Published online Jul 26, 2021. doi: 10.12998/wjcc.v9.i21.6081
Peer-review started: March 16, 2021
First decision: April 24, 2021
Revised: May 4, 2021
Accepted: May 19, 2021
Article in press: May 19, 2021
Published online: July 26, 2021
Special AT-rich sequence binding protein 2 (SATB2)-associated syndrome (SAS; OMIM 612313) is an autosomal dominant disorder. Alterations in the SATB2 gene have been identified as causative.
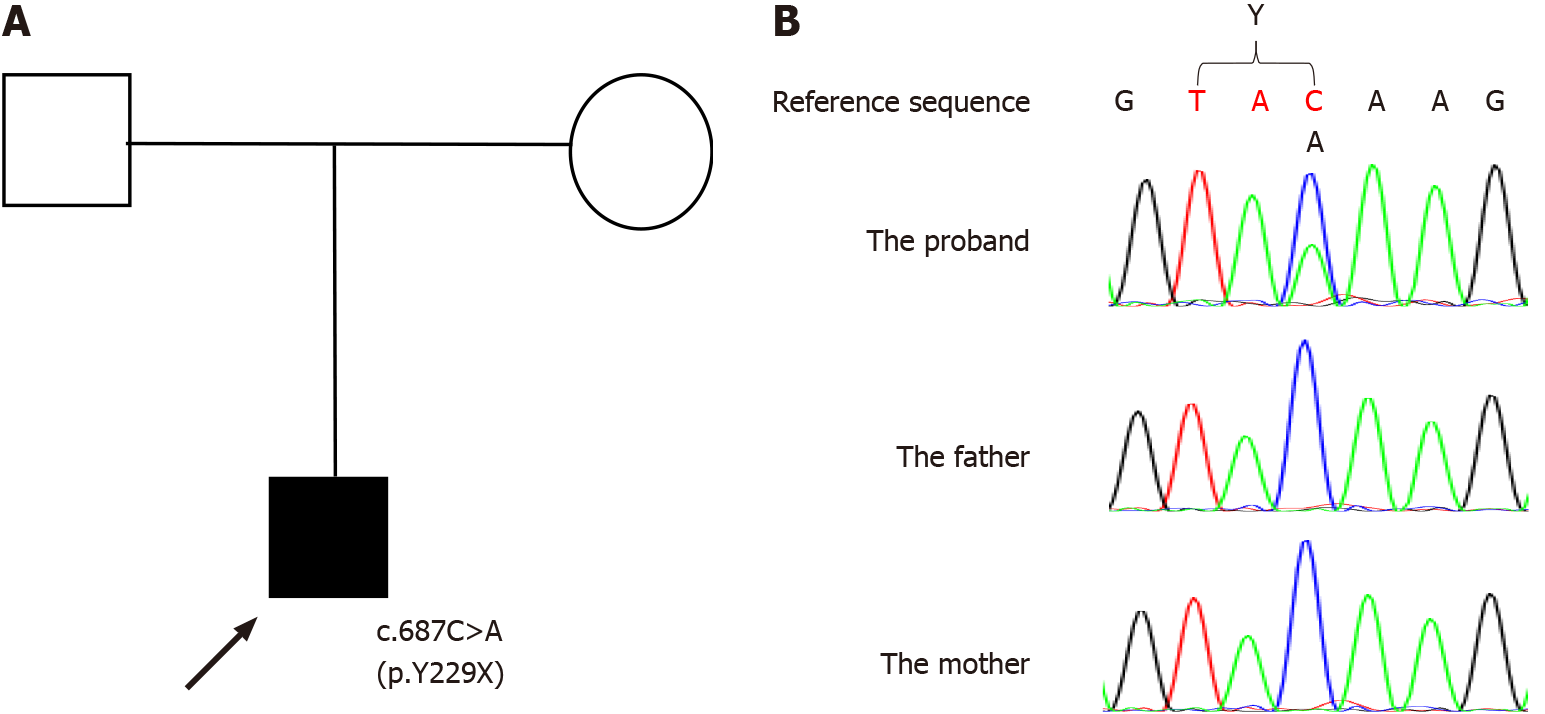
We report a case of a 13-year-old Chinese boy with lifelong global developmental delay, speech and language delay, and intellectual disabilities. He had short stature and irregular dentition, but no other abnormal clinical findings. A de novo heterozygous nonsense point mutation was detected by genetic analysis in exon 6 of SATB2, c.687C>A (p.Y229X) (NCBI reference sequence: NM_001172509.2), and neither of his parents had the mutation. This mutation is the first reported and was evaluated as pathogenic according to the guidelines from the American College of Medical Genetics and Genomics. SAS was diagnosed, and special education performed. Our report of a SAS case in China caused by a SATB2 mutation expanded the genotype options for the disease. The heterogeneous manifestations can be induced by complicated pathogenic involvements and functions of SATB2 from reviewed literatures: (1) SATB2 haploinsufficiency; (2) the interference of truncated SATB2 protein to wild-type SATB2; and (3) different numerous genes regulated by SATB2 in brain and skeletal development in different developmental stages.
