Published online Jul 26, 2021. doi: 10.12998/wjcc.v9.i21.5873
Peer-review started: February 3, 2021
First decision: March 14, 2021
Revised: March 22, 2021
Accepted: May 26, 2021
Article in press: May 26, 2021
Published online: July 26, 2021
Patients with lipopolysaccharide (LPS)-responsive beige-like anchor protein (LRBA) deficiency have a variety of clinical symptoms, but there is no apparent genotype–phenotype correlation, and patients carrying the same mutations may have different phenotypes. Therefore, it is not easy for doctors to make a decision regarding hematopoietic stem cell transplantation (HSCT) for LRBA-deficient patients. We hypothesized that there may be a protein–phenotype correlation to indicate HSCT for LRBA-deficient patients.
To report on three Chinese LRBA-deficient patients and determine the correlation between residual protein expression and disease phenotypes.
Clinical data of three Chinese LRBA-deficient patients were collected, and protein levels were detected by Western blot analysis. In addition, LRBA mutation information of another 83 previously reported patients was summarized.
All the major clinical findings indicated enteropathy, but patients 1 and 3 presented with more severe symptoms than patient 2. Endoscopy and histology indicated nonspecific colitis for patients 1 and 3 but Crohn's disease-like colitis for patient 2. Compound heterozygous mutations in LRBA were found in patient 1, and homozygous mutations in LRBA were found in patient 2 and patient 3. Only patient 2 responded well to traditional immunosuppressive treatment. Residual expression of the LRBA protein in patients 1 and 3 was very low, but in patient 2, a more than 0.5-fold in expression of the LRBA protein was found compared to that in the control. After HSCT, patient 1 had increased LRBA protein expression. We summarized the genetic information of 86 patients, and the mutations in patients 1 and 3 were novel mutations.
We described three Chinese LRBA-deficient patients, two of whom carried novel mutations. These patients had no genotype-phenotype correlations, but their residual LRBA protein expression might be associated with disease outcome and could be an indicator for HSCT.
Core Tip: Previous studies have showed that there is no apparent genotype–phenotype correlation for lipopolysaccharide-responsive beige-like anchor protein (LRBA) deficiency, but a protein–phenotype correlation may exist. In this study, we described three Chinese patients with LRBA deficiency. Although all their major clinical findings indicated enteropathy, they had different endoscopy findings and different response to the immunosuppressive treatment. Functional experiments revealed that a lack of LRBA protein expression may lead to worse disease outcomes and be an indicator for hematopoietic stem cell transplantation (HSCT). The results of this study will be valuable for the selection of an immunosuppressive treatment or HSCT for treating LRBA-deficient patients in the future.
- Citation: Tang WJ, Hu WH, Huang Y, Wu BB, Peng XM, Zhai XW, Qian XW, Ye ZQ, Xia HJ, Wu J, Shi JR. Potential protein–phenotype correlation in three lipopolysaccharide-responsive beige-like anchor protein-deficient patients. World J Clin Cases 2021; 9(21): 5873-5888
- URL: https://www.wjgnet.com/2307-8960/full/v9/i21/5873.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v9.i21.5873
Lipopolysaccharide-responsive beige-like anchor protein (LRBA) is a member of the PH-BEACH-WD40 (pleckstrin homology-beige and Chediak-Higashi-tryptophan aspartic acid dipeptide) protein family and participates in multiple cellular processes, including signal transduction, vesicular trafficking, transcriptional regulation, cytoskeleton assembly, chromatin dynamics, and apoptosis[1]. The LRBA gene is located on 4q31.3, contains 57 exons, and encodes a protein containing 2851 amino acids. The LRBA protein is a cytosolic protein that is expressed in several cell types, including hematopoietic, neural, gastrointestinal, and endocrine cells[2]. LRBA protein deficiency can cause dysregulation in activated T cells and circulating TFH (cTFH) cells due to marked Treg cell depletion and impaired Treg cell-mediated suppression caused by the large deficiency of cytotoxic T lymphocyte-associated antigen 4 (CTLA4) expression in residual Tregs[1,3-5]. LRBA deficiency is one of the most common autosomal recessive defects that cause common variable immunodeficiency (CVID), and it was first reported in 2012[6-8]. Patients with LRBA deficiency have a variety of clinical symptoms, including gastrointestinal (GI) symptoms, hypogammaglobulinemia, recurrent infections, lymphoproliferation, and autoimmune cytopenias (autoimmune hemolytic anemia (AIHA), immune thrombocytopenia (ITP), and neutropenia)[9-12].
To date, over 100 patients with LRBA deficiency have been reported, and most of these patients present with severe clinical symptoms that cannot be controlled by traditional immunosuppressive agents. However, there is no apparent genotype-phenotype correlation for LRBA deficiency; patients carrying the same mutations may have different phenotypes[3,13-16]. Therefore, it is not easy for doctors to make a decision regarding hematopoietic stem cell transplantation (HSCT) for LRBA-deficient patients. A recent study showed that there may be a protein-phenotype correlation[17].
LRBA deficiency is rarely reported in Chinese patients. In the present study, we reported three patients who were identified as carrying LRBA mutations. Functional experiments revealed that a lack of LRBA protein expression may lead to worse disease outcomes and be an indicator for HSCT.
Three individuals who were diagnosed with LRBA deficiency at our department were enrolled in this study. Clinical data, including complete blood counts, immunoglobulin levels, lymphocyte subsets, genetic mutations, and histopathological findings of biopsy specimens, were analyzed.
Whole-exome sequencing (WES) and analysis protocols were adapted for genetic analysis. Genomic DNA fragments of the patients and their parents were enriched for sequencing. The enriched DNA samples were indexed and sequenced on a HiSeq 2000 sequencer (Illumina, San Diego, CA, United States). Nucleotide changes observed in more than 5% of aligned reads were identified and reviewed by using NextGENe software (SoftGenetics, State College, PA, United States).
Missense mutations in LRBA from patients and their parents were confirmed by Sanger sequencing. Genomic DNA was extracted from peripheral blood using the Blood DNA Isolation Kit (Vazyme, DC111-01) according to the manufacturer’s instructions. PCR was carried out in a 25 μL reaction volume containing 70 ng of template DNA, 1 μL of each designed primer (10 µmol/L), 12.5 µL of Premix Taq (Takara, No: RR003A), and H2O. PCR was conducted using the following program: A denaturation step at 95 °C for 5 min; amplification via 35 cycles as follows: 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min; and a final extension step at 72 °C for 7 min. Then, the products were sent for purification and sequencing (ABI 3730xl, Jieli Company). Mutations were confirmed using SeqMan software.
Deletion mutations were confirmed by quantitative PCR in patient 1 and his parents. Briefly, Takara SYBR quantitative polymerase chain reaction (qPCR) Mix (RR820A), genomic DNA, and the primers were mixed, and qPCR was performed following the manufacturer’s instructions. The primer sequences are as follows: LRBA-exon 53 forward, 5’-gaatgcacaggaggcaaatc-3’ and reverse, 5’-ccagaagccacagacgaga-3’; LRBA-control forward, 5’-atcacaccagcagcattcag-3’ and reverse, 5’-tccacatggcttcctaaacc-3’.
LRBA protein analysis was performed on patients and their parents. Peripheral blood mononuclear cells (PBMCs) were lysed, and protein was extracted using RIPA buffer (Thermo Scientific 89900) according to the manufacturer’s instructions. Equal amounts of cell extracts were separated using 10% SDS polyacrylamide gels and then transferred to PVDF membranes at 100 V for 90 min. The membranes were blocked with 6% nonfat milk for 1 h and incubated overnight at 4 °C with primary antibodies against LRBA (Sigma-Aldrich, HPA023597), GAPDH (Protein-HRP, HRP-60004), and β-actin (Affinity, AF7018). After incubation with a secondary antibody, visualization was performed using ECL substrate (LumiBest ECL Substrate Solution Kit, Share-bio, Sb-wb011).
Quantity One software was used to analyze the gray value. The relative LRBA protein expression level was evaluated by the gray value of LRBA protein/the gray value of the internal reference protein. Then, the relative expression of LRBA in patient PBMCs was compared with that in the control group.
