Published online Dec 9, 2014. doi: 10.5497/wjp.v3.i4.162
Revised: October 15, 2014
Accepted: October 28, 2014
Published online: December 9, 2014
Protein kinases constitute a superfamily of therapeutic targets for a number of human and animal diseases that include more than 500 members accordingly to sequencing data of the human genome. The well characterized nature of protein kinases makes them excellent targets for drug development. Pharmacophore approaches have become one of the major tools in the area of drug discovery. Application of pharmacophore modeling approaches allows reducing of expensive overall cost associated with drug development project. Pharmacophore models are important functional groups of atoms in the proper spatial position for interaction with target protein. Various ligand-based and structure-based methods have been developed for pharmacophore model generation. Despite the successes in pharmacophore models generation these approaches have not reached their full capacity in application for drug discovery. In the following review, we summarize the published data on pharmacophore models for inhibitors of tyrosine protein kinases (EGFR, HER2, VEGFR, JAK2, JAK3, Syk, ZAP-70, Tie2) and inhibitors of serine/threonine kinases (Clk, Dyrk, Chk1, IKK2, CDK1, CDK2, PLK, JNK3, GSK3, mTOR, p38 MAPK, PKB). Here, we have described the achievements of pharmacophore modeling for protein kinase inhibitors, which provide key points for further application of generated pharmacophore hypotheses in virtual screening, de novo design and lead optimization.
Core tip: In the following review, we summarize the published data on pharmacophore models for inhibitors of tyrosine protein kinases (EGFR, HER2, VEGFR, JAK2, JAK3, Syk, ZAP-70, Tie2) and inhibitors of serine/threonine kinases (Clk, Dyrk, Chk1, IKK2, CDK1, CDK2, PLK, JNK3, GSK3, mTOR, p38 MAPK, PKB). Here, we have described the achievements of pharmacophore modeling for protein kinase inhibitors, which provide key points for further application of generated pharmacophore hypotheses in virtual screening, de novo design and lead optimization.
- Citation: Starosyla SA, Volynets GP, Bdzhola VG, Golub AG, Yarmoluk SM. Pharmacophore approaches in protein kinase inhibitors design. World J Pharmacol 2014; 3(4): 162-173
- URL: https://www.wjgnet.com/2220-3192/full/v3/i4/162.htm
- DOI: https://dx.doi.org/10.5497/wjp.v3.i4.162
Protein kinases are a group of enzymes which covalently modify proteins by adding phosphate groups from adenosine triphosphate (ATP) to serine, threonine or tyrosine residues and therefore, transduce a variety of signals in eukaryotic cells[1]. Kinases play a vital role in diverse cellular processes, functions, deregulations and now represent the second most important class of drug targets for pharmaceutical industry, after G-protein-coupled receptors[2]. Over the past decade about 20 drugs targeting kinases have been approved for clinical application, and much more are currently undergoing clinical studies[3].
Pharmacophore modeling is an important tool in drug development. A pharmacophore is the ensemble of steric and electronic features that is necessary to ensure the optimal supramolecular interactions with a specific biological target structure and to trigger (or to block) its biological response[4]. There are two approaches for pharmacophore construction-receptor-based methods that allow building pharmacophore models based on the interactions of ligands with receptors, and ligand-based methods allowing generation of pharmacophore models based on the training sets of active compounds.
The pharmacophore models are applicable for screening large compound libraries in silico for the search of new small molecule inhibitors because they allow select compounds exhibiting binding features complementary oriented to an active binding pocket[5].
In this review we discuss the published data on pharmacophore models for inhibitors of several tyrosine protein kinases and serine/threonine protein kinases (Table 1).
| Tyrosine protein kinases | |
| Epidermal growth factor receptor (EGFR; Erb-1; HER1 in humans) | Receptor |
| Human epidermal growth factor receptor 2 (HER2; erbB2; protooncogene Neu) | Receptor |
| Vascular endothelial growth factor receptor 2 | Receptor |
| Janus kinase 2 | Non-receptor |
| Janus kinase 3 | Non-receptor |
| Spleen tyrosine kinase | Non-receptor |
| Zeta-chain-associated protein kinase 70 | Non-receptor |
| TEK tyrosine kinase, endothelial | Receptor |
| Serine/threonine protein kinases | |
| Dual-specificity tyrosine-phosphorylation regulated kinase 1A | Non-receptor |
| Cdc2-like kinase | Non-receptor |
| Checkpoint kinase 1 | Non-receptor |
| Human inhibitor nuclear-factor κB kinase 2 | Non-receptor |
| Cyclin-dependent kinase 1 | Non-receptor |
| Cyclin-dependent kinase 2 | Non-receptor |
| Polo-like kinase | Non-receptor |
| c-Jun N-terminal kinase 3 | Non-receptor |
| Glycogen synthase kinase 3 | Non-receptor |
| Mammalian target of rapamycin | Non-receptor |
| p38 mitogen-activated protein kinase | Non-receptor |
| Protein kinase B | Non-receptor |
Protein tyrosine kinases (PTKs) are a family of enzymes that can transfer phosphate group from ATP to tyrosine amino acid residues of target proteins in cell. This covalent post-translational modification is a crucial event for regulation of various biological processes including growth, metabolism, differentiation and apoptosis. Recent advances have demonstrated that tyrosine kinases play significant role in development of different diseases suggesting PTKs as attractive targets in the search for therapeutic agents.
Tyrosine kinases are classified as receptor tyrosine kinases such as FGFR, epidermal growth factor receptor (EGFR), Vascular endothelial growth factor receptor (VEGFR), TEK tyrosine kinase, endothelial (Tie2) and non-receptor tyrosine kinases such as Spleen tyrosine kinase (Syk), Zeta-chain-associated protein kinase 70 (ZAP-70), ABL, SRC, FAK, Janus kinase (JAK)[6]. Each of receptor tyrosine kinases contains extracellular ligand-binding domain, transmembrane hydrophobic helix, and intracellular tyrosine protein kinase domain[7]. Non-receptor tyrosine kinases are cytosolic proteins, possessing considerable structural variability. The non-receptor tyrosine kinases have a kinase domain and often include several additional protein-protein interacting domains like SH2, SH3 and the PH domain[6].
The EGFR family comprises four cell surface receptors: HER1 (EGFR/erbB1), HER2 (erbB2), HER3 (erbB3) and HER4 (erbB4)[7]. Binding of specific ligands to three of these receptors causes their dimerization and activation. HER2 is called an “orphan receptor” because it does not interact with any ligand, but it dimerizes with other ligand-bound members of EGFR family[8].
HER1 (EGFR/erbB1): HER1 overexpression and overactivity are often associated with a wide range of cancers, including prostate, gastric, breast, colorectal, pancreatic, ovarian, lung cancers, head and neck squamous cell carcinoma and glioma[9]. Aberrant EGFR signaling has been implicated in psoriasis, eczema and atherosclerosis[10,11]. Therefore, the EGFR inhibitors can be used for the amelioration of these diseases.
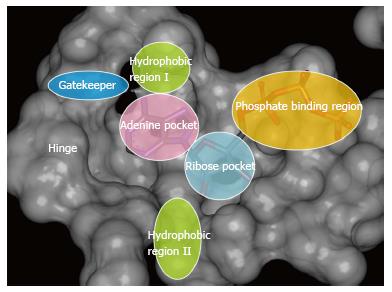
Furet et al[12] reported the first data concerning pharmacophore model for ATP-competitive inhibitors of EGFR. Accordingly to this Novartis pharmacophore hypothesis, the ATP-binding pocket in protein tyrosine kinases can be divided into five regions. In these five regions, three regions, namely, adenine region, sugar pocket, and hydrophobic region I, are primarily important to the binding affinity. Two other regions, hydrophobic region II and phosphate binding region are not of primary significance with respect to binding affinity, though they can be useful to enhance the inhibitor selectivity.
Also, pseudoreceptor model for EGFR was developed using a method FLARM. This model indicates the possible interactions between the receptor and ligand including two hydrogen bonds, one hydrophobic interaction and one sulfur-aromatic interaction, which are in accord with those in the Novartis pharmacophore model. Pharmacophore can be obtained according to the Novartis pharmacophore model and the pseudoreceptor model given by the FLARM method. 3D searching can then be done with the compound databases to find the lead compound of EGFR inhibitors[13].
HER2 (erbB2): HER2 has been demonstrated to play an essential role in the development and progression of about 25%-30% of human primary breast and ovarian cancers[14]. It was shown that the application of herceptin (a monoclonal antibody toward the HER2 receptor ectodomain) in combination with chemotherapy, leads to considerable regression of HER2-overexpressing metastatic breast tumors[15]. Therefore, the inhibition of HER2 has been considered a promising way of controlling malignant tumors.
Two groups of small molecules have been found to possess inhibitory activity toward HER2. A ligand-based approach was used for building HER2 pharmacophore model[16]. In the course of that work pharmacophore model generation was performed with Catalyst applying the Poling algorithm. From the calculated results, the best hypothesis bore good correlation with four features such as hydrogen bond donor, hydrogen bond acceptor, aliphatic and aromatic hydrophobic points. It seems that the formations of hydrogen bonds and the hydrophobic interactions are crucial for ligand binding.
VEGFR signaling regulates vascular development, angiogenesis, lymphangiogenesis[17] and has been involved in a wide range of human pathologies including cancer, atherosclerosis, and inflammatory diseases[18]. Therefore, VEGFR has been emerged as an attractive therapeutic target. The pharmacophore models for VEGFR inhibitors were reported in[18,19].
The ligand-based pharmacophore models were generated with Catalyst using the Poling algorithm and the “best conformational analysis” method. The best obtained hypothesis comprised four pharmacophore features: hydrogen bond donor, hydrogen bond acceptor, hydrophobic, and ring aromatic. The three-dimensional structure of ligand extracted from the crystal structure of 1YWN has been taken for shape query generation. The combined shape and hypothesis model was further used as a search query to screen Maybridge database. The query has been effectively performed to find one novel promising inhibitor of VEGFR kinase which possesses activity in cell lines[18].
Two structure-based pharmacophore models of VEGFR-2 kinase inhibitors were built using the SBF software. The first pharmacophoric hypothesis was based on the crystal structure of 1Y6A, with the selection hydrogen bonding interaction for Glu915, Cys917, and Asn921. The second pharmacophoric model was based on the crystal structure of 1YWN; the backbone amide-NH of Cys917 and Asp1044 were used as hydrogen bond donors; and the backbone carbonyl oxygen of Glu883 was the hydrogen bond acceptor. The results suggest the importance of the five features for pharmacophores: the presence of two hydrogen bond donors, one hydrogen bond acceptor and two hydrophobic groups. The screening accuracy was assessed using a series of known inhibitors[19].
JAK 2 and JAK 3: JAK2 and JAK3 are non-receptor protein tyrosine kinases involved in B-cell- and T-cell-mediated diseases[20]. The inhibition of these kinases can be a potential strategy for the treatment of lymphoid-derived disorders.
Pharmacophore models for JAK2 and JAK3 were generated with PHASE, a high-performance program module of Schrödinger for ligand-based drug design. PHASE provides six pharmacophoric features: hydrogen bond donor (D), hydrogen bond acceptor (A), positively charged (P), negatively charged (N), hydrophobic (H), and aromatic ring (R) features. Two ligand-based pharmacophore hypotheses were constructed for the dataset of inhibitor molecules of JAK2 and JAK3 to dig out the essential structural features required for inhibition of both enzymes. These models can be helpful for screening of novel molecules having inhibitory activity toward both enzymes. The best hypothesis of JAK2 was ADRR, indicating that JAK2 inhibitors have one hydrogen bond acceptor (A), one hydrogen bond donor (D) and two ring aromatic (R) features. The best model of JAK3 was ADDRR. Pearson correlation coefficient calculated for test set molecules demonstrated excellent predictive power of these hypotheses[20].
Syk and ZAP-70: Syk and ZAP-70 are cytoplasmic non-receptor tyrosine kinases which play critical roles in the intracellular signal transduction of hematopoietic cells[21]. Syk is a key mediator of immunoreceptor signalling in B-lymphocytes, mast cells, macrophages and neutrophils[22-24], and ZAP-70 in T-lymphocytes, basophils and natural killer cells[24,25]. Syk was shown to be an attractive drug target for therapy of type I hypersensitivity reactions such as allergic rhinitis, asthma, urticaria, anaphylaxis and autoimmune diseases such as multiple sclerosis, rheumatoid arthritis and systemic lupus erythematosus[23,26].
For a number of Syk and ZAP-70 small molecule inhibitors two reliable pharmacophore models were built with PHASE. The generated pharmacophore hypotheses combined with docking calculations were taken for further multi-step systematic virtual screening and finally 27 dual inhibitors of Syk and ZAP-70 were obtained as hits[24].
Also, 3D pharmacophore model of Syk inhibitors was developed by other authors applying HipHop and HypoRefine modules within Catalyst program package. Based on this model six compounds with good inhibitory potency against Syk were found[21].
Tie2: Tie2 is a receptor tyrosine protein kinase expressed almost exclusively in endothelial cells which plays an important role in blood vessel formation. This receptor negatively regulates the inflammatory response in endothelial cells, suppressing VEGF- and TNFα-induced expression of leukocyte adhesion molecules and procoagulant tissue factor[27]. Tie2 signaling also regulates pathologic angiogenesis, which includes tumor, psoriasis, choroidal neovascularization and rheumatoid arthritis angiogenesis[28]. The implication of Tie2 in pathologic angiogenesis makes this cellular receptor an attractive therapeutic target.
