Published online Jun 26, 2019. doi: 10.12998/wjcc.v7.i12.1383
Peer-review started: March 23, 2019
First decision: April 18, 2019
Revised: April 23, 2019
Accepted: May 1, 2019
Article in press: May 1, 2019
Published online: June 26, 2019
Periodontitis is a chronic inflammation of periodontal supporting tissue caused by local factors. Periodontal surgery can change the gene expression of peripheral blood mononuclear cells. However, little is known about the potential mechanism of surgical treatment for periodontitis.
To explore the potential molecular mechanism of surgical treatment for periodontitis.
First, based on the expression profiles of genes related to surgical treatment for periodontitis, a set of expression disorder modules related to surgical treatment for periodontitis were obtained by enrichment analysis. Subsequently, based on crosstalk analysis, we proved that there was a significant crosstalk relationship between module 3 and module 5. Finally, based on predictive analysis of multidimensional regulators, we identified a series of regulatory factors, such as endogenous genes, non-coding RNAs (ncRNAs), and transcription factors, which have potential regulatory effects on periodontitis.
A total of 337 genes related to surgical treatment for periodontitis were obtained, and 3896 genes related to periodontitis were amplified. Eight expression modules of periodontitis were obtained, involving the aggregation of 2672 gene modules. These modules are mainly involved in G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger, and adenylate cyclase-modulating G-protein coupled receptor signaling pathway. In addition, eight endogenous genes (including EGF, RPS27A, and GNB3) were screened by network connectivity analysis. Finally, based on this set of potential dysfunction modules, 94 transcription factors (including NFKB1, SP1, and STAT3) and 1198 ncRNAs (including MALAT1, CRNDE, and ANCR) were revealed. These core regulators are thought to be involved in the potential molecular mechanism of periodontitis after surgical treatment.
Based on the results of this study, we can show biologists and pharmacists a new idea to reveal the potential molecular mechanism of surgical treatment for periodontitis, and provide valuable reference for follow-up treatment programs.
Core tip: Based on the expression pattern and function of genes in peripheral blood mononuclear cells of periodontitis patients, key genes (including EGF, RPS27A, GNB3, etc.) were identified. Further, transcription factors (including NFKB1, SP1, STAT3, etc.,) and non-coding RNAs (including MALAT1, CRNDE, and ANCR) that significantly regulated gene co-expression modules were identified. Exploring the relationships between genes and regulators may reveal the potential molecular mechanism of surgical treatment for periodontitis.
- Citation: Ma JJ, Liu HM, Xu XH, Guo LX, Lin Q. Study on gene expression patterns and functional pathways of peripheral blood monocytes reveals potential molecular mechanism of surgical treatment for periodontitis. World J Clin Cases 2019; 7(12): 1383-1392
- URL: https://www.wjgnet.com/2307-8960/full/v7/i12/1383.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i12.1383
Periodontitis is a highly prevalent inflammatory disease in dental support tissues. It is caused by the growth of bacteria in the biofilm on the surface of teeth. As a mature periodontitis disease, it is characterized by progressive destruction of the supporting tissues of teeth. Periodontitis is a multifactorial disease. The susceptibility and severity of periodontitis are influenced by the polymorphism of immune response related genes, and are also determined by environmental and genetic factors[1,2]. Relevant studies have shown that periodontal diseases are associated with various systemic diseases, including cardiovascular diseases such as stroke and athero-sclerosis[3,4]. Many data show that periodontitis affects 20% of the world's population. The prevalence of periodontitis in adults is 35%, and it increases with age, reaches a peak in the 45-50 age group[5-7]. Periodontitis has a variety of risk factors, which may cause plaque, dental calculi, diabetes, and other diseases, and has multiple negative impacts on the quality of life of patients[5,8]. Therefore, many scholars actively explore the pathogenesis of periodontitis. It is well known that bacterial infection is the main cause of periodontal disease, and the bacterial colonization of actinomycetes and Porphyromonas gingivalis is the main cause of periodontitis[9,10]. On the one hand, the change of immune response in diabetic patients may cause periodontitis[11]. On the other hand, the severity of periodontal disease may also be related to the expression of multiple metalloproteinase genes in the oral mucosal epithelium[12]. Recent studies have shown that periodontitis, a chronic inflammatory oral infection, may have a profound impact on human health[13]. As far as the potential relationship between periodontitis and systemic diseases is concerned, the changes of immune cell function induced by periodontitis may cause lipid metabolism disorder through the me-chanism related to inflammatory cytokines, which has a serious negative impact on the health of whole body cells[13].