Global developmental delays are usually the initial presentations, and the diagnosis was challenging before other presentations occurred. Regular follow-up and genetic analysis can help to diagnose SAS early. Verification for genes affected by SATB2 mutations for heterogeneous manifestations may help to clarify the possible pathogenesis of SAS in the future.
Core tip: Our findings contribute to a growing list of special AT-rich sequence binding protein 2 (SATB2) mutations associated with SATB2-associated syndrome, which is a rare autosomal dominant disorder. The diagnosis was quite challenging when only developmental delays occurred without other manifestations. The heterogeneous manifestations can be induced by complicated pathogenic involvements and functions of SATB2 from reviewed literatures: (1) SATB2 haploinsufficiency; (2) The inter
- Citation: Zhu YY, Sun GL, Yang ZL. SATB2-associated syndrome caused by a novel SATB2 mutation in a Chinese boy: A case report and literature review. World J Clin Cases 2021; 9(21): 6081-6090
- URL: https://www.wjgnet.com/2307-8960/full/v9/i21/6081.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i21.6081
Special AT-rich sequence binding protein 2 (SATB2)-associated syndrome (SAS; OMIM 61231) is a multisystem disorder characterized by neurodevelopmental delays, behavioral issues, palatal and dental anomalies, craniofacial abnormalities, and frequent skeletal pathology[1]. It was first reported in a 16-year-old male in 1989 and termed Glass Syndrome at the time[2]. The frequency of SAS has been estimated in individuals with undiagnosed intellectual disability (ID)/developmental delay (DD) to be 0.24%-0.3%[3,4]. The diagnosis of SAS is likely to be inadequate and should be considered in children with ID, speech delay, cleft or high-arched palate, and abnormal dentition[5]. Up to July 2020, 194 individuals with SAS from more than 15 different countries had enrolled at a website for SAS (www.SATB2gene.com). The acronym has also been used to describe the signs: Severe speech anomalies, Abnormalities of the palate, Teeth anomalies, Behavioral issues with or without bone or brain magnetic resonance imaging (MRI) anomalies, and age of onset before 2 years of age (S.A.T.B.2.)[1].
For representative cases in a typical stage with the classic signs, the possible diagnosis is easy to consider. No specific laboratory tests are available for SAS, and non-specific elevation of alkaline phosphatase levels has been reported in some individuals with osteopenia[4,6]. Thus, early diagnosis is quite challenging in infants with only DD observed. The mean age at diagnosis has been reported to be 6.6 years (range: 1 wk to 34 years)[7].
Alterations in SATB2 have been identified as pathogenic causes, including contiguous deletions, intragenic deletions and duplications, translocations with secondary gene disruption, missense point mutations, and gonadal/somatic mosaicism[1,8-11]. SATB2, which is a kind of matrix attachment region–binding transcription factor and has a homeodomain, a CUTL domain, and two CUT domains, plays a key role in brain and skeletal development[12,13]. Human patients with SATB2 haploinsufficiency have shown some neurological symptoms, such as cognitive deficits, developmental delay, and developmental speech and language problem[14]. Here, we report a case of SAS confirmed by a novel nonsense mutation in SATB2 that is first reported here.
The proband (Figure 1A) was a 13-year-old Chinese boy with lifelong global DD. Brain development evaluations and genetic analysis were planned for the current medical visit after referring to local medical geneticists because the family hoped to have one more child.
There was no exposure to medications or alcohol during the pregnancy, and he was born spontaneously at 35 wk and in an incubator for 3 wk. He presented with delayed developmental milestones: unassisted sitting at about 1 year, unassisted standing at about 20 mo, and unassisted walking at about 2 years. He could speak only one word at 2 years old. He was admitted to the local hospital at that time, and brain MRI showed abnormal long T2 signals in the white matter, but electroencephalography was normal. He was diagnosed with cerebral palsy and brain dysplasia and did not receive any rehabilitation training before receiving special education from 6-years-old. He can eat and go to the toilet by himself but still speaks monosyllabic words and can hardly communicate with others. He experiences less sleep and is excitable.
He showed no other health problems in the past.
He is the only child of nonconsanguineous parents without marked family histories.
Upon physical examination, the patient weighed 28.3 kg (< 3rd percentile), his height was 140.5 cm (< 3rd percentile), and the occipitofrontal circumference was 50 cm (normal range). He was conscious and had a normal gait. He also presented irregular dentition but no special facial features. Patellar reflex was normal, and Babinski signs were negative. The hand-alternating movement test, heel-knee-tibia test, and intelligence quotient value could not be completed because of non-cooperation with the instructions.
Blood tests were normal, including ceruloplasmin, lactic acid, ammonia, and liver and kidney functions. Screening for genetic and metabolic diseases in the blood and urine was not suggestive of disease.