All the pharmacophore modeling calculations of type I and type II kinase inhibitors of Tie2 were performed with HipHop and HypoRefine modules within Catalyst program package. In connection with the lack of highly active type I protein kinase inhibitors and restricted their structural diversity, only qualitative HipHop pharmacophore models were generated for this type inhibitors of Tie2. The best hypothesis comprised five pharmacophore features, namely, hydrogen bond donor, hydrogen bond acceptor, general hydrophobic, hydrophobic aromatic and ring aromatic. For type II kinase inhibitors of Tie2, at the first step, a HipHop model was built with the aim to identify the common pharmacophore features which can be essential for potent inhibitors. Then, based on the information obtained from the HipHop hypothesis for type II kinase inhibitors, the quantitative pharmacophore models were created with the aid of HypoRefine module. The best HypoRefine hypothesis included two hydrogen bond donors, one hydrophobic aromatic, two general hydrophobic features, and two excluded volumes. The validation of this HypoRefine model with the test set method demonstrated good correlation between the experimental and estimated IC50 values, suggesting a good predictive power[29].
Serine/threonine protein kinases phosphorylate hydroxyl groups of serine or threonine residues of target proteins. Eukaryotic serine/threonine kinases can be classified into six groups: AGC, CaMK (for calcium-calmodulin dependent), CMGC (for CDK, MAP kinase, GSK and CDK-like), STE (homologs of STE11 and STE20), CK1 (for casein kinase-1), and TKL group (tyrosine kinase like). Accordingly to analysis of available structural data for members of each of the large groups it was revealed that the protein kinases possess similar architecture[30].
Dual-specificity tyrosine-phosphorylation regulated kinases (Dyrk) proteins are defined as dual-specificity protein kinases because they can phosphorylate serine, threonine and tyrosine residues. Dyrk1A has increased expression in Down Syndrown individuals and is implicated in the development of other pathologies, such as neurodegeneration, cardiac hypertrophy and bone homeostasis[31,32]. Hence, inhibition of Dyrk1A may have possible application as a therapeutical strategy for treatment of these diseases. Cdc2-like kinases (Clk) is implicated in the regulation of alternative splicing of mRNA isoforms, indicating that small molecule compounds able to modulate Clk activity may represent an important mechanism for the control of mRNA splicing[33].
Pharmacophore models of Dyrk1A and Clk4 inhibitors were built based on the structure of the five most active compounds. Both hypotheses are represented with AAARR, indicating they comprise three hydrogen bond acceptors and two hydrophobic groups. The models associated with Dyrk1A and Clk4 have pharmacophore features located at the similar positions, considering both active sets have common structural cores. For both models, two hydrogen bond acceptors and one hydrophobic group are mapped to the quinazoline ring, which is shared among all studied compounds. The other two features, or one hydrogen bond acceptor and one hydrophobic group, are mapped to the R3 substituent 1,3-benzodioxol, which is common for tested inhibitors[34].
Checkpoint kinase 1 (Chk1) is a serine/threonine protein kinase which plays an integral role in the regulation of cell cycle progression, normal cell division and is critical component for DNA damage response. The inhibition of Chk1 kinase has been shown to result in interrupting of the G2/M checkpoint, which would permit premature mitotic entry in the presence of DNA damage, leading to cell death. This suggests a potential therapeutic use of Chk1 inhibitors in cancer therapy[35].
All the pharmacophore modeling calculations for Chk1 were performed with Catalyst software package. The common pharmacophore features essential for promising Chk1 inhibitors were found with HipHop module. The best model involves four types of features, namely, hydrogen bond donor, hydrogen bond acceptor, ring aromatic and hydrophobic feature, indicating that the four types of features are important for potent Chk1 inhibitors[36].
Human inhibitor nuclear-factorκB kinase 2
The human inhibitor nuclear-factor κB kinase 2 (hIKK-2) is a serine/threonine protein kinase which belongs to the IKK complex and implicated in the activation of nuclear-factor κB transcription factor under inflammatory conditions. The inhibitors of hIKK-2 could have strong therapeutic potential for treatment of chronic inflammatory diseases.
The structure-based pharmacophore model for hIKK-2 was built by using LigandScout software based on the protein-ligand complexes which were obtained by the docking process of ATP-competitive inhibitors into active site of hIKK-2.
The ligand poses which satisfied the common pharmacophore features of protein kinase inhibitors necessary for interaction with ATP-binding site [ability to form hydrogen bonds with the amino acid residues in the hinge region (segment 96-99 in hIKK-2 sequence) and hydrophobic interactions with the hydrophobic cavity in the active site of hIKK-2 (for example, Val29, Lys44, Ile65 and Val152)] were taken as knowledge-based coherent. As a result of this analysis, 43 poses of the 21 hIKK-2 inhibitors were considered as knowledge-based coherent, and their corresponding sites (functional groups which form intermolecular interactions with the kinase domain of hIKK-2) were selected to generate structure-based pharmacophore model. This hypothesis comprised two hydrogen bond donors, one hydrogen bond acceptor and one hydrophobic group common to most of the 43 poses[37].
Cyclin-dependent kinase 1 (CDK1) is a serine/threonine protein kinase which plays a key role in promoting mitosis[38]. It was shown, that CDK1 inhibitors effectively blocked cell cycle progression in human tumor cell lines, indicating their potential clinical application as anticancer drugs[39].
A number of reliable binding hypotheses for CDK1 inhibitors were constructed with HypoGen module within Catalyst software package. HypoGen identifies a three-dimensional array of a maximum of five chemical features shared among the active ligands from training set providing relative alignment for each input molecule compatible with binding to target protein active site. The considered pharmacophore features can be hydrogen bond donors, hydrogen bond acceptors, aromatic planes, aliphatic, hydrophobic, positive and negative ionizable groups. The conformational flexibility of compounds from training set is modeled by generating multiple conformers covering a specified energy range for each input molecule. Successful pharmacophore models are complemented with exclusion spheres. Optimal sterically refined models obtained for CDK1 inhibitors were selected as search queries to screen the NCI, drugs and agrochemicals libraries. As a result, ten compounds demonstrated low micromolar activity toward CDK1, suggesting that generated pharmacophore hypothesis can be useful for search of potential anti-CDK1 agents[40].
Cyclin-dependent kinase 2 (CDK2) is important protein kinase for initiation of DNA synthesis in higher eukaryotes and is required for promoting the cell division cycle and for successful progression through S and G2 phases[41]. The importance of CDK2 for cell cycle progression has led to an active search of small molecule compounds inhibiting this enzyme as potential anticancer drugs.
Several ligand-based pharmacophore models for CDK2 small molecule inhibitors were generated independently with Catalyst by Hecker et al[42], Toba et al[43] and Vadivelan et al[44]. The multicomplex-based comprehensive pharmacophore map was built with LigandScout software by Zou et al[45]. It should be noted that during pharmacophore model construction Catalyst software takes into consideration only ligand information whereas LigandScout adds pharmacophore feature to the model when important interaction pattern between inhibitor and receptor is identified.
The authors of multicomplex-based comprehensive pharmacophore map compared their hypothesis with other reported CDK2 inhibitors models. It was revealed that each pharmacophore feature in the ligand-based models was mapped to the corresponding feature in comprehensive pharmacophore map suggesting that the last one includes more information over all three other models. Detailedly, during alignment of the final Hecker model and multicomplex-based comprehensive map it was shown that hydrogen bond acceptor feature in Hecker model was mapped to the feature of comprehensive map reflecting the interaction of small-molecule inhibitor with hinge region (A1); the hydrogen bond donor feature in Hecker model was matched to the feature representing the interaction with Gln131 (D5); the hydrophobic features were mapped to the features located at the solvent-accessible region (H1) and in the ribose-phosphate binding site (H3). The pharmacophore model generated by Toba et al[43] comprised two hydrogen bond donor features and three hydrophobic features. Each of these features was matched to the corresponding features D1, D5, H1, H2 and H3 of comprehensive pharmacophore map. The pharmacophore hypothesis constructed by Vadivelan et al[44] included two hydrogen bond acceptors, one hydrogen bond donor, and one hydrophobic feature. In comparison with comprehensive pharmacophore map, one of the hydrogen bond acceptor features was mapped to the feature that is located near Asp86 (A3), other one was matched to the feature representing the interaction with Lys33 (A4). The hydrogen bond donor feature and hydrophobic feature of Vadivelan model correspond to D1 and H3 features of multicomplex-based comprehensive pharmacophore map, respectively[45].
Polo-like kinases (PLKs) belong to a family of serine/threonine protein kinases and exist in four isoforms, namely, PLK1, PLK2, PLK3, and PLK4. The only one of these isoforms, PLK1, is shown to be implicated in the regulation of chromosome segregation, centrosome maturation, bipolar spindle formation and execution of cytokinesis[46]. The activity of PLK1 is increased in many tumor types, including lung, breast, colon, pancreatic, prostate and ovarian indicating its capability as a drug target[47].
The chemical feature-based pharmacophore models of PLK1 inhibitors were constructed by using HipHop and HypoGen modules within Catalyst program package. The best qualitative HipHop pharmacophore hypothesis contains seven features, namely, hydrophobic aromatic feature, two hydrophobic aliphatic moieties, three hydrogen bond acceptors and one hydrogen bond donor. The best quantitative HypoGen pharmacophore model, possessing the lowest rmsd value and the highest correlation coefficient, includes four features, namely, general hydrophobic, hydrophobic aliphatic, hydrogen bond acceptor and hydrogen bond donor. The results of validation for HypoGen pharmacophore model, which were obtained with the aid of the test set method, have demonstrated a really good correlation between the experimental and estimated IC50 values suggesting a good predictive ability[48].
The c-Jun N-terminal kinase 3 (JNK3) is a member of the mitogen-activated protein kinase (MAPK) family, which activates signaling pathways under environmental stress conditions[49]. JNK3 is expressed selectively in brain, heart, and testis[50]. It was shown, that JNK3 phosphorylates β-amyloid precursor protein, a conserved and ubiquitously expressed transmembrane glycoprotein involved in the development of Alzheimer’s disease[51]. Therefore, JNK3 appears to be an attractive therapeutic target for this neurodegenerative disease.
Pharmacophore models for JNK3 small-molecule inhibitors were generated with Catalyst software package. X-ray crystal structure of JNK3 (PDB ID: 2R9S) was taken for structure-based pharmacophore modeling and molecular docking simulations. A structure-based pharmacophore hypothesis was built using interaction generation module implemented in Discovery Studio. The most important interaction patterns were transformed into pharmacophore features, such as hydrophobes, hydrogen bond donors and hydrogen bond acceptors along with their direction vectors. The final refined structure-based pharmacophore model included hydrogen bond donor features with Lys68, Gly71, Ser72, Gln155, Met149 and hydrogen bond acceptors with Lys68, Gly71, Met149, Gln155 and three hydrophobic features.
The features obtained in the ligand-based pharmacophore model are well compared to the features of structure-based pharmacophore model. But, structure-based pharmacophore model had three additional features, not present in ligand-based pharmacophore hypothesis, which would be helpful for development of novel JNK3 inhibitors[49].
Glycogen synthase kinase 3 (GSK-3) is a serine/threonine protein kinase highly expressed in the nervous system, which regulates glycogen metabolism by insulin, and is involved in many different biological processes such as tumorigenesis, cell survival, and developmental patterning[52]. GSK-3 has recently emerged as a promising therapeutic target for the search of small molecule inhibitors which can be potential novel drug candidates for treatment of several human pathologies, including cancer, Alzheimer’s disease, stroke, bipolar disorders, type II diabetes and chronic inflammatory processes[53].
Pharmacophore models for GSK-3 inhibitors were constructed with the HypoGen module within the Catalyst software package based on list of 152 GSK-3 inhibitors. HypoGen allows automatic pharmacophore generation based on a library of at least 16 molecules with inhibitory activity toward proposed molecular target ranging over 4 orders of magnitude[5].
3D pharmacophore mapping methodology based on distance comparison technique was designed for the three GSK-3 inhibitors using DISCOtech™ module implemented in SYBYL 8.0. DISCOtech™ is a well established module in constructing pharmacophoric map. Taking into consideration a set of molecules which are characterized by the ability to interact with the same protein receptor, DISCOtech™ identifies features that could be components in a pharmacophore hypothesis. DISCOtech™ operates in distance space and can perform clique detection to build pharmacophore models on up to 300 conformers per molecule. Therefore, DISCOtech™ can be efficiently applied with at least 3-5 compounds to design reliable pharmacophore hypotheses[54].
Mammalian target of rapamycin (mTOR) is a ubiquitous serine/threonine protein kinase that regulates several important physiological functions like protein synthesis, metabolism, cell growth, proliferation, and autophagy. mTOR is also critical for a number of brain-specific mechanisms, such as synaptic plasticity, learning, and cortical development[55]. Recent studies have implicated mTOR to several human pathologies including cancer, diabetes, obesity, cardiovascular diseases and neurological disorders[56]. The pharmaceutical attractiveness of small molecule mTOR inhibitors coupled with the deficiency of crystallographic structural data for mTOR kinase domain, were starting point for development of ligand-based QSAR and pharmacophore models[57].
The Hip-Hop pharmacophore model was created with the Common Feature Pharmacophore Generation module implemented in Accelrys Discovery Studio 2.1. This pharmacophore hypothesis provides a geometrical representation of the features necessary for ligands to interact favorably with a receptor site and demonstrate biological activity. Hip-Hop identifies configurations or three-dimensional spatial arrangements of chemical features that are shared among all molecules in the set. Under the Common Feature Pharmacophore Generation protocol, were used four features such as hydrogen bond donor, hydrogen bond acceptor, ring aromatic, and hydrophobic to build the pharmacophore model.
The best pharmacophore hypothesis generated from 27 ATP-competitive inhibitors of mTOR comprised two hydrogen bond acceptors, one hydrophobic feature and one aromatic ring feature[58].
The p38 mitogen-activated protein (MAP) kinase (p38MAPK) is a serine/threonine protein kinase which plays a very important role in the pathophysiology of several inflammatory human diseases, such as, asthma, osteoarthritis and rheumatoid arthritis, a chronic obstructive pulmonary disease. Therefore, the inhibition of p38MAPK can be an effective strategy to prevent the development of these diseases[59].
Catalyst HypoGen pharmacophore approach was applied to obtain models for a collection of p38MAPK inhibitors[59,60]. Eight out of ten best hypotheses comprise the identical four features: one hydrogen bond acceptor, two hydrophobic aromatic and one hydrophobic feature, which indicates the stability of the models[59]. The obtained hypotheses are readily interpretable and can be applied for the rational discovery of new p38MAPK inhibitors[60].
The protein kinase B (PKB; Akt) family of serine/threonine kinases consists of three members: Akt1/PKBα, Akt2/PKBβ, and Akt3/PKBγ[61]. Akt is a central component in cell signaling pathways regulated by growth factors, cytokines, and other cellular stimuli. The activation of Akt leads to cell cycle progression (inhibiting apoptosis)[62]. Ligand-based pharmacophore model of Akt inhibitors was built using DISCOtech and GASP (genetic algorithm similarity program) module[63].
A crystal structure of Akt2 complexed with a known inhibitor (PDB ID: 3E8D) was taken for construction of structure-based pharmacophore hypothesis. The Interaction Generation protocol within DS program was used to create pharmacophoric features corresponding to all important interaction points at the ATP-binding pocket of Akt2. The obtained pharmacophore model consisted of seven pharmacophoric features, namely, hydrogen bond donor (HD), two hydrogen bond acceptors (HA1-2), and four hydrophobic groups (HY1-4), besides, eighteen exclusion volume spheres were also taken into account. HD is at the neighborhood of the carboxyl group of Asp293. HA1 is positioned to interact with the amino group of Ala232. HA2 is located near amino group of Phe294 and Asp293. Groups in accordance with these pharmacophoric features potentially able form hydrogen bonds with adjacent amino acid residues. HY1 is located in a hydrophobic pocket formed by Ala178, Met282 and Phe439. HY2 is situated in another hydrophobic pocket composed by Gly159, Gly161, Gly164 and Val166. HY3 is close to Lys181 and Met229 and HY3 is near to Phe294. There are short distances between HY4 and hydrophobic amino acids Phe163 and Lys181. Groups in accordance with these hydrophobic features may be involved in hydrophobic interactions with enzyme. Therefore, small-molecule compounds matching with some of these features may be potential inhibitors of Akt2[64].
The methods for pharmacophore model generation are divided in two categories: receptor-based and ligand-based. Receptor-based approaches can be used when the structure of molecular target is determined. In other case, ligand-based approaches can be applied for pharmacophore hypothesis generation.
During analysis of the pharmacophore approaches in protein kinase inhibitors design, it was revealed that despite of large amount of the structural data for protein kinases, the ligand-based approaches are more widely used for protein kinase pharmacophore model generation than the receptor-based. Ligand-based methods for pharmacophore elucidation include ALADDIN, DISCO, GERM, COMPASS, GASP, Catalyst HipHop, SCAMPI, Catalyst HypoGen, Phase, CLEW GAMMA, PARM, DANTE, etc.
3D-QSAR methods Catalyst HypoGen and Phase which use the known activity values of the small molecule inhibitors in the training set to build the hypothesis, are the most applied for protein kinase inhibitors pharmacophore models generation. These models include features common only for highly active compounds and also can contain excluded volumes (obtained based on the structure of inactive compounds), which couldn’t be occupied by inhibitors. The methods Catalyst HipHop, PharmaGist, DISCO can be used only for qualitative pharmacophore models generation which don’t take into consideration the information concerning activity of compounds. Qualitative pharmacophore model can be taken as a basis for further 3D-QSAR hypothesis generation.
The receptor-based methods of pharmacophore model elucidation are more rarely used for protein kinase pharmacophore design. These approaches can be useful for study of ligand-receptor interactions. The easiest way to build receptor-based 3D-pharmacophore model is to manually select key amino acid residues in the active site of enzyme, to determine the most important interactions with ligand and then based on this information to set pharmacophore features with specific properties. Other way to obtain 3D-pharmacophore model is to apply receptor-oriented algorithms which automatically evaluate the interactions between ligand and receptor. This modeling is performed with software packages Pharmer, Accelrys Discovery Studio, Schrödinger, MOE, LigandScout, etc.
The combination of receptor-based and ligand-based methods allows improve efficiency of novel bioactive compounds development[49].
The pharmacophore models, developed for search of novel inhibitors for tyrosine and serine/threonine protein kinases don’t have significant differences. For the best of our knowledge, the most protein kinase pharmacophore models are reported for inhibitors competing for the ATP-binding site and are in accord with the Novartis[12] and Traxler’s[65] pharmacophore models. Traxler’s pharmacophore model includes the same regions as Novartis pharmacophore model: adenine region, sugar pocket, hydrophobic region I, hydrophobic region II, phosphate binding region and also has one additional element - gatekeeper residue that plays an essential role in inhibitor binding and selectivity (Figure 1). In spite of the significant role of gatekeeper residue for inhibitor binding, in most cases the authors didn’t pay attention to this residue during pharmacophore model generation for protein kinase inhibitors.
The average number of pharmacophore features in the analyzed tyrosine and serine/threonine protein kinase inhibitors pharmacophore models is 4-5. Several models have more pharmacophore features but it should be noted that increasing of their number leads to improvement of model specificity which decreases ability of the model to identify hit compounds from diverse chemical classes.
The most pharmacophore hypotheses have 1-2 features (hydrogen bond acceptor and/or hydrogen bond donor) in adenine region. As a rule, these features are indicated as vectors directed to main chain of amino acid residue in the hinge region. Several pharmacophore models have additional aromatic or hydrophobic feature in adenine region.
Almost all pharmacophore models have hydrophobic or aromatic pharmacophore feature in hydrophobic pocket I. Several hypotheses also have hydrogen bond donor and/or hydrogen bond acceptor in this region which can additionally stabilize the inhibitor in ATP-binding pocket.
Only some models have hydrogen bond donor pharmacophore feature in sugar pocket. This region is not so important for binding affinity of inhibitors with protein kinase active site, as adenine region or hydrophobic pocket I.
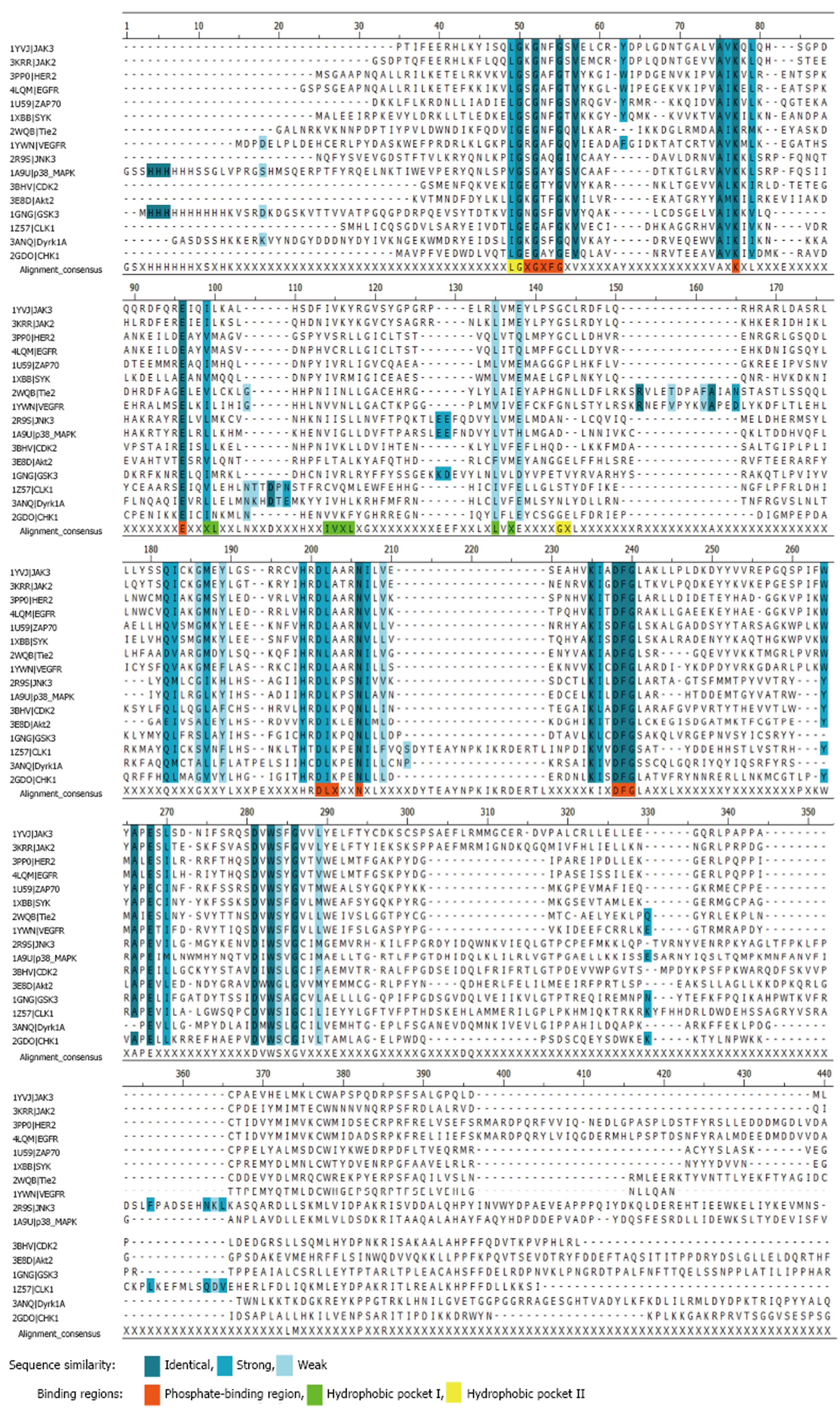
Phosphate-binding region has 1-2 pharmacophore features (hydrogen bond acceptor and/or hydrogen bond donor); hydrophobic pocket II has one hydrophobic or aromatic pharmacophore feature. We have performed multiple alignment of amino acid sequences for tyrosine and serine/threonine protein kinases for which pharmacophore models were reported. It was revealed that phosphate-binding regions are highly conservative in contrast to hydrophobic pockets I and II (Figure 2). Therefore, pharmacophore features located in phosphate-binding region don’t supply specificity for hit compounds, but correctly predicted hydrophobic or aromatic features in hydrophobic pockets I and II can be useful to improve the selectivity of inhibitors.
The most reported pharmacophore models were built for type I protein kinase inhibitors. Several pharmacophore hypotheses were also generated for type II protein kinase inhibitors. The models for both types are very similar, but in a case of type II protein kinase inhibitors, the model has pharmacophore features in the deep pocket of ATP-binding site.
In this article, the existing data concerning protein kinase inhibitors pharmacophore models were reviewed. Ligand-based and receptor-based methods can be equally applied for protein kinase pharmacophore models generation. The combination of these approaches can be useful for improving of efficiency of bioactive compounds design. With the help of the discovered pharmacophore models, the identification of possible protein kinase inhibitors can be much more accelerated.
P- Reviewer: Choi CY, Huang Y, Moens U S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Force T, Kuida K, Namchuk M, Parang K, Kyriakis JM. Inhibitors of protein kinase signaling pathways: emerging therapies for cardiovascular disease. Circulation. 2004;109:1196-1205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 100] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 2. | Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309-315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1668] [Cited by in F6Publishing: 1597] [Article Influence: 72.6] [Reference Citation Analysis (0)] |
| 3. | Cohen P, Alessi DR. Kinase drug discovery--what’s next in the field? ACS Chem Biol. 2013;8:96-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 293] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 4. | Wermuth CG, Ganellin CR, Lindberg P, Mitscher LA. Glossary of terms used in medicinal chemistry. Annu Rep Med Chem. 1998;33:385-395. [DOI] [Cited in This Article: ] |
| 5. | Taha MO, Bustanji Y, Al-Ghussein MA, Mohammad M, Zalloum H, Al-Masri IM, Atallah N. Pharmacophore modeling, quantitative structure-activity relationship analysis, and in silico screening reveal potent glycogen synthase kinase-3beta inhibitory activities for cimetidine, hydroxychloroquine, and gemifloxacin. J Med Chem. 2008;51:2062-2077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 71] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Paul MK, Mukhopadhyay AK. Tyrosine kinase - Role and significance in Cancer. Int J Med Sci. 2004;1:101-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 348] [Cited by in F6Publishing: 322] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 7. | Dowsett M, Cooke T, Ellis I, Gullick WJ, Gusterson B, Mallon E, Walker R. Assessment of HER2 status in breast cancer: why, when and how? Eur J Cancer. 2000;36:170-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 70] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | Brennan PJ, Kumagai T, Berezov A, Murali R, Greene MI. HER2/Neu: mechanisms of dimerization/oligomerization. Oncogene. 2002;21:328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 9. | Giaccone G. HER1/EGFR-targeted agents: predicting the future for patients with unpredictable outcomes to therapy. Ann Oncol. 2005;16:538-548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Jost M, Kari C, Rodeck U. The EGF receptor - an essential regulator of multiple epidermal functions. Eur J Dermatol. 2000;10:505-510. [PubMed] [Cited in This Article: ] |
| 11. | Dreux AC, Lamb DJ, Modjtahedi H, Ferns GA. The epidermal growth factor receptors and their family of ligands: their putative role in atherogenesis. Atherosclerosis. 2006;186:38-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 12. | Furet P, Caravatti G, Lydon N, Priestle JP, Sowadski JM, Trinks U, Traxler P. Modelling study of protein kinase inhibitors: binding mode of staurosporine and origin of the selectivity of CGP 52411. J Comput Aided Mol Des. 1995;9:465-472. [PubMed] [Cited in This Article: ] |
| 13. | Peng T, Pei J, Zhou J. 3D-QSAR and receptor modeling of tyrosine kinase inhibitors with flexible atom receptor model (FLARM). J Chem Inf Comput Sci. 2003;43:298-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 14. | Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707-712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4972] [Cited by in F6Publishing: 4931] [Article Influence: 140.9] [Reference Citation Analysis (0)] |
| 15. | Arteaga CL. Trastuzumab, an appropriate first-line single-agent therapy for HER2-overexpressing metastatic breast cancer. Breast Cancer Res. 2003;5:96-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Zhu LL, Hou TJ, Chen LR, Xu XJ. 3D QSAR analyses of novel tyrosine kinase inhibitors based on pharmacophore alignment. J Chem Inf Comput Sci. 2001;41:1032-1040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7:359-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2194] [Cited by in F6Publishing: 2264] [Article Influence: 125.8] [Reference Citation Analysis (0)] |
| 18. | Yu H, Wang Z, Zhang L, Zhang J, Huang Q. The discovery of novel vascular endothelial growth factor receptor tyrosine kinases inhibitors: pharmacophore modeling, virtual screening and docking studies. Chem Biol Drug Des. 2007;69:204-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Lee K, Jeong KW, Lee Y, Song JY, Kim MS, Lee GS, Kim Y. Pharmacophore modeling and virtual screening studies for new VEGFR-2 kinase inhibitors. Eur J Med Chem. 2010;45:5420-5427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 20. | Jasuja H, Chadha N, Kaur M, Silakari O. Dual inhibitors of Janus kinase 2 and 3 (JAK2/3): designing by pharmacophore- and docking-based virtual screening approach. Mol Divers. 2014;18:253-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 21. | Xie HZ, Li LL, Ren JX, Zou J, Yang L, Wei YQ, Yang SY. Pharmacophore modeling study based on known spleen tyrosine kinase inhibitors together with virtual screening for identifying novel inhibitors. Bioorg Med Chem Lett. 2009;19:1944-1949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 22. | Riccaboni M, Bianchi I, Petrillo P. Spleen tyrosine kinases: biology, therapeutic targets and drugs. Drug Discov Today. 2010;15:517-530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 80] [Article Influence: 5.7] [Reference Citation Analysis (1)] |
| 23. | Wong BR, Grossbard EB, Payan DG, Masuda ES. Targeting Syk as a treatment for allergic and autoimmune disorders. Expert Opin Investig Drugs. 2004;13:743-762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 123] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 24. | Kaur M, Kumari A, Bahia MS, Silakari O. Designing of new multi-targeted inhibitors of spleen tyrosine kinase (Syk) and zeta-associated protein of 70kDa (ZAP-70) using hierarchical virtual screening protocol. J Mol Graph Model. 2013;39:165-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 25. | Wang H, Kadlecek TA, Au-Yeung BB, Goodfellow HE, Hsu LY, Freedman TS, Weiss A. ZAP-70: an essential kinase in T-cell signaling. Cold Spring Harb Perspect Biol. 2010;2:a002279. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 259] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 26. | Mazuc E, Villoutreix BO, Malbec O, Roumier T, Fleury S, Leonetti JP, Dombrowicz D, Daëron M, Martineau P, Dariavach P. A novel druglike spleen tyrosine kinase binder prevents anaphylactic shock when administered orally. J Allergy Clin Immunol. 2008;122:188-194, 194.e1-3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Hughes DP, Marron MB, Brindle NP. The antiinflammatory endothelial tyrosine kinase Tie2 interacts with a novel nuclear factor-kappaB inhibitor ABIN-2. Circ Res. 2003;92:630-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 117] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 28. | Martin V, Liu D, Fueyo J, Gomez-Manzano C. Tie2: a journey from normal angiogenesis to cancer and beyond. Histol Histopathol. 2008;23:773-780. [PubMed] [Cited in This Article: ] |
| 29. | Xie QQ, Xie HZ, Ren JX, Li LL, Yang SY. Pharmacophore modeling studies of type I and type II kinase inhibitors of Tie2. J Mol Graph Model. 2009;27:751-758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 30. | Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. Substrate and docking interactions in Ser/Thr protein kinases. Chem Rev. 2007;107:5065-5081. [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 31. | Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011;25:449-462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 229] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 32. | Park J, Song WJ, Chung KC. Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol Life Sci. 2009;66:3235-3240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 145] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 33. | Mott BT, Tanega C, Shen M, Maloney DJ, Shinn P, Leister W, Marugan JJ, Inglese J, Austin CP, Misteli T. Evaluation of substituted 6-arylquinazolin-4-amines as potent and selective inhibitors of cdc2-like kinases (Clk). Bioorg Med Chem Lett. 2009;19:6700-6705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 34. | Pan Y, Wang Y, Bryant SH. Pharmacophore and 3D-QSAR characterization of 6-arylquinazolin-4-amines as Cdc2-like kinase 4 (Clk4) and dual specificity tyrosine-phosphorylation-regulated kinase 1A (Dyrk1A) inhibitors. J Chem Inf Model. 2013;53:938-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 35. | McNeely S, Beckmann R, Bence Lin AK. CHEK again: revisiting the development of CHK1 inhibitors for cancer therapy. Pharmacol Ther. 2014;142:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 129] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 36. | Chen JJ, Liu TL, Yang LJ, Li LL, Wei YQ, Yang SY. Pharmacophore modeling and virtual screening studies of checkpoint kinase 1 inhibitors. Chem Pharm Bull (Tokyo). 2009;57:704-709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 37. | Sala E, Guasch L, Iwaszkiewicz J, Mulero M, Salvadó MJ, Pinent M, Zoete V, Grosdidier A, Garcia-Vallvé S, Michielin O. Identification of human IKK-2 inhibitors of natural origin (part I): modeling of the IKK-2 kinase domain, virtual screening and activity assays. PLoS One. 2011;6:e16903. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 38. | Chow JP, Poon RY, Ma HT. Inhibitory phosphorylation of cyclin-dependent kinase 1 as a compensatory mechanism for mitosis exit. Mol Cell Biol. 2011;31:1478-1491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 39. | Chen S, Chen L, Le NT, Zhao C, Sidduri A, Lou JP, Michoud C, Portland L, Jackson N, Liu JJ. Synthesis and activity of quinolinyl-methylene-thiazolinones as potent and selective cyclin-dependent kinase 1 inhibitors. Bioorg Med Chem Lett. 2007;17:2134-2138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 34] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 40. | Al-Sha’er MA, Taha MO. Discovery of novel CDK1 inhibitors by combining pharmacophore modeling, QSAR analysis and in silico screening followed by in vitro bioassay. Eur J Med Chem. 2010;45:4316-4330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 41. | Hu B, Mitra J, van den Heuvel S, Enders GH. S and G2 phase roles for Cdk2 revealed by inducible expression of a dominant-negative mutant in human cells. Mol Cell Biol. 2001;21:2755-2766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 42. | Hecker EA, Duraiswami C, Andrea TA, Diller DJ. Use of catalyst pharmacophore models for screening of large combinatorial libraries. J Chem Inf Comput Sci. 2002;42:1204-1211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 52] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 43. | Toba S, Srinivasan J, Maynard AJ, Sutter J. Using pharmacophore models to gain insight into structural binding and virtual screening: an application study with CDK2 and human DHFR. J Chem Inf Model. 2006;46:728-735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 44. | Vadivelan S, Sinha BN, Irudayam SJ, Jagarlapudi SA. Virtual screening studies to design potent CDK2-cyclin A inhibitors. J Chem Inf Model. 2007;47:1526-1535. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 45. | Zou J, Xie HZ, Yang SY, Chen JJ, Ren JX, Wei YQ. Towards more accurate pharmacophore modeling: Multicomplex-based comprehensive pharmacophore map and most-frequent-feature pharmacophore model of CDK2. J Mol Graph Model. 2008;27:430-438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | Stewart HJ, Kishikova L, Powell FL, Wheatley SP, Chevassut TJ. The polo-like kinase inhibitor BI 2536 exhibits potent activity against malignant plasma cells and represents a novel therapy in multiple myeloma. Exp Hematol. 2011;39:330-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 47. | Chopra P, Sethi G, Dastidar SG, Ray A. Polo-like kinase inhibitors: an emerging opportunity for cancer therapeutics. Expert Opin Investig Drugs. 2010;19:27-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 48. | Wang HY, Cao ZX, Li LL, Jiang PD, Zhao YL, Luo SD, Yang L, Wei YQ, Yang SY. Pharmacophore modeling and virtual screening for designing potential PLK1 inhibitors. Bioorg Med Chem Lett. 2008;18:4972-4977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Kumar BV, Kotla R, Buddiga R, Roy J, Singh SS, Gundla R, Ravikumar M, Sarma JA. Ligand-based and structure-based approaches in identifying ideal pharmacophore against c-Jun N-terminal kinase-3. J Mol Model. 2011;17:151-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Kyriakis JM, Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92:689-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 873] [Cited by in F6Publishing: 979] [Article Influence: 81.6] [Reference Citation Analysis (0)] |
| 51. | Kimberly WT, Zheng JB, Town T, Flavell RA, Selkoe DJ. Physiological regulation of the beta-amyloid precursor protein signaling domain by c-Jun N-terminal kinase JNK3 during neuronal differentiation. J Neurosci. 2005;25:5533-5543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 97] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 52. | Martinez A, Castro A, Dorronsoro I, Alonso M. Glycogen synthase kinase 3 (GSK-3) inhibitors as new promising drugs for diabetes, neurodegeneration, cancer, and inflammation. Med Res Rev. 2002;22:373-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 245] [Cited by in F6Publishing: 249] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 53. | Dorronsoro I, Castro A, Martinez A. Inhibitors of glycogen synthase kinase-3: future therapy for unmet medical needs? Expert Opin Ther Patents. 2002;12:1527-1536. [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 54. | Khanfar MA, Asal BA, Mudit M, Kaddoumi A, El Sayed KA. The marine natural-derived inhibitors of glycogen synthase kinase-3beta phenylmethylene hydantoins: In vitro and in vivo activities and pharmacophore modeling. Bioorg Med Chem. 2009;17:6032-6039. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 55. | Wong M. Mammalian target of rapamycin (mTOR) pathways in neurological diseases. Biomed J. 2013;36:40-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 56. | Tsang CK, Qi H, Liu LF, Zheng XF. Targeting mammalian target of rapamycin (mTOR) for health and diseases. Drug Discov Today. 2007;12:112-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 310] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 57. | Khanfar MA, AbuKhader MM, Alqtaishat S, Taha MO. Pharmacophore modeling, homology modeling, and in silico screening reveal mammalian target of rapamycin inhibitory activities for sotalol, glyburide, metipranolol, sulfamethizole, glipizide, and pioglitazone. J Mol Graph Model. 2013;42:39-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 58. | Tanneeru K, Guruprasad L. Ligand-based 3-D pharmacophore generation and molecular docking of mTOR kinase inhibitors. J Mol Model. 2012;18:1611-1624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Sarma R, Sinha S, Ravikumar M, Kishore Kumar M, Mahmood SK. Pharmacophore modeling of diverse classes of p38 MAP kinase inhibitors. Eur J Med Chem. 2008;43:2870-2876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 60. | Xiao Z, Varma S, Xiao YD, Tropsha A. Modeling of p38 mitogen-activated protein kinase inhibitors using the Catalyst HypoGen and k-nearest neighbor QSAR methods. J Mol Graph Model. 2004;23:129-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 19] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 61. | Dillon RL, Muller WJ. Distinct biological roles for the akt family in mammary tumor progression. Cancer Res. 2010;70:4260-4264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 655] [Reference Citation Analysis (0)] |
| 62. | Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261-1274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4237] [Cited by in F6Publishing: 4568] [Article Influence: 268.7] [Reference Citation Analysis (0)] |
| 63. | Vyas VK, Ghate M, Goel A. Pharmacophore modeling, virtual screening, docking and in silico ADMET analysis of protein kinase B (PKB β) inhibitors. J Mol Graph Model. 2013;42:17-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 52] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 64. | Fei J, Zhou L, Liu T, Tang XY. Pharmacophore modeling, virtual screening, and molecular docking studies for discovery of novel Akt2 inhibitors. Int J Med Sci. 2013;10:265-275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 65. | Traxler P, Furet P. Strategies toward the design of novel and selective protein tyrosine kinase inhibitors. Pharmacol Ther. 1999;82:195-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 297] [Cited by in F6Publishing: 294] [Article Influence: 11.8] [Reference Citation Analysis (0)] |