INTRODUCTION
The global tumor statistics report released in 2018 shows that lung cancer is the malignant tumor with the highest morbidity and mortality in the world. The incidence of lung cancer accounts for 11.6% of the incidence of all tumors, and the mortality rate accounts for 18.4% of the deaths of all tumors[1]. Due to the late onset of clinical symptoms and limited screening procedures, a large number of patients are diagnosed as advanced[2]. Histologically, about 85% of new lung cancer cases are classified as non-small cell lung cancer (NSCLC), 10% are small cell lung cancer, and 5% are other variants[3]. Most NSCLC can be divided into three categories: squamous cell carcinoma, adenocarcinoma and large cell carcinoma[4]. Patients need the most accurate personalized treatment from doctors. Therefore, doctors need to obtain genomics, proteomics, immunohistochemistry and imaging data, in addition to histological, clinical and demographic information in order to develop precise treatment plans for patients. There are many factors, such as high cost of testing and treatment discontinuity, which will limit the timely access to data. This has aroused people’s interest in developing artificial intelligence.
Artificial intelligence (AI) is an important product of the rapid development of computer technology. It has a profound impact on the development of human society and the progress of science and technology through communication and cooperation with multidisciplinary and multifield, especially the organic combination with medicine, which is one of the most promising fields. John McCarthy first proposed the concept of AI: To develop machine software with human thinking mode, so that computers can think like humans[5]. Machine learning (ML) is a method to realize AI, which belongs to a subfield of AI. It analyzes and interprets data through machine algorithms, learns from it, and then makes decisions or predictions about something. Therefore, unlike manually writing software routines to complete specific tasks with a specific set of instructions, machines use a large number of data and algorithms to “train”, which give machines the ability to learn how to perform tasks. ML comes directly from the idea of the early artificial intelligence crowd. For many years, algorithm methods include decision tree learning, inductive logic programming, clustering, reinforcement learning and Bayesian network, etc. These algorithms allow information to be classified, predicted and segmented to provide insights that are difficult to obtain by the human eye or cognitive system.
Deep learning is a technology to realize ML. There are two key aspects in the description of advanced definition of deep learning: (1) A model composed of multilayer or multistage nonlinear information processing; and (2) A supervised or unsupervised learning method for feature representation at a higher and more abstract level[6]. There are many kinds of network learning models for deep learning, such as convolutional neural networks (CNN), recurrent neural networks, bi-directional long-term and short-term memory cyclic neural networks, multilayer neural networks, etc. Among them, the CNN is one of the representative algorithms of deep learning, which is a kind of feed forward neural networks with deep structure and convolution calculation. It consists of a series of layers. Each layer performs specific operations, such as convolution, pooling, loss calculation, etc. Each middle layer receives the output of the previous layer as its input and finally extracts the high-level abstraction through the fully connected layer. In the process of back propagation in the training stage, the weights of neural connection and kernel are optimized continuously. A CNN has the ability of representation learning, which can classify input information according to its hierarchical structure. Therefore, it is also called “translation invariant artificial neural network (ANN)”.
There are two main methods of data processing in ML: Supervised learning and unsupervised learning. Supervised learning specifically refers to the use of labeled data learning process to assist, so as to achieve learning objectives. The advantage is that the generalization ability of the machine itself can be given full play, and problems such as classification and regression can be effectively solved. Unsupervised learning does not need to be marked, and it explores the similarity between instances according to specific indicators and methods or the value relationship among features. The algorithms commonly used in unsupervised learning are as follows: Deep confidence network, automatic encoder, etc. The most important research problems of unsupervised learning include clustering, correlation analysis and dimension reduction. Other learning methods include reinforcement learning, which optimizes the model to get the best decision by giving different feedback to different choices in the iterative process, semisupervised learning that mixes supervised and unsupervised learning and transfer learning with models as an experiential training.
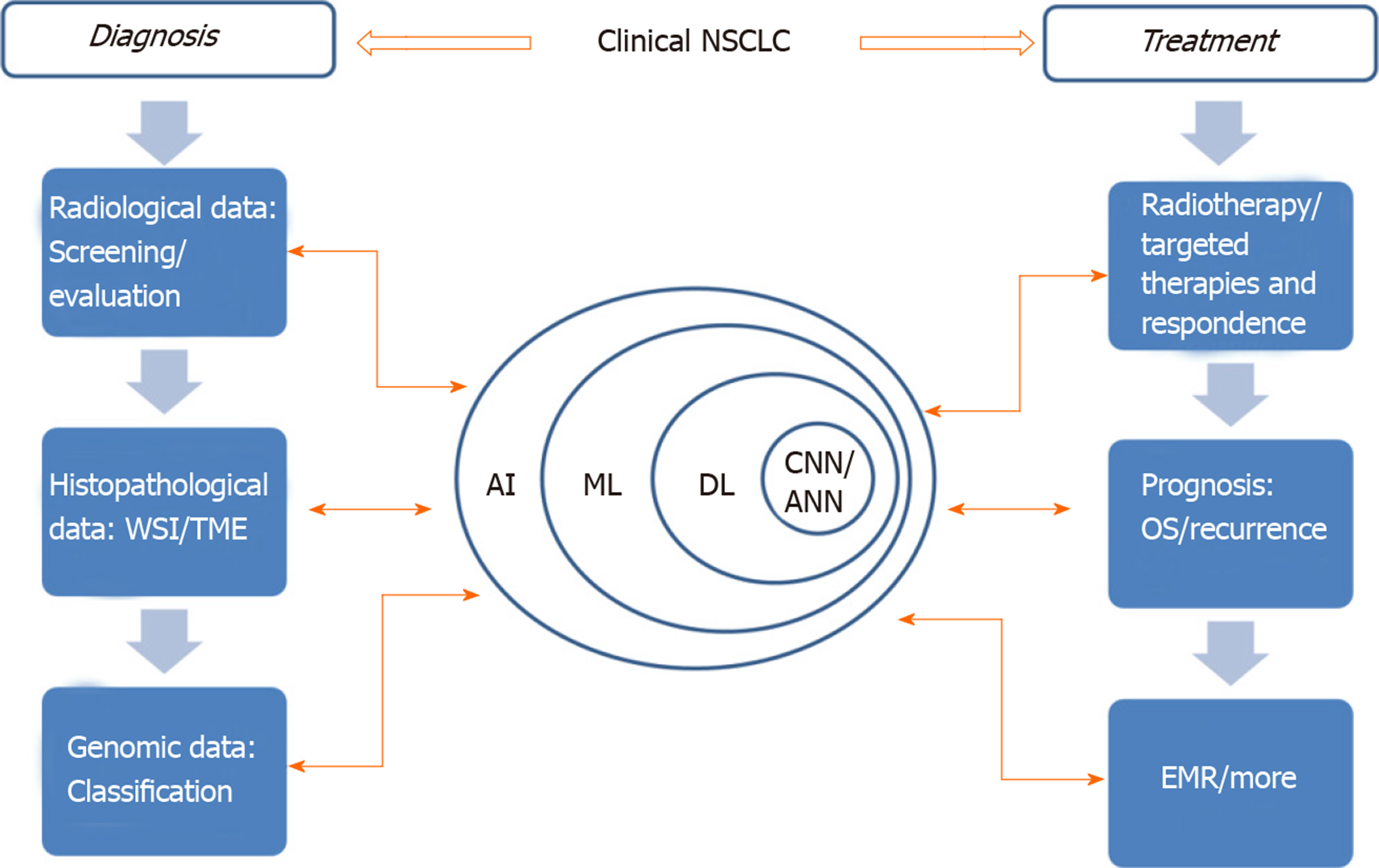
AI can improve patients’ treatment results, ameliorate patients’ treatment process and even mend medical management[7]. In view of the increasing application of AI in lung cancer treatment (Figure 1), this paper will review the AI applications being developed for NSCLC detection and treatment as well as the challenges facing clinical adaptability.
Figure 1 The application of artificial intelligence involved in clinical non-small cell lung cancer.
Learning process and application of AI in different fields are indicated by those two-way arrows. AI: Artificial intelligence; ANN: Artificial neural network; CNN: Convolutional neural networks; DL: Deep learning; EMR: Electronic medical record; ML: Machine learning; NSCLC: Non-small cell lung cancer; OS: Overall survival time; TME: Tumor microenvironment; WSI: Whole slide image.
APPLICATION OF AI IN SCREENING AND PRELIMINARY EVALUATION OF NSCLC
Pulmonary nodules are the early signs of lung cancer, which are of great significance for the diagnosis of early lung cancer. Early detection, early diagnosis and early treatment can improve the survival rate and prolong the survival time of patients. The national lung screening test showed that low-dose computed tomography (LDCT) screening was associated with a significant 20% reduction in overall mortality among current and previous high-risk smokers[8]. While conducting LDCT screening to detect patients with early-stage lung cancer, the number of health checkups, disease screenings and follow-up examinations is increasing. As a result, the workload of radiologists has multiplied. The increasing workload aggravates the fatigue of doctors, affects the quality of reading images and the accuracy of diagnosis results. The emergence of AI is just like a drop of sweet dew in a long drought for radiologists. AI can carry out self-learning and self-evolution under semi-supervision. At the same time as improving the accuracy of diagnosis, the time for doctors to read the images is greatly shortened, which solves the clinical needs well[9].
Most uncertain lung nodules were discovered by accident[10]. Every year, more than 1.5 million Americans are diagnosed with accidental detection of lung nodules[11]. Most of these nodules are benign granuloma and about 12% may be malignant[12]. Another potential hazard of lung cancer screening is the over diagnosis of slow-growing, inactive cancers. If left untreated, these cancers may not pose a threat. Therefore, over diagnosis must be identified and significantly reduced. Identifying the nature of pulmonary nodules by AI can effectively reduce the clinical work pressure as well as the long-term follow-up workload and ameliorate the psychological pressure of pulmonary nodule owners. In the field of cancer imaging, AI has found tremendous utility in three main clinical tasks: Detection, characterization and monitoring. In current clinical practice, imaging methods used to assess the presence of lung cancer include chest X-ray, computed tomography (CT) and positron emission tomography/computed tomography (PET/CT).
Chest X-ray is one of the most commonly used methods. The covering of the chest ribs on the lung field often affects the radiologists’ reading of the film and increases the missed diagnosis rate of the lung nodule shadow. von Berg et al[13] used a dual energy subtraction technology based on ANN to reduce the bone density shadow in the X-ray film, expose the lung nodule covered by the bone structure and improve the sensitivity and specificity of the radiologist in the diagnosis of lung nodules. Nam et al[14] recently developed an algorithm for detecting malignant pulmonary nodules on chest X-ray films based on deep learning and compared its performance with that of physicians, half of whom were radiologists. They used 43292 cases of chest X-ray data. The ratio of normal to pathological changes was 3.67. Using external validation data sets, they found that the area under the curve (AUC) of the developed algorithm was higher than 17 of the 18 doctors. When all doctors used this algorithm as the second reader, they found the improvement of nodule detection.
For lung cancer screening, the sensitivity and specificity of LDCT are much higher than that of general chest X-ray[15]. More than 200 thin-layer images can be reconstructed after high-resolution CT scanning or spiral CT scanning, which results in excessive reading of radiologists. Pulmonary nodules < 3 mm are more time-consuming and laborious. This has caused a considerable workload for radiologists in the traditional mode. Pulmonary nodule AI detection software is most sensitive to pulmonary nodules of 3-6 mm followed by nodules above 6 mm. Nodules of 3-6 mm are the most easily missed diagnosis by human vision[16]. After the application of AI, the daily working time can be halved without changing the inspection amount, and there will be no missed diagnosis due to excessive fatigue[17,18]. Detection refers to the positioning of objects of interest in X-rays or CTs and is collectively referred to as computer-aided detection[19]. In the early 2000s, methods of computer-aided detection for automatically detecting lung nodules on CT were based on traditional ML methods, such as support vector machines[20]. Computer-aided detection is used as an assistant in LDCT screening to find missed cancers and to detect brain metastases on MRI to improve radiological interpretation time while maintaining high detection sensitivity[21]. The computer-aided detection x system has been used for the diagnosis of pulmonary nodules by thin-layer CT[22].
Due to the simplicity of clinical implementation, size-based measurements such as the longest tumor diameter are widely used for staging and response assessment. However, size-based features and disease stages have limitations such as imprecise diagnosis. A preliminary work shows that AI can automatically quantify the radiographic characteristics of tumor phenotype, which has a significant prognosis for many types of cancer, including lung cancer[23]. Liu et al[24] combined a model of four semantic features (minor axis diameter, contour, concavity and texture) of quantitative scores. The accuracy of distinguishing malignant and benign nodules in lung cancer screening environment was 74.3%. In a separate study[25], semantic features were identified from small lung nodules (less than 6 mm) to predict the incidence of lung cancer in the context of lung cancer screening. The AUC of the final model was 0.930 based on the total score of emphysema, vascular attachment, nodal location, border definition and concavity. Paul et al[26] used a kind of pre-trained CNN after large-scale data training to detect lung cancer by extracting the features of CT images. They combined the extracted deep neural network features with the traditional quantitative features and obtained 90% accuracy (AUC: 0.935) by using the five best corrected linear unit features and five best traditional features extracted by vgg-f pre-trained CNN.
In recent years, the number of pure ground glass nodules (pGGN) has increased significantly. Judging its nature and making the treatment plan is very important. Qi et al[27] retrospectively analyzed the clinical follow-up data of 573 CT scans belonging to 110 patients with pGGNs from January 2007 to October 2018. The Dr. Wise system based on CNN was used to segment the initial CT scan and all subsequent CT scans automatically. Then, the diameter, density, volume, mass, volume doubling time and mass doubling time of pGGNs were calculated. Kaplan-Meier analyses with the log-rank test and Cox proportional hazards regression analysis were used to analyze the cumulative percentages of pGGN growth and identify risk factors for growth. It was found that persistent pGGNs showed a slow course. The 12-mo, 24.7-mo and 60.8-mo cumulative percentages of pGGN growth were 10%, 25.5% and 51.1%, respectively. Deep learning helps to clarify the natural history of pGGNs accurately. Those pGGNs with lobulated sign and larger initial diameter, volume and mass are more likely to grow up. Ardila et al[28] trained a deep learning algorithm on the NLST dataset, which came from 14851 patients and 578 of those patients developed lung cancer the following year. They tested the model on the first test data set of 6716 patients, and the AUC reached 94.4%. A part of 507 patients was compared with six radiologists. When a single CT is analyzed, the performance of the model was the same or higher than that of all radiologists.
The diagnosis of simultaneous or metachronous multiple pulmonary nodules is a new challenge for clinicians. In a retrospective study[29], a total of 53 patients with multiple pulmonary nodules, simultaneously or metachronously, were included. The coincidence rate of AI diagnosis and postoperative pathology to benign and malignant lesions was 88.8%. AI may represent a relevant diagnostic aid that can display more accurate and objective results when diagnosing multiple lung nodules. It may reduce the interpretation of results by displaying visual information directly to doctors and patients and the clinical status of multiple primary lung cancer patients. The time required and a reasonable follow-up and treatment plan may be more beneficial to the patient.
PET/CT using 18F-fluorodeoxyglucose (FDG) has been established as a great imaging method for the staging of patients with lung cancer[30]. Schwyzer et al[31] assessed whether machine learning would help detect lung cancer in FDG-PET imaging against the background of ultra-low-dose PET scans. The ANN was used to identify 3936 PET images, including images of lung tumors visible to the naked eye and image slices of patients without lung cancer. Based on clinical standard radiation dose PET images (PET 100%), 10% dose and 3.3% radiation dose (approximately 0.11 mSv), the diagnostic performance of the artificial neural network was evaluated. Their results indicated that even at very low effective radiation doses of 0.11 mSv, machine learning algorithms may contribute to fully automated lung cancer detection.
More and more new PET and single-photon emission computerized tomography tracers are used to explore various aspects of tumor biology, and hybrid multimodal imaging is increasingly used to provide multiparameter measurements. AI is needed to deal with the huge workload. According to reports[32], texture and color analysis of human FDG-PET images can be used to judge heterogeneity within tumors, thereby distinguishing NSCLC subtypes. Using support vector machine algorithm to extract texture and color features from FDG-PET images to differentiate histopathological tumor subtypes (squamous cell carcinoma and adenocarcinoma), the area under the receiver operating characteristic curve was 0.89. The use of the least absolute shrinkage and selection operator method[33] to derive radiographic descriptors of metastatic lymph nodes from FDG-PET images of patients with NSCLC has been found relate better with overall survival (OS) than the radiological data extracted from the primary tumor. Wang et al[34] made a comparison of ML methods for classifying NSCLC mediastinal lymph node metastasis from PET/CT images. A CNN and four ML methods (random forest, support vector machine, adaptive boosting and artificial neural networks) were used to classify mediastinal lymph node metastases of NSCLC. PET/CT images of 1397 lymph nodes were collected from 168 patients and were evaluated by the five methods with corresponding pathology analysis results as gold standard. The accuracy of CNN is 86%, which is not significantly different from the best ML method that uses standard diagnostic features or a combination of diagnostic features and texture features. CNN is more accurate than ML methods that simply use texture features.
APPLICATION OF AI IN HISTOPATHOLOGY OF NSCLC
In the differential diagnosis of lung cancer, it is necessary to classify the types or subtypes accurately. Because the hematoxylin-eosin (HE) stained full-scale whole slide image (WSI) is usually at the megapixel level, the much smaller image blocks (about 300 × 300 pixels) extracted from it are often used as training input. For example, Wang et al[35] trained a CNN model; each 300 × 300 pixel image block of lung adenocarcinoma WSIs stained by HE was classified as malignant or nonmalignant. The overall classification accuracy (malignant and nonmalignant) of the test set was 89.8%. This method can detect tumor rapidly when the tumor area is very small, which will greatly help pathologists in future clinical diagnoses. In the study reported by Teramoto et al[36], a deep CNN (DCNN) was developed for an automatic lung cancer classification scheme, which is a major deep learning technology. In the evaluation experiment, they used original database, including fine needle aspirate cytology images and HE stained WSIs and a graphics processing unit to train DCNN. First, the micro images were cropped and resampled to obtain the image with a resolution of 256 × 256 pixels. In order to prevent over fitting, the collected images were enhanced by rotation, flipping and filtering. The probability of three types of cancer was evaluated using the developed scheme, and its classification accuracy was evaluated using triple cross validation. In the results obtained, about 71% of the images were correctly classified, which is equivalent to the accuracy of cell technicians and pathologists.
The identification of early lung adenocarcinoma before operation, especially in the case of subcentimeter cancer, can provide important guidance for clinical decision making. Zhao et al[37] developed a 3D deep learning system based on 3D CNN and multitask learning. The deep learning system had better classification performance than radiologists. In terms of three-level weighted average F1 score, the model reached 63.3%, while the four radiologists reached 55.6%, 56.6%, 54.3% and 51.0%, respectively.
With tumor microenvironment increasingly considered as an important factor affecting tumor progression and immunotherapy response, tumor microenvironment for lung cancer has been studied in depth. Saltz et al[38] developed a CNN model to distinguish lymphocytes from necrotic or other tissues at the image spot level in multiple cancer types, including adenocarcinoma and small cell carcinoma of the lung. Then, by quantifying the spatial organization of lymphoid image plaques detected in WSIs, they reported the relationship between the distribution pattern, prognosis and lymphoid components of tumor infiltrated lymphocytes.
Lung cancer patients usually present with advanced, inoperable disease. Because the whole tumor specimen cannot be obtained, the size of the biopsy specimen obtained is usually very limited. It is difficult to distinguish squamous cell carcinoma and adenocarcinoma especially in poorly differentiated tumors because of their obscure histological features. ML in immunohistochemistry[39] was applied to establish a comprehensive and automatic diagnosis strategy for NSCLC biopsy specimen subtypes, which successfully solved this problem. Koh et al[40] described a comprehensive diagnostic strategy using a reliable and minimal immuno-histochemistry team for histopathological subtype analysis of NSCLC biopsy specimens. The team used two ML methods: Decision tree and support vector machines to learn from 30 small NSCLC biopsies with fuzzy morphology. The decision tree model showed that the highest accuracy of the combination of two markers (such as p63 and CK5/6) was about 72% except for three other markers (i.e. TTF-1, Napsin A and P40).
Wang et al[41] explored the correlation between the morphological features of the WSIs stained with HE and the NSCLC epidermal growth factor receptor (EGFR) mutation to achieve the purpose of predicting the risk of gene mutation. The results showed that the AUC of the EGFR mutation risk prediction model proposed in this paper can reach 72.4% on the test set, and the accuracy rate was 70.8%, suggesting a close relationship between morphological characteristics and EGFR mutations of NSCLC. Coudray et al[42] trained a DCNN (inception V3) to accurately and automatically classify the WSIs obtained from The Cancer Genome Atlas. Its performance was comparable to that of the pathologist, and the average AUC was 0.97. They trained the network to predict the ten most common mutations in lung adenocarcinoma and found that six genes (STK11, EGFR, FAT1, setbp1, KRAS and TP53) could be predicted by pathological images. In the nonexperimental population, AUC was 0.733-0.856. It suggested that deep learning models could help pathologists detect cancer subtypes or gene mutations.
APPLICATION OF AI IN GENOMIC CLASSIFICATION OF NSCLC
Various molecular abnormalities affecting oncogenes and tumor suppressor genes have been reported in NSCLC. It is so important to identify potential lung cancer genome subtypes that a specific targeted therapy was proposed. For example, mutations in EGFR or anaplastic lymphoma kinase (ALK) receptors are significant in NSCLC because they provide molecular targets for customized treatment regimens.
The gene expression profile of NSCLC subtype has been established by microarray[43,44]. Microarray data used to identify NSCLC genetic subtypes can be used to train ML algorithms to better understand genomic pathways. Yamamoto et al[45] screened 24 CT image traits performed in a training set of 59 patients, followed by random forest variable selection incorporating 24 CT traits plus six clinical-pathologic covariates to identify a radiomic predictor of ALK+ status. This predictor was then validated in an independent cohort (n = 113). Tests for accuracy and subset analyses were performed. It was found that ALK+ NSCLC had distinct characteristics at CT imaging that when combined with clinical covariate discriminated ALK+ from non-ALK tumors and could potentially identify patients with a shorter durable response to crizotinib.
With the commercialization of next generation sequencing technology and the improvement of the performance of these algorithms, clinicians will be able to better describe NSCLC based on genome data[46]. Duan et al[47] explored the application of the ANN model in the auxiliary diagnosis of lung cancer. They compared the effects of the back-propagation neural network with the Fisher discrimination model for lung cancer screening by combining the detection of four biomarkers, p16, RASSF1A and FHIT gene promoter methylation levels and the relative telomere length. The result of the back-propagation neural network AUC was higher than that of the Fisher discrimination analysis, which meant that the back-propagation neural network model for the prediction of lung cancer was better than Fisher discrimination analysis.
APPLICATION OF AI IN THERAPY OF NSCLC
Systemic treatment is needed in most stages of NSCLC; for example, those in stage II often need adjuvant radiotherapy and chemotherapy. The contour of organs at risk is an important but time-consuming part of radiotherapy treatment planning. Lustberg et al[48] analyzed the CT scan data of 20 patients with stage I-III NSCLC and compared the user adjusted contour and manual contour based on atlas and deep learning contour. It was found that the median time of manual contour drawing was 20 minutes. When using atlas-based contour drawing, a total of 7.8 minutes was saved, while the deep learning contour drawing saved 10 minutes. It showed that it was a feasible strategy for users to adjust the contour generated by the software, which could reduce the contour time of organs at risk in lung radiotherapy. Compared with the existing programs, deep learning shows encouraging results.
At present, targeted therapies[49] such as EGFR tyrosine kinase inhibitors, ALK inhibitors or angiogenesis inhibitors are used depending on the patients’ molecular status. The prediction of targeted therapy response is mainly accomplished by biopsy to analyze the status of the targeted mutation. AI prediction models can complement this by identifying the imaging phenotypes associated with mutation status. Support for this approach comes from quantitative imaging studies of patients with NSCLC treated with gefitinib. The results[50] showed that the mutation state of EGFR could be predicted by radiology. AI analysis of quantitative imaging data can also improve the assessment of response to targeted therapy. Bevacizumab (a monoclonal antibody against vascular endothelial growth factor)-treated NSCLC tumors had reduced FDG uptake and were found to have more patients responding to treatment (73% than 18%). In this study[51], both PET and CT were independent of OS (PET, P = 0.833; CT, P = 0.557).
The level of PD-L1 expression detected by immunohistochemistry is a key biomarker to identify whether NSCLC patients respond to the treatment of PD-1/PD-L1. The quantification of PD-L1 expression currently includes a pathologist’s visual estimate of the percentage of PD-L1 staining (tumor proportion score or TPS) in tumor cells. Kapil et al[52] proposed a new deep learning solution that can automatically and objectively grade PD-L1 expression for the first time in advanced NSCLC biopsy. Using a semisupervised approach and a standard full supervised approach, they integrated manual annotation for training and visual tumor proportion scores for quantitative evaluation by multiple pathologists. It was believed to be the first proof of concept study that showed that deep learning could accurately and automatically estimate the PD-L1 expression level and PD-L1 status of small biopsy samples.
Researchers have studied the use of ML in predicting treatment failure or death. For example, Jochems et al[53] studied ML methods for predicting early death in NSCLC patients after receiving therapeutic chemical radiation. Similarly, Zhou et al[54] used ML to predict the failure of stereotactic body radiotherapy in early NSCLC patients. Both groups used ML methods to establish the prognosis model of early mortality or treatment failure, which could be used to inform patients of treatment plan and optimize treatment. Kureshi et al[55] studied the role of multiple factors in predicting tumor response to EGFR-TKI therapy (erlotinib or gefitinib) in patients with advanced NSCLC.
APPLICATION OF AI IN PROGNOSIS OF NSCLC
Accurate classification, clinical stage, molecular subtype and therapies of NSCLC are all important because prognosis is closely related to these factors. Hsia et al[56] incorporated the clinical detection indicators and gene polymorphism detection results and predicted the prognosis of 75 lung cancer patients without indications of surgical treatment through the ANN model and made treatment plans accordingly. The actual average survival time of the patients was 12.44 ± 7.95 mo, while the ANN prediction result was 13.16 ± 1.77 mo with an accuracy of 86.2%. Zhu et al[57] successfully used DCNN to directly predict the survival time of patients from lung cancer pathological images. Another lung cancer study[58] showed that the prognosis of OS can be improved by adding genomic and radiological information to clinical models, thereby increasing the 95% confidence index from 0.65 (Noether P = 0.001) to 0.73 (P = 2 × 10-9), and the inclusion of radiation data led to a significant improvement in performance (P = 0.01).
Wang et al[59] proposed a computational histomorphometric image classifier using nuclear direction, texture, shape and tumor structure to predict the recurrence of early NSCLC diseases from digital HE tissue microarray slides. The results showed that the combination of these four features could predict the early recurrence of NSCLC, but it had nothing to do with clinical parameters such as gender, cancer stage and histological subtype. Yu et al[60] reported that Zernike shape characteristics of the nucleus could predict the recurrence of NSCLC adenocarcinoma and stage I squamous cell carcinoma.
In an article published in 2018, Saltz et al[38] described the use of CNN combined with pathologist’s feedback to automatically detect the spatial tissue of tumor infiltrating lymphocytes (TIL) in the tissue slide image of The Cancer Genome Atlas and found that this feature predicted the prognosis of 13 different cancer subtypes. In a related study, Corredor et al[61] showed the spatial arrangement of TIL clusters in early NSCLC, which was found by calculating the adjacent TILs and the prognosis of cancer cell nuclear recurrence risk compared with TIL density alone. The accuracy of the model in predicting recurrence was 82% and 75%, respectively, which proved to be an independent prognostic factor.
Blanc-Durand et al[62] trained a CNN in 189 NSCLC patients who received PET/CT examination. The subcutaneous adipose tissue, visceral adipose tissue and muscle weight were automatically segmented from the low-dose CT images. After a quintuple cross validation of a subset of 35 patients, body surface area was standardized as the anthropometric index extracted by deep learning. Cox risk regression analysis showed that body surface area normalized visceral adipose tissue/subcutaneous adipose tissue ratio was an independent predictor of progression free survival and OS in NSCLC patients.
Another study[63] evaluated the ability of CT radiomic features in patients with lung adenocarcinoma to predict distant metastasis. The phenotype of the primary tumor was quantified with 635 radiomic features in the pre-treatment CT scan. Univariate and multivariate analyses were performed using the consistency index to evaluate the efficacy of radiotherapy. Thirty-five radiomic features were used as prognostic indicators for distant metastasis (consistency index > 0.60, FDR < 5%) and 12 prognostic indicators. Notably, tumor volume was only a moderate prognostic indicator for distant metastasis in the discovery cohort (consistency index = 0.55, P = 2.77 × 10-5). This study suggested that radiomic features that capture the details of the tumor phenotype can be used as prognostic biomarkers for clinical factors such as distant metastasis.
APPLICATION IN ELECTRONIC MEDICAL RECORDS OF NSCLC
Electronic medical records (EMR) can be used in clinical diagnosis and treatment, medical insurance and scientific research. EMR is rich in information that can provide evidence of clinical diagnosis, treatment and data source of clinical research phenotype. In Wang et al[64]’s study, multiobjective ensemble deep learning, a dynamic integrated deep learning and adaptive model selection method based on multiobjective optimization, was developed. The information extracted from EMRs through analysis can better predict the treatment results than other conventional methods. According to accurate prognosis prediction, we can stratify the risk of treatment failure of lung cancer patients after radiotherapy. This method can help to design personalized treatment and follow-up plan and improve the survival rate of lung cancer patients after radiotherapy.
FUTURE CHALLENGES
It is one of the key directions of medical research in the information age to build a big database by collecting and integrating various biomics, clinical detection indicators and nonbiological environmental background data of patients. Effective analysis and interpretation of these data will be the top priority, and the integration and analysis of the existing massive information is precisely the biggest advantage of AI.
At present, the investment in AI in lung cancer and the entire medical field is huge, but there is still a certain distance from the actual clinical application. The lack of a high-quality standardized lung cancer clinical database is an important factor restricting AI’s use in lung cancer research. The deficiency of research sample size causes most prediction or diagnostic studies to not fully simulate the actual clinical environment, limiting the value of clinical applications. Studies[65] have pointed out that the current use of AI in the medical field, such as inadequacy of correct methods and evaluation criteria in ANN and the credibility of the results is questionable. In addition, in terms of social regulations, lack of common technical regulations on medical responsibility issues and information security issues exists.
In the future, major medical centers should take the lead to establish a multicenter standardized lung cancer clinical database as a world-class database in line with epidemiology and to develop an AI system that meets the clinical environment. Diagnosis, treatment and optimization of medical resources have positive significance. On the other hand, active promotion of AI-related system regulations, technical specification, audit systems to provide institutional support and corresponding constraints for the development of AI are needed. AI has promising prospects for lung cancer research in the future, but it is still full of challenges.
According to the accuracy stated, which is around 90%, misjudgment may happen in 10% of cases, which reflects a pitfall of AI. Therefore, in clinical work, AI must be placed in a subordinate position. It should exist as an assistant to clinicians and provide auxiliary information under the supervision of doctors to avoid mistakes as much as possible.
CONCLUSION
AI has become an indispensable method to solve complex problems in modern life. In this review, I introduced various attempts and applications of AI in clinical work of NSCLC patients. According to a large number of imaging, histology, genomics, EMR system and other data, doctors can accurately diagnose and treat NSCLC patients. It has been shown that AI is gradually becoming a powerful assistant for doctors. Oncologists, radiologists and surgeons should continue to integrate AI into the clinical treatment of NSCLC in order to provide more patients with accurate and personalized therapy. Over time, both patients and doctors will benefit from the combination of AI and clinical practice.