Published online Nov 27, 2013. doi: 10.5496/wjmg.v3.i4.34
Revised: September 6, 2013
Accepted: September 18, 2013
Published online: November 27, 2013
In order to provide the means for the design of novel rational anti-cancer drug therapies research efforts are concentrated on unravelling the molecular circuits which induce programmed cell death and block proliferation of cancer cells. Modern therapeutic strategies are based on the understanding of the complexity of physiological functions such as differentiation, development, immune responses, cell-cycle arrest, DNA damage repair, apoptosis, autophagy, energy metabolism, and senescence. It has become evident that this knowledge will provide the means to target the components of the pathways involved in these processes in a specific and selective manner thus paving the way for the development of effective and personalised anti-cancer therapies. Transcription is a crucial cellular process that regulates a multitude of physiological functions, which are essential in disease progression and cellular response to therapy. Transcription factors such as the p53 tumor suppressor and the hypoxia-inducible factor-α (HIF-α) are key players in carcinogenesis and cellular response to cancer therapies. Both of these transcription factors regulate gene expression of genes involved in cell death and proliferation, in some cases cooperating towards producing the same outcome and in some others mediating opposing effects. It is thus apparent that fine tuning of the activity of these transcription factors is essential to determine the cellular response to therapeutic regimens, in other words whether tumor cells will commit to apoptosis or evade engagement with the anti-proliferative effects of drugs leading to drug resistance. Our observations support the notion that the functional crosstalk between HIF-1α and p53 pathways and thus the fine tuning of their transcriptional activity is mediated by cofactors shared between the two transcription factors such as components of the p300 co-activator multiprotein complex. In particular, there is evidence to suggest that differential composition of the co-modulatory protein complexes associated with p53 and HIF-1α under diverse types of stress conditions differentially regulate the expression of distinct subsets of p53 and HIF-1α target genes involved in processes such as cell cycle arrest, apoptosis, chronic inflammation, and cellular energy metabolism thereby determining the cellular fate under particular types of micro-environmental stress.
Core tip: The results of our work endorse the notion that specific features determine targeting of transcription factors to distinct clusters of their target genes including the nature of the DNA binding sites found within the regulatory region of the promoter of each one of the target genes, the composition of the cofactor network associated with different transcription factors under diverse types of stress conditions and the precise posttranslational modifications of each one of the transcription factors linking characteristic PTM codes with discrete types of micro-environmental stress. These features are essential considerations for the design of effective therapeutics and individualised cancer treatment.
- Citation: Rajendran R, Krstic-Demonacos M, Demonacos C. Regulation of the cell fate by DNA damage and hypoxia. World J Med Genet 2013; 3(4): 34-40
- URL: https://www.wjgnet.com/2220-3184/full/v3/i4/34.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v3.i4.34
The transcriptional regulation of gene expression is a crucial mechanism by which cells maintain homeostasis, differentiate, survive and proliferate, respond to internal signals as well as those they receive from their surroundings, and adjust to local environmental conditions[1]. The transcription process is regulated mainly at two levels. One encompassing transcription factors and the transcriptional machinery, and the other involving chromatin which is the packaging structure of the DNA and consists of the four histone proteins H2A, H2B, H3 and H4 forming the nucleosome[2,3]. The two levels of regulation are connected to each other since access of the transcription machinery to the DNA is regulated by molecular modifications of the chromatin structure executed by remodelling reactions such as phosphorylation, methylation, and acetylation[4] which control the binding between transcription factors and DNA thereby selectively and specifically modulating gene expression of their target genes[5,6]. These modifications represent the so called ‘‘histone code’’[7], which is a type of encryption that indicates either open access (euchromatin structure) of transcription factors to the DNA and transcription initiation of the target genes or closed chromatin conformation (heterochromatin) and transcriptional repression[8,9]. In this respect transcriptional co-factors, which are proteins mediating histone modifications thus determining the open or closed chromatin conformation are of crucial importance in the activation or repression of gene expression and therefore for the cellular physiology[10-12].