Heparin sodium anticoagulated peripheral blood diluted with isometric phosphate-buffered saline (PBS) was slowly added to Ficoll-Paque Plus (GE Healthcare) and then centrifuged at 450 g at room temperature for 30 min. The PBMC layer was pipetted and washed twice with PBS. After centrifugation at 1500 rpm for 5 min, the PBMCs were stored until use in other experiments.
We searched PubMed using the terms "LRBA", "LRBA deficiency", "common variable immunodeficiency", and "primary immunodeficiency diseases". The first search date was January 1, 2012. The search was limited to full texts in English and human studies, and duplicate cases, asymptomatic cases, and cases with no genetic information were excluded. The reported genetic mutation information was summarized.
Statistics were analyzed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA, United States). Data are presented as the mean ± SD, and a t-test was used to analyze differences between groups. P < 0.05 was considered statistically significant.
Patient 1: A 5.4-year-old boy presented with severe chronic diarrhea and abdominal distention starting at 2 years of age (Table 1). Watery diarrhea often caused dehydration, hypokalemia, and failure to thrive, and the severe abdominal distention could only be alleviated after defecation. He required hospitalization several times since 2 years of age due to severe complications. During treatment courses, he developed recurrent upper respiratory infections, temporary hypothyroidism, and arthritis, and no infectious agents were found. He showed significant growth delay, with a weight of 9 kg (under the third percentile) and height of 75 cm at 3.7 years old (under the third percentile). Endoscopy and histology showed nonspecific colitis characterized by no crypt destruction and villus atrophy (Table 1). Despite changing medication and treatment, he did not show any improvement of his symptoms. He was the first child in his family.
| Patient 1 | Patient 2 | Patient 3 | ||
| Age of onset | 2.0 yr | 10.0 mo | < 1.0 yr | |
| Age of genetic | 3.9 | 2 | 3 | |
| Current age (yr) | 5.4 | 7.1 | 3.2 | |
| Sex | M | F | M | |
| Consanguinity | No | No | No | |
| Final outcome | Alive | Alive | Alive | |
| First symptoms | Diarrheal, abdominal distention | Diarrheal, hematochezia, abdomen pain | Diarrheal, growth delay | |
| Failure to thrive | (+) | (-) | (+) | |
| Other clinical findings | RURI, temporary hypothyroidism, neutropenia, urticaria, arthritis, growth delay | No | Elevated liver enzymes, renal pelvis | |
| Endoscopy findings | Non-specific colitis | Chron's-like disease | Non-specific colitis | |
| Infectious agents | No documented infectious agent | No documented infectious agent | Rotavirus, Klebsiella pneumoniae, Bacillus thuringiensis | |
| IgG (g/L) (RV: 4.95-12.85) | 12.7 | 8.7 | 4.4 | |
| IgA (g/L) (RV: 0.52-2.16) | 1.42 | 0.28 | 0.49 | |
| IgM (g/L) (RV: 0.65-2.01) | 0.8 | 1.01 | 2.04 | |
| White blood cells count (× 109) | 2.70-3.60 | 7.2 | 9.7 | |
| (RV: 4.0-10.0) | ||||
| Absolute lymphocyte | 2.00-2.80 | 3.6 | 3.36 | |
| Absolute neutrophil | 0.21-0.42 | 3.18 | 5.54 | |
| CD19+ B cells | (%) (RV: 14-21) | 12.78 | 18.63 | 12.03 |
| Count | 345.82 | 535.9 | 349.8 | |
| CD3+ T cells | (%) (RV: 64-73) | 76.97 | 69.66 | 59.69 |
| Count | 2083.5 | 2003.7 | 1735.4 | |
| CD3+CD4+ Th cells | (%) (RV: 29-36) | 39.86 | 37.37 | 33.32 |
| Count | 1078.83 | 1074.84 | 965.76 | |
| CD3+CD8+ Tc cells | (%) (RV: 24-34) | 32.04 | 31.4 | 24.38 |
| Count | 867.33 | 903.31 | 708.84 | |
| CD3-CD16+56+NK cells | (%) (RV: 11-23) | 8.88 | 10.34 | 26.98 |
| Count | 240.5 | 297.45 | 784.43 | |
| CD4/CD8 | 1.24 | 1.19 | 1.36 | |
| Immunosuppressive | / | Steroids, 5-ASA, AZA | Steroids | |
| HSCT | + | - | - | |
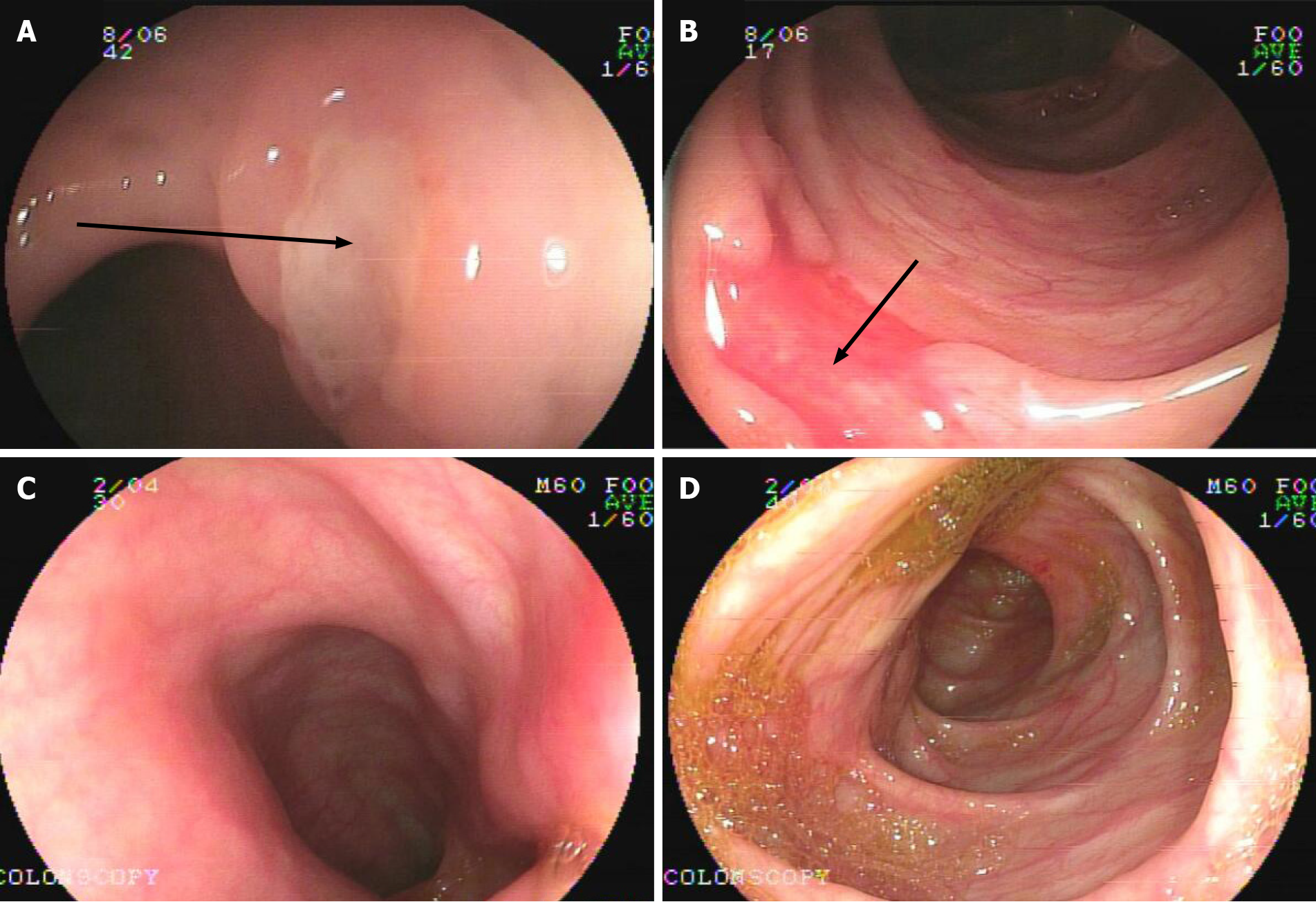
Patient 2: A 7.1-year-old girl presented with chronic diarrhea and hematochezia with recurrent abdominal pain at 10 mo old, and no infectious agents were found (Table 1). She had mucus and bloody stool 3-6 times a day, and her symptoms could be improved by antibiotics but persisted. During her treatment courses, no severe complications developed. Endoscopy and biopsies revealed chronic disease-like mucosal inflammation (Figure 1A and B). She was initially diagnosed with CD and had a healthy brother.
Patient 3: A 3.2-year-old boy was admitted to our department with complaints of chronic diarrhea and growth delay. He had watery, mucus stool 6–14 times a day since 1 year of age and required hospitalization several times for severe complications since then. Regular medicine, including antibiotics and formula changes, did not improve his symptoms. He showed significant growth delay, with a weight of 6.5 kg (under the third percentile) and height of 75 cm at 3.2 years old (under the third percentile). During treatment courses, he developed Rotavirus and Klebsiella pneumoniae infections, liver dysfunction, Bacillus thuringiensis sepsis, and temporary hypothyroidism. Left renal pelvis separation was found. Endoscopy and histology also showed nonspecific colitis characterized by no crypt destruction and villus atrophy.
For patient 1, the complete blood count showed neutropenia. Immune cell subclass analysis, which was performed at 3.7 years old, showed that the levels of IgG, IgA, and IgM were within the reference ranges for his age but a low CD19+ B cell count (Table 1). For patient 2, the complete blood count was normal, and immune cell subclass analysis, which was performed at 2 years old, showed that the IgA level was decreased (Table 1). For patient 3, an immunologic evaluation was performed at 3.2 years old; it was found that the complete blood count was normal, the IgG and IgA levels were slightly decreased, and the CD19+ B cell count and CD3+ T cell percentage were lower than the reference ranges for his age.
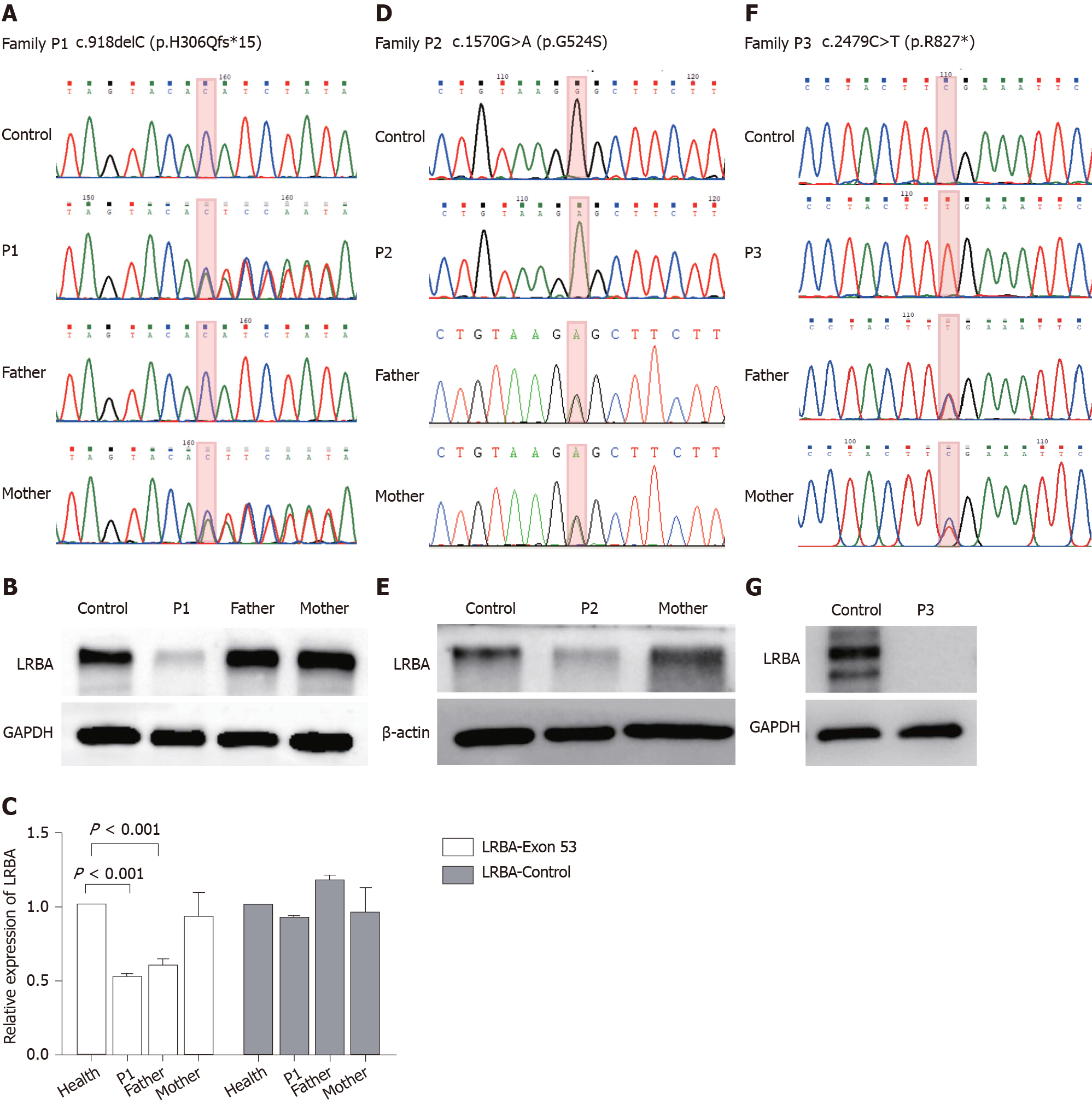
Genetic analysis of patient 1 revealed heterozygous c.918delc in exon 8 of LRBA. The patient’s mother was a heterozygous carrier, and his father showed no mutation (Figure 2A), but sequence alignment suggested that the father may have a large deletion. We performed qPCR to verify this hypothesis and found that both the child and father had a large deletion in exon 53 of LRBA (Figure 2C). To verify and confirm the genetic testing results, LRBA protein analysis was performed and showed that patient 1 had very low LRBA protein expression (Figure 2B), which was consistent with the WES results and symptoms.
Genetic analysis of patient 2 revealed homozygous c.1570G>A in exon 12 of LRBA, and both of her parents were heterozygous carriers (Figure 2D). Western blot analysis also confirmed that the patient had lower LRBA protein expression than that of the healthy controls (Figure 2E).
Genetic analysis of patient 3 revealed homozygous c.2479C>T in exon 21 of LRBA, and both of his parents were heterozygous carriers (Figure 2F). Western blot analysis also confirmed that the patient had lower LRBA protein expression than the healthy controls (Figure 2G).
Patient 1 showed no response or allergic reaction to immunosuppressive treatment. Because of the patient’s severe complications of chronic diarrhea and growth restriction, at 4.1 years old, he was treated with HSCT. To date, 1.3 years after HSCT, he is alive with no symptoms. He has been off immunosuppressive treatment for 8 mo with no signs of graft-vs-host disease (GVHD), and his health has improved markedly, with an increased growth rate (weight of 15 kg and height of 100 cm at 5.4 years old).
Patient 2 was treated with steroids to induce remission and 5-aminosalicylic acid and azathioprine to maintain remission. She was asymptomatic for 3 years and on normal food, with weight and height gain at normal rates. Endoscopy at 5 years old showed that there were no ulcers in her gut (Figure 1C and D).
Patient 3 still required hospitalization for dehydration, electrolyte disturbances, infections, and abdominal distention. Antibiotics and steroids were used, which seemed to partly improve his symptoms, and he defecated 2–5 times per day.
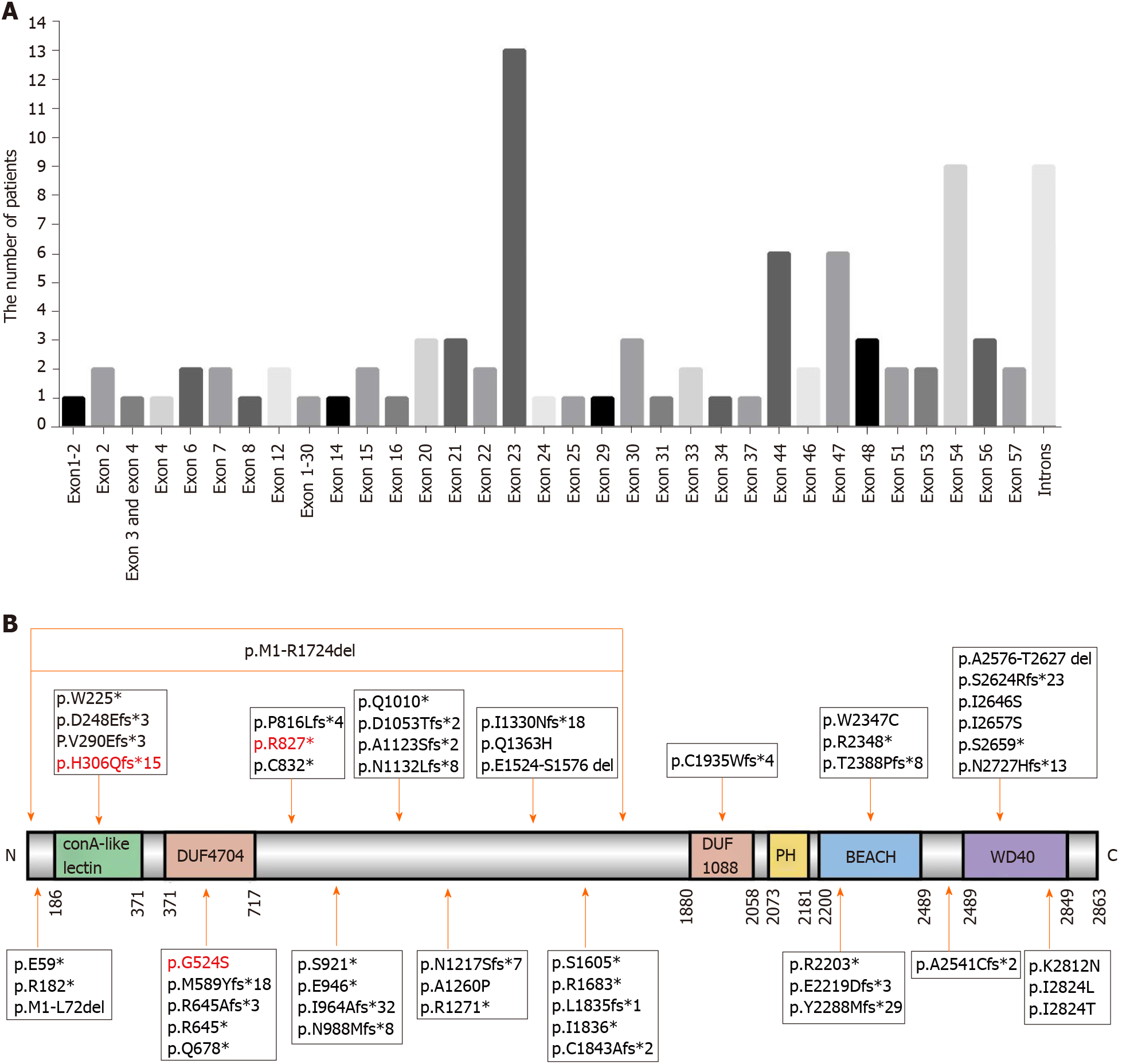
Finally, 83[2,6-9,11-14,16,18-43] patients with genetic information were reviewed, and their genetic mutation information together with that of our three patients are summarized in Table 2. A total of 54 mutations located in exons and 10 mutations located in introns were identified. Most patients carried mutations in exon 23 (13/85), exon 54 (9/85), introns (9/85), exon 44 (6/85), and exon 47 (6/85) (Figure 3A). The LRBA protein map shows the mutation sites (Figure 3B). The mutation sites in patient 1 (P84 in Table 2) and patient 3 (P86 in Table 2) were novel mutations, which are reported for the first time in this study. The mutation site in patient 2 (P85 in Table 2) was reported in another Chinese patient who carried compound heterozygous mutations[43]. This disease is more common in consanguineous families, and 85.4% (70 of 82) of patients who suffer from this disease were from consanguineous marriages, such as in Middle Eastern populations (Table 2). To date, seven Chinese patients with LRBA deficiency have been reported (Table 2).
| Patient | Ethnicity | CF | Affected exon/intron | Defect (cDNA) | Defect (protein) |
| P1[2,7] | Arab | Y | Exon 44 | c.6657_6658del | p.E2219Dfs*3 |
| P2[2,7] | Arab | Y | Exon 44 | c.6657_6658del | p.E2219Dfs*3 |
| P3 [2,7] | Arab | Y | Exon 44 | c.6657_6658del | p.E2219Dfs*3 |
| P4[2,7] | Arab | Y | Exon 44 | c.6657_6658del | p.E2219Dfs*3 |
| P5[2,7] | Arab | Y | Exon 44 | c.6657_6658del | p.E2219Dfs*3 |
| P6[2,6] | Sicilian | Y | Exon 30 | c.5047C>T | p.R1683* |
| P7[2,6] | Iranian | Y | Exon 2 | c.175G>T | p.E59* |
| P8[2,6,18] | Iranian | Y | Exons 1-2 | c.A1-G216del | p.M1-L72del |
| P9[2,8] | Pakistani | Y | Exons 1-30 | c.A1-G5171del | p.M1-R1724del |
| P10[2,9] | Arabian | Y | Exon 16 | c.2032C>T | p.Q678* |
| P11[2,9] | Lebanese | Y | Exon 7 | c.865_866delTG | p.V290Efs*3 |
| P12[2,19] | Kurdish | Y | Exon 48 | c.7162_7162delA | p.T2388Pfs*8 |
| P13[2,19] | Kurdish | Y | Exon 48 | c.7162_7162delA | p.T2388Pfs*8 |
| P14[2,20,21] | Turkish | Y | Exon 57; Exon 57 | c.A8470C; p.T8471C | p.I2824L; p.I2824T |
| P15[2] | NA | Y | Exon 33 | c.5505_5505delT | p.I1836* |
| P16[2] | North African | Y | Exon 44 | c.6607C>T | p.R2203* |
| P17[2] | Iranian | Y | Introns 8-9 | c.1014 + 1 C> T | |
| P18[2,12,14,22] | Iranian | Y | Exon 30 | c.C4814G | p.S1605* |
| P19[2] | Iranian | Y | Exon 4 | c.C544T | p.R182* |
| P20[2,12,14,22] | Iranian | Y | Intron 29-30 | c.4729 + 2insA | |
| P21[11] | Libyan | Y | Exon 20 | c.2445_2447delinsCC | p.P816Lfs*4 |
| P22[11] | Libyan | Y | Exon 20 | c.2445_2447delinsCC | p.P816Lfs*4 |
| P23[23] | Persian | Y | Exon 6 | c.743_744insAAGA | p.D248Efs*3 |
| P24[24] | Turkish | Y | Exon 48 | c.7162delA | p.T2388Pfs*8 |
| P25[25] | Egyptian | Y | Exon 46 | c.6862delT | p.Y2288Mfs*29 |
| P26[21,26] | Turkish | Y | Exon 33 | c.5505delT | p.I1836* |
| P27[27] | Caucasian | NA | Exon 23; Exon 54 | c.3647_3650delCTAA; c.7937T>G | p.N1217Sfs*7; p.I2646S |
| P28[28] | Omani | Y | Exon 23 | c.3811C>T | p.R1271* |
| P29[9,30] | Turkish | N | Exon 23; Exon 54 | c.3156_3156delT; c.7976C>A | p.D1053Tfs*2; p.S2659* |
| P30[16,21] | Turkish | Y | Exon 6 | c.675G > A | p.W225* |
| P31[30] | Moroccan | Y | Exon 47 | c.7042C>T | p.R2348* |
| P32[30] | Moroccan | Y | Exon 47 | c.7042C>T | p.R2348* |
| P33[30] | Omani | Y | Intron 34-35 | c.5581-1G>A | |
| P34[30] | Egyptian | Y | Exon 20 | c.2447_2447delC | p.P816Lfs*4 |
| P35[30] | Chinese | N | Intron 29-30 | c.(4729+1_4730-1)_(5171+1_ | |
| P36[22] | Pakistani | Y | Exon 24 | c.3988delinsAA | p.I1330Nfs*18 |
| P37[31] | Caucasian | Y | Exon 23 | c.2836_2839delGAAA | p.E946* |
| P38[32] | Arab | Y | Exon 54 | c.7970T>G | p.I2657S |
| P39[32] | Arab | Y | Exon 56 | c.8174_8175insCATG | p.N2727Hfs*13 |
| P40[32] | Arab | Y | Exon 53 | c.7869_7873delTTCTA | p.S2624Rfs*23 |
| P41[13] | Belarusian | N | Exon 22 | c.2762C>G | p.S921* |
| P42[13] | Belarusian | N | Exon 22 | c.2762C>G | p.S921* |
| P43[33] | NA | Y | Exon 21 | c.2496C>A | p.C832* |
| P44[33] | NA | Y | Exon 21 | c.2496C>A | p.C832* |
| P45[34] | NA | NA | NA | c.2450+1C >T | p.E789fs*792 |
| P46[35] | Moroccan | Y | Intron 20-21 | c.2450-3C>A | |
| P47[36] | NA | NA | Exon 15 | c.1931delinsCC | p.R645Afs*3 |
| P48[37] | Caucasian | N | Exon 23 | c.3366delinsAA | p. A1123Sfs*2 |
| P49[38] | Arab | Y | Intron 41-42 | c.6364-1G >C | |
| P50[16,39] | Turkish | Y | Exon 34 | c.5527delT | p.C1843Afs*2 |
| P51[16,39] | Turkish | Y | Exon 47 | c.7042C>T | p.R2348* |
| P52[16] | NA | Y | Exon 47 | c.7041G>T | p.W2347C |
| P53[16] | NA | Y | Intron 6–7 | IVS6+1delT | |
| P54[16] | NA | Y | Exon 31 | c.5504delT | L1835fs*1 |
| P55[21,32] | Turkish | Y | Exon 23 | c.2893_2900delinsGCCAGATA | p.I964Afs*32 |
| P56[16] | NA | Y | Exon 2 | c.175G>T | p.E59* |
| P657[16] | NA | Y | Exon 23 | c.2836_2839delGAAA | p.E946* |
| P58[2,6] | Arab | Y | Exon 54 | c.7970T>G | p.I2657S |
| P59[2,6] | Arab | Y | Exon 54 | c.7970T>G | p.I2657S |
| P60[2,9] | Lebanese | Y | Exon 7 | c.865_866delTG | p.V290Efs*3 |
| P61[2,32,40] | Arab | Y | Exon 54 | c.7970T>G | p.I2657S |
| P62[2,32,40] | Arab | Y | Exon 56 | c.8174_8175insCATG | p.N2727Hfs*13 |
| P63[2,32,40] | Arab | Y | Exon 56 | c.8174_8175insCATG | p.N2727Hfs*13 |
| P64[2,12,14,22] | Iranian | Y | Exon 30 | c.C4814G | p.S1605* |
| P65[2] | Lebanese | Y | Exon 23 | c.2963_2963delA | p.N988Mfs*8 |
| P66[2,12,14] | Iranian | Y | Intron 29-30 | c.4729+4dupA | |
| P67[41] | NA | Y | Exon 51 | c.7620_7621insT | p.A2541Cfs*2 |
| P68[41] | NA | Y | Exon 51 | c.7620_7621insT | p.A2541Cfs*2 |
| P69[25] | Egyptian | Y | Exon 46 | c.6862delT | p.Y2288Mfs*29 |
| P70[21] | Turkish | N | Exon 23; Exon 54 | c.3028G>A; c.7976G > C | p.Q1010*; p.S2659* |
| P71[30] | Omani | Y | Exon 23 | c.3811C>T | p.R1271* |
| P72[30] | Iranian | Y | Exon 14 | c.1764delinsTT | p.M589Yfs*18 |
| P73[22] | Turkish | Y | Intron 30-31 | c.5172-2A>G | |
| P74[32] | Arab | Y | Exon 54 | c.7970T>G | p.I2657S |
| P75[32] | Arab | Y | Exon 54 | c.7970T>G | p.I2657S |
| P76[16] | Turkish | N | Exon 37; Exon 47 | c.5805delT; c.7042C>T | p.C1935Wfs*4; |
| P77[16] | NA | Y | Exon 23 | c.3396_3397delCA | p.N1132Lfs*8 |
| P78[16] | Turkish | Y | Exon 47 | c.7042C>T | p.R2348* |
| P79[16] | NA | Y | Exon 23 | c.3811C>T | p.R1271* |
| P80[16] | NA | Y | Exon 3; Exon 4 | c.(501+1_502-1)_(733+1_734-1)del | P. G75_W183* |
| P81[42] | Chinese | NA | Exon 57; Exon 25 | c.8436G>C; c.4089A>T | p.K2812N; p.Q1363H |
| P82[43] | Chinese | N | Exon 15; Exon 29 | c.1933C>T; c.G4570-G4729del | p.R645*; p.E1524-S1576 del |
| P83[43] | Chinese | N | Exon 23; Exon 12 | c.3778G>C; c.1570G>A | p.A1260P; p.G524S |
| P84our patient | Chinese | N | Exon 8; Exon 53 | c.918delC; c.C7727-G7882 | p.H306Qfs*;15; p.A2576-T2627 |
| P85our patient | Chinese | N | Exon 12 | c.1570G>A | p.524G>S |
| P86our patient | Chinese | N | Exon 21 | c.2479C>T | p.Arg827* |
In this study, we described three Chinese patients with LRBA deficiency, and all their major clinical findings indicated enteropathy. Enteropathy is one of the most common early manifestations of LRBA deficiency[10,12,16,22], and it has been identified in 53.3%-88.2% of LRBA deficiency patients[2,12,16]. As previously reported, endoscopy findings can be divided into inflammatory bowel disease-like colitis or nonspecific colitis (characterized by no crypt destruction and villus atrophy, or completely normal and lymphocytic infiltration, resembling autoimmune colitis or celiac disease). Our three patients had different endoscopy findings; although patients 1 and 3 had more severe chronic diarrhea than patient 2, nonspecific colitis was found by endoscopy. IBD-like colitis was only found in patient 2 (1/3). Patients 1 and 3 also had different responses to immunosuppressive treatment; patient 2 responded well to treatment with traditional medicine, but patient 1 had no response to immunosuppressive treatment and, finally, had to undergo HSCT. It seemed that patient 3 also did not respond well to steroids.
It is not clear what caused the different endoscopy findings and different responses to immunosuppressive treatment. As previous studies have reported, there is no apparent genotype–phenotype correlation in LRBA deficiency, and our patients also seemed not to show a genotype–phenotype correlation. Both patients 2 and 3 carried homozygous mutations, but they had different clinical and endoscopy findings.
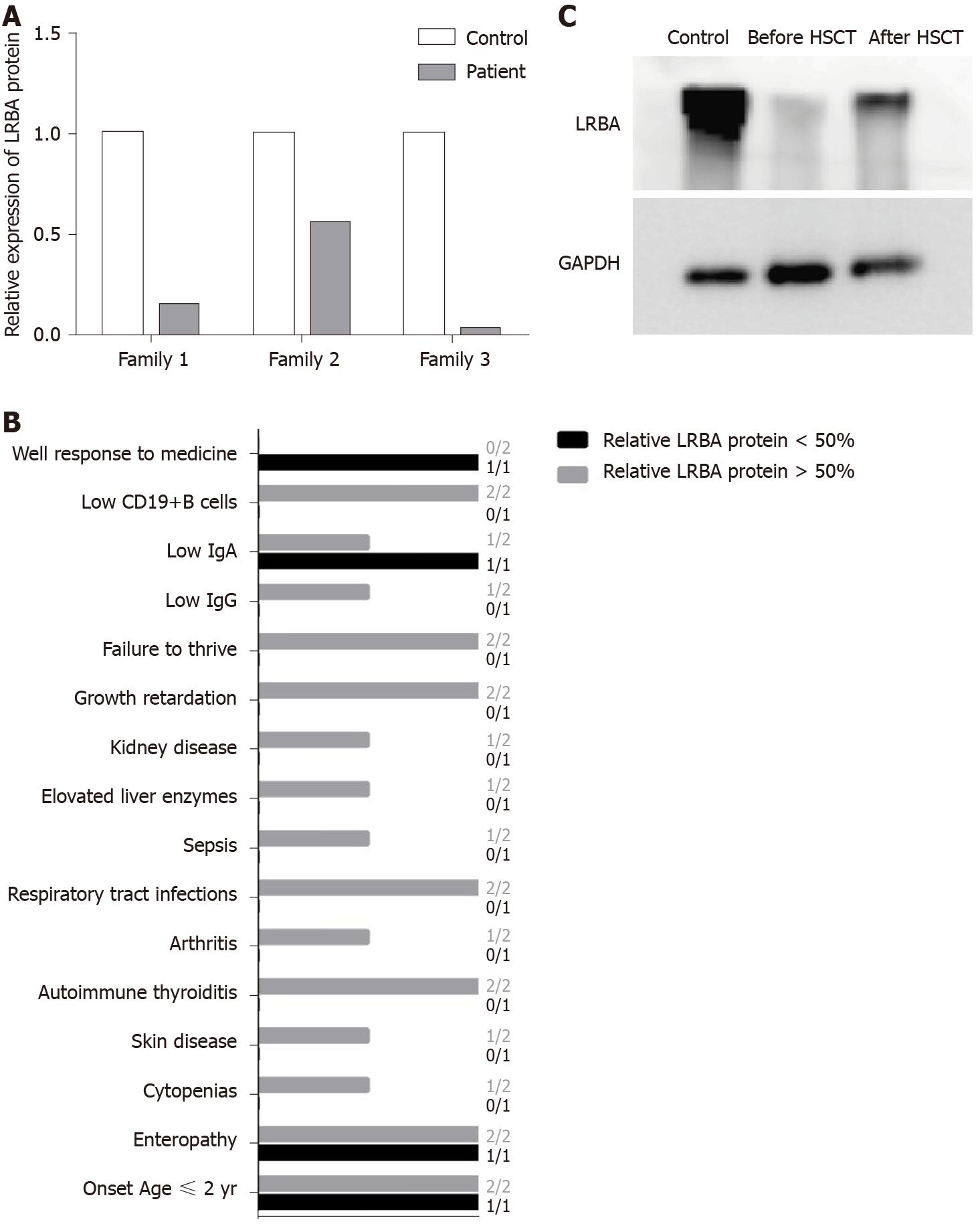
A recent study has shown a possible protein–phenotype correlation in LRBA deficiency[17], so we quantified the expression of residual LRBA protein. We found that patients 1 and 3 had very low protein expression compared with the controls (Figure 4A), but a more than 0.5-fold in expression of the LRBA protein was found in patient 2 compared to that in the control (Figure 4A). Patient 2 had fewer and weaker symptoms than patients 1 and 3 (Figure 4B), and only patient 2 responded well to traditional medicine (Figure 4B). After HSCT, patient 1 showed increased LRBA protein expression (Figure 4C). These results suggest that residual LRBA protein expression may be associated with disease outcome and that a lack of LRBA protein expression may indicate a worse disease outcome. HSCT has been suggested as the last available approach for LRBA patients with severe and complicated clinical manifestations who do not respond to conventional therapies[16,27]. However, because of complications and GVHD, it is better to have indicators to help doctors make a decision regarding HSCT. Our results suggest that the lack of LRBA protein expression may be an indicator for HSCT. When doctors treat LRBA-deficient patients, more protein information is needed. The results of this study will be valuable for the selection of an immunosuppressive treatment or HSCT for treating LRBA-deficient patients in the future.
We described three Chinese LRBA-deficient patients, two of whom carried novel mutations. Their major clinical findings indicated enteropathy, but they had different clinical phenotypes and different responses to immunosuppressive treatment. There may be a protein–phenotype correlation in LRBA deficiency, and residual LRBA protein expression may be associated with disease outcome and can be an indicator of HSCT.
There are limitations to this study. Only three patients were enrolled in this study, and the patients' LRBA protein levels were not quantified by flow cytometry and could not be compared among patients. We need to collect more data in the future and change our methods to improve the strength of our findings.
Patients with lipopolysaccharide-responsive beige-like anchor protein (LRBA) deficiency have a variety of clinical symptoms, and most patients present with severe clinical symptoms. However, it seems that there is no apparent genotype–phenotype correlation for LRBA deficiency. Therefore, it is not easy for doctors to make a decision regarding hematopoietic stem cell transplantation (HSCT) for LRBA-deficient patients. Therefore, we studied the protein–phenotype correlation in three LRBA-deficient patients and found that the lack of LRBA protein expression may indicate worse disease outcomes and be an indicator for HSCT.
The main motivation of this study was to study the protein–phenotype correlation in LRBA-deficient patients. The key problem to be solved is as follows: A lack of LRBA protein expression may indicate worse disease outcomes and be an indicator for HSCT.
The aim of this study was to identify potential protein–phenotype correlations in LRBA-deficient patients and look for indicators for HSCT. We hope that this study can provide some beneficial information to doctors regarding when HSCT should be considered in LRBA-deficient patients.
Whole-exome sequencing was adapted for genetic analysis in three LRBA-deficient patients, and their clinical data were analyzed. Protein was extracted from peripheral blood mononuclear cells of the three patients and their parents. Western blot was performed for protein analysis. Relative LRBA protein expression was determined for every patient and compared with the controls. Data are presented as the mean ± SD, and a t-test was used to analyze the differences. P < 0.05 was considered statistically significant.
The results showed that there may be a protein–phenotype correlation in LRBA deficiency, and residual LRBA protein expression may be associated with disease outcome and could be an indicator for HSCT. However, there are limitations to this study, and we need to collect more data in the future to strengthen our findings.
There may be a protein–phenotype correlation in LRBA deficiency, and residual LRBA protein expression may be associated with disease outcome and can be an indicator of HSCT.
In the future, we will collect more data from LRBA-deficient patients. It would be better to analyze the LRBA protein level by flow cytometry and compare the level among patients.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and Hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Nishizawa T S-Editor: Ma YJ L-Editor: Wang TQ P-Editor: Liu JH
| 1. | Azizi G, Yazdani R, Rae W, Abolhassani H, Rojas M, Aghamohammadi A, Anaya JM. Monogenic polyautoimmunity in primary immunodeficiency diseases. Autoimmun Rev. 2018;17:1028-1039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 2. | Alkhairy OK, Abolhassani H, Rezaei N, Fang M, Andersen KK, Chavoshzadeh Z, Mohammadzadeh I, El-Rajab MA, Massaad M, Chou J, Aghamohammadi A, Geha RS, Hammarström L. Spectrum of Phenotypes Associated with Mutations in LRBA. J Clin Immunol. 2016;36:33-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 116] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 3. | Azizi G, Mirshafiey A, Abolhassani H, Yazdani R, Ghanavatinejad A, Noorbakhsh F, Rezaei N, Aghamohammadi A. The imbalance of circulating T helper subsets and regulatory T cells in patients with LRBA deficiency: Correlation with disease severity. J Cell Physiol. 2018;233:8767-8777. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 4. | Lo B, Zhang K, Lu W, Zheng L, Zhang Q, Kanellopoulou C, Zhang Y, Liu Z, Fritz JM, Marsh R, Husami A, Kissell D, Nortman S, Chaturvedi V, Haines H, Young LR, Mo J, Filipovich AH, Bleesing JJ, Mustillo P, Stephens M, Rueda CM, Chougnet CA, Hoebe K, McElwee J, Hughes JD, Karakoc-Aydiner E, Matthews HF, Price S, Su HC, Rao VK, Lenardo MJ, Jordan MB. AUTOIMMUNE DISEASE. Patients with LRBA deficiency show CTLA4 Loss and immune dysregulation responsive to abatacept therapy. Science. 2015;349:436-440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 448] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 5. | Cepika AM, Sato Y, Liu JM, Uyeda MJ, Bacchetta R, Roncarolo MG. Tregopathies: Monogenic diseases resulting in regulatory T-cell deficiency. J Allergy Clin Immunol. 2018;142:1679-1695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 87] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 6. | Lopez-Herrera G, Tampella G, Pan-Hammarström Q, Herholz P, Trujillo-Vargas CM, Phadwal K, Simon AK, Moutschen M, Etzioni A, Mory A, Srugo I, Melamed D, Hultenby K, Liu C, Baronio M, Vitali M, Philippet P, Dideberg V, Aghamohammadi A, Rezaei N, Enright V, Du L, Salzer U, Eibel H, Pfeifer D, Veelken H, Stauss H, Lougaris V, Plebani A, Gertz EM, Schäffer AA, Hammarström L, Grimbacher B. Deleterious mutations in LRBA are associated with a syndrome of immune deficiency and autoimmunity. Am J Hum Genet. 2012;90:986-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 386] [Cited by in F6Publishing: 341] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 7. | Alangari A, Alsultan A, Adly N, Massaad MJ, Kiani IS, Aljebreen A, Raddaoui E, Almomen AK, Al-Muhsen S, Geha RS, Alkuraya FS. LPS-responsive beige-like anchor (LRBA) gene mutation in a family with inflammatory bowel disease and combined immunodeficiency.. J Allergy Clin Immunol. 2012;130:481-488.e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 178] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 8. | Burns SO, Zenner HL, Plagnol V, Curtis J, Mok K, Eisenhut M, Kumararatne D, Doffinger R, Thrasher AJ, Nejentsev S. LRBA gene deletion in a patient presenting with autoimmunity without hypogammaglobulinemia. J Allergy Clin Immunol. 2012;130:1428-1432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 9. | Charbonnier LM, Janssen E, Chou J, Ohsumi TK, Keles S, Hsu JT, Massaad MJ, Garcia-Lloret M, Hanna-Wakim R, Dbaibo G, Alangari AA, Alsultan A, Al-Zahrani D, Geha RS, Chatila TA. Regulatory T-cell deficiency and immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like disorder caused by loss-of-function mutations in LRBA. J Allergy Clin Immunol. 2015;135:217-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 175] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 10. | Gámez-Díaz L, August D, Stepensky P, Revel-Vilk S, Seidel MG, Noriko M, Morio T, Worth AJJ, Blessing J, Van de Veerdonk F, Feuchtinger T, Kanariou M, Schmitt-Graeff A, Jung S, Seneviratne S, Burns S, Belohradsky BH, Rezaei N, Bakhtiar S, Speckmann C, Jordan M, Grimbacher B. The extended phenotype of LPS-responsive beige-like anchor protein (LRBA) deficiency. J Allergy Clin Immunol. 2016;137:223-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 184] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 11. | Schreiner F, Plamper M, Dueker G, Schoenberger S, Gámez-Díaz L, Grimbacher B, Hilger AC, Gohlke B, Reutter H, Woelfle J. Infancy-Onset T1DM, Short Stature, and Severe Immunodysregulation in Two Siblings With a Homozygous LRBA Mutation. J Clin Endocrinol Metab. 2016;101:898-904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 12. | Azizi G, Abolhassani H, Mahdaviani SA, Chavoshzadeh Z, Eshghi P, Yazdani R, Kiaee F, Shaghaghi M, Mohammadi J, Rezaei N, Hammarström L, Aghamohammadi A. Clinical, immunologic, molecular analyses and outcomes of iranian patients with LRBA deficiency: A longitudinal study. Pediatr Allergy Immunol. 2017;28:478-484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 53] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 13. | Sharapova SO, Haapaniemi E, Sakovich IS, Rojas J, Gámez-Díaz L, Mareika YE, Guryanova IE, Migas AA, Mikhaleuskaya TM, Grimbacher B, Aleinikova OV. Novel LRBA Mutation and Possible Germinal Mosaicism in a Slavic Family. J Clin Immunol. 2018;38:471-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Azizi G, Abolhassani H, Habibi S, Rahamooz T, Mohammadi H, Jafarnezhad-Ansariha F, Mortazavi-Jahromi SS, Yazdani R, Aghamohammadi A. Two Faces of LRBA Deficiency in Siblings: Hypogammaglobulinemia and Normal Immunoglobulin Levels. J Investig Allergol Clin Immunol. 2018;28:48-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Azizi G, Jamee M, Yazdani R, Bagheri Y, Fayyaz F, Jadidi-Niaragh F, Abolhassani H, Aghamohammadi A. CTLA-4 Expression in CD4+ T Cells From Patients With LRBA Deficiency and Common Variable Immunodeficiency With No Known Monogenic Disease. J Investig Allergol Clin Immunol. 2018;28:422-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Cagdas D, Halaçlı SO, Tan Ç, Lo B, Çetinkaya PG, Esenboğa S, Karaatmaca B, Matthews H, Balcı-Hayta B, Arıkoğlu T, Ezgü F, Aladağ E, Saltık-Temizel İN, Demir H, Kuşkonmaz B, Okur V, Gümrük F, Göker H, Çetinkaya D, Boztuğ K, Lenardo M, Sanal Ö, Tezcan İ. A Spectrum of Clinical Findings from ALPS to CVID: Several Novel LRBA Defects. J Clin Immunol. 2019;39:726-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 17. | Tesch VK, Abolhassani H, Shadur B, Zobel J, Mareika Y, Sharapova S, Karakoc-Aydiner E, Rivière JG, Garcia-Prat M, Moes N, Haerynck F, Gonzales-Granado LI, Santos Pérez JL, Mukhina A, Shcherbina A, Aghamohammadi A, Hammarström L, Dogu F, Haskologlu S, İkincioğulları AI, Köstel Bal S, Baris S, Kilic SS, Karaca NE, Kutukculer N, Girschick H, Kolios A, Keles S, Uygun V, Stepensky P, Worth A, van Montfrans JM, Peters AMJ, Meyts I, Adeli M, Marzollo A, Padem N, Khojah AM, Chavoshzadeh Z, Avbelj Stefanija M, Bakhtiar S, Florkin B, Meeths M, Gamez L, Grimbacher B, Seppänen MRJ, Lankester A, Gennery AR, Seidel MG; Inborn Errors; Clinical, and Registry Working Parties of the European Society for Blood and Marrow Transplantation and the European Society for Immunodeficiencies. Long-term outcome of LRBA deficiency in 76 patients after various treatment modalities as evaluated by the immune deficiency and dysregulation activity (IDDA) score. J Allergy Clin Immunol. 2020;145:1452-1463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 18. | Aghamohammadi A, Mohammadi J, Parvaneh N, Rezaei N, Moin M, Espanol T, Hammarstrom L. Progression of selective IgA deficiency to common variable immunodeficiency. Int Arch Allergy Immunol. 2008;147:87-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 19. | Seidel MG, Hirschmugl T, Gamez-Diaz L, Schwinger W, Serwas N, Deutschmann A, Gorkiewicz G, Zenz W, Windpassinger C, Grimbacher B, Urban C, Boztug K. Long-term remission after allogeneic hematopoietic stem cell transplantation in LPS-responsive beige-like anchor (LRBA) deficiency. J Allergy Clin Immunol 2015; 135: 1384-90. e1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 20. | Serwas NK, Kansu A, Santos-Valente E, Kuloğlu Z, Demir A, Yaman A, Gamez Diaz LY, Artan R, Sayar E, Ensari A, Grimbacher B, Boztug K. Atypical manifestation of LRBA deficiency with predominant IBD-like phenotype. Inflamm Bowel Dis. 2015;21:40-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 21. | Kostel Bal S, Haskologlu S, Serwas NK, Islamoglu C, Aytekin C, Kendirli T, Kuloglu Z, Yavuz G, Dalgic B, Siklar Z, Kansu A, Ensari A, Boztug K, Dogu F, Ikinciogullari A. Multiple Presentations of LRBA Deficiency: a Single-Center Experience. J Clin Immunol. 2017;37:790-800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 22. | Mozdarani H, Kiaee F, Fekrvand S, Azizi G, Yazdani R, Zaki-Dizaji M, Mozdarani S, Nosrati H, Abolhassani H, Aghamohammadi A. G2-lymphocyte chromosomal radiosensitivity in patients with LPS responsive beige-like anchor protein (LRBA) deficiency. Int J Radiat Biol. 2019;95:680-690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 23. | Shokri S, Nabavi M, Hirschmugl T, Aghamohammadi A, Arshi S, Bemanian MH, Fallahpour M, Molatefi R, Rekabi M, Eslami N, Ahmadian J, Darabi K, Sedighi GR, Monajemzadeh M, Modaresi M, Parvaneh N, Boztug K, Rezaei N. LPS-Responsive Beige-Like Anchor Gene Mutation Associated With Possible Bronchiolitis Obliterans Organizing Pneumonia Associated With Hypogammaglobulinemia and Normal IgM Phenotype and Low Number of B Cells. Acta Med Iran. 2016;54:620-623. [PubMed] [Cited in This Article: ] |
| 24. | Tesi B, Priftakis P, Lindgren F, Chiang SC, Kartalis N, Löfstedt A, Lörinc E, Henter JI, Winiarski J, Bryceson YT, Meeths M. Successful Hematopoietic Stem Cell Transplantation in a Patient with LPS-Responsive Beige-Like Anchor (LRBA) Gene Mutation. J Clin Immunol. 2016;36:480-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Bakhtiar S, Ruemmele F, Charbit-Henrion F, Lévy E, Rieux-Laucat F, Cerf-Bensussan N, Bader P, Paetow U. Atypical Manifestation of LPS-Responsive Beige-Like Anchor Deficiency Syndrome as an Autoimmune Endocrine Disorder without Enteropathy and Immunodeficiency. Front Pediatr. 2016;4:98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Sari S, Dogu F, Hwa V, Haskologlu S, Dauber A, Rosenfeld R, Polat M, Kuloglu Z, Kansu A, Dalgic B, Ikinciogullari A. A Successful HSCT in a Girl with Novel LRBA Mutation with Refractory Celiac Disease. J Clin Immunol. 2016;36:8-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 27. | Bakhtiar S, Gámez-Díaz L, Jarisch A, Soerensen J, Grimbacher B, Belohradsky B, Keller KM, Rietschel C, Klingebiel T, Koletzko S, Albert MH, Bader P. Treatment of Infantile Inflammatory Bowel Disease and Autoimmunity by Allogeneic Stem Cell Transplantation in LPS-Responsive Beige-Like Anchor Deficiency. Front Immunol. 2017;8:52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 28. | Al Sukaiti N, AbdelRahman K, AlShekaili J, Al Oraimi S, Al Sinani A, Al Rahbi N, Cho V, Field M, Cook MC. Agammaglobulinaemia despite terminal B-cell differentiation in a patient with a novel LRBA mutation. Clin Transl Immunology. 2017;6:e144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Azizi G, Abolhassani H, Yazdani R, Mohammadikhajehdehi S, Parvaneh N, Negahdari B, Mohammadi J, Aghamohammadi A. New therapeutic approach by sirolimus for enteropathy treatment in patients with LRBA deficiency. Eur Ann Allergy Clin Immunol. 2017;49:235-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 30. | Johnson MB, De Franco E, Lango Allen H, Al Senani A, Elbarbary N, Siklar Z, Berberoglu M, Imane Z, Haghighi A, Razavi Z, Ullah I, Alyaarubi S, Gardner D, Ellard S, Hattersley AT, Flanagan SE. Recessively Inherited LRBA Mutations Cause Autoimmunity Presenting as Neonatal Diabetes. Diabetes. 2017;66:2316-2322. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | Bratanič N, Kovač J, Pohar K, Trebušak Podkrajšek K, Ihan A, Battelino T, Avbelj Stefanija M. Multifocal gastric adenocarcinoma in a patient with LRBA deficiency. Orphanet J Rare Dis. 2017;12:131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Shamriz O, Shadur B, NaserEddin A, Zaidman I, Simanovsky N, Elpeleg O, Kerem E, Reiter J, Stepensky P. Respiratory manifestations in LPS-responsive beige-like anchor (LRBA) protein-deficient patients. Eur J Pediatr. 2018;177:1163-1172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Eren Akarcan S, Edeer Karaca N, Aksu G, Aykut A, Yilmaz Karapinar D, Cetin F, Aydinok Y, Azarsiz E, Gambineri E, Cogulu O, Ulusoy Severcan E, Alper H, Kutukculer N. Two male siblings with a novel LRBA mutation presenting with different findings of IPEX syndrome. JMM Case Rep. 2018;5:e005167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Besnard C, Levy E, Aladjidi N, Stolzenberg MC, Magerus-Chatinet A, Alibeu O, Nitschke P, Blanche S, Hermine O, Jeziorski E, Landman-Parker J, Leverger G, Mahlaoui N, Michel G, Pellier I, Suarez F, Thuret I, de Saint-Basile G, Picard C, Fischer A, Neven B, Rieux-Laucat F, Quartier P; Members of the French reference center for pediatric autoimmune cytopenias (CEREVANCE). Pediatric-onset Evans syndrome: Heterogeneous presentation and high frequency of monogenic disorders including LRBA and CTLA4 mutations. Clin Immunol. 2018;188:52-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 35. | De Bruyne M, Bogaert DJ, Venken K, Van den Bossche L, Bonroy C, Roels L, Tavernier SJ, van de Vijver E, Driessen A, van Gijn M, Gámez-Diaz L, Elewaut D, Grimbacher B, Haerynck F, Moes N, Dullaers M. A novel LPS-responsive beige-like anchor protein (LRBA) mutation presents with normal cytotoxic T lymphocyte-associated protein 4 (CTLA-4) and overactive TH17 immunity. J Allergy Clin Immunol. 2018;142:1968-1971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 36. | Fernández KS, Antony R, Kumar A. Patients with "ALPS-like phenotype" diagnosed with immune dysregulation due to LRBA deficiency. Pediatr Blood Cancer. 2019;66:e27558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 37. | Soler-Palacín P, Garcia-Prat M, Martín-Nalda A, Franco-Jarava C, Rivière JG, Plaja A, Bezdan D, Bosio M, Martínez-Gallo M, Ossowski S, Colobran R. LRBA Deficiency in a Patient With a Novel Homozygous Mutation Due to Chromosome 4 Segmental Uniparental Isodisomy. Front Immunol. 2018;9:2397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 38. | Rajpurkar M, Buck S, Lafferty J, Wakeling E, Ravindranath Y, Savaşan S. Acquired Pure Red Cell Aplasia and Acquired Amegakaryocytic Thrombocytopenia Associated With Clonal Expansion of T-Cell Large Granular Lymphocytes in a Patient With Lipopolysaccharide-responsive Beige-like Anchor (LRBA) Protein Deficiency. J Pediatr Hematol Oncol. 2019;41:e542-e545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 39. | Okur FV, Kuskonmaz B, Cagdas D, Tezcan I, Uckan-Cetinkaya D. Bone marrow transplantation with Favorable outcome in three patients with LPS-responsive beige-like anchor (LRBA) deficiency. Clin Immunol. 2019;203:162-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 40. | Revel-Vilk S, Fischer U, Keller B, Nabhani S, Gámez-Díaz L, Rensing-Ehl A, Gombert M, Hönscheid A, Saleh H, Shaag A, Borkhardt A, Grimbacher B, Warnatz K, Elpeleg O, Stepensky P. Autoimmune lymphoproliferative syndrome-like disease in patients with LRBA mutation. Clin Immunol. 2015;159:84-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 41. | Lévy E, Stolzenberg MC, Bruneau J, Breton S, Neven B, Sauvion S, Zarhrate M, Nitschké P, Fischer A, Magérus-Chatinet A, Quartier P, Rieux-Laucat F. LRBA deficiency with autoimmunity and early onset chronic erosive polyarthritis. Clin Immunol. 2016;168:88-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 42. | Li R, Zheng Y, Li Y, Zhang R, Wang F, Yang D, Ma Y, Mu X, Cao Z, Gao Z. Common Variable Immunodeficiency with Genetic Defects Identified by Whole Exome Sequencing. Biomed Res Int. 2018;2018:3724630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Liu L, Wang Y, Dong X, Lin L, Sun J, Wang X. A report of two cases with LRBA gene mutaiton. Zhongguo Xunzheng Erke Zazhi. 2019;14:25-29. [DOI] [Cited in This Article: ] |