After understanding some of the pathogenesis of periodontitis, scholars around the world are actively studying the corresponding treatment mechanisms and inhibition methods. For example, according to van Winkelhoff et al[14], mechanical periodontal therapy combined with metronidazole and amoxicillin is effective for subgingival inhibition of actinomycete infection in patients with severe periodontitis. In addition, triclosan and copolymer toothpaste were found to be able to effectively control the development of plaque and gingivitis, thereby inhibiting the occurrence of periodontitis[15]. Relevant studies have shown that periodontitis is essentially an inflammatory reaction in periodontal attachment caused by bacteria[16]. Periodontal infection is not only related to chronic inflammation, but also to oxidative stress in severe periodontitis and its treatment[17]. YKL-40, as a chitin-binding glycoprotein, may be an inflammatory marker of periodontitis, which has been confirmed by Kido et al[18]. On the one hand, as one of the biomarkers of periodontitis, pulp-like cell-1 may play an important role in the association between periodontal infection and systemic inflammatory response[19]. On the other hand, the increased inflammatory response of higher serum C-reactive protein level may be as important as the promotion of periodontitis[20].
Previous data show that periodontal therapy can change the gene expression pattern of peripheral blood monocytes[21]. Therefore, we can think that the gene expression of peripheral blood mononuclear cells also has a feedback regulation effect on periodontal therapy. Therefore, in our study, we proposed a comprehensive analysis method based on the gene expression patterns and functional pathways of peripheral blood mononuclear cells. It combined co-expression analysis and enrichment analysis of functional pathways, and focused on predicting a series of core non-coding RNAs (ncRNAs) and transcription factors to reveal the potential molecular mechanism of surgical treatment for periodontitis.
First, the gene expression profiles of 3896 related genes in periodontitis were analyzed by weighted gene co-expression network analysis (WGCNA)[22] and the gene modules of co-expression were searched. Second, the weighted value of correlation coefficient was used to calculate the correlation coefficient between any two genes (Person Coefficient) by taking the N power of the correlation coefficient. The connection between genes in the network obeys scale-free networks, which makes the algorithm more biologically meaningful. Then, a hierarchical clustering tree was constructed by the correlation coefficient between genes. Different branches of the clustering tree represent different gene modules, and different colors represent different modules.
First, Python was used to program the human protein interaction network (score > 950) in String to generate 1000 random networks[23] while keeping the network size and the degree of each node unchanged. Second, according to the number of interaction pairs between significant modules obtained by random network statistics, the number of interaction pairs between modules was compared with the number of interaction pairs in random background. When the number of interaction pairs between modules was larger than that of interaction pairs in random background, it was called crosstalk. The method of calculating significant crosstalk was as follows: Under the background of random network, the number of interaction pairs between modules in N random networks was larger than that in real networks. The formula of calculating P-value is: P = n/N (in this work, N = 1000). When a P-value was <0.05, there was a crosstalk interaction among modules. Then, Cytoscape[24] was used to display the significant crosstalk to visually observe the complex regulatory relationship between modules. In addition, Cytoscape was used for display and network analysis (including connectivity calculation). Finally, the genes with the greatest connectivity was screened and considered as the core molecule to regulate the progress of the module and identified as the intrinsic gene. These endogenous genes may represent potential key molecules in the effect of surgical treatment on periodontitis.
Exploring the function and signal pathway of genes is often an effective means to study the molecular mechanism of disease, while the function and pathway of module gene participation can characterize the dysfunction mechanism of modules in the process of disease occurrence. Therefore, we used R language Cluster profiler package[25] to analyze the enrichment of GO function (pvalue Cutoff = 0.01, qvalueCutoff = 0.01) and KEGG pathway (pvalueCutoff = 0.05, qvalueCutoff = 0.2), respectively. According to the function and pathway of module gene participation, it was identified as a functional disorder module related to periodontitis.
First, we downloaded all human transcription factor target data in TRRUST V2 database[26], and obtained 116 interaction pairs of 94 transcription factors. Then, human ncRNA-protein data (score > 0.5) were downloaded from RAID 2.0 database[27], and 1678 interaction pairs involving 1198 ncRNAs were obtained. Then, pivot analysis based on these interaction data was performed to identify the regulatory effects of these transcription factors and ncRNAs on the modules. Pivot analysis refers to searching for at least two interacting drivers with the module in the target pair and calculating the significance of the interaction between the driver and the module according to the hypergeometric test. Transcription factors and ncRNAs with a P-value < 0.01 were the pivots of the significant regulatory module. Finally, the pivots were identified as the core pivots by statistical analysis.
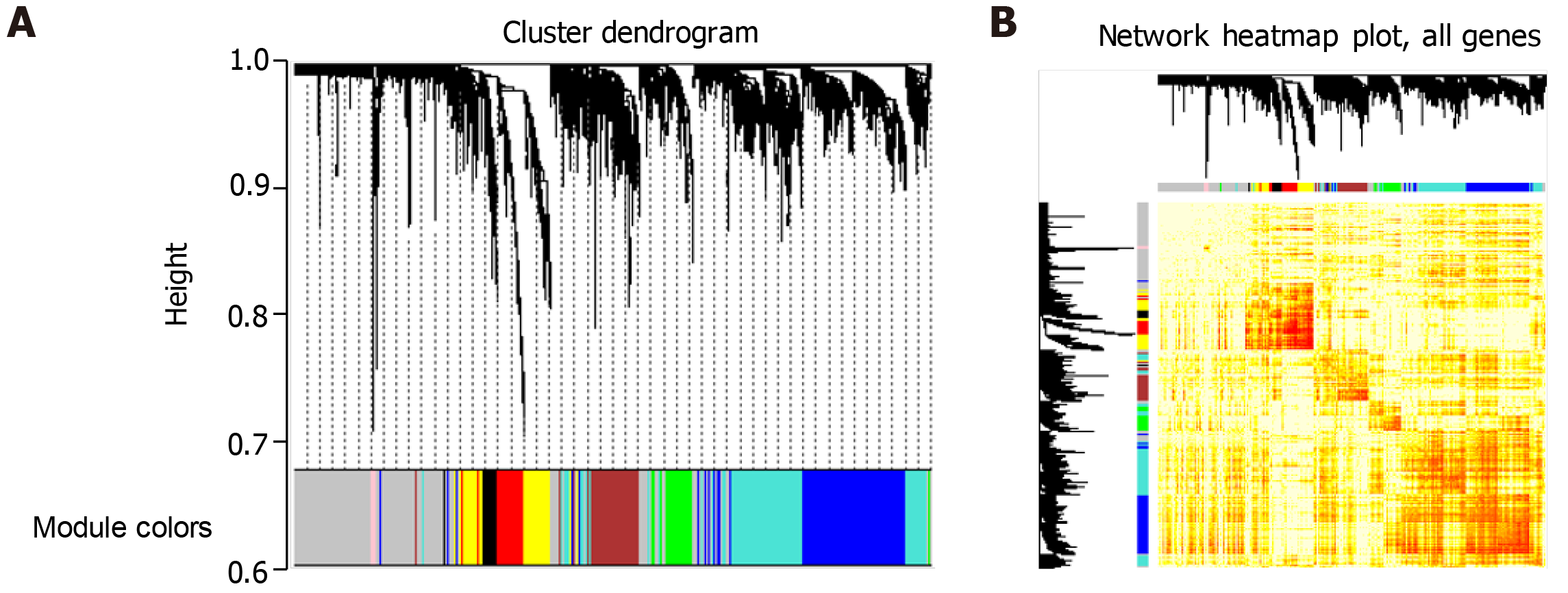
Because the regulatory mechanism of periodontitis-related genes and the synergistic relationship between genes are not clear, we conducted in-depth research. First, a total of 337 genes related to surgical treatment of periodontitis was integrated into several databases. Second, we amplified 337 related genes and got 3896 related genes. We analyzed the WGCNA co-expression of the amplified genes and observed the expression of these genes in 15 patients with periodontitis. We found that eight groups of genes expressed synergistically in the group based on cohesion. Finally, these eight groups of genes were identified as eight functional modules, involving 2672 clusters of module genes (Figure 1). These functional modules may participate in different functions and pathways, representing different regulatory mechanisms to mediate the occurrence and development of periodontitis.
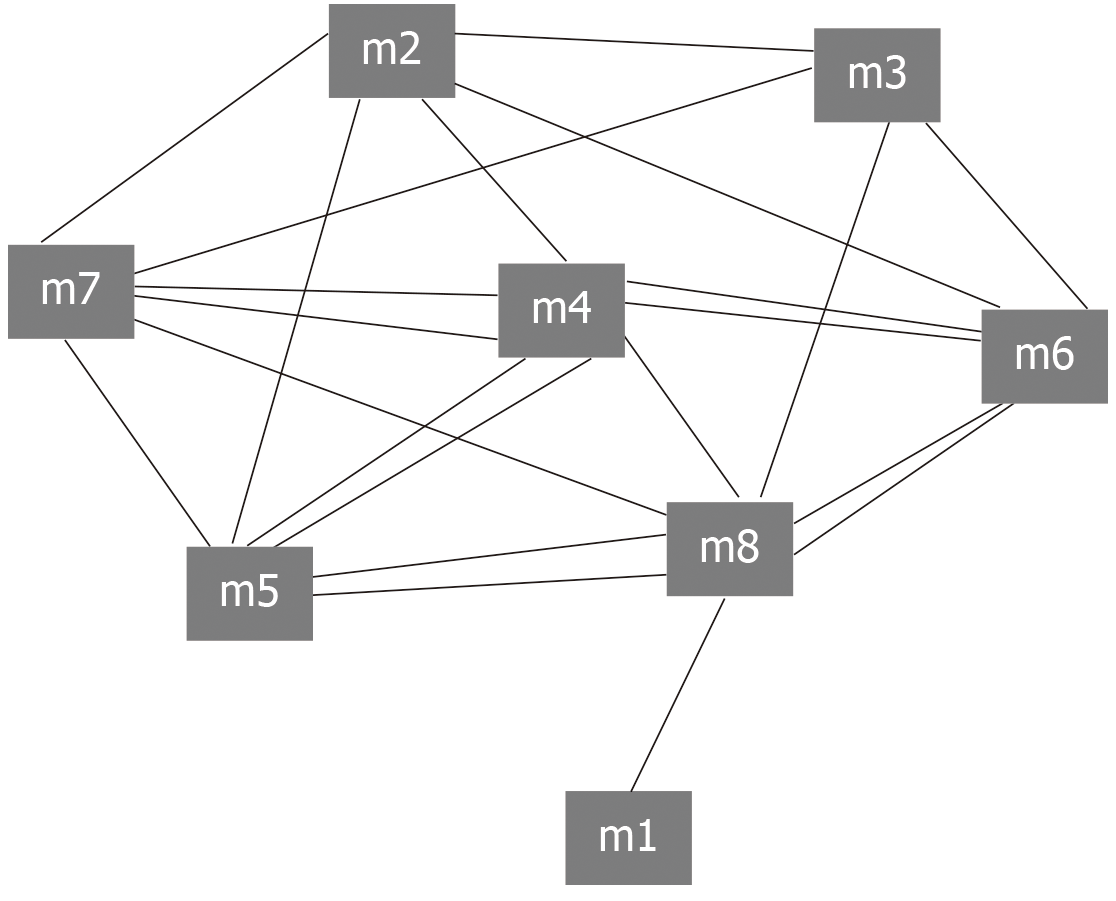
The intergenic regulation has always been complex and changeable. Similarly, the relationships between modules are complex and diverse. The interaction between modules is the pivot to maintain the relationship between modules and the whole world, and is also the key to exploring the function of modules. Exploring the crosstalk relationship between modules is helpful to deepen our understanding of the potential dysfunction mechanism of module regulation of periodontitis. Therefore, we conducted crosstalk analysis among modules based on the relationship between module genes. The results showed that there was a complex interaction between modules 2, 3, 4, 5, 6, 7, and 8, while other modules had simple crosstalk (Figure 2). This crosstalk may represent the driving force between modules, which together regulate the potential molecular mechanism of peripheral blood monocytes in the surgical treatment of periodontitis.
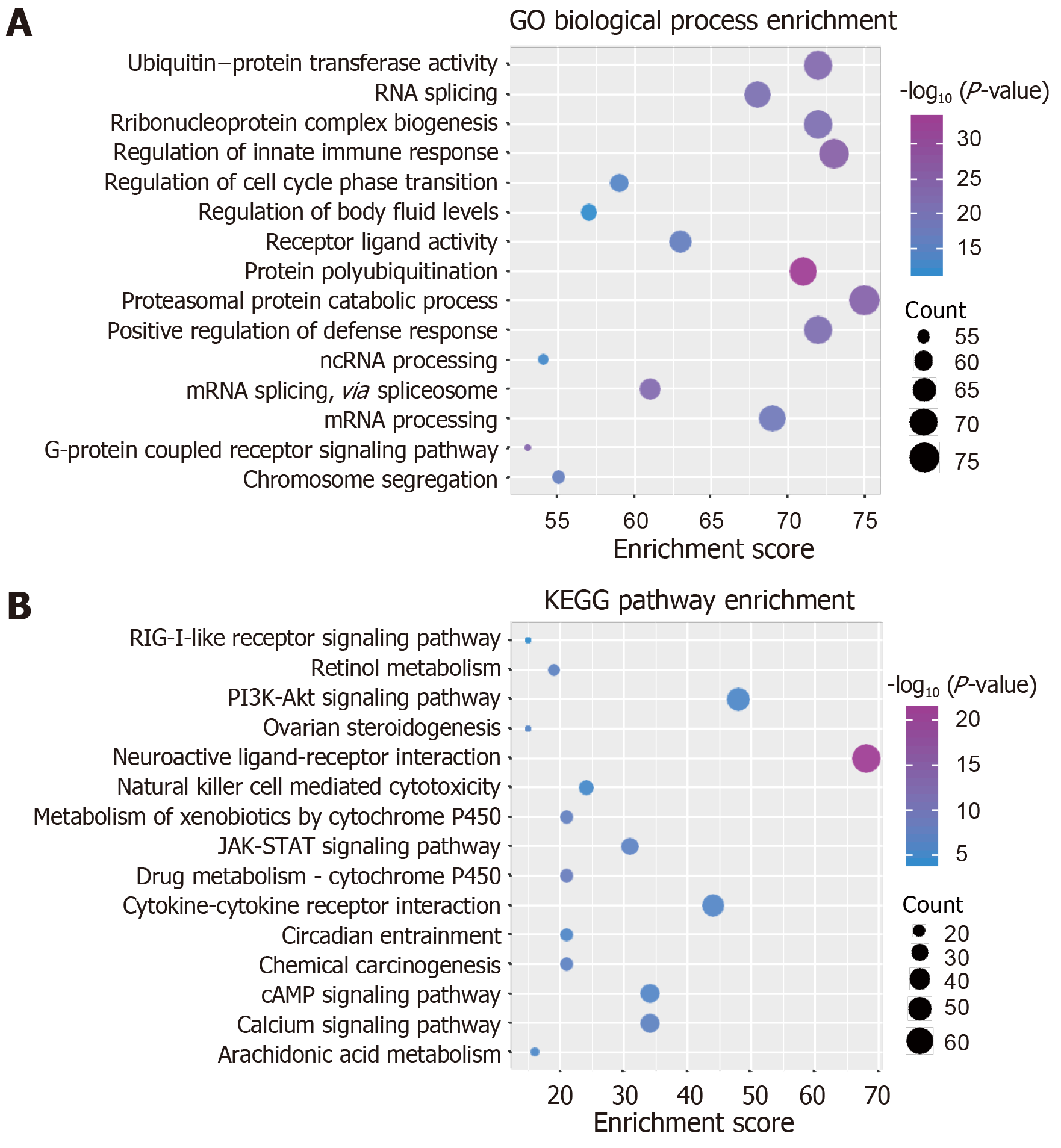
Studying the function and pathway of gene involvement is an important means to identify its mediated pathogenesis. In order to study the possible dysfunction caused by module gene disorder, we analyzed the enrichment of function and pathway of each module. The results showed that most of the functional modules were enriched in periodontitis-related functions and pathways. GO function and KEGG pathway enrichment analyses were carried out on the eight functional modules. A total of 45887 functions and 1796 KEGG pathway enrichment results were obtained. Among them, there are 5950 molecular functions, 3976 cell components, and 35691 biological processes in which these genes participate (Figure 3, Table 1). It is worth noting that G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger, and adenylate cyclase-modulating G-protein coupled receptor signaling pathway, which are significantly involved, may be identified as the core signaling pathways involved in the potential disorder mechanism of periodontitis. From the above data, we can find that these signaling pathways may be closely related to the molecular mechanism of periodontitis in the surgical treatment of peripheral blood monocytes.
| ID | Description | P-value | Count |
| GO:0007631 | Feeding behavior | 1.49E-13 | 26 |
| GO:0070371 | ERK1 and ERK2 cascade | 2.32E-13 | 46 |
| GO:0070372 | Regulation of ERK1 and ERK2 cascade | 5.73E-13 | 44 |
| GO:0050878 | Regulation of body fluid levels | 2.12E-12 | 57 |
| GO:0097485 | Neuron projection guidance | 2.44E-12 | 38 |
| GO:0007411 | Axon guidance | 8.41E-12 | 37 |
| GO:0035567 | Non-canonical Wnt signaling pathway | 2.61E-11 | 28 |
| GO:0007218 | Neuropeptide signaling pathway | 3.08E-11 | 23 |
| GO:2000027 | Regulation of organ morphogenesis | 4.06E-11 | 36 |
| GO:0019932 | Second-messenger-mediated signaling | 5.57E-11 | 43 |
| GO:0009410 | Response to xenobiotic stimulus | 5.72E-11 | 38 |
| GO:0051047 | Positive regulation of secretion | 8.82E-11 | 46 |
| GO:1903532 | Positive regulation of secretion by cell | 9.27E-11 | 44 |
| GO:0019373 | Epoxygenase P450 pathway | 1.21E-10 | 11 |
| GO:1905330 | Regulation of morphogenesis of an epithelium | 2.75E-10 | 29 |
| hsa04080 | Neuroactive ligand-receptor interaction | 7.10E-22 | 68 |
| hsa00982 | Drug metabolism - cytochrome P450 | 5.57E-09 | 21 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 1.62E-08 | 21 |
| hsa04020 | Calcium signaling pathway | 4.82E-08 | 34 |
| hsa00830 | Retinol metabolism | 4.97E-08 | 19 |
| hsa05204 | Chemical carcinogenesis | 6.96E-08 | 21 |
| hsa04630 | JAK-STAT signaling pathway | 9.51E-08 | 31 |
| hsa04060 | Cytokine-cytokine receptor interaction | 3.93E-07 | 44 |
| hsa04024 | cAMP signaling pathway | 3.95E-07 | 34 |
| hsa04913 | Ovarian steroidogenesis | 4.34E-07 | 15 |
| hsa04713 | Circadian entrainment | 1.21E-06 | 21 |
| hsa04151 | PI3K-Akt signaling pathway | 2.21E-06 | 48 |
| hsa00590 | Arachidonic acid metabolism | 2.89E-06 | 16 |
| hsa04650 | Natural killer cell mediated cytotoxicity | 6.05E-06 | 24 |
| hsa04724 | Glutamatergic synapse | 6.17E-06 | 22 |
Further, our study of 2672 modular genes revealed that there were also target interactions among these genes. In-depth study of gene interaction within the module was then performed to calculate its connectivity and screen each module with the highest connectivity of the intrinsic genes. Eight endogenous genes including EGF, RPS27A, and GNB3 were screened from eight dysfunction modules. In addition, besides the module-driven genes, there were also key regulatory factors such as ncRNAs and transcription factors, which also play an indispensable role in the potential molecular mechanism of periodontitis. Scientific prediction of ncRNAs regulating dysfunction module genes is helpful for us to explore the post-transcriptional regulation mechanism of periodontitis. To this end, pivot analysis based on the targeting relationship between ncRNAs and genes was carried out to predict ncRNA regulators causing module dysfunction. We identified 198 ncRNAs that had significant regulatory effects on modules, involving 1678 ncRNA-module interaction pairs. Statistical analysis of the results showed that MALAT1 had a significant regulatory relationship with six dysfunction modules and may play an important role in the potential molecular mechanism of periodontitis, so it was identified as the core ncRNA. ANCR also plays an important role in the molecular mechanism of periodontitis. In addition, other ncRNAs may also play a role in driving the potential molecular mechanism of periodontitis. Similarly, transcription factors are equally important in regulating gene transcription. Many studies have shown that disordered expression of transcription factors may lead to the occurrence of various diseases. The dysfunction of periodontitis is also closely related to the dysfunction of transcription factors, which is also reflected in the regulation of dysfunction module by transcription factors. Therefore, we used pivot analysis to predict the module according to the regulation of genes by transcription factors. The results showed that there were 94 transcription factors involving 116 pivot-module interaction pairs. Statistical analysis of these transcription factors showed that NFKB1, SP1, and STAT3 had significant regulatory effects on 6 and 4 dysfunction modules, respectively, and they are likely to participate in the potential molecular mechanism of surgical treatment for periodontitis. These data suggest that transcription factors may play an important role in the molecular mechanism of surgical treatment of periodontitis. These transcription factors, which have significant regulatory effects on multiple dysfunction modules, have been identified as the core transcription factors driving the development of periodontitis.
It is well known that periodontitis is a multifactorial disease. Studies have shown that some clinical variability of periodontitis may be caused by genetic factors[28]. As a high-risk inflammatory disease, periodontitis is considered a potential risk factor for systemic diseases such as cardiovascular diseases[20]. Therefore, scholars around the world are actively exploring the detailed and specific pathogenesis and treatment mechanism of periodontitis. In this study, we investigated the gene expression patterns and functional pathways of peripheral blood mononuclear cells to reveal the potential molecular mechanism of the effect of surgical treatment on periodontitis. We proposed a comprehensive approach, which first integrated 3896 genes related to periodontitis and carried out co-expression analysis. Finally, eight functional modules with synergistic expression were identified, involving 2672 module gene clusters. Based on the results of functional enrichment, we also obtained that these modules are significantly involved in G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger, and adenylate cyclase-modulating G-protein coupled receptor signaling pathway. Among them, adenylate cyclase has been found to be significantly involved in many signaling pathways, which also indicates that adenylate cyclase plays an important role in the signaling pathway in periodontitis. According to the research of Petrovich et al[29], the decrease of adenylate cyclase activity increases with the severity of periodontal pathological process, so it may play an important role in the pathogenesis of periodontitis. In addition, we also found that fibroblasts are also an important functional pathway involved in periodontitis, which is involved in the derivation of osteoclasts, contributing to the loss of attachment and destruction of periodontal ligaments and leading to periodontitis[30].
On the other hand, we identified the transcription factors involved in these dysfunctional modules for periodontitis-related genes, and obtained 94 transcription factors involving 116 pivot-module interaction pairs. Statistical analysis of these transcription factors revealed that NFKB1 had significant regulatory effects on five functional modules, which may be the core transcription factor involved in the potential molecular mechanism of surgical treatment for periodontitis. NFKB1 has been found to be involved in immune responses, cell proliferation, and other processes. NFKB1 is an important molecule involved in osteoclast differentiation, so inhibition of its growth can be used to treat periodontitis and various bone and joint diseases [31]. Both SP1 and STAT3 have regulatory effects on the four functional modules. Transcription factor SP1 has been found in a large number of cases of periodontitis, indicating that it plays an important role in causing periodontitis[1]. The expression of STAT3 was inhibited in gingival epithelial cells of patients with diabetic periodontitis. Therefore, down-regulation of STAT3 signaling could improve diabetic periodontitis to some extent, which was confirmed by Wang et al[32]. These tran-scription factors can be considered as core transcription factors regulating the potential molecular mechanism of surgical treatment for periodontitis.
In addition, ncRNAs have been considered as important regulators of disease occurrence and development. We conducted pivot analysis based on the targeting relationship between ncRNAs and genes. The prediction results showed that there were 1198 ncRNAs that had significant regulatory effects on modules, including 1678 ncRNA-module interaction pairs. Statistical analysis of the results showed that MALAT1 had a significant regulatory relationship with six dysfunction modules, which were considered as key regulators of periodontitis. According to Ziebolz et al[33], MALAT1 can participate in the pathogenesis of periodontitis through a variety of pathways, including cytokine-cytokine receptor, adhesion molecule, and chemokine signal transduction pathway[2]. In addition, ANCR has been found to play an im-portant role in regulating the osteogenic differentiation of periodontitis. In that study, ANCR has been found to regulate two functional modules[33]. At the same time, we screened a series of genes with the greatest connectivity, which were considered as the core driving molecules and identified as intrinsic genes. In total, there were eight endogenous genes, which may represent the potential key disorders in periodontitis treated by surgery. Analysis of these eight endogenous genes showed that EGF is an inhibitory factor for cell apoptosis[34]. According to the research results of many scholars, it can be shown that EGF has the effect of bone catabolism, which may stimulate osteoclast formation and tooth movement, and has a certain regulatory effect on the occurrence and development of periodontitis[35].
Based on the results of this study, we obtained several key functional modules of surgical treatment for periodontitis. These modules provide many proven periodontitis-related genes and candidate factors to be tested, which provide a theoretical basis for further study of periodontitis. We have also predicted a series of potential regulatory factors; these core regulatory factors may become the focus of future research on periodontitis.
Periodontal diseases are associated with various systemic diseases, including cardiovascular diseases such as stroke and atherosclerosis, which seriously affect the quality of life for patients.
At present, little is known about the potential mechanism of surgical treatment for periodontitis.
To explore the mechanism of surgical treatment for periodontitis based on the gene expression of peripheral blood mononuclear cells.
Co-expression analysis, enrichment analysis, crosstalk analysis, and pivot regulatory factor prediction
A total of 337 genes related to periodontitis were clustered into 8 co-expression modules. These genes are mainly involved in G-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger, and adenylate cyclase-modulating G-protein coupled receptor signaling pathway. In addition, 94 transcription factors (including NFKB1, SP1, and STAT3) and 1198 ncRNAs (including MALAT1, CRNDE, and ANCR) regulatory module genes were identified.
The key factors we have identified affect the recovery of periodontitis after surgery through a variety of biological processes and signaling pathways
The results of this study provide a new theoretical basis and individualized treatment direction for follow-up treatment.
Manuscript source: Unsolicited manuscript
Specialty type: Medicine, Research and Experimental
Country of origin: China
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Vieyra JP, Gurav AN S-Editor: Dou Y L-Editor: Wang TQ E-Editor: Wu YXJ
| 1. | Larsson L, Thorbert-Mros S, Rymo L, Berglundh T. Interleukin-10 genotypes of the -1087 single nucleotide polymorphism influence sp1 expression in periodontitis lesions. J Periodontol. 2011;82:1376-1382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 2. | Peng W, Deng W, Zhang J, Pei G, Rong Q, Zhu S. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem Biophys Res Commun. 2018;503:815-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 3. | Schenkein HA, Sabatini R, Koertge TE, Brooks CN, Purkall DB. Anti-cardiolipin from periodontitis patients induces MCP-1 production by human umbilical vein endothelial cells. J Clin Periodontol. 2013;40:212-217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Behle JH, Papapanou PN. Periodontal infections and atherosclerotic vascular disease: an update. Int Dent J. 2006;56:256-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | van Winkelhoff AJ, Winkel EG, Vandenbroucke-Grauls CM. [Periodontitis: a hidden chronic infection]. Ned Tijdschr Geneeskd. 2001;145:557-563. [PubMed] [Cited in This Article: ] |
| 6. | Schaefer AS, Richter GM, Nothnagel M, Laine ML, Rühling A, Schäfer C, Cordes N, Noack B, Folwaczny M, Glas J, Dörfer C, Dommisch H, Groessner-Schreiber B, Jepsen S, Loos BG, Schreiber S. A 3' UTR transition within DEFB1 is associated with chronic and aggressive periodontitis. Genes Immun. 2010;11:45-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 7. | Horning GM, Hatch CL, Lutskus J. The prevalence of periodontitis in a military treatment population. J Am Dent Assoc. 1990;121:616-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Preshaw PM, Alba AL, Herrera D, Jepsen S, Konstantinidis A, Makrilakis K, Taylor R. Periodontitis and diabetes: a two-way relationship. Diabetologia. 2012;55:21-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 782] [Cited by in F6Publishing: 825] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 9. | Kumar RS, Prakash S. Impaired neutrophil and monocyte chemotaxis in chronic and aggressive periodontitis and effects of periodontal therapy. Indian J Dent Res. 2012;23:69-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Novak MJ, Novak KF. Early-onset periodontitis. Curr Opin Periodontol. 1996;3:45-58. [PubMed] [Cited in This Article: ] |
| 11. | Kranthi J, Sudhakar K, Kulshrestha R, Raju PK, Razdan A, Srinivasa Ts. Comparison of the serum immunoglobulin IgM level in diabetic and nondiabetic patients with chronic periodontitis. J Contemp Dent Pract. 2013;14:814-818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 12. | Kinoshita N, Awano S, Yoshida A, Soh I, Ansai T. Periodontal disease and gene-expression levels of metalloendopeptidases in human buccal mucosal epithelium. J Periodontal Res. 2013;48:606-614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Iacopino AM, Cutler CW. Pathophysiological relationships between periodontitis and systemic disease: recent concepts involving serum lipids. J Periodontol. 2000;71:1375-1384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 151] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | van Winkelhoff AJ, Tijhof CJ, de Graaff J. Microbiological and clinical results of metronidazole plus amoxicillin therapy in Actinobacillus actinomycetemcomitans-associated periodontitis. J Periodontol. 1992;63:52-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 196] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 15. | Seymour GJ, Palmer JE, Leishman SJ, Do HL, Westerman B, Carle AD, Faddy MJ, West MJ, Cullinan MP. Influence of a triclosan toothpaste on periodontopathic bacteria and periodontitis progression in cardiovascular patients: a randomized controlled trial. J Periodontal Res. 2017;52:61-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 16. | Tucker R. Periodontitis and pregnancy. J R Soc Promot Health. 2006;126:24-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 17. | D'Aiuto F, Nibali L, Parkar M, Patel K, Suvan J, Donos N. Oxidative stress, systemic inflammation, and severe periodontitis. J Dent Res. 2010;89:1241-1246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 218] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 18. | Kido J, Bando Y, Bando M, Kajiura Y, Hiroshima Y, Inagaki Y, Murata H, Ikuta T, Kido R, Naruishi K, Funaki M, Nagata T. YKL-40 level in gingival crevicular fluid from patients with periodontitis and type 2 diabetes. Oral Dis. 2015;21:667-673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Bostanci N, Oztürk VÖ, Emingil G, Belibasakis GN. Elevated oral and systemic levels of soluble triggering receptor expressed on myeloid cells-1 (sTREM-1) in periodontitis. J Dent Res. 2013;92:161-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 20. | Pejcic A, Kesic LJ, Milasin J. C-reactive protein as a systemic marker of inflammation in periodontitis. Eur J Clin Microbiol Infect Dis. 2011;30:407-414. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Papapanou PN, Sedaghatfar MH, Demmer RT, Wolf DL, Yang J, Roth GA, Celenti R, Belusko PB, Lalla E, Pavlidis P. Periodontal therapy alters gene expression of peripheral blood monocytes. J Clin Periodontol. 2007;34:736-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10254] [Cited by in F6Publishing: 12661] [Article Influence: 791.3] [Reference Citation Analysis (0)] |
| 23. | Hopf TA, Green AG, Schubert B, Mersmann S, Schärfe CPI, Ingraham JB, Toth-Petroczy A, Brock K, Riesselman AJ, Palmedo P, Kang C, Sheridan R, Draizen EJ, Dallago C, Sander C, Marks DS. The EVcouplings Python framework for coevolutionary sequence analysis. Bioinformatics. 2018;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 116] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 24. | Carlin DE, Demchak B, Pratt D, Sage E, Ideker T. Network propagation in the cytoscape cyberinfrastructure. PLoS Comput Biol. 2017;13:e1005598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 25. | Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11591] [Cited by in F6Publishing: 16462] [Article Influence: 1371.8] [Reference Citation Analysis (0)] |
| 26. | Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380-D386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1155] [Cited by in F6Publishing: 969] [Article Influence: 161.5] [Reference Citation Analysis (0)] |
| 27. | Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li Y, Liu L, Hou P, Cui T, Tan P, Hu Y, Zhang T, Huang Y, Li X, Yu J, Wang D. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45:D115-D118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 156] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 28. | Grigoriadou ME, Koutayas SO, Madianos PN, Strub JR. Interleukin-1 as a genetic marker for periodontitis: review of the literature. Quintessence Int. 2010;41:517-525. [PubMed] [Cited in This Article: ] |
| 29. | Petrovich IuA, Podorozhnaia RP, Genesina TI, Beloklitskaia GF. [Adenylate cyclase and guanylate cyclase in the saliva of healthy persons and in periodontitis]. Stomatologiia (Mosk). 1991;30-33. [PubMed] [Cited in This Article: ] |
| 30. | Beklen A, Al-Samadi A, Konttinen YT. Expression of cathepsin K in periodontitis and in gingival fibroblasts. Oral Dis. 2015;21:163-169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 31. | Jimi E, Fukushima H. [NF-κB signaling pathways and the future perspectives of bone disease therapy using selective inhibitors of NF-κB]. Clin Calcium. 2016;26:298-304. [PubMed] [Cited in This Article: ] |
| 32. | Wang Q, Li H, Xie H, Fu M, Guo B, Ding Y, Li W, Yu H. 25-Hydroxyvitamin D3 attenuates experimental periodontitis through downregulation of TLR4 and JAK1/STAT3 signaling in diabetic mice. J Steroid Biochem Mol Biol. 2013;135:43-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Li S, Liu X, Li H, Pan H, Acharya A, Deng Y, Yu Y, Haak R, Schmidt J, Schmalz G, Ziebolz D. Integrated analysis of long noncoding RNA-associated competing endogenous RNA network in periodontitis. J Periodontal Res. 2018;53:495-505. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Vaahtokari A, Aberg T, Thesleff I. Apoptosis in the developing tooth: association with an embryonic signaling center and suppression by EGF and FGF-4. Development. 1996;122:121-129. [PubMed] [Cited in This Article: ] |
| 35. | Alves JB, Ferreira CL, Martins AF, Silva GA, Alves GD, Paulino TP, Ciancaglini P, Thedei G, Napimoga MH. Local delivery of EGF-liposome mediated bone modeling in orthodontic tooth movement by increasing RANKL expression. Life Sci. 2009;85:693-699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |