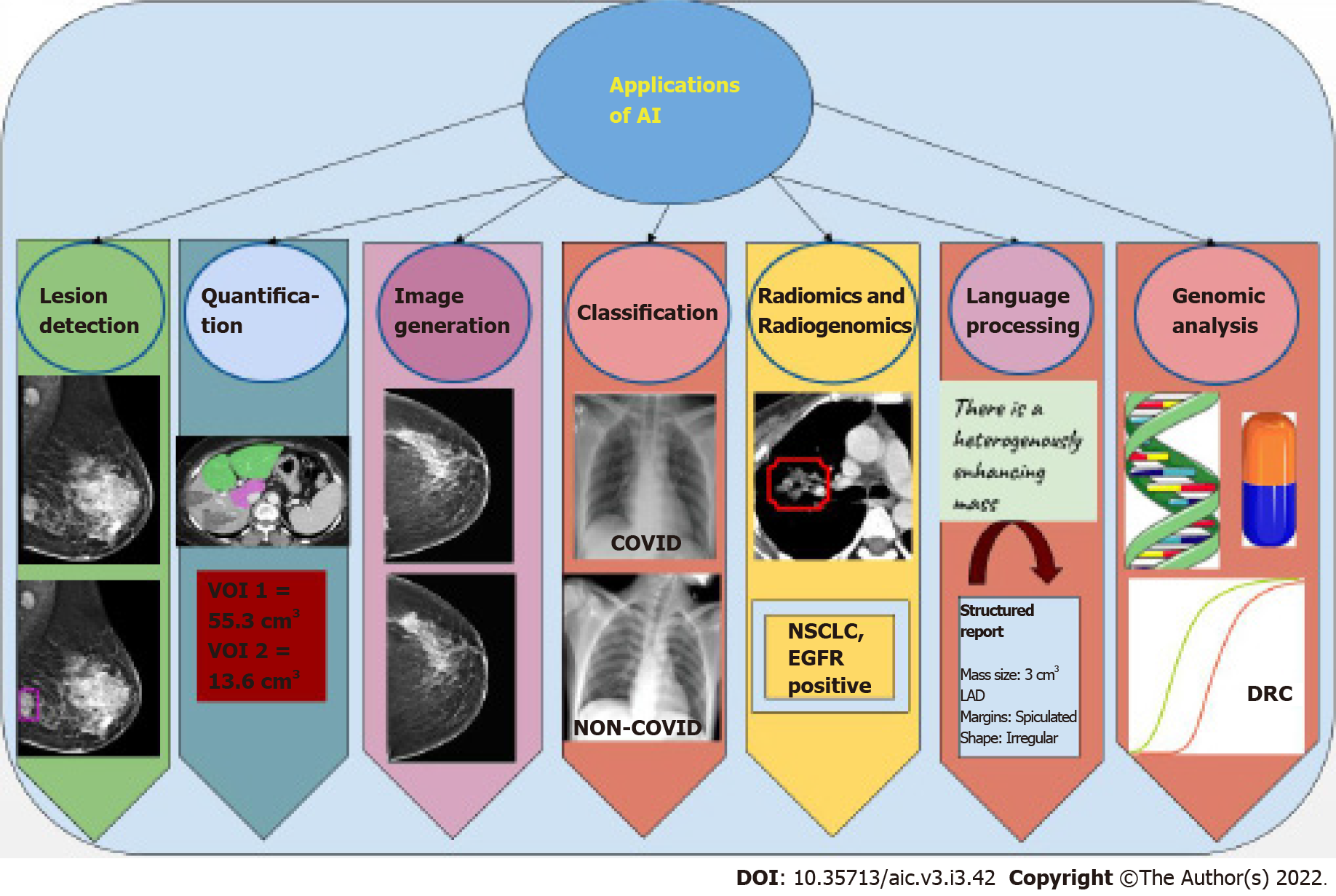
Most applications of AI in oncology are currently in the field of radiology and pathology, given the abundant digital data available in these fields. These tasks may be classified into specific categories (Figure 4). For readers wishing to foray into the field, an explanation of each kind of task, as well as data requirements and some examples of applications of these tasks is given below.
A classification task is one in which the AI algorithm classifies each image as belonging to one of several target categories. These categories are given at the level of image or patient. For instance, whether a particular mammogram has cancer or not.
Data requirement: Training the network requires input images (mammograms in the above example), and an image level ground truth label (presence or absence of cancer in the above case). These are relatively easy to obtain if reports are available in a digital format, since automated extraction of diagnosis from free-text reports may be performed. Usually thousands of such images are required for training. Large public datasets of labelled natural images exist, such as “ImageNet” with over 14 million images[7], and several classification networks trained on these databases also exist, such as Alexnet, Inception and ResNet. These networks trained on these large public databases can be adapted to the medical domain, a process called “transfer learning”.
Classification tasks can be evaluated by calculating the area under the receiver operating characteristic curve (AUROC) and by drawing a confusion matrix from which accuracy of classification can be calculated.
Applications: Some examples of classification tasks include breast density categorization on mammograms[8], detection of stroke on head computed tomography (CT) in order to prioritize their reading[9], and prioritising chest radiographs based on presence of pneumothorax in them[10].
Advantages: The most advantageous use of classification networks is for triage. These can be used to classify images that need urgent attention, or those that need a re-look by a reporting radiologist, pathologist, or ophthalmologist. This helps to reduce workload and effectively divert resources where required.
Disadvantages: When a classification task is performed by an algorithm, it simply classifies an image into a certain category, say ‘benign’ or ‘malignant’ for a mammogram, or ‘COVID’ or ‘non-COVID’ for a chest radiograph. It does not indicate which part of the image is used for classification, or indeed, if multiple lesions are present, which lesion is classified. This translates to reduced ‘explainability’ of such a model, where the results cannot be understood logically.
A detection task is one in which the network would predict the presence as well as location of a lesion on an image. Unlike a classification task, which is performed at the image or patient level, the detection task is performed at the lesion level. For example, if the network draws a box around a cancer on a mammogram, the task is a detection task.
Data requirement: Training requires images as input, and the ground truth needs to be provided as a box (called a bounding box) around each lesion, with their labels mentioned. This would typically have to be done prospectively, as this is not performed in the routine work-flow of most departments. In the above example, each mammogram would have to be annotated with bounding boxes by an expert radiologist (usually by multiple radiologists to avoid missing/misclassifying lesions), and each box would have to be assigned a label (as benign/malignant or with a BIRADS score, depending on what output is expected). Several publicly available datasets such as the COCO dataset[11] exist for natural images, with several networks trained on these datasets for object detection (such as RCNN, faster-RCNN and YOLO).
Detection tasks are evaluated by calculating the intersection over union between a predicted box and a ground truth box, that is, by calculating how close a predicted box is in comparison to the ground truth box. All boxes over a certain cut-off are considered a correct prediction. A free-response operating curve is drawn and sensitivity of the network at specific false positivity rates can be computed and compared.
Applications: The most prominent applications in oncology are detection of nodules on chest radiographs[12] and CT scans of the lungs[13-15], and detection of masses and calcifications on mammography[16].
A lesion segmentation task essentially involves classifying each pixel in the image as belonging to a certain category. So unlike a classification task (image or patient level) or detection task (lesion level), a segmentation task is performed at the pixel level. For instance, classification of each pixel of a CT image of the liver as background liver or a lesion would result in demarcating the exact margins of a lesion. The volume of these pixels may then be calculated to give the volume of the right lobe and left lobe of the liver separately.
Data requirement: Here, exact hand annotation of the lesion in question by the expert is required. This involves drawing an exact boundary demarcating the exact lesion in each section of the scan. Since this is routinely performed for radiotherapy planning, such data may be leveraged for building relevant datasets. Datasets like the COCO dataset exist with pixel level annotations for natural images.
These algorithms are evaluated with segmentation accuracy, IOU with the ground truth annotations (described in the previous section), or Dice scores[17].
Advantages: There is a tremendous advantage to the use of AI for segmentation, particularly quantification, in terms of increasing throughput and reducing the man-hours required for these tasks. In some cases such as quantification of extent of emphysema, which is particularly tedious for human operators, ready acceptance of AI may be found.
Applications: Automated liver volume calculations (liver volumetry) is an important application which can significantly reduce the time of the radiologist spent in the process[18,19]. Segmentation of cerebral vessels to perform flow calculations[20] and segmentation of ischemic myocardial tissue[21] are other such applications.
Image generation refers to the network “drawing” an image, based on images that it has seen. For instance, if a network is trained with low dose CT and corresponding high resolution CT images, it may learn to faithfully draw the high-resolution CT image, given the low dose CT. The most successful neural network to perform this task is called a generative adversarial network (GAN), first described by Ian Goodfellow[22]. This involves training two CNNs- a generator, which draws the image, and a discriminator, which determines whether a given image is real or generated. The two CNNs are trained simultaneously, with each trying to get better than the other.
Data requirement: This kind of network is usually trained in an “unsupervised” manner, that is, no ground truth is required. Therefore, no expert time is required in annotating these images. Only curated datasets of a particular kind of images are required.
This kind of network is difficult to evaluate, since no objective measure is typically defined. Evaluation by human eyes is generally considered the best.
Applications: GAN has found use in several interesting and evolving applications. This includes CT and magnetic resonance imaging (MRI) reconstruction techniques to improve spatial resolution while reducing the radiation dose or time of acquisition, respectively. GANs can also be trained to correct or remove artifacts from images[23]. An interesting application of GAN has been in generating images of a different modality, given an image of a certain modality. An example is generation of a positron emission tomography (PET) image from a CT image[24], a brain MRI image from a brain CT image[25], or a T2 weighted image from T1 weighted image[26].
Advantages: An interesting application of GAN has been used for simulation training for diagnostic imaging[27,28]. Students may be trained to recognise a wide variety of pathology using the synthetic images generated from these networks. This may be particularly important in certain scenarios such as detecting masses in dense breasts.
Disadvantages: These networks seem to possess a supra-human capability. The generated images cannot be verified for authenticity of texture or indeed even representation and thus may lead to an inherent mistrust of ‘synthetic’ images.
Natural language processing (NLP) refers to understanding of natural human language. While processing structured information is relatively easy, most data in the real world is locked up in the form of sentences in natural language. For example, understanding what is written in radiology reports would require processing of free-text, and this task is called NLP.
Data requirement: Large publically available datasets such as the “Google blogger corpus” (text) and “Spoken Wikipedia corpuses” (spoken language) are available, over which networks can be trained to understand natural language. However, large medical corpuses with reports pertaining to specific tasks are needed for tackling specific medical problems. With more robust electronic medical records (EMR), integrated Hospital and Radiology Information Systems (HIS and RIS), as well as recorded medical transcripts, this field is likely to grow rapidly.
Applications: The applications of NLP range from extraction of clinical information from reports and EMR to train deep neural networks, to designing chatbots for conversing with patients.
Predictive modelling has been at the core of medical practice for decades. While initial attempts were centered at developing scoring systems, or metrics that could be calculated from a few lab parameters, predictive modelling can be much more complex today because of the number of variables that ML systems can analyse.
A simple example of such a model is the “cholesterol ratio” (total cholesterol/HDL) which is used to estimate the risk of cardiac disease. As our models are capable of processing many variables, in fact capable of processing whole images, predictive models can be much more nuanced. Radiomics and radiogenomics are in fact an extension of the same, built to predict survival, response to therapy, or future risk of cancer, with more complex feature extraction and analysis from radiology images.
Data requirement: Building such models requires longitudinal data. Simple ML models would require lesser data in comparison to DL models. The amount of data required essentially depends upon which level ML is used at. For instance, if lesion segmentation is performed manually, feature extraction is performed with routine textural features, and feature selection is performed by means of traditional tools such as simple clustering or principle component analysis, then ML model would only use these selected features to make the desired prediction, and the amount of data required is relatively small. However, if DL is used end-to-end, the data requirement is much higher.
Predictive models are also assessed through AUROC and confusion matrices from which accuracy of prediction can be calculated.
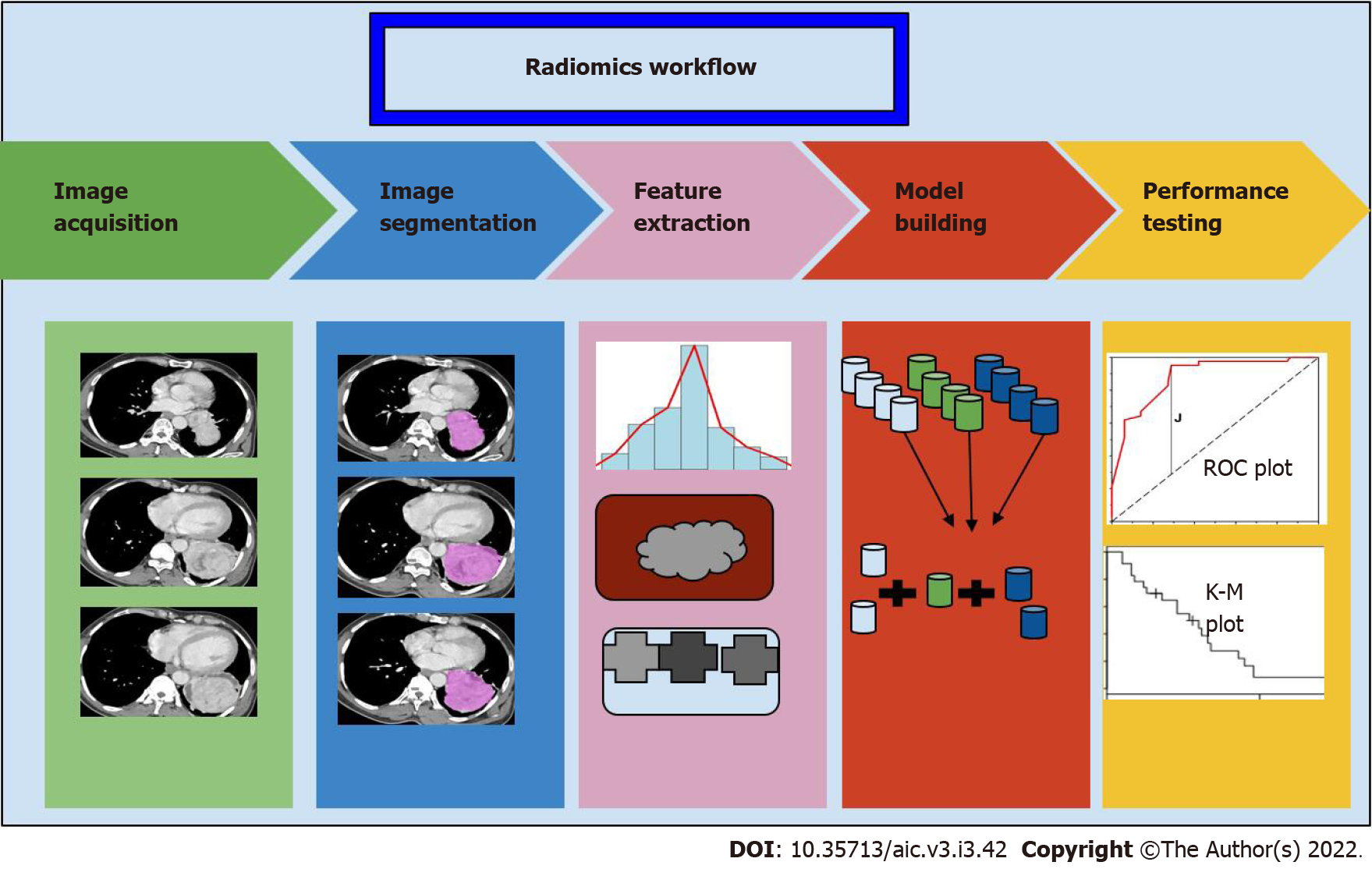
Radiomics involve four steps: (1) Segmentation; (2) Extraction of features; (3) Selection of features; and (4) model building for prediction (Figure 5). Segmentation involves drawing a margin around a lesion. This may be performed by an expert manually, or automatically. Features of the lesion are then defined. These may be semantic, that is defined by an expert, such as tissue heterogeneity, spiculated margins, or quantitative features (such as mean, median, histogram analysis, and filter-extracted features). This may yield several 100 features, of which overlapping features should be removed before analysis. Subsequently, a few selected features may then be fed into a ML model along with the outcomes that are to be predicted. ML or DL may also be applied at the initial stages, for segmentation and feature extraction itself, rather than at the last step.
Applications: Predictive models are extremely useful in oncology. Studies have shown that features extracted from images can be used to predict the response to various kinds of therapies. Morshid et al[29] and Abajian et al[30] showed good accuracy in predicting response to transarterial chemoembolisation. Studies have also correlated the imaging features extracted with genomic information, for example, several studies have shown that imaging features can accurately predict EGFR mutation status in patients with lung cancer[31-34]. Segal et al[35] showed that 28 CT texture features could decode 78% of the genes expressed in hepatocellular carcinoma. More recent work also shows that DL models can predict future risk of development of cancer. Eriksson et al[36] studied a model that identified women at a high likelihood of developing breast cancer within 2 years based on the present mammogram. All these pave the way towards more personalised management of patients with cancer.
The next generation of personalised medicine is undoubtedly ‘genomic medicine’, wherein not just targeted therapy but also diagnostic procedures are tailored as per the genetic make-up of an individual.
In addition, there is a growing effort towards population based studies for pooling of large scale genomic data and understanding the relationship between genomics, clinical phenotype, metabolism, and such domains. The challenges with these techniques are the huge amounts of data obtained from a single cycle, and the computational requirements in its processing and analysis. Thus, both ML and DL are ideally suited to deal with each step of the process starting from genome sequencing to data processing and interpretation.
For instance, a DL model that combined both histological and genomic data in patients with brain tumors to predict the overall survival, was able to show non-inferiority compared to human experts[37].
Data requirement: The essential in this field is not the data, but rather the ability to process the data. Since the human genome contains approximately 3 billion base pairs and thousands of genes, the data becomes extremely high dimensional. CNNs and recurrent neural networks (RNNs) have been proven to be the best approach to evaluate multiple DNA fragments in parallel, similar to the approach used in next generation sequencing (NGS)[38]. RNN models have also been used to perform microRNA and target prediction from gene expression data[39].
Applications: In addition to the applications detailed above, AI has also found use in variant identification, particularly Google’s ‘deep variant’ which has shown superior performance to existing methods despite not being trained on genomic data[40]. Other studies have also used ML to identify disease biomarkers and predict drug response[41,42].
Much of medical care today is moving away from patients, with focus shifting towards interpreting digital data in the form of blood reports, imaging data, pathology reports, genomic information, etc. The sheer amount of data has rendered face to face patient care less important, as synthesizing this information takes significant time and effort.
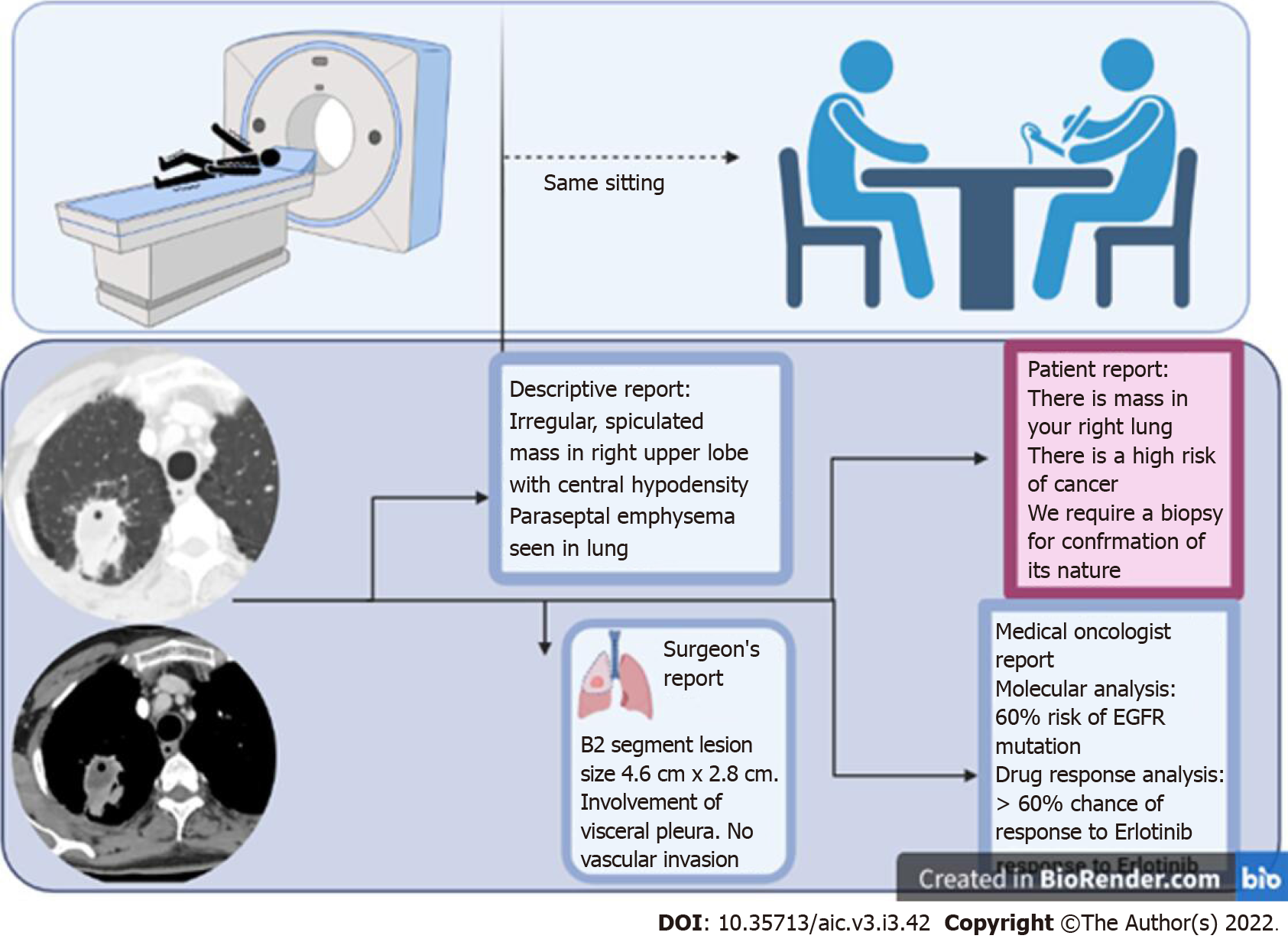
ML and DL have, however, ushered in a new era with endless possibilities. For instance, in a field like radiology, where AI is likely to have maximum impact, the onco-radiology reporting room of the future is likely to be dramatically different from where we are currently. AI, by reducing the amount of time spent in preparing a report, may pre-prepare images and sample reports, allowing a radiologist to spend time with the patient, examine the clinical files, and provide the report immediately after the examination (unlike in current practice where a radiologist sees the images, never meets the patient, and gives them a report about 24 h later). This report can potentially be transcribed into several reports simultaneously - for instance a patient friendly report, in easy to understand non-medical language, a physician report with important sections and lesions marked on the image, and a traditional descriptive radiology report. In fact, the radiology report is likely to have much more information than currently considered possible, including the possibility of a particular mutation, possibility of response to a particular therapy, and even reconstructed images translated to different modalities which may help determine the most important site of biopsy.
While Amara’s law for new technology may well apply (which says that any new technology is overestimated early on, and underestimated later[43]), the potential of AI and the vistas that it opens up cannot be ignored. As the technology evolves, many of the changes it brings about will enable a leap towards the era of personalised medicine (Figure 6).
AI thus holds great potential. The most significant advantage of AI rests in the fact that since it is data-driven, it holds the potential to derive inferences from very large databases, in a short span of time. It brings with it the possibility to standardize clinical care, reduce interpretation times, and improve accuracy of diagnosis, and may help enable patient centricity in cancer care.
Like any new technology, however, AI must be used with care and only after thorough clinical tests. The most significant disadvantage derives from the fact that it is a “black-box”, with little explainability. Little is known about the reasons behind the decisions taken by neural networks, making it imperative for the decisions to be seen and approved by human experts.
In summary, there is tremendous scope of AI in cancer care, particularly in the image related tasks. With the development of neural networks capable of performing complex tasks, the era of personalised medicine seems a reality with AI. Thus, judicious use must be encouraged to maximise the long term benefits that outlive the initial enthusiasm of discovery.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): D, D
Grade E (Poor): 0
P-Reviewer: Cabezuelo AS, Spain; Tanabe S, Japan; Wang R, China A-Editor: Yao QG, China S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Dembrower K, Wåhlin E, Liu Y, Salim M, Smith K, Lindholm P, Eklund M, Strand F. Effect of artificial intelligence-based triaging of breast cancer screening mammograms on cancer detection and radiologist workload: a retrospective simulation study. Lancet Digit Health. 2020;2:e468-e474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 91] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 2. | Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. Radiographics. 2017;37:505-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 647] [Cited by in F6Publishing: 653] [Article Influence: 93.3] [Reference Citation Analysis (0)] |
| 3. | Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep Learning: A Primer for Radiologists. Radiographics. 2017;37:2113-2131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 511] [Cited by in F6Publishing: 598] [Article Influence: 99.7] [Reference Citation Analysis (0)] |
| 4. | Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234-1248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1505] [Cited by in F6Publishing: 1342] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 5. | Choudhery S, Gomez-Cardona D, Favazza CP, Hoskin TL, Haddad TC, Goetz MP, Boughey JC. MRI Radiomics for Assessment of Molecular Subtype, Pathological Complete Response, and Residual Cancer Burden in Breast Cancer Patients Treated With Neoadjuvant Chemotherapy. Acad Radiol. 2022;29 Suppl 1:S145-S154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 27] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 6. | Chen X, Tong X, Qiu Q, Sun F, Yin Y, Gong G, Xing L, Sun X. Radiomics Nomogram for Predicting Locoregional Failure in Locally Advanced Non-small Cell Lung Cancer Treated with Definitive Chemoradiotherapy. Acad Radiol. 2022;29 Suppl 2:S53-S61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 7. | Deng J, Dong W, Socher R, Li-Jia Li, Kai Li, Li Fei-Fei. ImageNet: A large-scale hierarchical image database, 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, IEEE, 2009; 248–255. [DOI] [Cited in This Article: ] |
| 8. | Wu N, Geras KJ, Shen Y, Su J, Kim SG, Kim E, Wolfson S, Moy L, Cho K. Breast density classification with deep convolutional neural networks. ArXiv171103674 Cs Stat 2017. [DOI] [Cited in This Article: ] |
| 9. | Karthik R, Menaka R, Johnson A, Anand S. Neuroimaging and deep learning for brain stroke detection - A review of recent advancements and future prospects. Comput Methods Programs Biomed. 2020;197:105728. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Taylor AG, Mielke C, Mongan J. Automated detection of moderate and large pneumothorax on frontal chest X-rays using deep convolutional neural networks: A retrospective study. PLoS Med. 2018;15:e1002697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 107] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 11. | Lin T-Y, Maire M, Belongie S, Bourdev L, Girshick R, Hays J, Perona P, Ramanan D, Zitnick CL, Dollár P. Microsoft COCO: Common Objects in Context. ArXiv14050312 Cs, 2015. [DOI] [Cited in This Article: ] |
| 12. | Nam JG, Park S, Hwang EJ, Lee JH, Jin KN, Lim KY, Vu TH, Sohn JH, Hwang S, Goo JM, Park CM. Development and Validation of Deep Learning–based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology. 2019;290:218-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 234] [Cited by in F6Publishing: 289] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 13. | Hamidian S, Sahiner B, Petrick N, Pezeshk A. 3D Convolutional Neural Network for Automatic Detection of Lung Nodules in Chest CT. Proc SPIE Int Soc Opt Eng. 2017;10134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Guo W, Li Q. High performance lung nodule detection schemes in CT using local and global information. Med Phys. 2012;39:5157-5168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Setio AA, Ciompi F, Litjens G, Gerke P, Jacobs C, van Riel SJ, Wille MM, Naqibullah M, Sanchez CI, van Ginneken B. Pulmonary Nodule Detection in CT Images: False Positive Reduction Using Multi-View Convolutional Networks. IEEE Trans Med Imaging. 2016;35:1160-1169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 792] [Cited by in F6Publishing: 489] [Article Influence: 61.1] [Reference Citation Analysis (0)] |
| 16. | Abdelhafiz D, Yang C, Ammar R, Nabavi S. Deep convolutional neural networks for mammography: advances, challenges and applications. BMC Bioinformatics. 2019;20:281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 17. | Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in Auto-Segmentation. Semin Radiat Oncol. 2019;29:185-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 204] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 18. | Winkel DJ, Weikert TJ, Breit HC, Chabin G, Gibson E, Heye TJ, Comaniciu D, Boll DT. Validation of a fully automated liver segmentation algorithm using multi-scale deep reinforcement learning and comparison versus manual segmentation. Eur J Radiol. 2020;126:108918. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 19. | Winther H, Hundt C, Ringe KI, Wacker FK, Schmidt B, Jürgens J, Haimerl M, Beyer LP, Stroszczynski C, Wiggermann P, Verloh N. A 3D Deep Neural Network for Liver Volumetry in 3T Contrast-Enhanced MRI. Rofo. 2021;193:305-314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 20. | Chen L, Xie Y, Sun J, Balu N, Mossa-Basha M, Pimentel K, Hatsukami TS, Hwang JN, Yuan C. Y-net: 3D intracranial artery segmentation using a convolutional autoencoder. ArXiv171207194 Cs Eess, 2017. [DOI] [Cited in This Article: ] |
| 21. | Chen C, Qin C, Qiu H, Tarroni G, Duan J, Bai W, Rueckert D. Deep Learning for Cardiac Image Segmentation: A Review. Front Cardiovasc Med. 2020;7:25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 395] [Cited by in F6Publishing: 282] [Article Influence: 70.5] [Reference Citation Analysis (0)] |
| 22. | Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative Adversarial Networks. ArXiv14062661 Cs Stat, 2014. [DOI] [Cited in This Article: ] |
| 23. | Harms J, Lei Y, Wang T, Zhang R, Zhou J, Tang X, Curran WJ, Liu T, Yang X. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. 2019;46:3998-4009. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 132] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 24. | Ben-Cohen A, Klang E, Raskin SP, Soffer S, Ben-Haim S, Konen E, Amitai MM, Greenspan H. Cross-modality synthesis from CT to PET using FCN and GAN networks for improved automated lesion detection. Eng Appl Artif Intell. 2019;78:186-194. [DOI] [Cited in This Article: ] |
| 25. | Jin CB, Kim H, Liu M, Jung W, Joo S, Park E, Ahn YS, Han IH, Lee JI, Cui X. Deep CT to MR Synthesis Using Paired and Unpaired Data. Sensors (Basel). 2019;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 26. | Ourselin S, Alexander DC, Westin CF, Cardoso MJ. Preface. 24th International Conference, IPMI 2015, Sabhal Mor Ostaig, Isle of Skye, UK, June 28 - July 3, 2015. Proceedings. Inf Process Med Imaging. 2015;24:V-VII. [PubMed] [Cited in This Article: ] |
| 27. | Kutter O, Shams R, Navab N. Visualization and GPU-accelerated simulation of medical ultrasound from CT images. Comput Methods Programs Biomed. 2009;94:250-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Vitale S, Orlando JI, Iarussi E, Larrabide I. Improving realism in patient-specific abdominal ultrasound simulation using CycleGANs. Int J Comput Assist Radiol Surg. 2020;15:183-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Morshid A, Elsayes KM, Khalaf AM, Elmohr MM, Yu J, Kaseb AO, Hassan M, Mahvash A, Wang Z, Hazle JD, Fuentes D. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol Artif Intell. 2019;1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 30. | Abajian A, Murali N, Savic LJ, Laage-Gaupp FM, Nezami N, Duncan JS, Schlachter T, Lin M, Geschwind JF, Chapiro J. Predicting Treatment Response to Intra-arterial Therapies for Hepatocellular Carcinoma with the Use of Supervised Machine Learning-An Artificial Intelligence Concept. J Vasc Interv Radiol. 2018;29:850-857.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 106] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 31. | Li S, Ding C, Zhang H, Song J, Wu L. Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys. 2019;46:4545-4552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 32. | Digumarthy SR, Padole AM, Gullo RL, Sequist LV, Kalra MK. Can CT radiomic analysis in NSCLC predict histology and EGFR mutation status? Medicine (Baltimore). 2019;98:e13963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 33. | Rizzo S, Petrella F, Buscarino V, De Maria F, Raimondi S, Barberis M, Fumagalli C, Spitaleri G, Rampinelli C, De Marinis F, Spaggiari L, Bellomi M. CT Radiogenomic Characterization of EGFR, K-RAS, and ALK Mutations in Non-Small Cell Lung Cancer. Eur Radiol. 2016;26:32-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 157] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 34. | Tu W, Sun G, Fan L, Wang Y, Xia Y, Guan Y, Li Q, Zhang D, Liu S, Li Z. Radiomics signature: A potential and incremental predictor for EGFR mutation status in NSCLC patients, comparison with CT morphology. Lung Cancer. 2019;132:28-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 35. | Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675-680. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 400] [Cited by in F6Publishing: 399] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 36. | Eriksson M, Czene K, Strand F, Zackrisson S, Lindholm P, Lång K, Förnvik D, Sartor H, Mavaddat N, Easton D, Hall P. Identification of Women at High Risk of Breast Cancer Who Need Supplemental Screening. Radiology. 2020;297:327-333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 37. | Mobadersany P, Yousefi S, Amgad M, Gutman DA, Barnholtz-Sloan JS, Velázquez Vega JE, Brat DJ, Cooper LAD. Predicting cancer outcomes from histology and genomics using convolutional networks. Proc Natl Acad Sci U S A. 2018;115:E2970-E2979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 426] [Cited by in F6Publishing: 455] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 38. | D’Agaro E. Artificial intelligence used in genome analysis studies. EuroBiotech J. 2018;2:78-88. [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Lee B, Baek J, Park S, Yoon S. deepTarget: End-to-end Learning Framework for microRNA Target Prediction using Deep Recurrent Neural Networks, Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, New York, NY, USA, Association for Computing Machinery, 2016; 434–442. [DOI] [Cited in This Article: ] |
| 40. | Poplin R, Chang PC, Alexander D, Schwartz S, Colthurst T, Ku A, Newburger D, Dijamco J, Nguyen N, Afshar PT, Gross SS, Dorfman L, McLean CY, DePristo MA. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36:983-987. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 453] [Cited by in F6Publishing: 483] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 41. | Zafeiris D, Rutella S, Ball GR. An Artificial Neural Network Integrated Pipeline for Biomarker Discovery Using Alzheimer's Disease as a Case Study. Comput Struct Biotechnol J. 2018;16:77-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 42. | Madhukar NS, Elemento O. Bioinformatics Approaches to Predict Drug Responses from Genomic Sequencing. Methods Mol Biol. 2018;1711:277-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 43. | Ratcliffe S. Oxford Essential Quotations, Oxford University Press. 2016. [Cited in This Article: ] |