The next generation sequencing-based trio whole exome sequencing plus copy number variation sequencing was performed for genetic analysis (the process was performed by Beijing Chigene Translational Medicine Research Center). The EDTA-treated peripheral blood were collected with informed consent of the patient’s guardians. The DNA was extracted using the Blood Genome Column Medium Extraction Kit (Kangweishiji, China) according to the manufacture instructions and was subjected to quality control using Qubit 2.0 fluorimeter and electrophoresis with 0.8% agarose gel for further protocol. The whole exome library was constructed using xGen Exome Research Panel v1.0 (IDT, Iowa, United States) that consists of 429826 individually synthesized and quality-controlled probes, which targets 39 Mb protein-coding region (19396 genes) of the human genome and covers 51 Mb of end-to-end tiled probe space. High-throughput sequencing was performed by Illumina NovaSeq 6000 series sequencer (PE150). A de novo heterozygous nonsense point mutation in exon 6 in SATB2, c.687C>A (p.Y229X) (NCBI reference sequence: NM_001172509.2) was detected in the proband. Direct sequencing validated the detected missense mutations. PCR was performed to amplify the exon with the detected mutation and its neighboring introns using the forward primer: 5’-TAGTTTTAAGGGAGCCAACTAGGA-3’ and the reverse primer: 5’-TCAGTTTACAGGAGAGTGACTGTTT-3’. PCR products were purified using micro concentrating centrifugal filter columns (Millipore, Bedford, MA, United States), sequenced by ABI 3730XL (Applied Biosystems, Forster City, CA, United States), and analyzed by DNASTAR software.
The proband had the mutation that neither of his parents had (Figure 1B). The detected heterozygous mutation c.687C>A (p.Y229X) is near the end of coding region for the CUTL domain and leads to a stop codon. We think the mutation c.687C>A (p.Y229X) leads to an abnormal transcription and then a truncated protein, like a nearby mutation c.715C>T (p.R239X) that had been reported to lead to an abnormal transcription and a truncated protein[3,15,16]. We did not investigate the mutation in control groups, but the minor allele frequency for the mutation was not found in the dbSNP database (https://www.ncbi.nlm.nih.gov/snp/), 1000 Genomes Project[17], Virtual Chinese Genome Database[18], or Genome Variation Map[19]. The effect of the mutation was evaluated according to the guidelines of the American College of Medical Genetics and Genomics[20], and the mutation was considered pathogenic (Table 1). The mutation is the first reported for SAS.
| Weight for pathopoiesis | Pathogenic criterion for the mutation | |||
| Functional evaluation | PVS1 (very strong) | Null variant in SATB2 where loss of function is a known mechanism of the disease | ||
| PM2 (moderate) | Absence in population database | |||
| PP3 (supporting) | Multiple lines of computational evidence support a deleterious effect on the gene/gene production | |||
| PolyPhen-2 | SIFT | MutationTaster | ||
| Probably damaging | Damaging | Disease causing | ||
| Final evaluation | Pathogenic | |||
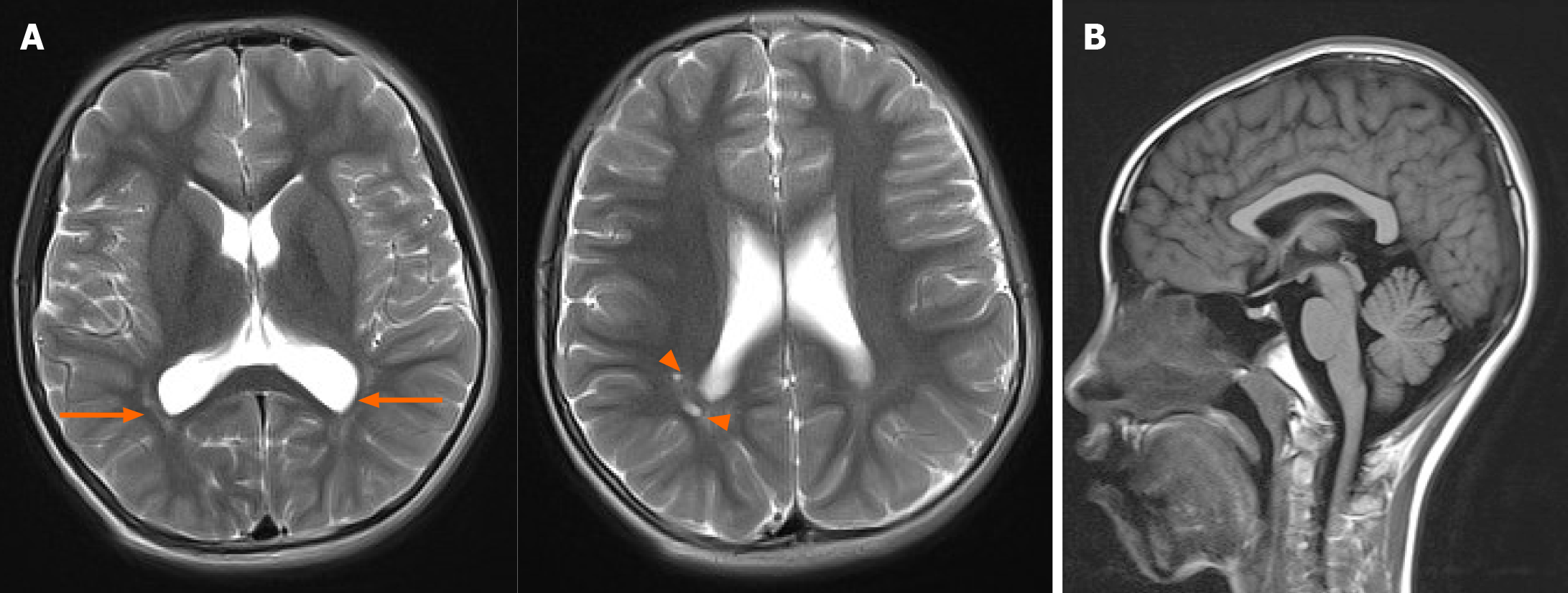
An electroencephalography was normal, but brain MRI revealed multiple small cystic lesions in the white matter near the posterior horns of the right lateral ventricle and long T2 signals in the white matter adjacent to the bilateral posterior horns of the lateral ventricles (Figure 2A).
SAS was diagnosed.
After diagnosis, special education in local welfare institutions and symptomatic treatment were performed (for example: oral chloral hydrate for sleeping problems if necessary).
At 6 mo follow-up, no speech improvements were observed.
For SAS patients, preterm birth was reported quite rarely[1,2]. The first reported patient was delivered at 32 wk[2]. But SATB2 is not a candidate gene for spontaneously preterm birth for now. For our case, he was delivered at 35 wk without obvious causes, and we did not find associations between the preterm birth and SATB2. Many neurodevelopmental problems are associated with prematurity compared to full term infants, including DD, language problems, and poorer academic performance in the future. Some developmental problems may be corrected if interventions were performed early[21]. Differentiating underlying causes for DD early in premature infants can help to make more accurate treatment maneuvers. SAS includes a constellation of manifestations that do not occur simultaneously or necessarily (Table 2).
| Signs for SAS | Reported frequency (%) | |
| Demographics | Gender (M:F) | 3:2 |
| Age < 4 yr | 31 | |
| Age 4-10 yr | 37 | |
| Age 10-18 yr | 19 | |
| Adults | 13 | |
| Severe speech anomalies | DD/ID | 100 |
| Speech delay | 95 | |
| Abnormalities of the palate | Cleft palate, high-arched palate, and bifid uvula | 76 |
| Micrognathia | 42 | |
| Teeth anomalies | Abnormal upper central incisors | 36 |
| Dental crowding | 36 | |
| Hypodontia | 16 | |
| Delayed primary dentition | 6 | |
| Diastema | 4 | |
| Behavioral issues with or without bone or brain MRI anomalies | Feeding difficulties | 39 |
| Growth restriction | 34 | |
| Enlarged ventricles | 12 | |
| Agenesis of corpus callosum | 5 | |
| Age of onset before 2 yr | — | — |
All SAS patients have DD/ID, 95% have speech delay[22], 82% have a limited vocabulary under ten words or absent speech by 10-years-old, and their first words are spoken at a median of 24 mo of age (range 10-144 mo)[7]. In three patients from Japan, meaningful words had not been acquired at 5-years-old[23]. The process usually did not regress after gaining some abilities. Only one case with an 8.6-Mb deletion of 2q32.2-q33.1, including SATB2, was reported to have ID regress from mild to severe and speech from poor to absent between the ages of 6 and 12 years[24]. For our case, DD and limited speech were considered to be caused by the prematurity, and it was hoped that he would develop to normal in the future when he was referred to the hospital in the first few years. Therefore, we think the attention to the prematurity history may be one cause for the delayed diagnosis for the patient.
Seventy-six percent of individuals with SAS have palatal abnormalities such as cleft palate, high-arched palate, and bifid uvula. In 42% of SAS individuals, micrognathia is found[22]. For our case, no such abnormalities were found.
Thirty-six percent of individuals with SAS have abnormal upper central incisors. In addition, 36% have dental crowding, 16% have hypodontia, 6% have delayed primary dentition, and/or 4% have diastema[22]. Delayed permanent root formation, delayed or missing second bicuspids, and malformed teeth have been found on radiography[25]. Multiple odontomas have been reported as an occasional manifestation in SAS patients[26]. In our case, dental crowding and irregular dentation were found, and he still did not grow any permanent teeth.
A broad spectrum of behavioral issues has been reported, including pleasant personality, irritability, hyperactivity, sleep problems, sensory issues, stereotyped repetitive movements, and Rett syndrome-like phenotypes[3,7,22,27,28]. Nonspecific brain abnormalities detected by MRI are found in half of affected individuals, including ventricle enlargements, thin corpus callosum, enlarged perivascular spaces, and abnormal myelinization in white matter[7,22]. Some Pectus, finger, or spine deformities have been reported in a few affected individuals, some with concurrent osteopenia[8,22]. In our case, the patient slept less, and brain MRI indicated periventricular leukomalacia and poor myelinization, which is relatively common in subjects with a history of preterm or neonatal respiratory distress syndrome, and showed normal corpus callosum without obvious physical abnormalities (Figure 2B).
Individuals with SAS fail to reach normal developmental milestones from infancy. Some other signs may exist from infancy, such as mild but nonspecific facial dysmorphism, hypotonia, feeding difficulties, clinical seizures, and abnormal muscle strength[22]. Growth restriction, severe heart defects, small or undescended testicles, inguinal hernias, hypospadias, and thin skin or hair have been reported in several cases with large deletions including SATB2[22]. In our case, the patient had short stature, though genetic analysis did not reveal large gene deletions.
According to the gene database for SAS (https://satb2gene.com/pros-molecular-data/), 194 variants have been identified, and single nucleotide variants causing a premature stop codon were the most frequent type (46%) followed by missense variants (25%), large deletions > 1 Mb (19%), and small deletions (12%). According to the published data, 120 unique variants have been identified in SATB2, and the most common ones were single nucleotide variants inducing the occurrence of a stop codon (42.5%), and next were the missense variants (25.8%). About 74.2% of missense variants are located in CUT1, CUT2, or homeodomain DNA protein domains[12]. The mutation detected in our case (c.687C>A (p.Y229X)) is a nonsense point mutation that has not been reported previously and located in the CUTL domain.
The phenotypes are heterogeneous according to the fragment deletion size and locations of mutations in SATB2 [12]. For example, abnormal myelination and/or white matter abnormalities were relatively common (26%) in patients with pathogenic nonsense, missense, and frameshift variants, while abnormalities in heart, testicles, skin, or hair were reported in patients with large fragment deletions[7,22]. In addition, even the same genotypes of p.R239* that have been detected in different individuals have phenotypes that were not identical (from Genotype-Phenotype database at https://satb2gene.com/pros-molecular-data/).
The reasons for the heterogeneous manifestations for SAS can be the complicated pathogenic involvements and functions of SATB2. SATB2 is a kind of matrix attachment region-binding transcription factor, with high levels of expression in the brain, including the cerebral cortex and spinal cord, and plays a role in central nervous system development[29]. SATB2 haploinsufficiency can be the cause for intellectual disability. Mutant SATB2 protein appears functionally inactive because of the disrupted associations with chromatin or matrix[3,14]. The oligomerization of the N-terminal domain of SATB1, another SATB family protein, is critical for DNA-binding affinity[30]. The truncated protein with left N-terminus and CUTL domain in a patient with a heterozygous nonsense mutation of c.715C>T (p.R239X) has been shown to localize to the nucleus and interferes with the normal activity of wild-type SATB2 protein by forming a dimer with wild-type SATB2[16]. In our case, the mutation is near the reported mutation of c.715C>T (p.R239X), induces a stop codon at the end of the CUTL domain, and can leave an unaffected N-terminus and oligomerization domain of SATB2. This may disturb the DNA binding and gene regulation like the reported mutation of c.715C>T (p.R239X).
SATB2 is involved in transcription regulation and chromatin remodeling. SATB2 can activate UPF3B transcription, and defects in UPF3B in humans can induce cognitive deficits and craniofacial dysmorphisms[16]. SATB2 regulates a transcriptional network of multiple key determinants for skeletal development by activating or repressing DNA bound protein or enhancing the activity of other DNA binding proteins[13]. Three gene sets related to or regulated by SATB2 have been analyzed at different stages of development; the sets include many genes contributing to schizophrenia and educational attainment[31]. Other research has shown that SATB2 interacts with different protein networks at developing and adult stages in the cortex, indicating that the SATB2 function shifts in the cortex over a lifetime[13]. In the mouse, Satb2 regulates regionalization of the retrosplenial cortex by controlling Nr4a2 and Ctip2 during development[32], and loss of Satb2 in cortex and hippocampus contributed to abnormal behavior[33]. Satb2 was involved in the specification of upper-layer neuron specification in the neocortex in mouse by regulating expression of specific genes[34]. In the developing cerebral cortex, Satb2 regulated the formation of corticocortical connections by repressing the expression of Ctip2, which is a transcription factor involved in the extension of subcortical projections from cortical neurons[35,36].
The SATB2 function shift at different developmental stages and complex interactions with related proteins may be involved in the pathogenesis of variable manifestations in SAS patients.
Diagnosis for SAS can be quite challenging in infants with only DD, hypotonia, feeding difficulties, and palatal issues. Cerebral palsy may be considered at this time, especially for those who were preterm or had neonatal asphyxia, but nonprogressing motor developmental abnormalities during follow-up are the main characteristic of cerebral palsy[37]. Leukodystrophy is usually the initial diagnosis for those with some neurological manifestations and abnormal findings in white matter on brain MRI and can occur at different ages, from newborn to adult. The initial clinical manifestation of leukodystrophies is often nonspecific, such as motor impairment, cognitive impairment, and seizures. Further diagnosis of leukodystrophies constitutes many kinds of rare heritable disorders and is challenging in most cases[38]. Angelman syndrome and related disorders can be considered during infancy, which are characterized by severe cognitive disability, motor dysfunction, speech impairment, and hyperactivity, but inappropriate happy demeanor may help diagnose[20,39]. Dental and behavioral problems over time may indicate SAS during follow-up. KBG syndrome manifests dental abnormalities (macrodontia of upper central incisors), DD, and other characteristic features that help in diagnosis, including distinctive craniofacial features, such as triangular face, broadened nasal bridge, thin upper lip, and synophrys[20,40].
Today, next generation sequencing technologies, especially whole exome sequencing and whole genome sequencing, are available for clinical genetic diagnosis and differential diagnosis of heritable diseases, especially for distinct differential diagnosis[41]. Cerebral palsy and leukodystrophy are clinical diagnoses, but the etiologies vary and are not fully known. Approximately one-third of cases do not have traditional risk factors, such as prematurity, hypoxic–ischemic injury, placental insufficiency, and prenatal infection, and genetic mutations may be involved in the pathogenesis of a substantial proportion of cases of cerebral palsy[37]. Distinct diagnoses of leukodystrophies have significantly increased due to genetic diagnostic tests[38]. Genetic testing of UBE3A for Angelman syndrome and ANKRD11 for KBG syndrome can help to differentiate the diseases from SAS[39,40].
No specific therapy or guidelines are available for SAS, and regular evaluations after diagnosis and treatment of symptoms are recommended. Specialists in different fields deal with sleep disturbances, seizures, feeding difficulties, behavioral issues, scoliosis, tibial bowing, and joint contractures, among others. Developmental support/special education is referred for DD/ID[20]. Prompts for restructuring oral muscular phonetic targets and aggressive speech therapy (3 ×/wk) at 2-years-old have shown improvements in speech over time, and some communication devices, such as picture exchange communication system, may provide additional help for SAS patients[1].
We confirmed SAS by the genetic analysis and collected the clinical features. A new variant was found. Because of phenotypic heterogeneity, limited physical findings, and lack of specific laboratory tests, SAS can be difficult to diagnose in infants with DD but no other signs. Surveillance should focus on speech development, dental status, and behavior issues. Next generation sequencing can help differentiate underlying diseases and predict the appropriate prognosis, and we recommend genetic testing for children who show delays over 6 mo compared with normal development milestones in gross movement and/or speech and language development in clinical practice. In future studies, verification for different gene sets that are affected by SATB2 mutations for heterogeneous manifestations may help to clarify the possible pathogenesis of SAS.
The authors thank the family for participating and supporting this study.
Manuscript source: Unsolicited manuscript
Specialty type: Genetics and heredity
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Strainiene S S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Wang LYT
| 1. | Zarate YA, Fish JL. SATB2-associated syndrome: Mechanisms, phenotype, and practical recommendations. Am J Med Genet A. 2017;173:327-337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 68] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 2. | Glass IA, Swindlehurst CA, Aitken DA, McCrea W, Boyd E. Interstitial deletion of the long arm of chromosome 2 with normal levels of isocitrate dehydrogenase. J Med Genet. 1989;26:127-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Bengani H, Handley M, Alvi M, Ibitoye R, Lees M, Lynch SA, Lam W, Fannemel M, Nordgren A, Malmgren H, Kvarnung M, Mehta S, McKee S, Whiteford M, Stewart F, Connell F, Clayton-Smith J, Mansour S, Mohammed S, Fryer A, Morton J; UK10K Consortium; Grozeva D, Asam T, Moore D, Sifrim A, McRae J, Hurles ME, Firth HV, Raymond FL, Kini U, Nellåker C, Ddd Study, FitzPatrick DR. Clinical and molecular consequences of disease-associated de novo mutations in SATB2. Genet Med. 2017;19:900-908. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Zarate YA, Steinraths M, Matthews A, Smith WE, Sun A, Wilson LC, Brain C, Allgove J, Jacobs B, Fish JL, Powell CM, Wasserman WW, van Karnebeek CD, Wakeling EL, Ma NS. Bone health and SATB2-associated syndrome. Clin Genet. 2018;93:588-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Döcker D, Schubach M, Menzel M, Munz M, Spaich C, Biskup S, Bartholdi D. Further delineation of the SATB2 phenotype. Eur J Hum Genet. 2014;22:1034-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Boone PM, Chan YM, Hunter JV, Pottkotter LE, Davino NA, Yang Y, Beuten J, Bacino CA. Increased bone turnover, osteoporosis, progressive tibial bowing, fractures, and scoliosis in a patient with a final-exon SATB2 frameshift mutation. Am J Med Genet A. 2016;170:3028-3032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Zarate YA, Smith-Hicks CL, Greene C, Abbott MA, Siu VM, Calhoun ARUL, Pandya A, Li C, Sellars EA, Kaylor J, Bosanko K, Kalsner L, Basinger A, Slavotinek AM, Perry H, Saenz M, Szybowska M, Wilson LC, Kumar A, Brain C, Balasubramanian M, Dubbs H, Ortiz-Gonzalez XR, Zackai E, Stein Q, Powell CM, Schrier Vergano S, Britt A, Sun A, Smith W, Bebin EM, Picker J, Kirby A, Pinz H, Bombei H, Mahida S, Cohen JS, Fatemi A, Vernon HJ, McClellan R, Fleming LR, Knyszek B, Steinraths M, Velasco Gonzalez C, Beck AE, Golden-Grant KL, Egense A, Parikh A, Raimondi C, Angle B, Allen W, Schott S, Algrabli A, Robin NH, Ray JW, Everman DB, Gambello MJ, Chung WK. Natural history and genotype-phenotype correlations in 72 individuals with SATB2-associated syndrome. Am J Med Genet A. 2018;176:925-935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 8. | Kaiser AS, Maas B, Wolff A, Sutter C, Janssen JW, Hinderhofer K, Moog U. Characterization of the first intragenic SATB2 duplication in a girl with intellectual disability, nearly absent speech and suspected hypodontia. Eur J Hum Genet. 2015;23:704-707. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 9. | Liedén A, Kvarnung M, Nilssson D, Sahlin E, Lundberg ES. Intragenic duplication--a novel causative mechanism for SATB2-associated syndrome. Am J Med Genet A. 2014;164A:3083-3087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Grelet M, Mortreux J, Alazard E, Sigaudy S, Philip N, Missirian C. SATB2-associated syndrome: first report of a gonadal and somatic mosaicism for an intragenic copy number variation. Clin Dysmorphol. 2019;28:205-210. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 11. | Qian Y, Liu J, Yang Y, Chen M, Jin C, Chen P, Lei Y, Pan H, Dong M. Paternal Low-Level Mosaicism-Caused SATB2-Associated Syndrome. Front Genet. 2019;10:630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Zarate YA, Bosanko KA, Caffrey AR, Bernstein JA, Martin DM, Williams MS, Berry-Kravis EM, Mark PR, Manning MA, Bhambhani V, Vargas M, Seeley AH, Estrada-Veras JI, van Dooren MF, Schwab M, Vanderver A, Melis D, Alsadah A, Sadler L, Van Esch H, Callewaert B, Oostra A, Maclean J, Dentici ML, Orlando V, Lipson M, Sparagana SP, Maarup TJ, Alsters SI, Brautbar A, Kovitch E, Naidu S, Lees M, Smith DM, Turner L, Raggio V, Spangenberg L, Garcia-Miñaúr S, Roeder ER, Littlejohn RO, Grange D, Pfotenhauer J, Jones MC, Balasubramanian M, Martinez-Monseny A, Blok LS, Gavrilova R, Fish JL. Mutation update for the SATB2 gene. Hum Mutat. 2019;40:1013-1029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 13. | Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Fariñas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971-986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 381] [Cited by in F6Publishing: 392] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 14. | Cera I, Whitton L, Donohoe G, Morris DW, Dechant G, Apostolova G. Genes encoding SATB2-interacting proteins in adult cerebral cortex contribute to human cognitive ability. PLoS Genet. 2019;15:e1007890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 15. | Leoyklang P, Suphapeetiporn K, Siriwan P, Desudchit T, Chaowanapanja P, Gahl WA, Shotelersuk V. Heterozygous nonsense mutation SATB2 associated with cleft palate, osteoporosis, and cognitive defects. Hum Mutat. 2007;28:732-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 16. | Leoyklang P, Suphapeetiporn K, Srichomthong C, Tongkobpetch S, Fietze S, Dorward H, Cullinane AR, Gahl WA, Huizing M, Shotelersuk V. Disorders with similar clinical phenotypes reveal underlying genetic interaction: SATB2 acts as an activator of the UPF3B gene. Hum Genet. 2013;132:1383-1393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 17. | 1000 Genomes Project Consortium. , Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10217] [Cited by in F6Publishing: 9980] [Article Influence: 1108.9] [Reference Citation Analysis (0)] |
| 18. | Ling Y, Jin Z, Su M, Zhong J, Zhao Y, Yu J, Wu J, Xiao J. VCGDB: a dynamic genome database of the Chinese population. BMC Genomics. 2014;15:265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 19. | Song S, Tian D, Li C, Tang B, Dong L, Xiao J, Bao Y, Zhao W, He H, Zhang Z. Genome Variation Map: a data repository of genome variations in BIG Data Center. Nucleic Acids Res. 2018;46:D944-D949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 20. | Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13374] [Cited by in F6Publishing: 17945] [Article Influence: 1993.9] [Reference Citation Analysis (0)] |
| 21. | Hee Chung E, Chou J, Brown KA. Neurodevelopmental outcomes of preterm infants: a recent literature review. Transl Pediatr. 2020;9:S3-S8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 22. | Zarate YA, Kaylor J, Fish J, Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Mirzaa G, Amemiya A. SATB2-Associated Syndrome. 1993;. [PubMed] [Cited in This Article: ] |
| 23. | Yamada M, Uehara T, Suzuki H, Takenouchi T, Yoshihashi H, Suzumura H, Mizuno S, Kosaki K. SATB2-associated syndrome in patients from Japan: Linguistic profiles. Am J Med Genet A. 2019;179:896-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Gregoric Kumperscak H, Krgovic D, Vokac NK. Specific behavioural phenotype and secondary cognitive decline as a result of an 8.6 Mb deletion of 2q32.2q33.1. J Int Med Res. 2016;44:395-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 25. | Scott J, Adams C, Beetstra S, Zarate YA. SATB2-associated syndrome (SAS) and associated dental findings. Spec Care Dentist. 2019;39:220-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 26. | Kikuiri T, Mishima H, Imura H, Suzuki S, Matsuzawa Y, Nakamura T, Fukumoto S, Yoshimura Y, Watanabe S, Kinoshita A, Yamada T, Shindoh M, Sugita Y, Maeda H, Yawaka Y, Mikoya T, Natsume N, Yoshiura KI. Patients with SATB2-associated syndrome exhibiting multiple odontomas. Am J Med Genet A. 2018;176:2614-2622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 27. | Cotton AP, Gokarakonda S, Caffrey AR, Zarate YA, Kumar N. Behavioral phenotype and sleep problems in SATB2-associated syndrome. Dev Med Child Neurol. 2020;62:827-832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Lee JS, Yoo Y, Lim BC, Kim KJ, Choi M, Chae JH. SATB2-associated syndrome presenting with Rett-like phenotypes. Clin Genet. 2016;89:728-732. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur J Neurosci. 2005;21:658-668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 184] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 30. | Wang Z, Yang X, Chu X, Zhang J, Zhou H, Shen Y, Long J. The structural basis for the oligomerization of the N-terminal domain of SATB1. Nucleic Acids Res. 2012;40:4193-4202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Whitton L, Apostolova G, Rieder D, Dechant G, Rea S, Donohoe G, Morris DW. Genes regulated by SATB2 during neurodevelopment contribute to schizophrenia and educational attainment. PLoS Genet. 2018;14:e1007515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 32. | Zhang L, Song NN, Zhang Q, Mei WY, He CH, Ma P, Huang Y, Chen JY, Mao B, Lang B, Ding YQ. Satb2 is required for the regionalization of retrosplenial cortex. Cell Death Differ. 2020;27:1604-1617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Zhang Q, Huang Y, Zhang L, Ding YQ, Song NN. Loss of Satb2 in the Cortex and Hippocampus Leads to Abnormal Behaviors in Mice. Front Mol Neurosci. 2019;12:33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 34. | Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, Sestan N, Molnár Z, Tarabykin V. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 457] [Cited by in F6Publishing: 481] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 35. | Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Fariñas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364-377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 457] [Cited by in F6Publishing: 470] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 36. | Leone DP, Heavner WE, Ferenczi EA, Dobreva G, Huguenard JR, Grosschedl R, McConnell SK. Satb2 Regulates the Differentiation of Both Callosal and Subcerebral Projection Neurons in the Developing Cerebral Cortex. Cereb Cortex. 2015;25:3406-3419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 37. | Fahey MC, Maclennan AH, Kretzschmar D, Gecz J, Kruer MC. The genetic basis of cerebral palsy. Dev Med Child Neurol. 2017;59:462-469. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 38. | Ashrafi MR, Amanat M, Garshasbi M, Kameli R, Nilipour Y, Heidari M, Rezaei Z, Tavasoli AR. An update on clinical, pathological, diagnostic, and therapeutic perspectives of childhood leukodystrophies. Expert Rev Neurother. 2020;20:65-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 38] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 39. | Margolis SS, Sell GL, Zbinden MA, Bird LM. Angelman Syndrome. Neurotherapeutics. 2015;12:641-650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 40. | Morel Swols D, Foster J 2nd, Tekin M. KBG syndrome. Orphanet J Rare Dis. 2017;12:183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 41. | Zarate YA, Perry H, Ben-Omran T, Sellars EA, Stein Q, Almureikhi M, Simmons K, Klein O, Fish J, Feingold M, Douglas J, Kruer MC, Si Y, Mao R, McKnight D, Gibellini F, Retterer K, Slavotinek A. Further supporting evidence for the SATB2-associated syndrome found through whole exome sequencing. Am J Med Genet A. 2015;167A:1026-1032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |