Published online Jun 28, 2020. doi: 10.35713/aic.v1.i1.1
Peer-review started: May 20, 2020
First decision: June 4, 2020
Revised: June 9, 2020
Accepted: June 12, 2020
Article in press: June 12, 2020
Published online: June 28, 2020
Cancer is a major public health problem worldwide. Current predictions suggest that 13 million people will die each year from cancer by 2030. Thus, new ideas are urgently needed to change paradigms in the global fight against cancer. Over the last decades, artificial intelligence (AI) emerged in the field of cancer research as a new and promising discipline. Although emerging, a great potential is appreciated in AI to improve cancer diagnosis and prognosis, as well as to identify relevant therapeutics in the current era of personalized medicine. Developing pipelines connecting patient-generated health data easily translatable into clinical practice to assist clinicians in decision making represents a challenging but fascinating task. AI algorithms are mainly fueled by multi omics data which, in the case of cancer research, have been largely derived from international cancer programs, including The Cancer Genome Atlas (TCGA). Here, I briefly review some examples of supervised and unsupervised big data derived from TCGA programs and comment on how AI algorithms have been applied to improve the management of patients with cancer. In this context, Artificial Intelligence in Cancer journal was specifically launched to promote the development of this discipline, by serving as a forum to publish high-quality basic and clinical research articles in various fields of AI in oncology.
Core tip: Artificial intelligence (AI) emerged in the field of cancer research as a new and promising discipline to improve the management of patients with cancer, including more accurate and fastest diagnosis to facilitate the therapeutic decision. AI models are mainly fueled by multi omics data. Integrating omics data and clinical data of patients represents a challenging but fascinating task.
- Citation: Coulouarn C. Artificial intelligence and omics in cancer. Artif Intell Cancer 2020; 1(1): 1-7
- URL: https://www.wjgnet.com/2644-3228/full/v1/i1/1.htm
- DOI: https://dx.doi.org/10.35713/aic.v1.i1.1
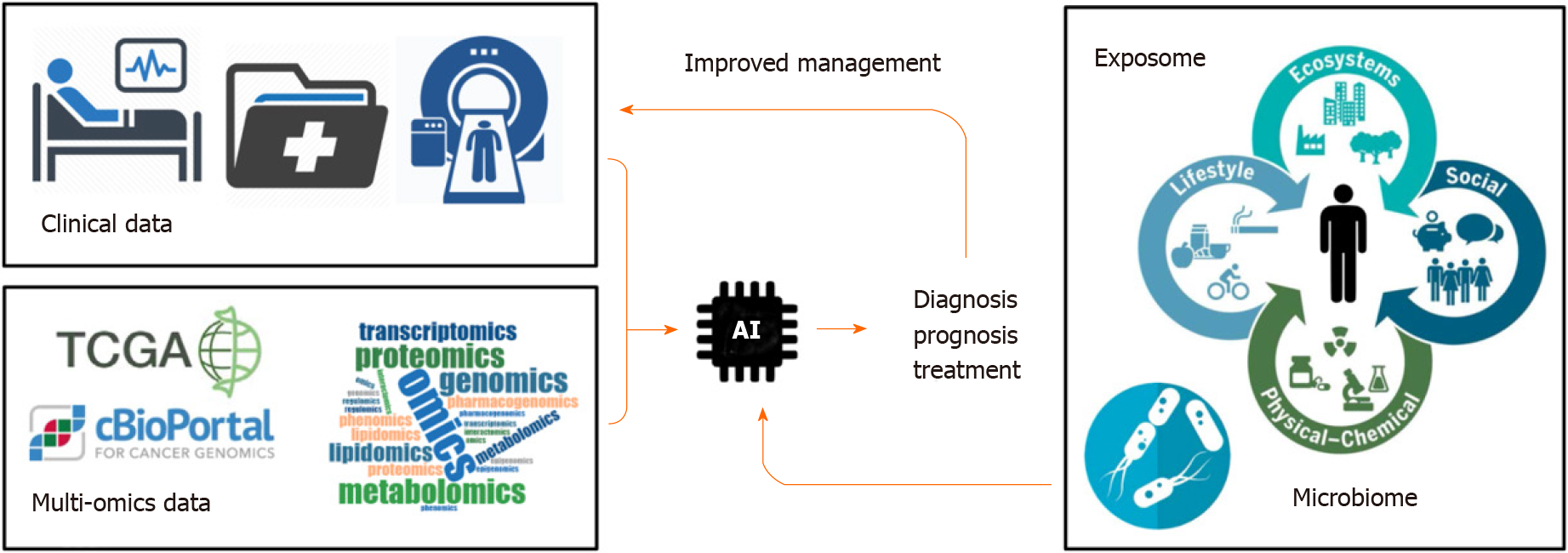
Cancer is a public health problem worldwide[1]. Predictions suggest that 13 million people will die each year from cancer by 2030[2]. Tumor heterogeneity represents an important obstacle to establish efficient therapeutic strategies. Over the last decades, large-scale pan-genomic studies allowed to address tumor heterogeneity in multiple cancers and to provide a landscape of alterations occurring at multiple levels in tumor cells (e.g. at DNA, RNA and protein levels). Thus, international consortia have been initiated, including The Cancer Genome Atlas (TCGA) and its landmark cancer genomics program, which molecularly characterized over 84000 cases from 67 primary sites so far (https://portal.gdc.cancer.gov). Accordingly, TCGA and other cancer programs generated over 2.5 petabytes of genomic, epigenomic, transcriptomic, and proteomic data. This explosive growth of data represented a major driving force to develop innovative artificial intelligence (AI) methods, including deep learning algorithms, capable of analyzing large and multifaceted datasets in an integrated and comprehensive way[3]. By using algorithms that imitate the thinking process, deep learning allows computational models that are composed of multiple processing layers to learn representations of data with multiple levels of abstraction and to discover intricate structure in large data sets[4]. These automated methods, popularized in the society by image or speech recognition algorithms, are now moving into the field of health, including cancer research. Indeed, innovative algorithms are developed to extract meaningful genomic patterns and to translate this conceptual basic information into clinical applications, notably to improve cancer diagnosis, prognosis prediction and treatment efficacy (Figure 1). Here, I briefly review some examples of supervised and unsupervised big data derived from TCGA programs and comment on how AI algorithms have been applied to improve the management of patients with cancer.
TCGA programs represented a major advance in the field of cancer research, allowing both supervised analysis of specific cancers and unsupervised analysis of pan-cancer datasets. Thus, supervised comparative and comprehensive analyses that distinguished clinically relevant molecular subtypes were reported in several cancers, including gastrointestinal (GI) cancers[5], gynecologic and breast cancers[6], pancreatic[7] or liver[8] cancers. Unsupervised analyses have been also performed using pan-cancer datasets. By analyzing mutation profiles, copy-number changes, gene fusions, mRNA expression, and DNA methylation in 9125 tumors profiled by TCGA, a detailed landscape of oncogenic pathway alterations was notably charted in 33 cancer types. Tumors were stratified into 64 subtypes, and patterns of co-occurrence and mutual exclusivity alterations were identified using SELECT, a method that infers conditional selection dependencies between alterations from occurrence patterns[9]. Importantly, using dedicated knowledge base of clinically actionable alterations, it was shown that 57% of tumors had at least one alteration potentially targetable and 30% of tumors had multiple targetable alterations, indicating opportunities for combination therapy[9]. This type of information will be crucial in the current area of cancer precision medicine to develop effective combination therapies that address or prevent resistance to initially successful single agent therapies. Pan-cancer supervised analyses were also performed to highlight frequent alterations in key signaling pathways involved in cancer progression. transforming growth factor beta (TGFβ) is a pleiotropic cytokine that harbors a functional duality in cancer, i.e. exhibiting tumor suppressive features at early stages but switching toward pro-metastatic activities at late tumor stages[10]. Interestingly, genetic alterations in TGFβ signaling, affecting mostly metastatic-associated genes, were observed in 39% of pan-cancer TCGA cases, and were particularly enriched in GI cancers[11]. Specific algorithms have been also used to characterize the immune tumor microenvironment across 33 cancer types analyzed by TCGA. By integrating major immunogenomics methods, including analysis of genomic profiles, hematoxylin and eosin stained tumor sections and deconvolution analysis of mRNA sequencing (mRNA-seq) data, six immune subtypes were characterized, spanning multiple tumor types, with potential therapeutic and prognostic implications for cancer management[12]. Interestingly, one so-called TGFβ dominant subtype, displayed the highest TGFβ signature and a high lymphocytic infiltrate. This observation is particularly relevant with the emergence of effective immunotherapies, including the recent development of an innovative immuno-therapeutic that simultaneously blocks the PD-L1 checkpoint protein and the TGFβ signaling pathway[13].
From a basic point of view, several efforts have been made also to integrate multi omics data and to provide a better understanding of tumor biology. As an example, a deep learning-based predictive model using deep denoising auto-encoder and multi-layer perceptron was developed to quantitatively capture how genetic and epigenetic alterations correlate with directionality of gene expression in liver cancer[14]. Similarly, an innovative one-class logistic regression machine-learning algorithm was used to identify stemness features associated with oncogenic dedifferentiation[15]. Interestingly, an unanticipated correlation of cancer stemness with immune checkpoint expression and infiltrating immune cells was highlighted in the tumor microenvironment[15]. The analysis of gene regulatory networks from available omics data is a challenging task given that biological data is prone to different kinds of noise and ambiguity. Soft computing tools, such as fuzzy sets, evolutionary strategies, and neurocomputing, have been found to be helpful in providing low-cost, acceptable solutions in the presence of various types of uncertainties[16].
Cancer diagnosis using deep learning has been recently reviewed[17]. Soft computing techniques also provided solutions for cancer, regarding diagnosis, prediction, inference and classification[18,19,20]. The approaches are mainly based on segmentation processes using convolutional neural networks (CNN) in clinical images notably acquired from computed tomography (CT) and magnetic resonance imaging (MRI). AI allows integrating quantitative, multiparametric and functional imaging data to automatically recognize complex patterns and to provide quantitative, rather than qualitative, assessments of radiographic characteristics[21]. A classification of skin lesions using a single CNN, trained end-to-end from images directly, using only pixels and disease labels as inputs, nicely illustrates the interest and the power of AI algorithms[22]. Indeed, a CNN trained using a dataset of 129450 clinical images (2032 different cases) was capable of classifying skin lesions with a level of competence comparable to dermatologists[22]. By helping clinicians in characterizing early benign and/or malignant lesions, AI recently emerged as the next step towards precision pathology. Screening programs for early detection of colorectal cancer (CRC) have been shown to reduce mortality in multiple studies. Thus, a machine learning-based algorithm (MeScore) was trained to predict the occurrence of CRC and to identify a group of individuals at a high risk for CRC. Remarkably, MeScore can help identifying individuals in the population who would benefit most from CRC screening, including those with no clinical signs or symptoms of CRC[23]. In another study, a total of 1970 whole slide images of 731 cases of nasopharyngeal carcinoma were divided into training, validation and testing sets. A CNN model was trained to classify images into three categories: Chronic nasopharyngeal inflammation, lymphoid hyperplasia and nasopharyngeal carcinoma. Remarkably, the model equals the senior pathologist when considered in terms of accuracy, specificity, sensitivity, area under the curve and consistency[24]. Thus, this couple of examples suggests that deep learning algorithms could potentially assist pathologists in clinical practice by providing a second opinion and thus increasing consistency on the diagnosis.
Gene expression profiling has been extensively used to derive prognostic signatures in multiple types of cancers. However, these signatures are usually derived from a single type of omics data (e.g. mRNA, miRNA, lncRNA profiling). Integration of multifaceted datasets with different levels of information appears relevant to better reflect the biology of a specific tumor. Accordingly, integrated genome-wide epigenetic and multi omics analyses using AI entered in the era of precision medicine with the burst of data generated over the last decades[25]. Thus, a deep learning multi omics model integrating RNA-seq, miRNA-seq, and methylation data from TCGA, was reported to robustly predict survival of patients with liver cancer[26]. A more aggressive subtype was associated with frequent TP53 inactivation mutations, higher expression of stemness markers, and activated WNT and AKT signaling pathways[26]. Pathway-based biomarker identification with crosstalk analysis has been also reported in liver cancer for efficiently differentiating patients into moderate or aggressive risk subtypes with significant differences in terms of survival[27]. Besides, deep-learning algorithms based on whole slide histological images were reported to predict prognosis of patients with liver cancer. By using a training set made of 390 slides from 206 tumors and a validating set made of 342 slides from 328 patients, a model was built for predicting the survival of patients after surgical resection of hepatocellular carcinoma[28]. Notably, the study highlights the importance of pathologist/machine interactions for the construction of deep-learning algorithms[28]. By processing 5202 digital pathology images from 13 cancer types, a deep-learning model established tumor-infiltrating lymphocytes maps correlated with molecular data, tumor subtypes, immune profiles and patient survival[29]. The application of deep learning in cancer prognosis has been shown to be equivalent or better than current approaches, as recently reviewed[30].
Deep learning-based analysis of multi omics data finds its natural place for the development of personalized therapies in cancer, notably by linking molecular actionable alterations with specific drugs already developed for these alterations or through a drug repositioning process (also referred to as drug repurposing). Deep learning models also enable large scale virtual screening of compound databases for predictive activity profiling against targets important for multiple cancers. Such large scale screening facilitate the quick and cost-effective repurposing of existing drugs[31]. By using a pharmacogenomics database of 1001 cancer cell lines, deep neural networks were trained for predicting drug response and their performance was assessed on multiple clinical cohorts[32]. By integrating RNA-seq, copy number, and mutations from 33 different cancer types (TCGA PanCanAtlas project), a deep learning model was shown to successfully predict RAS activation across cancer types and to identify phenocopying variants (e.g. NF1 loss). The model represents a useful tool to predict response to MEK inhibitors and identify the best responders[33]. Specific algorithms for drug repurposing have been also developed, based notably on linking gene expression profiles of tumors with gene signatures of bioactive molecules. Thus, the L1000 Connectivity Map is a library of gene expression signatures established in cell lines after pharmacologic or genetic (knockdown or over-expression) perturbation (approximately 20000 compounds, 4500 knockdowns, and 3000 over-expressions)[34]. This approach has been successfully used to propose epigenetic modulators (e.g. HDAC inhibitors) as relevant innovative therapeutics to target several hallmarks of liver cancer[35]. Using the same approach, anthelminthic drugs were also identified as potential therapeutic candidates in liver cancer[36]. Thus, combined with a robust stratification of human tumors, AI would help predicting response to individual therapy. Although translation between research and clinical practice requires to fully addressing the question of the reproducibility and interpretability of the developed algorithms, there is no doubt that AI will positively impact clinical decision-making, providing a more personalized management of patients[37]. Another aspect that needs to be fully appraised is the regulatory issue for AI technologies, including clinically approved algorithms (Software as Medical Devices, SaMD), e.g. in terms of personal data sharing[38].
Over the last decades, cancer genomic programs generated a large amount of multi omics data. This information fueled the development of innovative algorithms to extract meaningful information possibly translatable into clinical practices. AI emerged only recently in the field of cancer research. However, specific studies demonstrated already the possibility of AI to improve diagnosis and prognosis of patients with cancer and to develop innovative targeted therapeutics. Although, the actual algorithms are fueled mainly with omics data and clinical images (e.g. genetic, epigenetic, transcriptomic, proteomic, metabolomics profiles, CT scan, MRI), they pave the way for future models that will also integrate personalized clinical information related to lifestyle of each patient, including environmental exposure (exposome) or microbiome composition that may influence response to treatment[39] (Figure 1). As a promising future direction, research on exposome, genetic factors, microbiome, immunity, and molecular tissue biomarkers is needed using AI and omics technologies. This field referred to as molecular pathological epidemiology (MPE) aims at investigating those factors in relation to molecular pathologies and clinical outcomes by means of computational analyses. Thus, MPE represents a promising area of investigation to better understand how a particular exposure influences the carcinogenic and pathologic process[40,41].
In this context, Artificial Intelligence in Cancer journal was specifically launched to promote the development of this discipline, by serving as a forum to publish high-quality basic and clinical research articles in various fields of AI in oncology.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: European Association for the Study of the Liver; European Network for the Study of Cholangiocarcinoma; and International Lactation Consultant Association.
Specialty type: Oncology
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A, A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hu B, Jurman G, Liu Y, Ogino S, Santos-García G S-Editor: Wang JL L-Editor: A E-Editor: Liu JH
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50857] [Article Influence: 8476.2] [Reference Citation Analysis (44)] |
| 2. | The Lancet. GLOBOCAN 2018: counting the toll of cancer. Lancet. 2018;392:985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 3. | Landhuis E. Deep learning takes on tumours. Nature. 2020;580:551-553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 4. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36149] [Cited by in F6Publishing: 17263] [Article Influence: 1918.1] [Reference Citation Analysis (0)] |
| 5. | Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, Kim J, Chatila W, Akbani R, Kanchi RS, Rabkin CS, Willis JE, Wang KK, McCall SJ, Mishra L, Ojesina AI, Bullman S, Pedamallu CS, Lazar AJ, Sakai R; Cancer Genome Atlas Research Network, Thorsson V, Bass AJ, Laird PW. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell. 2018;33:721-735.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 318] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 6. | Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, Liu Y, Fan H, Shen H, Ravikumar V, Rao A, Schultz A, Li X, Sumazin P, Williams C, Mestdagh P, Gunaratne PH, Yau C, Bowlby R, Robertson AG, Tiezzi DG, Wang C, Cherniack AD, Godwin AK, Kuderer NM, Rader JS, Zuna RE, Sood AK, Lazar AJ, Ojesina AI, Adebamowo C, Adebamowo SN, Baggerly KA, Chen TW, Chiu HS, Lefever S, Liu L, MacKenzie K, Orsulic S, Roszik J, Shelley CS, Song Q, Vellano CP, Wentzensen N; Cancer Genome Atlas Research Network, Weinstein JN, Mills GB, Levine DA, Akbani R. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 2018;33:690-705.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 362] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 7. | Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017;32:185-203.e13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1247] [Cited by in F6Publishing: 1153] [Article Influence: 164.7] [Reference Citation Analysis (0)] |
| 8. | Cancer Genome Atlas Research Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell. 2017;169:1327-1341.e23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1175] [Cited by in F6Publishing: 1608] [Article Influence: 229.7] [Reference Citation Analysis (1)] |
| 9. | Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, Chakravarty D, Daian F, Gao Q, Bailey MH, Liang WW, Foltz SM, Shmulevich I, Ding L, Heins Z, Ochoa A, Gross B, Gao J, Zhang H, Kundra R, Kandoth C, Bahceci I, Dervishi L, Dogrusoz U, Zhou W, Shen H, Laird PW, Way GP, Greene CS, Liang H, Xiao Y, Wang C, Iavarone A, Berger AH, Bivona TG, Lazar AJ, Hammer GD, Giordano T, Kwong LN, McArthur G, Huang C, Tward AD, Frederick MJ, McCormick F, Meyerson M; Cancer Genome Atlas Research Network, Van Allen EM, Cherniack AD, Ciriello G, Sander C, Schultz N. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321-337.e10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1916] [Cited by in F6Publishing: 1702] [Article Influence: 283.7] [Reference Citation Analysis (0)] |
| 10. | Papoutsoglou P, Louis C, Coulouarn C. Transforming Growth Factor-Beta (TGFβ) Signaling Pathway in Cholangiocarcinoma. Cells. 2019;8:960. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Korkut A, Zaidi S, Kanchi RS, Rao S, Gough NR, Schultz A, Li X, Lorenzi PL, Berger AC, Robertson G, Kwong LN, Datto M, Roszik J, Ling S, Ravikumar V, Manyam G, Rao A, Shelley S, Liu Y, Ju Z, Hansel D, de Velasco G, Pennathur A, Andersen JB, O'Rourke CJ, Ohshiro K, Jogunoori W, Nguyen BN, Li S, Osmanbeyoglu HU, Ajani JA, Mani SA, Houseman A, Wiznerowicz M, Chen J, Gu S, Ma W, Zhang J, Tong P, Cherniack AD, Deng C, Resar L; Cancer Genome Atlas Research Network, Weinstein JN, Mishra L, Akbani R. A Pan-Cancer Analysis Reveals High-Frequency Genetic Alterations in Mediators of Signaling by the TGF-β Superfamily. Cell Syst. 2018;7:422-437.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 118] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 12. | Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS; Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The Immune Landscape of Cancer. Immunity. 2018;48:812-830.e14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3025] [Cited by in F6Publishing: 3110] [Article Influence: 518.3] [Reference Citation Analysis (0)] |
| 13. | Lan Y, Zhang D, Xu C, Hance KW, Marelli B, Qi J, Yu H, Qin G, Sircar A, Hernández VM, Jenkins MH, Fontana RE, Deshpande A, Locke G, Sabzevari H, Radvanyi L, Lo KM. Enhanced preclinical antitumor activity of M7824, a bifunctional fusion protein simultaneously targeting PD-L1 and TGF-β. Sci Transl Med. 2018;10:eaan5488. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 355] [Article Influence: 71.0] [Reference Citation Analysis (0)] |
| 14. | Seal DB, Das V, Goswami S, De RK. Estimating gene expression from DNA methylation and copy number variation: A deep learning regression model for multi-omics integration. Genomics. 2020;112:2833-2841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J, Omberg L, Gevaert O, Colaprico A, Czerwińska P, Mazurek S, Mishra L, Heyn H, Krasnitz A, Godwin AK, Lazar AJ; Cancer Genome Atlas Research Network, Stuart JM, Hoadley KA, Laird PW, Noushmehr H, Wiznerowicz M. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell. 2018;173:338-354.e15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 705] [Cited by in F6Publishing: 1151] [Article Influence: 230.2] [Reference Citation Analysis (0)] |
| 16. | Mitra S, Das R, Hayashi Y. Genetic networks and soft computing. IEEE/ACM Trans Comput Biol Bioinform. 2011;8:94-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 17. | Munir K, Elahi H, Ayub A, Frezza F, Rizzi A. Cancer Diagnosis Using Deep Learning: A Bibliographic Review. Cancers (Basel). 2019;11:1235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 128] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 18. | Gambhir S, Malik SK, Kumar Y. Role of Soft Computing Approaches in HealthCare Domain: A Mini Review. J Med Syst. 2016;40:287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 19. | Bhatia A, Mago V, Singh R. Use of soft computing techniques in medical decision making: A survey. Proceedings of the 2014 IEEE International Conference on Advances in Computing, Communications and Informatics (ICACCI); 2014 Sep 24-27; New Delhi, India. IEEE. 2014:1131-1137. [DOI] [Cited in This Article: ] |
| 20. | Yardimci A. A survey on use of soft computing methods in medicine. In: de Sá JM, Alexandre LA, Duch W, Mandic D, editors. Artificial Neural Networks â ICANN 2007 - Lecture Notes in Computer Science, vol 4669. Berlin, Heidelberg: Springer, 2007: 69-79. [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 21. | Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1552] [Cited by in F6Publishing: 1365] [Article Influence: 227.5] [Reference Citation Analysis (2)] |
| 22. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5683] [Cited by in F6Publishing: 4645] [Article Influence: 663.6] [Reference Citation Analysis (0)] |
| 23. | Kinar Y, Akiva P, Choman E, Kariv R, Shalev V, Levin B, Narod SA, Goshen R. Performance analysis of a machine learning flagging system used to identify a group of individuals at a high risk for colorectal cancer. PLoS One. 2017;12:e0171759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 24. | Diao S, Hou J, Yu H, Zhao X, Sun Y, Lambo RL, Xie Y, Liu L, Qin W, Luo W. Computer-Aided Pathological Diagnosis of Nasopharyngeal Carcinoma Based on Deep Learning. Am J Pathol. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 25. | Hamamoto R, Komatsu M, Takasawa K, Asada K, Kaneko S. Epigenetics Analysis and Integrated Analysis of Multiomics Data, Including Epigenetic Data, Using Artificial Intelligence in the Era of Precision Medicine. Biomolecules. 2019;10:62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 26. | Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24:1248-1259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 472] [Article Influence: 78.7] [Reference Citation Analysis (0)] |
| 27. | Fa B, Luo C, Tang Z, Yan Y, Zhang Y, Yu Z. Pathway-based biomarker identification with crosstalk analysis for robust prognosis prediction in hepatocellular carcinoma. EBioMedicine. 2019;44:250-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 28. | Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, Pronier E, Laurent A, Amaddeo G, Regnault H, Sommacale D, Ziol M, Pawlotsky JM, Mulé S, Luciani A, Wainrib G, Clozel T, Courtiol P, Calderaro J. Predicting survival after hepatocellular carcinoma resection using deep-learning on histological slides. Hepatology. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 29. | Saltz J, Gupta R, Hou L, Kurc T, Singh P, Nguyen V, Samaras D, Shroyer KR, Zhao T, Batiste R, Van Arnam J; Cancer Genome Atlas Research Network, Shmulevich I, Rao AUK, Lazar AJ, Sharma A, Thorsson V. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018;23:181-193.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 581] [Cited by in F6Publishing: 495] [Article Influence: 82.5] [Reference Citation Analysis (0)] |
| 30. | Zhu W, Xie L, Han J, Guo X. The Application of Deep Learning in Cancer Prognosis Prediction. Cancers (Basel). 2020;12:603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 31. | Issa NT, Stathias V, Schürer S, Dakshanamurthy S. Machine and deep learning approaches for cancer drug repurposing. Semin Cancer Biol. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 94] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 32. | Sakellaropoulos T, Vougas K, Narang S, Koinis F, Kotsinas A, Polyzos A, Moss TJ, Piha-Paul S, Zhou H, Kardala E, Damianidou E, Alexopoulos LG, Aifantis I, Townsend PA, Panayiotidis MI, Sfikakis P, Bartek J, Fitzgerald RC, Thanos D, Mills Shaw KR, Petty R, Tsirigos A, Gorgoulis VG. A Deep Learning Framework for Predicting Response to Therapy in Cancer. Cell Rep. 2019;29:3367-3373.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 33. | Way GP, Sanchez-Vega F, La K, Armenia J, Chatila WK, Luna A, Sander C, Cherniack AD, Mina M, Ciriello G, Schultz N; Cancer Genome Atlas Research Network, Sanchez Y, Greene CS. Machine Learning Detects Pan-cancer Ras Pathway Activation in The Cancer Genome Atlas. Cell Rep. 2018;23:172-180.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 34. | Duan Q, Flynn C, Niepel M, Hafner M, Muhlich JL, Fernandez NF, Rouillard AD, Tan CM, Chen EY, Golub TR, Sorger PK, Subramanian A, Ma'ayan A. LINCS Canvas Browser: interactive web app to query, browse and interrogate LINCS L1000 gene expression signatures. Nucleic Acids Res. 2014;42:W449-W460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 198] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 35. | Allain C, Angenard G, Clément B, Coulouarn C. Integrative Genomic Analysis Identifies the Core Transcriptional Hallmarks of Human Hepatocellular Carcinoma. Cancer Res. 2016;76:6374-6381. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 36. | Chen B, Garmire L, Calvisi DF, Chua MS, Kelley RK, Chen X. Harnessing big 'omics' data and AI for drug discovery in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2020;17:238-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 37. | Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, Ferrero E, Agapow PM, Zietz M, Hoffman MM, Xie W, Rosen GL, Lengerich BJ, Israeli J, Lanchantin J, Woloszynek S, Carpenter AE, Shrikumar A, Xu J, Cofer EM, Lavender CA, Turaga SC, Alexandari AM, Lu Z, Harris DJ, DeCaprio D, Qi Y, Kundaje A, Peng Y, Wiley LK, Segler MHS, Boca SM, Swamidass SJ, Huang A, Gitter A, Greene CS. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface. 2018;15:20170387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1148] [Cited by in F6Publishing: 764] [Article Influence: 127.3] [Reference Citation Analysis (0)] |
| 38. | Walradt T, Glissen Brown JR, Alagappan M, Lerner HP, Berzin TM. Regulatory considerations for artificial intelligence technologies in GI endoscopy. Gastrointest Endosc. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 39. | Jim HSL, Hoogland AI, Brownstein NC, Barata A, Dicker AP, Knoop H, Gonzalez BD, Perkins R, Rollison D, Gilbert SM, Nanda R, Berglund A, Mitchell R, Johnstone PAS. Innovations in research and clinical care using patient-generated health data. CA Cancer J Clin. 2020;70:182-199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 40. | Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 403] [Cited by in F6Publishing: 436] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 41. | Ogino S, Nowak JA, Hamada T, Milner DA, Nishihara R. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu Rev Pathol. 2019;14:83-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 154] [Article Influence: 30.8] [Reference Citation Analysis (0)] |