The detailed understanding of the regulation of gene expression has provided the means to comprehend how aberrant regulation of the transcriptional events can lead to disease[13]. The role of DNA binding transcription factors and their modulators, of the non-coding RNAs, as well as the effects of epigenetic changes on the structure of the chromatin on transcription regulation and the impact of these events on the cellular physiology has been elucidated for many different diseases, for example diabetes[14] cardiovascular disease[15], neurological disorders[16], rheumatoid arthritis[17] cancer[18] and conditions such as obesity[19] and ageing[20]. Transcriptional regulation is not only important to understand the initiation, development, and prognosis of the disease but it is also imperative in predicting the cellular response to therapeutic modalities[21-24].
A characteristic example of the importance of the transcription process in the outcome of the disease and the cellular response to drug treatment has been demonstrated by the function of the transcription factor and tumor suppressor protein p53[25]. p53 is a transcription factor responding alternatively to diverse types of stress conveying different signals in a manner dependent on the type of stress[26] by modulating gene expression of specific subsets of its target genes involved in vital and sometimes contradicting cellular functions such as cell cycle control[27], apoptosis[28], senescence[29,30], autophagy[31], DNA damage repair[32,33], and tumor energy metabolism[34]. It is worth noting that more than 90% of p53 mutations in human cancers occur in its DNA binding domain[35] hampering the ability of this transcription factor to bind to DNA and transactivate its transcription target genes and emphasising the importance of transcription in oncogenesis[36]. Under mild stress conditions p53 facilitates cell survival by activating a set of genes involved in cell cycle arrest and DNA damage repair[37]. In prolonged stress or irreversible DNA damage p53 activates programmed cell death[38]. Post-translational modifications of p53 including ubiquitination, phosphorylation, methylation and acetylation are also very important in the regulation of its protein stability and transcription target selectivity[39].
Hypoxia is an important pathophysiological state found mainly in solid tumors since the rapid growth of cancer tissues is associated with vascularisation deficiency, and therefore low oxygen availability which reaches levels below 5%[40]. Hypoxic conditions give rise to the expression of genes encoding proteins which promote angiogenesis, invasion and metastasis, and enhanced glycolytic metabolism[41-45]. Major contributing factors to the cellular and systemic adaptation in response to hypoxic conditions are primarily the hypoxia-inducible factors (HIFs)[45,46]. HIF-1 is a transcription factor that regulates the induction of various genes facilitating adaptation and survival of cells in low oxygen conditions such as erythropoietin[47] vascular endothelial growth factor[48] glucose transporters, and glycolytic enzymes[49,50].
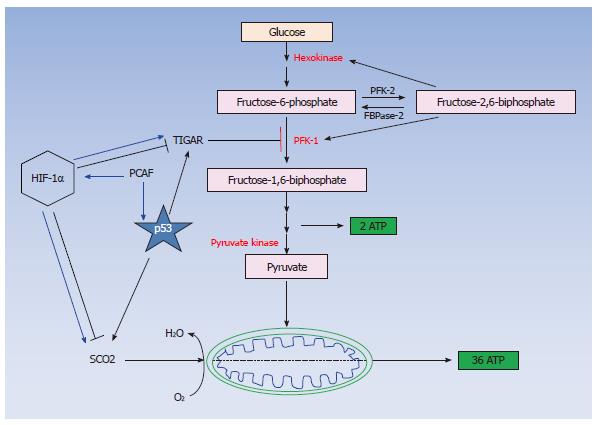
The functional crosstalk between HIF-1α and p53 pathways at several levels has been extensively studied[51-53] and indicated that under certain conditions p53 and HIF-1α co-operate in inducing apoptosis whereas they exert opposing functions in G1 cell cycle arrest[54,55]. p53 has also been shown to be stabilized in hypoxia mimicking conditions in a HIF-1α dependent manner[56] although its transcriptional activity is attenuated in hypoxia since it is incapable to induce the expression of its transcription targets including pro-apoptotic members of the Bcl-2 family under these conditions[57]. Although the molecular mechanisms involved have not yet been clearly elucidated, it appears that both p53 and HIF-1α regulate cellular energy production pathways by modulating the gene expression of glucose transporters and enzymes involved in glycolysis and oxidative phosphorylation. In particular, the glucose transporter GLUT-1 is downregulated by p53 and upregulated by HIF-1α[58-60] and similarly hexokinase 2 is upregulated by mutated p53[61,62], and induced by HIF-1α[63]. These contradicting observations are due at least in part to the differential interactions of p53 and HIF-1α with their common co-activators or co-repressors[64-67].
The p300/CBP transcriptional coactivator assembles a number of diverse cofactor proteins into multicomponent complexes[68] and is itself involved in the regulation of the transcriptional activity of both HIF-1 and p53[69,70]. The steroid receptor coactivator 1 is a component of the p300/CBP complex[71] and another common cofactor shared between HIF-1[72] and p53[73]. In addition, the nuclear receptor coactivator TIF2 interacts with HIF-1 to potentiate its transcriptional activity[74], although it inhibits p53 transcription potential when fused with the acetyltransferase MOZ associated with acute myeloid leukaemia[75].
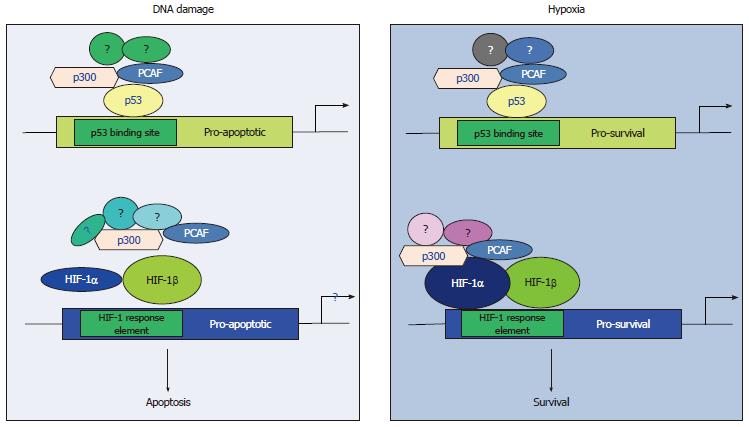
Our studies investigating the crosstalk between p53 and HIF-1α[64,65,76] have elucidated an additional molecular mechanism explaining the inability of p53 to activate its pro-apoptotic targets in hypoxia and implicate p300/CBP associated factor (PCAF) in the fine-tuning of the transcriptional activity and protein stability of both p53 and HIF-1α in DNA damage and hypoxic conditions. PCAF is a common cofactor for both p53 and HIF-1α[64,67] and is recruited to the transcriptional complex of the one or the other transcription factor in a tissue and type of stress dependent manner (Figure 1) determining the pathway of energy production (Figure 2) and the cellular fate under diverse stress conditions[64,65] providing an additional evidence for the importance of the co-activator function in determining the cell fate under hypoxia by modulating both p53 and HIF-1α responses.
The therapeutic activity of many anti-cancer agents depends on their ability to specifically and selectively induce apoptotic pathways in cancer cells. Radioactivity and chemotherapeutic drugs mediate their pro-apoptotic effects through the induction of pro-apoptotic pathways regulated by transcription factors such as the tumor suppressor protein p53. The tumor suppressor p53 signalling pathway is a highly regulated process involving a cascade of events, mediated among other pathways by various transcriptional co-factors such as the p300, and other p300 associated factors such as the tetratricopeptide domain 5 and PCAF[77,78]. These co-factors regulate the p53 transcriptional activity and protein stability by acetylating different lysine residues in its C-terminal region and in this way they contribute to the p53 mediated cellular adaptation to diverse types of stress[79,80]. In addition, it has become clear from the studies investigating the molecular mechanisms of the regulation of HIF-1α protein stability and transcriptional activity that p300 is required for the trans-activation of HIF-1α and that there is competition for limiting amounts of this cofactor in hypoxia between HIF-1α and p53[69,81-83].
Poor response or resistance to anti-cancer chemotherapeutics by hypoxic tumors has been evidenced and it is attributed to the lack of vascular system that would allow efficient drug delivery to these tumors[84]. Likewise, radiation therapy requires oxygen radicals for efficient production of DNA strand breaks, and thus hypoxic tumor microenvironment contributes to radioresistance[44,84]. Furthermore, repression of the p53 transcriptional activity and inability of this transcription factor to induce its pro-apoptotic targets in hypoxic conditions is an additional mechanism conferring drug resistance to hypoxic tumors[85].
Our observations have provided evidence supporting the view that distinct subpopulations of transcription co-activator complexes as well as differential posttranslational modifications determine the transcriptional target selectivity of both p53 and HIF-1α under diverse micro-environmental conditions[64,65,76] resulting in the expression of distinct subsets of genes, which carry out different functions, in a type of stress dependent manner. This distinction in the transcriptional cofactors’ function can be interpreted in a variety of ways. Firstly, transcription cofactors might facilitate the recruitment of different transcription factors to distinct regions of the genome[86] thus allowing different transcription factors to carry out specialised functions determining the cellular fate (survival or apoptosis) (Figure 1). Secondly, differences in the structure of the promoter between the different targets of various transcription factors could be responsible for preferential binding of particular subsets of these targets by alternatively posttranslationally modified transcription factors. For example, PCAF dependent acetylation of either p53 or HIF-1α is a mechanism by which these transcription factors distinguish between their pro-survival or pro-apoptotic target promoters[64] or glycolytic or oxidative phosphorylation inducers (Figure 2)[65,87-89].
To substantiate this hypothesis we are currently using genome wide ChIP-seq approaches to uncover the specific transcriptional circuitries that determine the specificity and target selectivity of several transcription factors including p53, glucocorticoid receptor, estrogen receptor, HIF-1α and NF-KB which play very important roles in carcinogenesis. The ultimate aim of this investigation is to acquire essential knowledge that will guide the identification of new transcriptional targets in the DNA damage response and low oxygen availability networks and thus facilitate the development of selective therapeutics for potential personalized cancer therapeutics.
P- Reviewer: Maluf SW S- Editor: Zhai HH L- Editor: A E- Editor: Zheng XM
| 1. | Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29:1305-1312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 477] [Cited by in F6Publishing: 416] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 2. | Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Genet Dev. 1998;8:140-146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 364] [Cited by in F6Publishing: 390] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46:43-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 159] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 4. | Li J, Lin Q, Yoon HG, Huang ZQ, Strahl BD, Allis CD, Wong J. Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. Mol Cell Biol. 2002;22:5688-5697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 108] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | Luo RX, Dean DC. Chromatin remodeling and transcriptional regulation. J Natl Cancer Inst. 1999;91:1288-1294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 92] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 6. | Mazzio EA, Soliman KF. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics. 2012;7:119-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 7. | Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6156] [Cited by in F6Publishing: 5912] [Article Influence: 246.3] [Reference Citation Analysis (0)] |
| 8. | Kim TM, Park PJ. Advances in analysis of transcriptional regulatory networks. Wiley Interdiscip Rev Syst Biol Med. 2011;3:21-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | van Steensel B. Chromatin: constructing the big picture. EMBO J. 2011;30:1885-1895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 105] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 10. | Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 282] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 11. | Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145-1155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 361] [Cited by in F6Publishing: 359] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 12. | Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761-772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 535] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 13. | Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237-1251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1049] [Cited by in F6Publishing: 936] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 14. | Lamhamedi-Cherradi SE, Zheng S, Hilliard BA, Xu L, Sun J, Alsheadat S, Liou HC, Chen YH. Transcriptional regulation of type I diabetes by NF-kappa B. J Immunol. 2003;171:4886-4892. [PubMed] [Cited in This Article: ] |
| 15. | Kuwahara K, Nakao K. New molecular mechanisms for cardiovascular disease: transcriptional pathways and novel therapeutic targets in heart failure. J Pharmacol Sci. 2011;116:337-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 16. | Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr Genomics. 2009;10:154-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 159] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 17. | Li G, Han N, Li Z, Lu Q. Identification of transcription regulatory relationships in rheumatoid arthritis and osteoarthritis. Clin Rheumatol. 2013;32:609-615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Yu H, Tu K, Wang YJ, Mao JZ, Xie L, Li YY, Li YX. Combinatorial network of transcriptional regulation and microRNA regulation in human cancer. BMC Syst Biol. 2012;6:61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 19. | Cummings DE, Schwartz MW. Genetics and pathophysiology of human obesity. Annu Rev Med. 2003;54:453-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 263] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 20. | Roy AK, Oh T, Rivera O, Mubiru J, Song CS, Chatterjee B. Impacts of transcriptional regulation on aging and senescence. Ageing Res Rev. 2002;1:367-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 63] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 21. | Kufe D, Weichselbaum R. Radiation therapy: activation for gene transcription and the development of genetic radiotherapy-therapeutic strategies in oncology. Cancer Biol Ther. 2003;2:326-329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 47] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Li X, Zhang J, Gao H, Vieth E, Bae KH, Zhang YP, Lee SJ, Raikwar S, Gardner TA, Hutchins GD. Transcriptional targeting modalities in breast cancer gene therapy using adenovirus vectors controlled by alpha-lactalbumin promoter. Mol Cancer Ther. 2005;4:1850-1859. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 23. | Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med. 2002;53:615-627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1854] [Cited by in F6Publishing: 1853] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 24. | Lynch JT, Rajendran R, Xenaki G, Berrou I, Demonacos C, Krstic-Demonacos M. The role of glucocorticoid receptor phosphorylation in Mcl-1 and NOXA gene expression. Mol Cancer. 2010;9:38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527-2532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 265] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 26. | Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 425] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 27. | Amundson SA, Myers TG, Fornace AJ. Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene. 1998;17:3287-3299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 283] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 28. | Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077-4085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 810] [Cited by in F6Publishing: 1011] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 29. | Feng Z, Lin M, Wu R. The Regulation of Aging and Longevity: A New and Complex Role of p53. Genes Cancer. 2011;2:443-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 30. | Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013;Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 471] [Cited by in F6Publishing: 729] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 31. | Contreras AU, Mebratu Y, Delgado M, Montano G, Hu CA, Ryter SW, Choi AM, Lin Y, Xiang J, Chand H. Deacetylation of p53 induces autophagy by suppressing Bmf expression. J Cell Biol. 2013;201:427-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Meek DW. The p53 response to DNA damage. DNA Repair (Amst). 2004;3:1049-1056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 241] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 33. | Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotechnol. 2012;2012:170325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 174] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 34. | Ma W, Sung HJ, Park JY, Matoba S, Hwang PM. A pivotal role for p53: balancing aerobic respiration and glycolysis. J Bioenerg Biomembr. 2007;39:243-246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 35. | Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1046] [Cited by in F6Publishing: 1276] [Article Influence: 91.1] [Reference Citation Analysis (0)] |
| 36. | Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 459] [Cited by in F6Publishing: 470] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 37. | Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413-431. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2178] [Cited by in F6Publishing: 2306] [Article Influence: 153.7] [Reference Citation Analysis (0)] |
| 38. | Benchimol S. p53-dependent pathways of apoptosis. Cell Death Differ. 2001;8:1049-1051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 143] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 39. | Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 385] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 40. | Subarsky P, Hill RP. The hypoxic tumour microenvironment and metastatic progression. Clin Exp Metastasis. 2003;20:237-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 284] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 41. | Semenza GL. Angiogenesis in ischemic and neoplastic disorders. Annu Rev Med. 2003;54:17-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 292] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 42. | Dachs GU, Chaplin DJ. Microenvironmental control of gene expression: implications for tumor angiogenesis, progression, and metastasis. Semin Radiat Oncol. 1998;8:208-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 79] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 43. | Le QT, Denko NC, Giaccia AJ. Hypoxic gene expression and metastasis. Cancer Metastasis Rev. 2004;23:293-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 183] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 44. | Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 822] [Cited by in F6Publishing: 808] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 45. | Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587-4602. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 328] [Cited by in F6Publishing: 344] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 46. | Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev. 1998;8:588-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 814] [Cited by in F6Publishing: 845] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 47. | Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271:32529-32537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1226] [Cited by in F6Publishing: 1280] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 48. | Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604-4613. [PubMed] [Cited in This Article: ] |
| 49. | Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct Cis-acting sequences. J Biol Chem. 1995;270:29083-29089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 384] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 50. | Firth JD, Ebert BL, Pugh CW, Ratcliffe PJ. Oxygen-regulated control elements in the phosphoglycerate kinase 1 and lactate dehydrogenase A genes: similarities with the erythropoietin 3’ enhancer. Proc Natl Acad Sci USA. 1994;91:6496-6500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 386] [Cited by in F6Publishing: 417] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 51. | Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14:34-44. [PubMed] [Cited in This Article: ] |
| 52. | Blagosklonny MV, An WG, Romanova LY, Trepel J, Fojo T, Neckers L. p53 inhibits hypoxia-inducible factor-stimulated transcription. J Biol Chem. 1998;273:11995-11998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 215] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 53. | Safronova O, Morita I. Transcriptome remodeling in hypoxic inflammation. J Dent Res. 2010;89:430-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 54. | Goda N, Dozier SJ, Johnson RS. HIF-1 in cell cycle regulation, apoptosis, and tumor progression. Antioxid Redox Signal. 2003;5:467-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 90] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 55. | Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448-2460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 428] [Cited by in F6Publishing: 410] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 56. | An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 611] [Cited by in F6Publishing: 620] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 57. | Zhao Y, Chen XQ, Du JZ. Cellular adaptation to hypoxia and p53 transcription regulation. J Zhejiang Univ Sci B. 2009;10:404-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519-9525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 512] [Cited by in F6Publishing: 543] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 59. | Chen Z, Lu W, Garcia-Prieto C, Huang P. The Warburg effect and its cancer therapeutic implications. J Bioenerg Biomembr. 2007;39:267-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 228] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 60. | Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627-2633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 468] [Cited by in F6Publishing: 525] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 61. | Mathupala SP, Heese C, Pedersen PL. Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J Biol Chem. 1997;272:22776-22780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 182] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 62. | Shen L, Sun X, Fu Z, Yang G, Li J, Yao L. The fundamental role of the p53 pathway in tumor metabolism and its implication in tumor therapy. Clin Cancer Res. 2012;18:1561-1567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 63. | Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381-7393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 432] [Cited by in F6Publishing: 475] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 64. | Xenaki G, Ontikatze T, Rajendran R, Stratford IJ, Dive C, Krstic-Demonacos M, Demonacos C. PCAF is an HIF-1alpha cofactor that regulates p53 transcriptional activity in hypoxia. Oncogene. 2008;27:5785-5796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 65. | Rajendran R, Garva R, Ashour H, Leung T, Stratford I, Krstic-Demonacos M, Demonacos C. Acetylation mediated by the p300/CBP-associated factor determines cellular energy metabolic pathways in cancer. Int J Oncol. 2013;42:1961-1972. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 66. | Melvin A, Rocha S. Chromatin as an oxygen sensor and active player in the hypoxia response. Cell Signal. 2012;24:35-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 67. | Lim JH, Lee YM, Chun YS, Chen J, Kim JE, Park JW. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864-878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 556] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 68. | Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 693] [Cited by in F6Publishing: 682] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 69. | Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858-861. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1118] [Cited by in F6Publishing: 1100] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 70. | Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202-1209. [PubMed] [Cited in This Article: ] |
| 71. | Yao TP, Ku G, Zhou N, Scully R, Livingston DM. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626-10631. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 333] [Cited by in F6Publishing: 352] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 72. | Ruas JL, Poellinger L, Pereira T. Role of CBP in regulating HIF-1-mediated activation of transcription. J Cell Sci. 2005;118:301-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 76] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 73. | Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 361] [Cited by in F6Publishing: 390] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 74. | Liu W, Shen SM, Zhao XY, Chen GQ. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int J Biochem Mol Biol. 2012;3:165-178. [PubMed] [Cited in This Article: ] |
| 75. | Kindle KB, Troke PJ, Collins HM, Matsuda S, Bossi D, Bellodi C, Kalkhoven E, Salomoni P, Pelicci PG, Minucci S. MOZ-TIF2 inhibits transcription by nuclear receptors and p53 by impairment of CBP function. Mol Cell Biol. 2005;25:988-1002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 76. | Andreou K, Rajendran R, Krstic-Demonacos M, Demonacos C. Regulation of CXCR4 gene expression in breast cancer cells under diverse stress conditions. Int J Oncol. 2012;41:2253-2259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 77. | Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1617] [Cited by in F6Publishing: 1606] [Article Influence: 94.5] [Reference Citation Analysis (0)] |
| 78. | Demonacos C, Krstic-Demonacos M, Smith L, Xu D, O’Connor DP, Jansson M, La Thangue NB. A new effector pathway links ATM kinase with the DNA damage response. Nat Cell Biol. 2004;6:968-976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 79. | Grossman SR. p300/CBP/p53 interaction and regulation of the p53 response. Eur J Biochem. 2001;268:2773-2778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 185] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 80. | Demonacos C, Krstic-Demonacos M, La Thangue NB. A TPR motif cofactor contributes to p300 activity in the p53 response. Mol Cell. 2001;8:71-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 60] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 81. | Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002;111:709-720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 544] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 82. | Seo HW, Kim EJ, Na H, Lee MO. Transcriptional activation of hypoxia-inducible factor-1alpha by HDAC4 and HDAC5 involves differential recruitment of p300 and FIH-1. FEBS Lett. 2009;583:55-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 83. | Lawless MW, O’Byrne KJ, Gray SG. Oxidative stress induced lung cancer and COPD: opportunities for epigenetic therapy. J Cell Mol Med. 2009;13:2800-2821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 84. | Brown JM. Tumor hypoxia in cancer therapy. Methods Enzymol. 2007;435:297-321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 177] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 85. | Sermeus A, Michiels C. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2011;2:e164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 204] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 86. | Pan Y, Nussinov R. Lysine120 interactions with p53 response elements can allosterically direct p53 organization. PLoS Comput Biol. 2010;6:e1000878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 87. | Chen S, Sang N. Histone deacetylase inhibitors: the epigenetic therapeutics that repress hypoxia-inducible factors. J Biomed Biotechnol. 2011;2011:197946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 88. | Chen R, Xu M, Hogg RT, Li J, Little B, Gerard RD, Garcia JA. The acetylase/deacetylase couple CREB-binding protein/Sirtuin 1 controls hypoxia-inducible factor 2 signaling. J Biol Chem. 2012;287:30800-30811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 89. | Mortuza R, Chen S, Feng B, Sen S, Chakrabarti S. High glucose induced alteration of SIRTs in endothelial cells causes rapid aging in a p300 and FOXO regulated pathway. PLoS One. 2013;8:e54514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 148] [Article Influence: 13.5] [Reference Citation Analysis (0)] |