Published online Feb 24, 2022. doi: 10.5306/wjco.v13.i2.135
Peer-review started: April 5, 2021
First decision: July 6, 2021
Revised: July 11, 2021
Accepted: January 13, 2022
Article in press: January 13, 2022
Published online: February 24, 2022
Breast cancer is the most common female cancer and a major cause of morbidity and mortality. Progress in breast cancer therapeutics has been attained with the introduction of targeted therapies for specific sub-sets. However, other subsets lack targeted interventions and thus there is persisting need for identification and characterization of molecular targets in order to advance breast cancer thera
To analyze the role of lesions in neurotrophic receptor tyrosine kinase (NTRK) genes in breast cancers.
Analysis of publicly available genomic breast cancer datasets was performed for identification and characterization of cases with fusions and other molecular abnormalities involving NTRK1, NTRK2 and NTRK3 genes.
NTRK fusions are present in a small number of breast cancers at the extensive GENIE project data set which contains more than 10000 breast cancers. These cases are not identified as secretory in the database, suggesting that the histologic characterization is not always evident. In the breast cancer The Cancer Genome Atlas (TCGA) cohort the more common molecular lesion in NTRK genes is amplification of NTRK1 observed in 7.9% of breast cancers.
Neurotrophin receptors molecular lesions other than fusions are observed more often than fusions. However, currently available NTRK inhibitors are effective mainly for fusion lesions. Amplifications of NTRK1, being more frequent in breast cancers, could be a viable therapeutic target if inhibitors efficacious for them become available.
Core Tip: Molecular lesions in neurotrophic receptor tyrosine kinase (NTRK) receptors have been brought to the forefront of cancer therapy with the introduction of specific inhibitors which are effective in cancers with fusions involving the family of receptors. In breast cancer fusions involving the NTRK receptors are rarely seen and concern exclusively the secretory sub-type. In non-secretory breast carcinomas amplifications are observed in a minority of cases.
- Citation: Stravodimou A, Voutsadakis IA. Neurotrophic receptor tyrosine kinase family members in secretory and non-secretory breast carcinomas. World J Clin Oncol 2022; 13(2): 135-146
- URL: https://www.wjgnet.com/2218-4333/full/v13/i2/135.htm
- DOI: https://dx.doi.org/10.5306/wjco.v13.i2.135
Breast cancer is the most common neoplasm in women and a significant cause of morbidity and mortality. In United States alone, an estimated 284000 cases of breast cancer will occur in 2021, with more than 44000 resulting deaths[1]. Breast cancers represent about 30% of all cancers diagnosed in women. Despite recent advances in breast cancer therapies produced by an improved understanding of molecular pathogenesis, the disease remains difficult to treat when metastatic. It is currently well established that breast cancer does not represent a single entity, but several sub-types exist with implications for prognosis and treatment[2]. The most frequent sub-type is estrogen receptor (ER) positive and negative for the epidermal growth factor receptor (EGFR) family receptor 2, known as human EGFR receptor 2 (HER2) and is further divided in a more indolent disease corresponding with the genomic luminal A sub-type and a more aggressive, less estrogen-dependent disease aligning with the genomic luminal B classification[3]. Another breast cancer sub-type is positive for the HER2 receptor, while the triple negative sub-type is negative for both ER and HER2, as well as the receptor for Progesterone, PR, which is positive in most ER positive cases. Treatments for each sub-type has evolved to differ significantly and therapeutic decisions in breast cancer depend on the sub-type. HER2 positive cancers, for example, are treated with monoclonal antibodies, small molecule kinase inhibitors and antibody drug conjugates targeting HER2[4].
Several other receptor tyrosine kinases have been implicated in cancer pathogenesis and progression and treatments targeting some of them exist. These include the angiogenesis kinase receptor vascular endothelial growth factor receptor (VEGFR) and its ligand VEGF, EGFR, fibroblast growth factor receptor (FGFR) and c-Met, the receptor for hepatocyte growth factor (HGF). In breast cancer, besides HER2, no receptor tyrosine kinase inhibitors have been approved. Recently, inhibitors of the neurotrophic kinase family of tyrosine kinase receptors have become available and confirmed to be effective in cancers with fusions involving these receptors[5]. These fusions are observed in the majority of rare histologic types of cancers such as mammary analogue secretory carcinomas of the salivary glands and secretory breast cancers but are exceedingly rare in more common histologic subtypes of cancers[6]. The current analysis examines fusions and other molecular lesions of neurotrophin receptors in secretory and non-secretory carcinomas of the breast using publicly available genomic data.
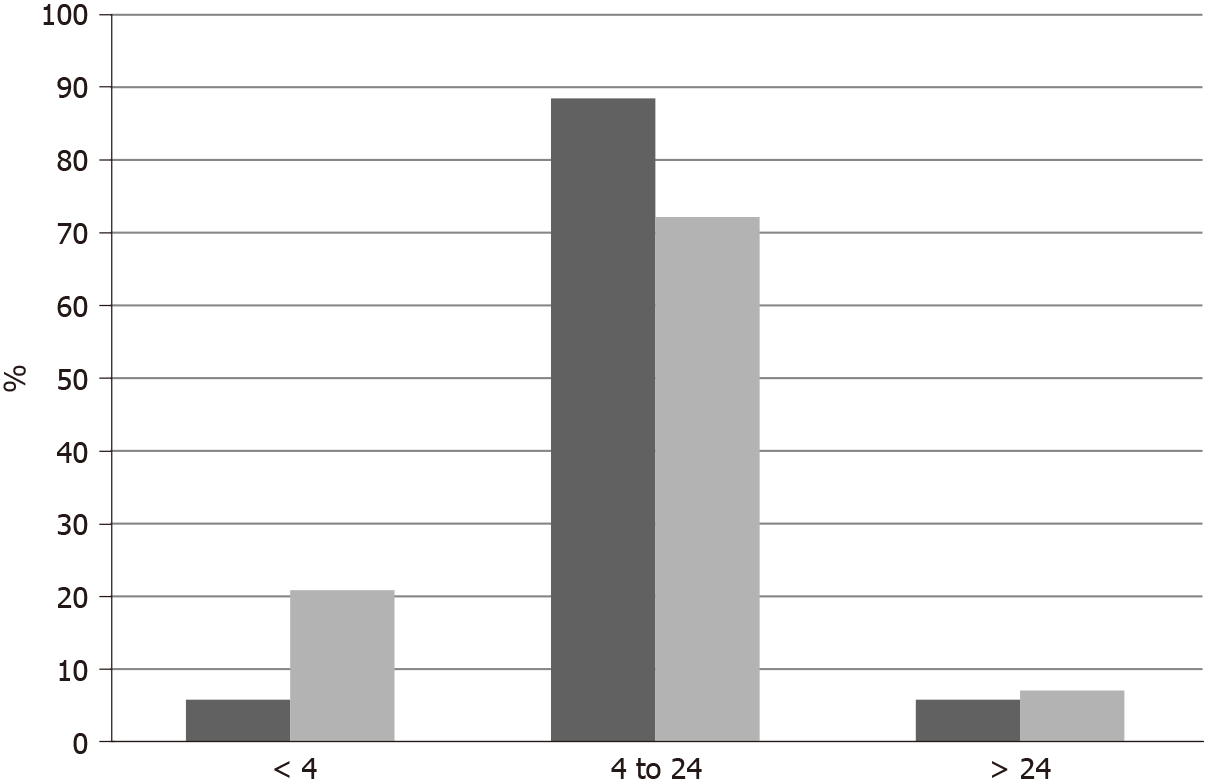
Published genomic studies of molecular lesions in neurotrophic receptor tyrosine kinases (NTRKs) in breast cancer were interrogated in the cBioportal platform (http://www.cbioportal.org). cBioportal is a platform freely available to investigators containing molecular studies and corresponding clinical data[7,8]. The platform allows for a multi-dimensional interrogation of genomic data from publicly available studies. cBioportal also provides the opportunity to associate data of molecular lesions (mutations, fusions, copy number alterations and mRNA hyperexpression or hypo-expression) of any gene of interest in studies from the Cancer Genome Atlas (TCGA) and other studies with patient clinical characteristics and survival outcomes[7]. The current analysis is based on the TCGA breast cancer study and on the project GENIE study, both included in the cBioportal and providing open access to their data[9,10]. The analysis of copy number alterations (CNAs) in TCGA is performed with the algorithm Genomic Identification of Significant Targets in Cancer (GISTIC) [11]. In GISTIC, putative amplification of a given gene is defined as a score of 2 or above. TCGA provides an aneuploidy score (AS) as a measure of chromosomal instability of each sample. AS is calculated as the sum of the number of chromosome arms in each sample that have copy number alterations (gains or losses). A chromosome arm is considered copy number altered, either gained or lost, if there is a somatic copy number alteration in more than 80% of the length of the arm as calculated by an algorithm called ABSOLUTE based on Affymetrix 6.0 SNP arrays[12]. Chromosomal arms with somatic copy number alterations in 20% to 80% of the arm length are considered not evaluable and chromosomal arms with somatic copy number alterations in less than 20% of the arm length are considered not altered. mRNA expression grids in cBioportal are constructed and normalized using the RSEM (RNA-Seq by Expectation Maximization) algorithm[13].
Expression of NTRK proteins in breast cancer were evaluated using publicly available data from the Human Protein Atlas (www.proteinatlas.org), a database of protein expressions in human normal and neoplastic tissues[14]. The Human Protein Atlas contains a semi-quantitative immunohistochemistry-based evaluation of the expression of proteins of interest.
The effect of mRNA expression level of NTRK1 gene on survival of breast cancer patients was examined with data derived from the online publicly available platform Kaplan Meier Plotter
The Fisher’s exact test or the χ2 test and the t test, respectively, are used to compare categorical and continuous data. Kaplan-Meier survival curves were compared using the Log Rank test. All statistical comparisons were considered significant if P < 0.05.
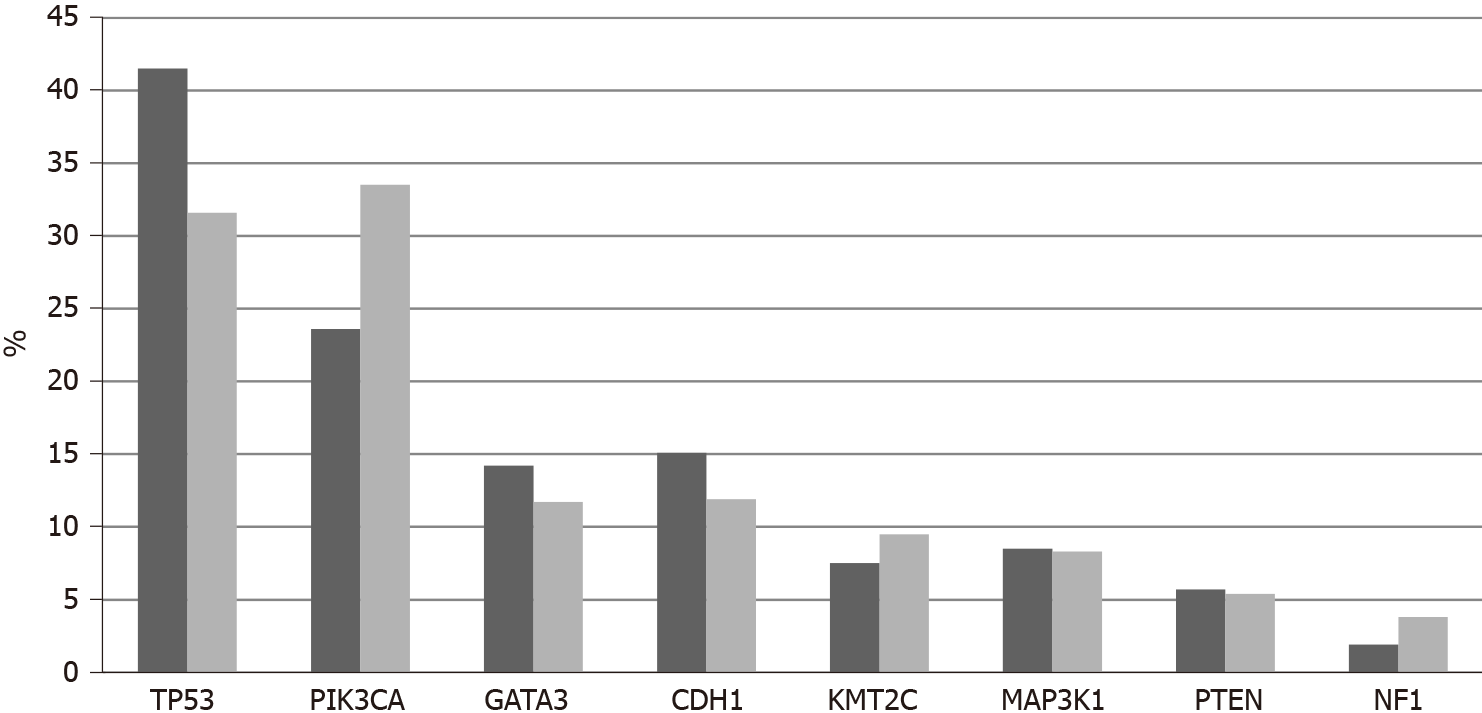
Among 11,886 patients with breast cancer in the project GENIE cohort, 27 patients (0.22%) had fusions in one of the three NTRK genes. Two male patients were included among the 27 NTRK fusion positive breast cancer patients. Most patients with information for race were White, while 2 patients were Black and one was Asian. Histologically, 25 patients were diagnosed with ductal carcinomas or breast carcinomas not otherwise specified and one patient had an invasive lobular carcinoma. Interestingly, only one case was diagnosed as juvenile secretory carcinoma, in a male patient. A total of 32 fusions involving the 3 NTRK genes were present in the 27 patients (5 patients had more than one different fusions). The most frequent fusions involved NTRK3, in 16 samples, followed by NTRK1 in 13 samples and NTRK2 in 3 samples. Fusions in seven cases were intragenic and a variety of partners were involved in other fusions. Recurring partner genes include ETV6 with NTRK3 and LMNA with NTRK1. Frequent mutations observed in breast cancers such as in TP53, PIK3CA and GATA3 genes are also observed in cases with NTRK gene fusions. with a frequency not statistically significant different compared with cases without NTRK fusions. Similarly, common amplifications in breast cancers of CCND1 at 11q13.3, ERBB2 at 17q12, NSD3 at 8p11.23 and c-MYC at 8q24.21 are encountered in cases with NTRK fusions in frequencies comparable to cancers without NTRK fusions.
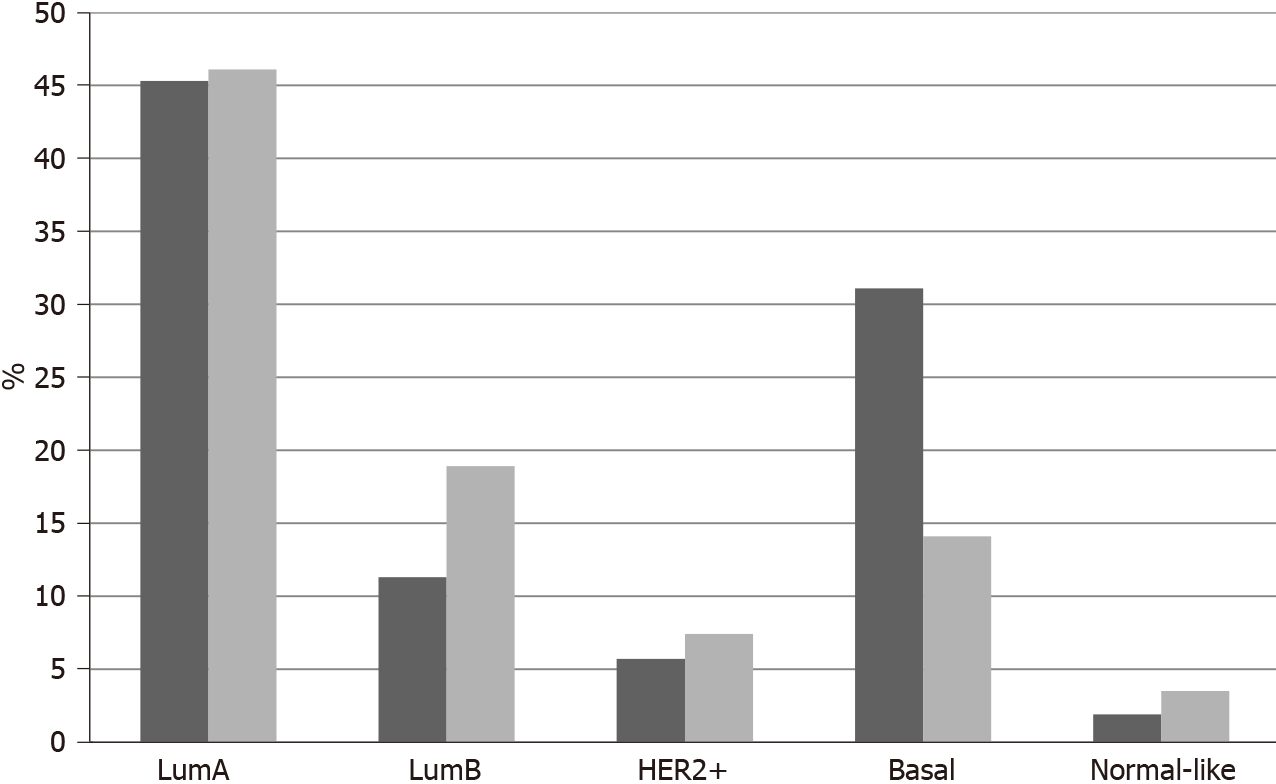
In TCGA breast cancer cohort, no fusions involving the 3 NTRK genes were observed. The most common molecular lesions were amplifications that were observed in 9.8% of patients, most commonly in NTRK1 (7.9%) and more rarely in NTRK3 (1.9%) and in NTRK2 (0.2%). NTRK amplified cancers have a distribution of histologic types (ductal, lobular, mixed, other) that is similar to non-amplified cases. NTRK amplified cancers are basal more commonly than NTRK non-amplified cancers (31.1% vs 14.1%,
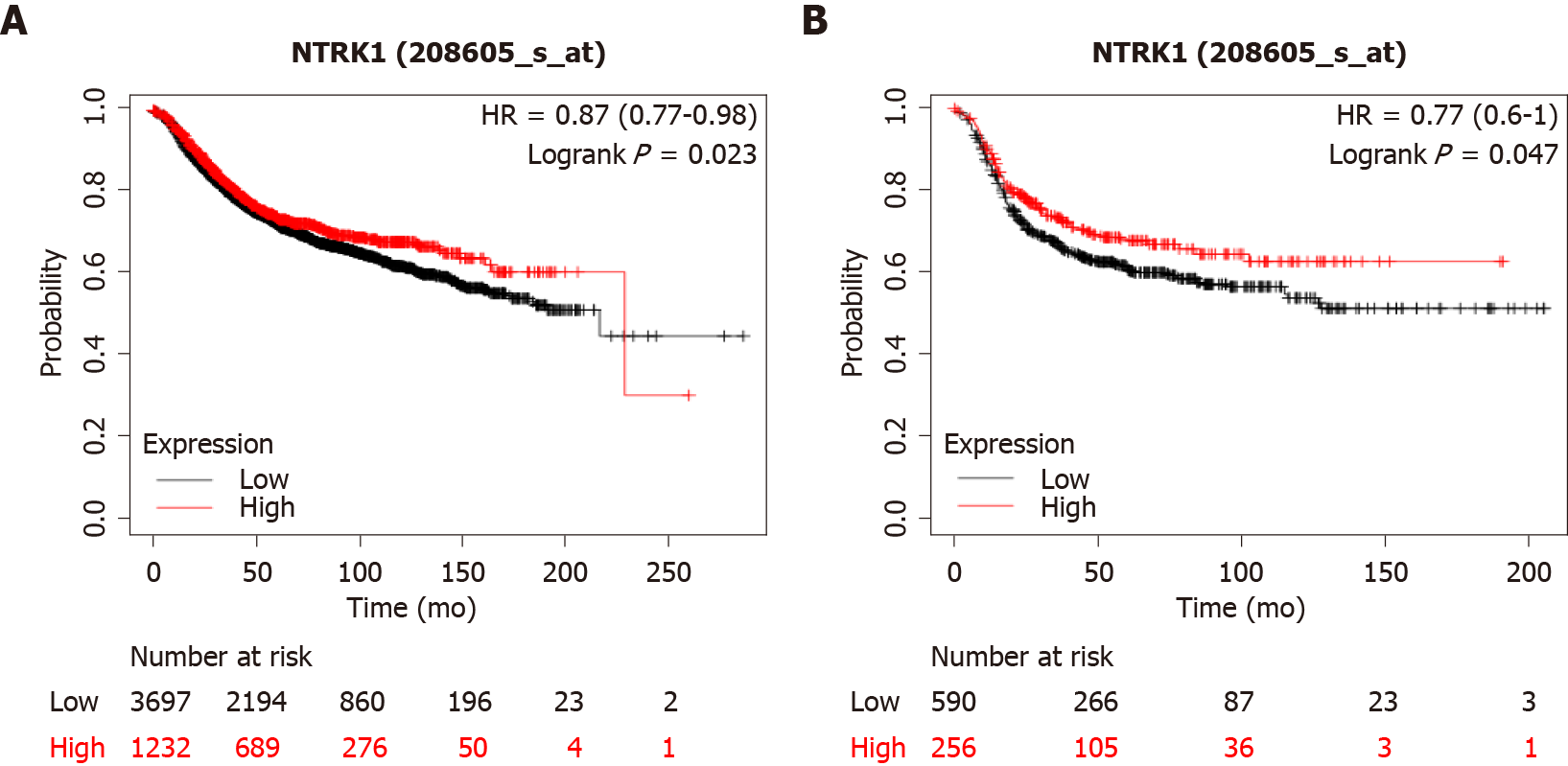
In breast cancer cases with amplifications of NTRK genes, expression of the respective mRNAs is not increased, except in rare cases. However, breast cancers with increased NTRK1 mRNA expression at the upper quartile tend to have a better overall survival (OS) than cancers with low NTRK1 mRNA expression, falling in the three lower quartiles (Figure 4A). This holds true also for an analysis restricted to basal breast cancers (Figure 4B).
Protein expression of TrkA is present in most breast cancers as evaluated by immunohistochemistry. The Human Protein Atlas has found moderate to high expression of the protein in 72.7% (8 of 11 samples) and 63.6% (7 of 11 samples) using two different antibodies (HPA035799 rabbit polyclonal antibody from Sigma-Aldrich and CAB004606 mouse monoclonal antibody from Santa Cruz Biotechnology). One and three additional cases have shown a low intensity of staining. In contrast, almost all cases of breast cancer examined showed low or absent staining for TrkB and TrkC in the Human Protein Atlas.
The human neurotrophin system consists of four ligands, nerve growth factor (NGF), brain derived neurotrophic factor (BDNF), neurotrophin 3 (NT-3) and neurotrophin 4/5 (NT-4/5), and three Trk receptors, TrkA, TrkB, TrkC, as well as two additional non-Trk receptors, p75NTR and sortilin[16]. NGF is the ligand for receptor TrkA, encoded by gene NTRK1, BDNF and NT-4/5 are the ligands for TrkB encoded by gene NTRK2 and NT-5 is the ligand for TrkC, encoded by gene NTRK3. NT-5 can also ligate the two other Trk receptors. All four ligands ligate the common neurotrophin receptor p75NTR, which also serves as a receptor for the precursor forms of the four ligands pro-NGF, pro-BDNF, pro-NT-3 and pro-NT-4/5 and possesses an intracellular death domain[17]. All precursor forms ligate, additionally, sortilin, also known as neurotensin receptor 3 (NTSR3). The three Trk kinase receptors have a common structure with an extracellular part consisting of two cysteine-rich domains separated by three leucine-rich repeats. Closer to the cell membrane, the three Trk receptors possess two immunoglobulin-like domains. The intracellular part of the receptors holds the tyrosine kinase domain with five conserved tyrosines, three of which are phosphorylated when the receptors are activated by ligand binding. Downstream of receptors activation, the MAPK and PI3K/Akt kinase cascades are activated transmitting signals of proliferation and apoptosis inhibition. The physiologic role of neurotrophins signaling is best described in the nervous system, both in peripheral nervous system and the brain[18]. In peripheral nervous system, NGF signaling stimulates neurite growth and prevents apoptosis. NGF is also functional in brain where BDNF is also present and involved in signal transduction through binding and induction of phosphorylation of TrkB resulting in neuron survival and differentiation[18,19].
Similarities of downstream signaling between the neurotrophin receptors and the EGFR family of tyrosine kinase receptors, which are well-known oncogenes, have led to the exploration of the role of Trk receptors in cancer[18,20]. Expression of receptor TrkA is observed in breast cancer cell lines and in breast cancer tissues, where it is more prominent than surrounding normal breast epithelium[21]. TrkA signaling enhances breast cancer cell proliferation and anchorage independent growth. Moreover, TrkA inhibits apoptosis and anoikis in metastatic sites. In breast cancer cells, activation of TrkA transmits signals through the MAPK pathway consistent with the physiologic circuit of the receptor[18]. Pro-NGF derived from cancer cells also binds TrkA and signals in an autocrine manner[22]. TrkA ligation promotes interaction with another receptor kinase EphA2 and downstream activation of Src and Akt kinases, resulting in phosphorylation and activation of these kinases and stimulation of cell growth[22]. In contrast, pharmacologic or small interfering RNA inhibition of TrkA or EphA2 decreases tumor growth and metastases of breast cancer in an in vivo mouse model. Immunohistochemical studies showed that Pro-NGF is expressed at higher levels in breast cancer tissues compared with normal breast tissues and benign lesions[23]. Receptor p75NTR is also expressed in breast cancer cells and leads to increased proliferation and inhibition of apoptosis through activation of transcription factor NF-κB[24]. p75NTR interacts with NF-κB through mediator TRADD (TNFR Associated Death Domain) when MCF-7 breast cancer cells are exposed to NGF and absence of TRADD promotes apoptosis in the same conditions[24].
BDNF and its receptor TrkB are expressed in breast cancer cells and promote cell survival and proliferation[25,26]. Knock down of BDNF or pharmacologic inhibition of TrkB reduced the viability of breast cancer cells in vitro. TrkB is expressed in cancer stem cells with the CD44 positive phenotype derived from triple negative breast cancers and participates in a paracrine circuit ligated by BDNF derived from differentiated breast cancer cells[27]. BDNF stimulation induces stemness core factor KLF4 and promotes stemness phenotype and stem cell renewal. In addition, TrkB expression predicts relapse of localized triple negative breast cancers[27]. Related to stemness, epithelial to mesenchymal transition (EMT) is a process that promotes cancer cell metastasis[28]. TrkB is a target of micro-RNA miR-200c, which protects against EMT and preserves breast cancer cell epithelial phenotype, further supporting the role of TrkB in breast cancer promotion[29].
Further interest in neurotrophin family receptors in cancer was stimulated by the discovery of rare fusions involving the receptor genes NTRK1, NTRK2 and NTRK3 that act as oncogenic drivers and can be neutralized by NTRK inhibitors[30]. Fusions involving most frequently NTRK3 were confirmed to be the underlying molecular mechanism of secretory mammary carcinomas but were absent in other more common breast cancer histologies[30,31]. The carcinogenic mechanism of the fusion involves deregulated activation of the RAS-MAPK and PI3K-Akt pathways that are the physiologic targets of neurotrophic kinases signaling[32]. In contrast, these fusions do not affect the differentiation programs of cells, as suggested by their presence in diverse cancers, in addition to secretory breast carcinomas, including nephromas, infant fibrosarcomas, mammary analogue secretory carcinomas of the salivary glands and acute myeloblastic leukemias[33,34].
Breast secretory carcinoma is a rare type of breast cancer malignancy, accounting for 0.15% to 0.2% of all breast cancers[35]. It has generally an indolent clinical behavior, with late local recurrence and prolonged survival, even with lymph node involvement, that occurs at nearly 30% of cases[36]. It was initially described by Mc Divitt and Stewart as a variation of mammary carcinoma in the pediatric population, also called “juvenile carcinoma”[37]. In 1980, Tavassoli and Norris[38] reported several cases presenting at variable ages and recommended to replace the previous definition with the term “secretory carcinoma”. Secretory breast cancer displays a particular phenotype related to the presence of large amounts of intracellular and extracellular secretory material. Although it has been described as a sub-family of basal-like triple negative breast cancers with cytokeratin 5/6 expression and no expression of either hormone receptors or HER2, this tumor has a unique immunophenotypic profile among triple negative breast carcinomas. It stains diffusely for mammaglobin and gross cystic disease fluid protein (GCDFP) and displays strong and diffuse SOX10 and S-100 Labeling[39,40]. The hallmark genetic alteration of this cancer is a t(12;15) translocation, resulting in the ETV6-NTRK3 gene fusion which is pathognomonic, detected in more than 90% of secretory breast carcinomas. The same translocation is also identified in the mammary analog secretory carcinoma, its salivary gland counterpart[30,39,41]. Given that NTRK3 protein expression is not detected in breast epithelial cells physiologically, immunohistochemical analysis using pan-Trk antibodies has been shown to constitute a sensitive and specific approach for the detection of NTRK3 rearrangements in breast secretory carcinomas[42]. Absence of Trk receptor expression in non-secretory breast cancer was also observed in a micro-array study of 339 breast cancer patients, using a pan-Trk rabbit monoclonal antibody[43]. However, since TrkA is expressed in a sub-set of non-secretory breast cancers, as shown in the human Protein Atlas, the intensity of staining should be taken into consideration and confirmatory molecular detection of fusions should be standard practice.
Patients with tumors that harbor NTRK fusions as a specific molecular alteration may benefit from treatment with selective tyrosine kinase inhibitors or antagonistic monoclonal antibodies. There is accumulating evidence for the efficacy of Trk inhibitors in the control of the disease in these patients. Two Trk inhibitors, larotrectinib and entrectinib, are both approved by the American Food and Drug Administration (FDA) and the European Medicines Agency (EMA). Larotrectinib is a selective inhibitor of the Trk proteins (including TrkA, TrkB, and TrkC) approved for the treatment of locally advanced and metastatic solid tumors with NTRK fusions, in both adult and pediatric cancer patients. Its efficacy and safety have been studied in three multicenter, phase 1/2, open-label, single-arm clinical trials[44-46]. An objective response rate (ORR) of 78% [95% confidence interval (CI): 71%-84%] regardless of histology, age, and type of NTRK fusion was shown in the joint analysis of these studies. The median progression-free survival (PFS) was 36.8 months [95%CI: 25.7 mo - not estimated (NE)] with 90% (95%CI: 75%-90%) of patients being alive at 1 year. A specific adult population analysis, that included 5% with breast cancers, has showed an ORR of 71% (95%CI: 62%-79%), a median PFS of 25.8 mo (15.2 mo to NE), and 87% of patients alive at 1 year. The most frequent fusion transcripts were NTRK3 (54%) and NTRK1 (43%), and only 3% had NTRK2[47]. Impressive responses have also been observed in locally advanced disease highlighting the potential utility of Trk inhibition as neoadjuvant therapy in the non-metastatic setting. The most common side effects were fatigue (30%), constipation (27%), cough (27%), elevation of liver enzymes (24%), dizziness (25%) and nausea (24%), mainly grade 1-2. Only 2% of patients had to discontinue treatment and 16% had grade 3-4 toxicity related to treatment, predominantly elevation of transaminases (3%), decreased neutrophil count (2%) and anemia (2%).
Entrectinib, a multikinase inhibitor of the TrkA, TrkB, TrkC, ROS1, and ALK proteins was tested in three clinical trials, two phase 1 and one "basket" phase 2 trial in the adult population with tumors harboring NTRK fusions[48]. Breast cancer represented the 11% of the tumors in these trials. In the analysis of the first 54 patients included, an ORR of 59% (95%CI: 45%-72%), a PFS of 11.2 mo (95%CI: 8.0-14.9 mo), and an OS of 23.9 mo (95%CI: 16.8 mo to NE) were observed. Entrectinib crosses the blood-brain barrier and has showed activity at the CNS level. In the 22% of patients with brain metastases the intracranial ORR was 55% (95%CI: 23%-83%)[48,49]. The activity of Trk inhibitors in the CNS is crucial because cancers that harbor NTRK fusions can present with primary or metastatic intracranial disease. The tolerability of the inhibitors was acceptable with the most frequent adverse events being anemia (10%), weight gain (14%), dyspnea, and asthenia (25%)[50,51]. The majority of adverse events encountered were grade 1-2. These side effects are manageable with drug dose modifications.
Despite the marked efficacy of Trk inhibitors and, in many cases, long-lasting responses, resistance may occur leading to progression. Several potential mechanisms of resistance have been described, including through the development of mutations in the NTRK genes, mutations of mitogen-activated protein kinase (MAPK) pathway genes such as BRAF (V600E) and KRAS (G12D), and the amplification of MET[52]. Razavi et al[53] identified lesions in the MAPK pathway that were responsible for resistance to endocrine therapy. The same findings were supported by Ross et al[54], who aimed at characterizing kinase fusions within a large cohort of 4857 patients with advanced breast cancer. In total, 56% with fusion-positive breast cancers had a history of previous endocrine therapy and none of the fusion-positive breast cancer samples harbored ESR1 hotspot mutations. Two patients with acquired LMNA-NTRK1 fusions and metastatic disease received larotrectinib and demonstrated clinical benefit[54]. The kinase fusions, even if they are rare in breast cancer, they are enriched in hormone-resistant, metastatic carcinomas and mutually exclusive with ESR1 mutations. These data expand the spectrum of genetic alterations activating MAPK signaling that can substitute for ESR1 mutations in this setting. Thus, molecular testing in metastatic breast cancer at progression after endocrine therapy should include fusion testing, especially in case of absence of ESR1 hotspot alterations. Specific mutations in NTRK1 producing substitutions at position p.G595R and p.G667C have been described as associated with resistance to entrectinib in a patient with colon cancer bearing the LMNA-NTRK1 rearrangement[55].
Second-generation Trk inhibitors such as selitrectinib and repotrectinib, have been developed, and have shown activity in these patients, in both adult and pediatric populations, in the first in human dose escalation clinical trials[56,57]. Currently there are ongoing trials evaluation the safety and efficacy of these drugs in solid tumors. Inspired by the resistance of other kinases, dual blocking TRKs or combination inhibitors could be a new treatment approach. The inhibitor foretinib was able to inhibit xenografts with the LMNA-NTRK1 fusion that had become resistance to entrectinib after acquiring the above mentioned mutation p.G667C, suggesting that mutations producing resistance to first generation inhibitors may still be sensitive to second generation inhibitors in development[58].
Regarding NTRK gene amplification, it was shown to result in TRK overexpression in a sub-set of breast cancers[59]. Among the 1250 analyzed tumor specimens, NRTK amplification was detected in 2.2% cases, representing 14.8% of cases with TrkA protein overexpression. The efficacy of TRK inhibitors in tumors harboring NTRK gene amplification is not well characterized. These patients were not included in the trials of TRK inhibitors. Interestingly, two patients with NTRK gene amplification have been described in whom larotrectinib has marked a durable antitumor activity[60,61] suggesting that these therapies may have a role in a sub-set of amplified patients, who remain to be identified. Regarding patients with mutations as a primary abnormality in Trk proteins (as opposed to the discussed above secondary mutations in patients with fusions), TRK inhibitors have not shown activity[61]. It has been suggested that these mutations are not driver lesions in these cancers. However, it is possible that a subset of these mutations are indeed oncogenic[62]. In various hematologic malignancies including AML, ALL, CML and myelofibrosis, NTRK2 and NTRK3 mutations were observed in about 5% of patients and cells bearing some of them were inhibited by entrectinib in vitro[63].
The current study confirms the rarity of NTRK fusions in breast cancer with a prevalence rate of less than 1%. Amplifications of NTRK1 are more common and are associated with basal cancers and TP53 mutations which are most common in this breast cancer type. Although amplification of the NTRK1 gene does not always lead to mRNA over-expression, TrkA protein is expressed in many breast cancers and thus can be a therapeutic target. In contrast to our results, another study that examined expression of TrkA at the protein level suggested that the receptor is more often expressed in HER2 positive cancers compared to luminal and basal carcinomas[64]. This study also confirmed discrepancies between gene dosage and protein expression. Clearly, transcription and post-transcription regulations are critical for TrkA expression in breast cancer.
Breast cancer is a common female cancer. It constitutes a major cause of morbidity and mortality. Progress in breast cancer therapeutics has been attained with the introduction of targeted therapies for specific sub-sets. However, other breast cancer subsets lack targeted therapeutics.
Personalized therapies for breast cancer have the potential to improve outcomes. These therapies take consideration of specific molecular abnormalities that could be targeted for optimal results.
This article aims to analyze the landscape of neurotrophic receptor tyrosine kinase (NTRK) abnor
An analysis of publicly available genomic breast cancer datasets was performed for identification and characterization of cases with fusions and other molecular abnormalities involving NTRK1, NTRK2 and NTRK3 genes.
NTRK fusions are rare in breast cancers as a whole. They are present in a small number of breast cancers at the extensive GENIE project data set which contains more than 10000 breast cancers. These cases are not identified as secretory in the database.
NTRK lesions other than fusions are more common than fusions in breast cancers. However, confirmation of efficacy for the currently available inhibitors exist only for NTRK fusions.
Amplifications of NTRK receptor genes are more common than fusions in breast cancers. Inhibitors effective for interfering with the activity of amplified NTRK receptors could advance therapeutics in this subset of breast cancers.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hou L S-Editor: Gao CC L-Editor: A P-Editor: Gao CC
| 1. | Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8287] [Cited by in F6Publishing: 10043] [Article Influence: 3347.7] [Reference Citation Analysis (2)] |
| 2. | Russnes HG, Lingjærde OC, Børresen-Dale AL, Caldas C. Breast Cancer Molecular Stratification: From Intrinsic Subtypes to Integrative Clusters. Am J Pathol. 2017;187:2152-2162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 150] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 3. | Stravodimou A, Voutsadakis IA. The Future of ER+/HER2- Metastatic Breast Cancer Therapy: Beyond PI3K Inhibitors. Anticancer Res. 2020;40:4829-4841. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 4. | Voutsadakis IA. HER2 in stemness and epithelial-mesenchymal plasticity of breast cancer. Clin Transl Oncol. 2019;21:539-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Garrido P, Hladun R, de Álava E, Álvarez R, Bautista F, López-Ríos F, Colomer R, Rojo F. Multidisciplinary consensus on optimising the detection of NTRK gene alterations in tumours. Clin Transl Oncol. 2021;23:1529-1541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 6. | Perreault S, Chami R, Deyell RJ, El Demellawy D, Ellezam B, Jabado N, Morgenstern DA, Narendran A, Sorensen PHB, Wasserman JD, Yip S. Canadian Consensus for Biomarker Testing and Treatment of TRK Fusion Cancer in Pediatric Patients. Curr Oncol. 2021;28:346-366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 7. | Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8187] [Cited by in F6Publishing: 9923] [Article Influence: 902.1] [Reference Citation Analysis (0)] |
| 8. | Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685-D690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 805] [Cited by in F6Publishing: 747] [Article Influence: 53.4] [Reference Citation Analysis (0)] |
| 9. | Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8940] [Cited by in F6Publishing: 8654] [Article Influence: 721.2] [Reference Citation Analysis (0)] |
| 10. | Printz C. AACR releases large cancer genomic data set from project GENIE. Cancer. 2017;123:1685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 11. | Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007-20012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 836] [Cited by in F6Publishing: 800] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 12. | Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ; Cancer Genome Atlas Research Network, Cherniack AD, Beroukhim R, Meyerson M. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018;33:676-689.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 546] [Cited by in F6Publishing: 560] [Article Influence: 93.3] [Reference Citation Analysis (0)] |
| 13. | Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10832] [Cited by in F6Publishing: 12114] [Article Influence: 931.8] [Reference Citation Analysis (0)] |
| 14. | Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Pontén F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7696] [Cited by in F6Publishing: 8811] [Article Influence: 979.0] [Reference Citation Analysis (0)] |
| 15. | Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322-49333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 582] [Cited by in F6Publishing: 725] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 16. | Hondermarck H. Neurotrophins and their receptors in breast cancer. Cytokine Growth Factor Rev. 2012;23:357-365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Tajbakhsh A, Mokhtari-Zaer A, Rezaee M, Afzaljavan F, Rivandi M, Hassanian SM, Ferns GA, Pasdar A, Avan A. Therapeutic Potentials of BDNF/TrkB in Breast Cancer; Current Status and Perspectives. J Cell Biochem. 2017;118:2502-2515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 18. | Bradshaw RA, Chalkley RJ, Biarc J, Burlingame AL. Receptor tyrosine kinase signaling mechanisms: Devolving TrkA responses with phosphoproteomics. Adv Biol Regul. 2013;53:87-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609-642. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1840] [Cited by in F6Publishing: 1848] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 20. | Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2737] [Cited by in F6Publishing: 2704] [Article Influence: 150.2] [Reference Citation Analysis (0)] |
| 21. | Lagadec C, Meignan S, Adriaenssens E, Foveau B, Vanhecke E, Romon R, Toillon RA, Oxombre B, Hondermarck H, Le Bourhis X. TrkA overexpression enhances growth and metastasis of breast cancer cells. Oncogene. 2009;28:1960-1970. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 152] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 22. | Lévêque R, Corbet C, Aubert L, Guilbert M, Lagadec C, Adriaenssens E, Duval J, Finetti P, Birnbaum D, Magné N, Chopin V, Bertucci F, Le Bourhis X, Toillon RA. ProNGF increases breast tumor aggressiveness through functional association of TrkA with EphA2. Cancer Lett. 2019;449:196-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 23. | Demont Y, Corbet C, Page A, Ataman-Önal Y, Choquet-Kastylevsky G, Fliniaux I, Le Bourhis X, Toillon RA, Bradshaw RA, Hondermarck H. Pro-nerve growth factor induces autocrine stimulation of breast cancer cell invasion through tropomyosin-related kinase A (TrkA) and sortilin protein. J Biol Chem. 2012;287:1923-1931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 24. | El Yazidi-Belkoura I, Adriaenssens E, Dollé L, Descamps S, Hondermarck H. Tumor necrosis factor receptor-associated death domain protein is involved in the neurotrophin receptor-mediated antiapoptotic activity of nerve growth factor in breast cancer cells. J Biol Chem. 2003;278:16952-16956. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 58] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 25. | Yang X, Martin TA, Jiang WG. Biological influence of brain-derived neurotrophic factor on breast cancer cells. Int J Oncol. 2012;41:1541-1546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 26. | Cornelio DB, DE Farias CB, Prusch DS, Heinen TE, Dos Santos RP, Abujamra AL, Schwartsmann G, Roesler R. Influence of GRPR and BDNF/TrkB signaling on the viability of breast and gynecologic cancer cells. Mol Clin Oncol. 2013;1:148-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Yin B, Ma ZY, Zhou ZW, Gao WC, Du ZG, Zhao ZH, Li QQ. The TrkB+ cancer stem cells contribute to post-chemotherapy recurrence of triple-negative breast cancers in an orthotopic mouse model. Oncogene. 2015;34:761-770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 28. | Voutsadakis IA. The network of pluripotency, epithelial-mesenchymal transition, and prognosis of breast cancer. Breast Cancer (Dove Med Press). 2015;7:303-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Howe EN, Cochrane DR, Richer JK. Targets of miR-200c mediate suppression of cell motility and anoikis resistance. Breast Cancer Res. 2011;13:R45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 30. | Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367-376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 712] [Cited by in F6Publishing: 641] [Article Influence: 29.1] [Reference Citation Analysis (1)] |
| 31. | Kim J, Kim S, Ko S, In YH, Moon HG, Ahn SK, Kim MK, Lee M, Hwang JH, Ju YS, Kim JI, Noh DY, Park JH, Rhee H, Han W. Recurrent fusion transcripts detected by whole-transcriptome sequencing of 120 primary breast cancer samples. Genes Chromosomes Cancer. 2015;54:681-691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 32. | Tognon C, Garnett M, Kenward E, Kay R, Morrison K, Sorensen PH. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 2001;61:8909-8916. [PubMed] [Cited in This Article: ] |
| 33. | Euhus DM, Timmons CF, Tomlinson GE. ETV6-NTRK3--Trk-ing the primary event in human secretory breast cancer. Cancer Cell. 2002;2:347-348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 34. | Alves LDB, de Melo AC, Farinha TA, de Lima Araujo LH, Thiago LS, Dias FL, Antunes HS, Amaral Eisenberg AL, Santos Thuler LC, Cohen Goldemberg D. A systematic review of secretory carcinoma of the salivary gland: where are we? Oral Surg Oral Med Oral Pathol Oral Radiol. 2021;132:e143-e152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 35. | Lakhamni SR, Ellis IO, Schnitt SJ, Tan PH, van de Vijver MJ. WHO classification of the Breast. International Agency for Research on Cancer, Lyon, France 2012. [cited 23 February 2021]. In: International Collaboration on Cancer Reporting [Internet]. Available from: www.iccr-cancer.org. [Cited in This Article: ] |
| 36. | Horowitz DP, Sharma CS, Connolly E, Gidea-Addeo D, Deutsch I. Secretory carcinoma of the breast: results from the survival, epidemiology and end results database. Breast. 2012;21:350-353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 37. | McDivitt RW, Stewart FW. Breast carcinoma in children. JAMA. 1966;195:388-390. [PubMed] [Cited in This Article: ] |
| 38. | Tavassoli FA, Norris HJ. Secretory carcinoma of the breast. Cancer. 1980;45:2404-2413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 39. | Del Castillo M, Chibon F, Arnould L, Croce S, Ribeiro A, Perot G, Hostein I, Geha S, Bozon C, Garnier A, Lae M, Vincent-Salomon A, MacGrogan G. Secretory Breast Carcinoma: A Histopathologic and Genomic Spectrum Characterized by a Joint Specific ETV6-NTRK3 Gene Fusion. Am J Surg Pathol. 2015;39:1458-1467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 40. | Diallo R, Schaefer KL, Bankfalvi A, Decker T, Ruhnke M, Wülfing P, Jackisch C, Luttges J, Sorensen PH, Singh M, Poremba C. Secretory carcinoma of the breast: a distinct variant of invasive ductal carcinoma assessed by comparative genomic hybridization and immunohistochemistry. Hum Pathol. 2003;34:1299-1305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 60] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Skálová A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, Starek I, Geierova M, Simpson RH, Passador-Santos F, Ryska A, Leivo I, Kinkor Z, Michal M. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599-608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 673] [Cited by in F6Publishing: 660] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 42. | Harrison BT, Fowler E, Krings G, Chen YY, Bean GR, Vincent-Salomon A, Fuhrmann L, Barnick SE, Chen B, Hosfield EM, Hornick JL, Schnitt SJ. Pan-TRK Immunohistochemistry: A Useful Diagnostic Adjunct For Secretory Carcinoma of the Breast. Am J Surg Pathol. 2019;43:1693-1700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 43. | Remoué A, Conan-Charlet V, Bourhis A, Flahec GL, Lambros L, Marcorelles P, Uguen A. Non-secretory breast carcinomas lack NTRK rearrangements and TRK protein expression. Pathol Int. 2019;69:94-96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, Turpin B, Dowlati A, Brose MS, Mascarenhas L, Federman N, Berlin J, El-Deiry WS, Baik C, Deeken J, Boni V, Nagasubramanian R, Taylor M, Rudzinski ER, Meric-Bernstam F, Sohal DPS, Ma PC, Raez LE, Hechtman JF, Benayed R, Ladanyi M, Tuch BB, Ebata K, Cruickshank S, Ku NC, Cox MC, Hawkins DS, Hong DS, Hyman DM. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med. 2018;378:731-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1577] [Cited by in F6Publishing: 1672] [Article Influence: 278.7] [Reference Citation Analysis (0)] |
| 45. | Hong DS, DuBois SG, Kummar S, Farago AF, Albert CM, Rohrberg KS, van Tilburg CM, Nagasubramanian R, Berlin JD, Federman N, Mascarenhas L, Geoerger B, Dowlati A, Pappo AS, Bielack S, Doz F, McDermott R, Patel JD, Schilder RJ, Tahara M, Pfister SM, Witt O, Ladanyi M, Rudzinski ER, Nanda S, Childs BH, Laetsch TW, Hyman DM, Drilon A. Larotrectinib in patients with TRK fusion-positive solid tumours: a pooled analysis of three phase 1/2 clinical trials. Lancet Oncol. 2020;21:531-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 335] [Cited by in F6Publishing: 522] [Article Influence: 130.5] [Reference Citation Analysis (0)] |
| 46. | McDermott R, van Tilburg CM, Farago AF, Kummar S, Tan DSW, Albert CM, Berlin J, Lassen UN, Doz F, Geoerger B, Mascarenhas L, Federman N, Reeves JA, Dima L, Brega N, De La Cuesta E, Laetsch TW, Hong DS, Drilon A. Survival benefits of larotrectinib in an integrated database of patients with TRK fusion cancer. Ann Oncol. 2020;31 (suppl_4):S1034-S1051. [Cited in This Article: ] |
| 47. | Drilon AE, Farago AF, Shao-Weng Tan D, Kummar S, McDermott RS, Berlin J, Patel JD, Brose MS, Leyvraz S, Tahara M, Solomon BM, Reeves JA, Fellous MM, Brega N, Childs BH, Lassen UN, Hong DS. Activity and safety of larotrectinib in adult patients with TRK fusion cancer: An expanded data set. J Clin Oncol. 2020;38:3610. [Cited in This Article: ] |
| 48. | Demetri GD, Paz-Ares L, Farago AF, Liu SV, Chawla SP, Tosi D, Kim ES, Blakely CM, Krauss JC, Sigal D, Bazhenova L, John T, Besse B, Wolf J, Seto T, Chow-Maneval E, Multani PS, Johnson A, Simmons B, Doebele RC. Efficacy and safety of entrectinib in patients with NTRK fusion-positive tumours: Pooled analysis of STARTRK-2, STARTRK-1, and ALKA-372–001. Ann Oncol. 2018;29:ix173-ix178. [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 49. | Rolfo C, Dziadziuszko R, Doebele RC, Demetri G, Simmons B, Huang X, Ye C, Paz-Ares L. Updated efficacy and safety of entrectinib in Clinical and Translational Oncology 1 3 patients with NTRK fusion-positive tumors: Integrated analysis of STARTRK-2, STARTRK-1 and ALKA-372-001. Ann Oncol. 2019;30:v180. [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 50. | Doebele RC, Drilon A, Paz-Ares L, Siena S, Shaw AT, Farago AF, Blakely CM, Seto T, Cho BC, Tosi D, Besse B, Chawla SP, Bazhenova L, Krauss JC, Chae YK, Barve M, Garrido-Laguna I, Liu SV, Conkling P, John T, Fakih M, Sigal D, Loong HH, Buchschacher GL Jr, Garrido P, Nieva J, Steuer C, Overbeck TR, Bowles DW, Fox E, Riehl T, Chow-Maneval E, Simmons B, Cui N, Johnson A, Eng S, Wilson TR, Demetri GD; trial investigators. Entrectinib in patients with advanced or metastatic NTRK fusion-positive solid tumours: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21:271-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 574] [Cited by in F6Publishing: 850] [Article Influence: 170.0] [Reference Citation Analysis (0)] |
| 51. | Han SY. TRK Inhibitors: Tissue-Agnostic Anti-Cancer Drugs. Pharmaceuticals (Basel). 2021;14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | Cocco E, Schram AM, Kulick A, Misale S, Won HH, Yaeger R, Razavi P, Ptashkin R, Hechtman JF, Toska E, Cownie J, Somwar R, Shifman S, Mattar M, Selçuklu SD, Samoila A, Guzman S, Tuch BB, Ebata K, de Stanchina E, Nagy RJ, Lanman RB, Houck-Loomis B, Patel JA, Berger MF, Ladanyi M, Hyman DM, Drilon A, Scaltriti M. Resistance to TRK inhibition mediated by convergent MAPK pathway activation. Nat Med. 2019;25:1422-1427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 53. | Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, Penson A, Shen R, Pareja F, Kundra R, Middha S, Cheng ML, Zehir A, Kandoth C, Patel R, Huberman K, Smyth LM, Jhaveri K, Modi S, Traina TA, Dang C, Zhang W, Weigelt B, Li BT, Ladanyi M, Hyman DM, Schultz N, Robson ME, Hudis C, Brogi E, Viale A, Norton L, Dickler MN, Berger MF, Iacobuzio-Donahue CA, Chandarlapaty S, Scaltriti M, Reis-Filho JS, Solit DB, Taylor BS, Baselga J. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell. 2018;34:427-438.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 464] [Cited by in F6Publishing: 568] [Article Influence: 94.7] [Reference Citation Analysis (0)] |
| 54. | Ross DS, Liu B, Schram AM, Razavi P, Lagana SM, Zhang Y, Scaltriti M, Bromberg JF, Ladanyi M, Hyman DM, Drilon A, Zehir A, Benayed R, Chandarlapaty S, Hechtman JF. Enrichment of kinase fusions in ESR1 wild-type, metastatic breast cancer revealed by a systematic analysis of 4854 patients. Ann Oncol. 2020;31:991-1000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 55. | Russo M, Misale S, Wei G, Siravegna G, Crisafulli G, Lazzari L, Corti G, Rospo G, Novara L, Mussolin B, Bartolini A, Cam N, Patel R, Yan S, Shoemaker R, Wild R, Di Nicolantonio F, Bianchi AS, Li G, Siena S, Bardelli A. Acquired Resistance to the TRK Inhibitor Entrectinib in Colorectal Cancer. Cancer Discov. 2016;6:36-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 233] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 56. | Drilon A, Nagasubramanian R, Blake JF, Ku N, Tuch BB, Ebata K, Smith S, Lauriault V, Kolakowski GR, Brandhuber BJ, Larsen PD, Bouhana KS, Winski SL, Hamor R, Wu WI, Parker A, Morales TH, Sullivan FX, DeWolf WE, Wollenberg LA, Gordon PR, Douglas-Lindsay DN, Scaltriti M, Benayed R, Raj S, Hanusch B, Schram AM, Jonsson P, Berger MF, Hechtman JF, Taylor BS, Andrews S, Rothenberg SM, Hyman DM. A Next-Generation TRK Kinase Inhibitor Overcomes Acquired Resistance to Prior TRK Kinase Inhibition in Patients with TRK Fusion-Positive Solid Tumors. Cancer Discov. 2017;7:963-972. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 288] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 57. | Drilon A, Ou SI, Cho BC, Kim DW, Lee J, Lin JJ, Zhu VW, Ahn MJ, Camidge DR, Nguyen J, Zhai D, Deng W, Huang Z, Rogers E, Liu J, Whitten J, Lim JK, Stopatschinskaja S, Hyman DM, Doebele RC, Cui JJ, Shaw AT. Repotrectinib (TPX-0005) Is a Next-Generation ROS1/TRK/ALK Inhibitor That Potently Inhibits ROS1/TRK/ALK Solvent- Front Mutations. Cancer Discov. 2018;8:1227-1236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 285] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 58. | Nishiyama A, Yamada T, Kita K, Wang R, Arai S, Fukuda K, Tanimoto A, Takeuchi S, Tange S, Tajima A, Furuya N, Kinoshita T, Yano S. Foretinib Overcomes Entrectinib Resistance Associated with the NTRK1 G667C Mutation in NTRK1 Fusion-Positive Tumor Cells in a Brain Metastasis Model. Clin Cancer Res. 2018;24:2357-2369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 59. | Lee SI, Kim NDK, Lee S-H, Kim ST, Park SH, Park JO, Park YS, Lim HY, Kang WK, Park WY, Bang HJ, Kim KM, Park K, Lee J. NTRK gene amplification in patients with metastatic cancer. Precis Future Med. 2017;1:129-137. [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 60. | Hempel D, Wieland T, Solfrank B, Grossmann V, Steinhard J, Frick A, Hempel L, Eberl T, Gaumann A. Antitumor Activity of Larotrectinib in Esophageal Carcinoma with NTRK Gene Amplification. Oncologist. 2020;25:e881-e886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 61. | Hong DS, Bauer TM, Lee JJ, Dowlati A, Brose MS, Farago AF, Taylor M, Shaw AT, Montez S, Meric-Bernstam F, Smith S, Tuch BB, Ebata K, Cruickshank S, Cox MC, Burris HA 3rd, Doebele RC. Larotrectinib in adult patients with solid tumours: a multi-centre, open-label, phase I dose-escalation study. Ann Oncol. 2019;30:325-331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 62. | Nanda N, Fennell T, Low JA. Identification of tropomyosin kinase receptor (TRK) mutations in cancer. J Clin Oncol. 2015;33 (suppl 15):1553. [Cited in This Article: ] |
| 63. | Joshi SK, Qian K, Bisson WH, Watanabe-Smith K, Huang A, Bottomly D, Traer E, Tyner JW, McWeeney SK, Davare MA, Druker BJ, Tognon CE. Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood. 2020;135:2159-2170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 64. | Griffin N, Marsland M, Roselli S, Oldmeadow C, Attia J, Walker MM, Hondermarck H, Faulkner S. The Receptor Tyrosine Kinase TrkA Is Increased and Targetable in HER2-Positive Breast Cancer. Biomolecules. 2020;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |