Published online Aug 26, 2017. doi: 10.4331/wjbc.v8.i3.163
Peer-review started: February 1, 2017
First decision: March 8, 2017
Revised: April 28, 2017
Accepted: May 12, 2017
Article in press: May 15, 2017
Published online: August 26, 2017
To identify and characterize the effect of phosphorylation on the subcellular localization of Ankrd54.
HEK293T cells were treated with calyculin A, staurosporin or phorbol 12-myristate 13-acetate (PMA). Cells were transfected with eGFP-tagged Ankrd54 with or without Lyn tyrosine kinase (wild-type, Y397F mutant, or Y508F mutant). The subcellular localization was assessed by immunofluorescence imaging of cells, immunoblotting of subcellular fractionations. The phosphorylation of Ankrd54 was monitored using Phos-tagTM gel retardation. Phosphorylated peptides were analysed by multiple-reaction-monitoring (MRM) proteomic analysis.
Activation of PKC kinases using PMA promoted nuclear export of Ankrd54 and correlated with increased Ankrd54 phosphorylation, assayed using Phos-tagTM gel retardation. Co-expression of an active form of the PKCδ isoform specifically promoted both phosphorylation and cytoplasmic localization of Ankrd54, while PKCδ, Akt and PKA did not. Alanine mutation of several serine residues in the amino-terminal region of Ankrd54 (Ser14, Ser17, Ser18, Ser19) reduced both PMA induced cytoplasmic localization and phosphorylation of Ankrd54. Using MRM proteomic analysis, phosphorylation of the Ser18 residue of Ankrd54 was readily detectable in response to PMA stimulation. PMA stimulation of cells co-expressing Ankrd54 and Lyn tyrosine kinase displayed increased co-immunoprecipitation and enhanced co-localization in the cytoplasm.
We identify phosphorylation by PKCδ as a major regulator of nuclear-cytoplasmic shuttling of Ankrd54, and its interaction with the tyrosine kinase Lyn.
Core tip: Ankrd54 is a nuclear-cytoplasmic shuttling adaptor that interacts with Lyn, Btk, Txk, and HCLS1. Activation of PKC kinases promoted nuclear export and phosphorylation of Ankrd54, and increased interaction and cytoplasmic co-localization with Lyn. Co-expression of an active form of the PKCδ isoform specifically promoted both phosphorylation and cytoplasmic localization of Ankrd54. Alanine mutation of several serine residues in the amino-terminal region of Ankrd54 reduced both phorbol 12-myristate 13-acetate induced cytoplasmic localization and phosphorylation of Ankrd54. These results identify PKCδ as a major regulator of nuclear-cytoplasmic shuttling and interaction of Ankrd54 with Lyn, through its phosphorylation of at least the Ser18 residue.
- Citation: Samuels AL, Louw A, Zareie R, Ingley E. Control of nuclear-cytoplasmic shuttling of Ankrd54 by PKCδ. World J Biol Chem 2017; 8(3): 163-174
- URL: https://www.wjgnet.com/1949-8454/full/v8/i3/163.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v8.i3.163
Protein nuclear-cytoplasmic compartmentalization and the shuttling of proteins between the nucleus and cytoplasm is regulated at multiple levels and is vitally important for many cellular processes (e.g., transcription). During interphase, when the nuclear envelope exists, the nuclear pore complex (NPC) controls the nuclear entry and exit of many proteins and protein complexes[1]. While proteins/complexes less than approximately 40 kDa in size can typically diffuse through the NPC opening, larger proteins rely on an active process to traverse the NPC and enter/exit the nucleus. The primary control mechanism for most proteins passing through the NPC is the exposure of a nuclear localization sequence (NLS) and/or a nuclear export sequence (NES)[2]. The classical NLS is enriched in basic residues and mediates interaction with an importin α/βcomplex, which then guides the NLS containing cargo through the NPC, and it is released into the nucleus upon RanGTP binding[3]. Proteins exiting the nucleus typically display a leucine-rich NES which interacts with Crm1 in the presence of RanGTP and mediates their export through the NPC. Many proteins that actively shuttle between the nucleus and cytoplasm contain both NLS and NES motifs[4].
Posttranslational modifications of nuclear-cytoplasmic shuttling proteins have been shown to regulate the exposure and interaction of NLS and NES motifs with the importin α/β and Crm1 complexes, respectively[5]. Phosphorylation appears to be a major regulator and mediator of both NLS and NES activity, with examples of phosphorylation promoting nuclear import (e.g., STATs and Erk1/2) as well as nuclear export (e.g., NFAT)[6-8].
Previously we identified Ankrd54 (also called Liar) as a novel nuclear-cytoplasmic (containing functional NLS and NES motifs) shuttling adaptor of the Src family tyrosine kinase Lyn and other signalling molecules (e.g., HCLS1), that regulates down-stream signalling in erythropoietin-responsive erythroid cells[9]. Ankrd54 was also identified as an adaptor for the Tec family tyrosine kinases Btk and Txk[10]. Both these studies illustrated that Ankrd54 interacts with the SH3 domains of these kinases/signalling molecules via its ankyrin repeats and this interaction can regulate the nuclear-cytoplasmic compartmentalization of Lyn/Btk[9,10]. While we identified that erythropoietin stimulation of erythroid cells promoted nuclear speckle accumulation of both endogenous Ankrd54 and Lyn[9], the molecular mechanism controlling Ankrd54 nuclear-cytoplasmic shuttling remained unclear. Consequently, we investigated the role of phosphorylation in regulating Ankrd54 localization to nuclear/cytoplasmic compartments and identified an amino-terminal serine cluster that is required for phorbol ester (PMA)-stimulated cytoplasmic accumulation of Ankrd54, mediated by the serine/threonine kinase PKCδ isoform. Intriguingly, tyrosine phosphorylation of PKCδ by Src family kinases activates the serine/threonine kinase, which could then promote cytosolic accumulation of Ankrd54 and cytoplasmic interaction with Lyn.
HEK293T cells were maintained in Delbocco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum (FCS). Polyclonal rabbit antibodies were raised against murine Ankrd54 using the purified full-length protein fused to GST, as the antigen, and high titer serum was purified on protein-A beads as described previously[9]. Additional antibodies used for immunoblotting were anti-Lyn (Santa-Cruz Biotechnology, sc-15 and sc-7274); anti-β-actin (AC-15, ab6276, Abcam, Cambridge, United Kingdom); anti-Histone H3 (H0164, Sigma-Aldrich); and anti-14-3-3ζ (ab51129, Abcam). TRITC-labeled phalloidin (P1591, Sigma-Aldrich) was used to visualize filamentous actin (F-actin). Phorbol 12-myristate 13-acetate (PMA, P8139), staurosporine (S4400) and leptomycin B (L2913) were purchased from Sigma-Aldrich, calyculin A (#9902) was purchased from Cell Signaling Technologies, and Phos-tag™-Acrylamide TM (ALL-107) was purchased from Wako Laboratory Chemicals (Japan).
All plasmid constructs were generated by site-directed mutagenesis using oligonucleotides (sequences available upon request), subcloning in frame into the appropriate vector and confirmation by sequencing. Murine Lyn, LynY397F and LynY508F expressing plasmids have previously been described[11]. Full-length murine Ankrd54 cDNA was generated by incorporation of a 5’RACE product and a cDNA isolated from a cDNA library using site-directed mutagenesis as previously described[9]. eGFP-tagged Ankrd54 was generated by subcloning murine Ankrd54 into the mammalian expression vector pEGFPC1 (Invitrogen, Carlsbad, CA, United States). This eGFP-Ankrd54 expression construct was then used as a substrate for site-directed mutagenesis to generate the NLS (Ankrd54KKRL111AALA, Ankrd54RR98AA, Ankrd54D98-114) and NES (Ankrd54LSL289ASA) mutants as previously described[9]. The S14A/S17A/S18A/S19A mutant was generated by the same procedure using oligonucleotides that simultaneously introduced these serine-alanine mutations in one reaction. The mCherry-SH3 domain fusion was generated by subcloning the SH3 domain from murine Lyn cDNA into the pmCherry-C1 (Invitrogen) vector as an in-frame C-terminal fusion of mCherry. The Akt mammalian expression vectors were a kind gift from Professor Brian A. Hemmings (FMI, Switzerland) and have been previously described[12]. The PKC mammalian expression vectors for PKCα and PKCδ isoforms were a generous gift from Professor Jae-Won Soh (Inha University, South Korea) and have been described previously[13,14]. Expression vectors for the catalytic domain of cAMP-dependent protein kinase (PKA) were a kind gift from Professor G. Stanley McKnight (University of Washington, Seattle, WA, United States) and have previously been described[15].
Cells (HEK293T) were transiently transfected using Lipofectamine 2000 (Invitrogen) and harvested 24-48 h post transfection. Protein lysates were prepared in 1% nonidet P40, 0.1% SDS, 150 mmol/L NaCl, 50 mmol/L Tris-HCl pH 8.0, 1 mmol/L EDTA, 1 mmol/L EGTA, 25 mmol/L sodium fluoride, 25 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 × protease inhibitor cocktail (Complete, ROCHE) or 1 mmol/L benzamidine/1 mmol/L phenylmethylsulphoyl fluoride (PMSF)/1 μg/mL aprotinin, for 30 min on ice, followed by centrifugation at 10000 × g for 20 min. Protein concentration was estimated using the BioRad Dc protein assay according to manufacturer’s instructions (BioRad), using bovine serum albumin as a standard.
For immunoprecipitation, 5 mg of total protein from clarified cell lysates were incubated with specific antibodies (10 μg) for 2 h at 4 °C, collected with protein G-Sepharose beads (Sigma-Aldrich) for 16 h (4 °C) before washing in lysis buffer, and subsequent analysis by SDS-PAGE.
For separation of nuclear and cytoplasmic compartments subcellular fractionation was performed. Cells were washed (twice) with cold PBS, and lysed in cell lysis buffer (20 mmol/L Tris-HCl pH 8, 0.5% NP40, 10 mmol/L NaCl, 3 mmol/L MgCl2 supplemented with protease inhibitors) for 30 min on ice with vigorous pipetting every 5 min. Nuclei were sedimented by centrifugation at 5000 × g for 5 min at 4 °C and the supernatant (cytoplasmic fraction) was collected. The nuclear pellet was washed 3 times in cell lysis buffer to remove cytoplasmic traces and the nuclei were lysed in nuclear lysis buffer (50 mmol/L Tris-HCl, 1% SDS) on ice for 20 min followed by brief sonication to ensure nuclei rupture. Nuclear debri was collected by centrifugation at 12500 × g for 10 min at 4 °C and the supernatant (nuclear fraction) was collected. Nuclear and cytoplasmic fractions were stored at -80 °Cprior to analysis by SDS-PAGE and Western blotting.
After probing with specific primary antibodies, proteins were revealed using either secondary antibody coupled to horseradish peroxidase (GE Healthcare or Cell Signaling Technology,) and detection by enhanced chemiluminescence (GE Healthcare), or with fluorescently labeled secondary antibodies and an Odyssey scanner (LI-COR Biosciences, Lincoln, NE, United States). Quantitation of immunoblot analysis was performed using on Odyssey scanned blots using Image Studio Light (V3.1.4, LI-COR Biosciences).
Glass coverslips (No. 1.5H, Marienfeld) were coated with either poly-L-lysine (Sigma-Aldrich) or serum before seeding cells (HEK293T). At the end of specific cell treatments of specific time points, coverslips were washed with PBS (37 °C) then fixed in 4% paraformaldehyde/PBS for 15 min at 37 °C, followed by permeabilization with 0.5% triton X-100 in PBS (5 min). Coverslips were then either processed immediately or stored at -80 °C with minimal PBS media. Coverslips were blocked for 2 h at 25 °C in 3% BSA/PBS/Tween-20 (0.1%), then incubated with primary antibody diluted in blocking buffer at 4 °C for 16 h, then washed 3 times in PBS/Tween-20 (15 min) before incubation with fluorophore-conjugated secondary antibodies (Alexa-Fluor-labeled-488/594, Life-Technologies), with Hoechst 33342 at 0.01 μg/mL, for 1 h at 25 °C, then washed in PBS/Tween-20 before mounting in Vectashield (H-1000, Vector Labs). For some experiments TRITC-labeled phalloidin (P1951, Sigma-Aldrich) at 50 μg/mL was added to the secondary antibody/Hoechst solution to visualize filamentous actin (F-actin). For experiments where only transfected fluorescent protein was being analysed then permeabilized cells were incubated directly with Hoechst with or without TRITC-phalloidin. For cells co-transfected with eGFP-Ankrd54 and Lyn expressing plasmids, Lyn was detected using anti-Lyn antibodies (Sant-Cruz Biotechnology, sc-15) and Alexa-Fluor-546 donkey anti-rabbit IgG (ThermoFisher Scientific). Fluorescent microscopy was performed using three different instruments: An MRC 1024 UV laser scanning confocal microscope (Bio-Rad) using a Nikon Diaphot 300 microscope fitted with a 40 ×/1.15 oil objective and analysed using Confocal Assistant (v4.02, Bio-Rad); an × 60 plan-Apo 1.40 oil objective on an inverted Elcips-Ti microscope (Nikon, Japan), fitted with a CoolSNAP HQ2 CCD camera (Photometrics), images analysed using NIS Elements 4.x (Nikon); or on a DeltaVision Elite system and analysed with softWoRx suite 2.0 (GE Healthcare). Generally, images were processed as z-stacks (deconvolution software used for the non-confocal systems) and maximum image projections generated. Quantitation of subcellular localization was performed on 50-100 individual cells for each experiment by analyzing 10 nuclear and 10 cytoplasmic sub-compartments per individual cell. Ratios of nuclear:cytoplasmic amounts of fluorescence (eGFP channel) were then collated. For statistical analysis experiments were repeated as three biological replicates and analysed by students t-test or ANOVA with two-tailed analysis, with orthogonal comparisons (using Prism v7.0b.154, Graphpad). Data are presented as mean ± SD, P < 0.05, P < 0.01, ns P > 0.05. The statistical methods of this study were reviewed by Michael Phillips from the Harry Perkins Institute of Medical Research.
Protein bands obtained following SDS-PAGE were reduced and alkylated using DTT and iodoacetamide before digestion with chymotrypsin at pH approximately 8.0. Peptides were extracted as previously described[16] and analysed by electrospray ionisation mass spectrometry using an Ultimate 3000 nano HPLC system (Dionex) coupled to a 4000 Q TRAP mass spectrometer (Applied Biosystems). Peptides were resolved using a C18 PepMap100 column (LC Packings) running a linear gradient of water/acetonitrile/with 0.1% formic acid. Multiple reaction monitoring (MRM) transitions for the putative phosphopeptides (flanking Ser18 of ANKKRD54) were generated using the Skyline software[17]. Both singly-phosphorylated and nonphosphorylated forms of the peptides, with one optional missed cleavage, were targeted. A non-phosphorylated peptide (sequence: QQDVEPRDEL, corresponding to amino acid 72-81) was also included in the MRM experiment as an internal normalization control. Spectra were imported back into the Skyline software for interpretation. Peak heights from each sample were divided by the relative peak height of the internal normalization control in the same sample to obtain normalized quantities. Normalized values associated with each hypermass were summed to obtain an overall quantity indicator for that hypermass. The overall hypermass quantities were then divided over the maximum observed for that hypermass to obtain the overall percentage for the hypermass.
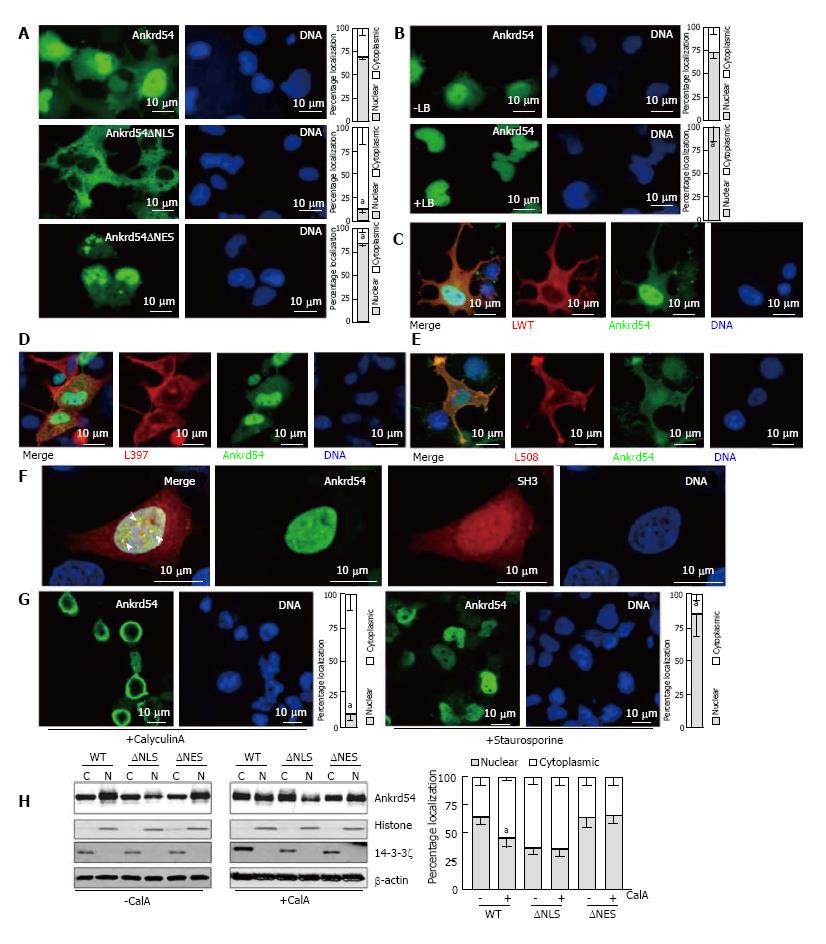
We have previously shown that myc/HA-tagged and untagged Ankrd54 contain functional NLS and NES sequences[9]. For this study, we generated an N-terminal eGFP fusion of Ankrd54 and confirmed that the NLS and NES were still functional in this fusion construct (Figure 1A). Further, the Crm1 inhibitor Leptomycin B promoted strong nuclear accumulation of eGFP-Ankrd54 (Figure 1B) as has been observed with myc/HA-tagged Ankrd54 previously[9]. Interaction of eGFP-Ankrd54 and Lyn was also analogous to that previously delineated with the strongest co-localization seen with constitutively active (Y508F) Lyn and minimal with inactive (Y397F) Lyn (Figure 1C-E).
In erythroid cells Lyn and Ankrd54 both co-localize to nuclear speckles after erythropoietin stimulation[9], while when overexpressed in HEK293 cells active Lyn and Ankrd54 co-localize in the cytoplasm/plasma-membrane (Figure 1E). To ascertain if Ankrd54 could co-localize with Lyn in the nucleus via their Ankyrin repeat-SH3 interaction, we co-expressed an mCherry fusion of the SH3 domain of Lyn with eGFP-tagged Ankrd54 (Figure 1F). Here eGFP-Ankrd54 was predominantly nuclear and localized to nuclear speckles. The SH3 domain of Lyn localized to both the cytoplasm and nucleus with stronger accumulation in the nucleus and specifically within Ankrd54 co-localized nuclear speckles (Figure 1F).
Having established that the eGFP fusion of Ankrd54 retained functional NLS and NES, localization dynamics and Lyn/SH3 domain interactions, we then sought to delineate what signals could influence its nuclear-cytoplasmic shuttling. Modulation of phosphorylation using the phosphatase inhibitor CalyculinA and the kinase inhibitor Staurosporine produced profound effects on the subcellular localization of eGFP-Ankrd54 (Figure 1G and H). Immunofluorescence microscopy revealed that activation of kinases through inhibition of phosphatases with CalyculinA promoted cytoplasmic accumulation of eGFP-Ankrd54, while inhibition of kinases with Staurosporine significantly increased nuclear accumulation (Figure 1G). Biochemical subcellular fractionation and immunoblot analysis also showed that CalyculinA significantly promoted cytoplasmic accumulation of eGFP-Ankrd54 (Figure 1H). Further, addition of CalyculinA was unable to influence the compartmentalization of either NLS or NES deletion mutants of eGFP-Ankrd54, which accumulate in the cytoplasm or nucleus, respectively (Figure 1H).
Identification of PKCδ as mediating phosphorylation and cytoplasmic accumulation of Ankrd54
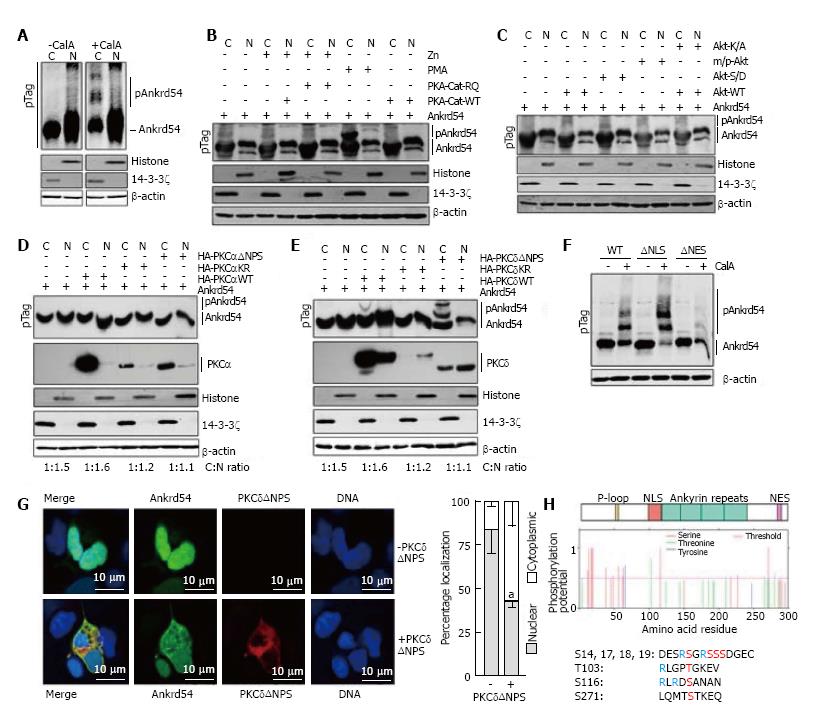
Having established that global phosphorylation is important for NES mediated cytoplasmic accumulation of Ankrd54 (Figure 1G and H), we then investigated the direct effect of phosphorylation on Ankrd54 subcellular localization. Analysis of phosphorylation databases (PhosphoSitePlus®, Cell Signaling Technologies; PhosphoNET, Kinexus Bioinformatics Corporation, Canada; and PHOSIDA, Max Planck Institute of Biochemistry, Germany), indicated that Ankrd54 is a phosphoprotein with several residues identified in mass spectrometric data. Utilizing Phos-Tag™ SDS-PAGE to detect phosphorylation-mediated mobility shifted Ankrd54, we discovered that CalyculinA promoted phosphorylation of Ankrd54 in the cytoplasmic fraction (Figure 2A). Using this Phos-Tag mobility shift assay we then analysed the ability of co-expressing candidate kinases (predicted from database searches, PhosphoNET, PHOSIDA) and kinase activators for their ability to mimic the CalyculinA effect. While co-expression of constitutively active forms of Akt or the catalytic subunit of PKA failed to promote a band-shift of Ankrd54, the addition of the PKC activator PMA did produce a clear phospho-Ankrd54 shifted band (Figure 2B and C). Consequently, we then tested the ability of two widely expressed PMA activated PKC isoforms (a conventional PKC, PKCα; and a novel PKC, PKCδ) to promote cytoplasmic phosphorylation of Ankrd54 (Figure 2D and E). This clearly demonstrated that PKCδ could promote phosphorylation of cytoplasmic Ankrd54, especially when expressed as a constitutively active form (ΔNPS, N-terminal pseudo-substrate domain deletion), while PKCδ failed to promote Ankrd54 phosphorylation (Figure 2D and E). Further, quantitation of the PKCδNPS promoted phospho-Ankrd54 revealed that PKCδ also promoted cytoplasmic accumulation of Ankrd54 (Cytoplasmic:nuclear ratio; Ankrd54 1.0:1.5; Ankrd54 + PKCδΔNPS 1.0:0.3). In addition, the CalyculinA mediated band-shift of cytoplasmic Ankrd54 was enhanced with NLS deleted, but strongly reduced in NES deleted mutants of Ankrd54 (Figure 2F). Consequently, we then ascertained the effect of co-expressing active PKCδ (PKCδΔNPS) with Ankrd54 on the subcellular localization of Ankrd54 by immunofluorescent microscopy (Figure 2G). This revealed, in agreement with the biochemical analysis (Figure 2E), that PKCδ significantly promotes cytoplasmic accumulation of Ankrd54.
Utilizing predictive algorithms (NetPho3.1)[18,19] and interpretation of potential site of PKC phosphorylation (Figure 2H), identified four regions of potential phosphorylation; an amino-terminal cluster of serines (Ser14, Ser17, Ser18, Ser19), Thr103, Ser116 and Ser271. The C-terminal site Ser271, close to the NES motif, showed low probability of PKC phosphorylation due to a lack of N-terminal basic residues within the site. The Thr103 and Ser116 sites are located within the NLS motif (Thr103) and immediately C-terminal (Serh116) to the NLS motif, and the N-terminal cluster (Ser14, Ser17, Ser18, Ser19) all have potential for PKC-medicated phosphorylation.
While we identified several potential PKC phosphorylation sites, only the N-terminal Ser14/Ser17/Ser18/Ser19 cluster has been robustly detected in mass-spectrometry datasets (PhosphositePlus®, PhosphoNET, PHOSIDA), in addition to several phosphorylated residues located close to the P-loop (Thr31, Ser38, Ser44, Ser54, Ser58, Ser63), with the Thr103, Ser116 and Ser271 motifs not robustly detected in these global phosphorylation analysis datasets. Consequently, we focused our initial attention on the amino-terminal Ser14/Ser17/Ser18/Ser19 cluster and its phosphorylation in response to PMA treatment. Using MRM of singly phosphorylated peptides encompassing the N-terminal serine cluster (Table 1) we identified this region as showing strong correlation of its phosphorylation with exposure of cells to PMA. Significantly, this included an inverse correlation of PMA stimulation with the non-phosphorylated peptide.
| Target peptide | m/z | Charge state | Transition | Normalized quantity | Overall percentage | ||
| +PMA | -PMA | +PMA | -PMA | ||||
| Non-phosphorylated S18 peptide | 936.078 | 3+ | AATGGGADDESRSGRSSSDGECAVAPEPL.y4+ | 4.2E+02 | 1.9E+03 | 26% | 91% |
| AATGGGADDESRSGRSSSDGECAVAPEPL.y2+ | 8.5E+02 | 2.5E+03 | |||||
| AATGGGADDESRSGRSSSDGECAVAPEPL.y15+++ | 2.6E+02 | 1.1E+03 | |||||
| 702.310 | 4+ | AATGGGADDESRSGRSSSDGECAVAPEPL.y4+ | ND | ND | ND | ND | |
| AATGGGADDESRSGRSSSDGECAVAPEPL.y2+ | ND | ND | |||||
| 826.871 | 4+ | AATGGGADDESRSGRSSSDGECAVAPEPLAEAGGL.y6+ | 2.0E+03 | 3.2E+03 | 61% | 100% | |
| AATGGGADDESRSGRSSSDGECAVAPEPLAEAGGL.y2+ | 2.0E+03 | 3.2E+03 | |||||
| Normalization Control (Q72-L81) | 614.794 | 2 | QQDVEPRDEL.y8+ | 1 | 1 | 94% | 100% |
| QQDVEPRDEL.y6+ | 1 | 1 | |||||
| QQDVEPRDEL.y5+ | 1 | 1 | |||||
| Singly-phosphorylated S18 peptide | 962.733 | 3+ | AATGGGADDESRSGRSSSDGECAVAPEPL(p).y4+ | 1.3E+04 | 7.0E+03 | 70% | 44% |
| AATGGGADDESRSGRSSSDGECAVAPEPL(p).y2+ | 1.1E+03 | 1.6E+03 | |||||
| AATGGGADDESRSGRSSSDGECAVAPEPL(p).y15+++ | |||||||
| 722.302 | 4+ | AATGGGADDESRSGRSSSDGECAVAPEPL(p).y4+ | 5.5E+03 | 2.4E+03 | 71% | 38% | |
| AATGGGADDESRSGRSSSDGECAVAPEPL(p).y2+ | 5.8E+02 | 9.0E+02 | |||||
| 846.862 | 4+ | AATGGGADDESRSGRSSSDGECAVAPEPLAEAGGL(p).y6+ | ND | ND | 100% | 83% | |
| AATGGGADDESRSGRSSSDGECAVAPEPLAEAGGL(p).y2+ | 1.1E+03 | 8.8E+02 | |||||
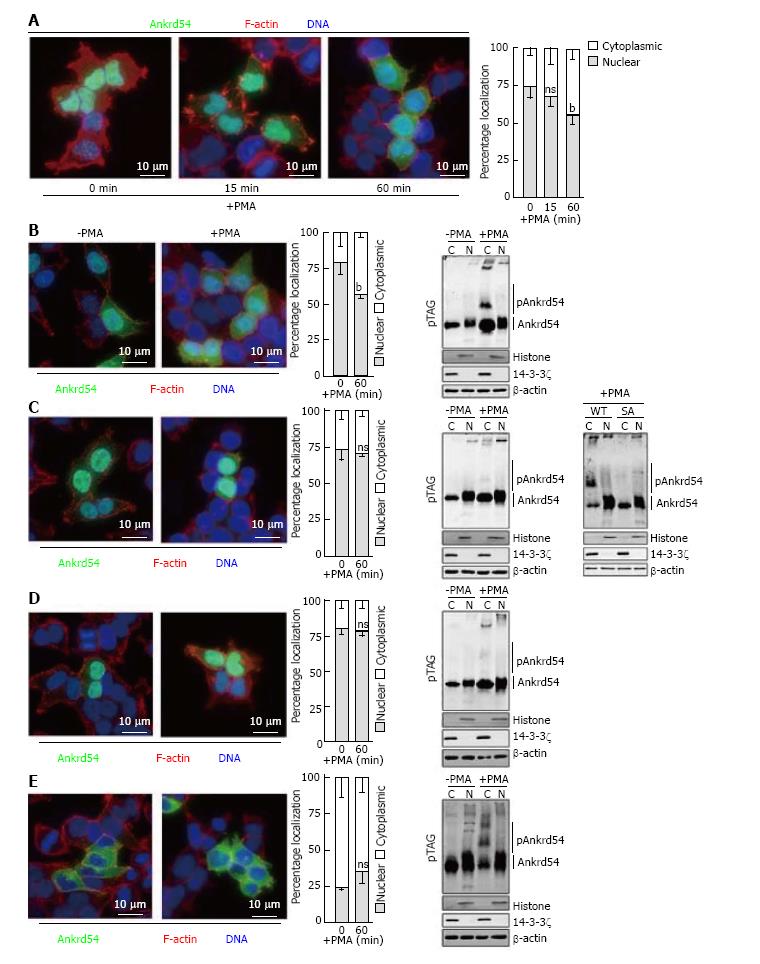
Stimulation of eGFP-Ankrd54 transfected HEK293 cells with PMA promotes significant cytoplasmic accumulation of Ankrd54 by 1 h post phorbol ester addition, assayed by immunofluorescent microscopy (Figure 3A). Consequently, we used this time point of PMA stimulation to analyse the effect of mutagenesis of the N-terminal serine cluster (S14A, S17A, S18A, S19A) on cytoplasmic accumulation of Ankrd54 (Figure 3B-E). While wild-type Ankrd54 (Figure 3B) showed significant cytoplasmic accumulation after PMA stimulation in addition to phosphorylation (assayed by Phos-Tag® SDS-PAGE), the S14/17/18/19A mutant failed to show significant alteration to its predominantly nuclear subcellular localization, and failed to show a robust phosphorylation mobility shift (Figure 3C). Importantly, direct in-gel comparison of wild-type and the SA mutant of Ankrd54 showed a strong reduction in the band shift upon PMA stimulation of mutant Ankrd54 (Figure 3C, far right panel). Interestingly, mutagenesis of the NES motif (promoting nuclear localization) also disrupted the PMA-mediated cytoplasmic accumulation and phosphorylation of Ankrd54 (Figure 3D), while the NLS motif mutant (predominantly cytoplasmic) showed no significant further cytoplasmic accumulation upon PMA stimulation, but did show strong PMA-mediated phosphorylation (Figure 3E).
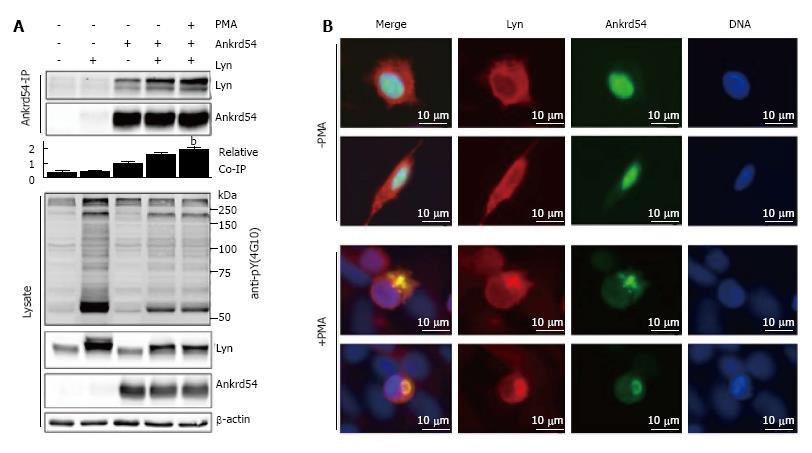
Stimulation of eGFP-Ankrd54 and Lyn co-transfected HEK293 cells with PMA for 1 h promotes interaction of Lyn and Ankrd54, assayed by immunoblotting of Ankrd54 immunoprecipitates (Figure 4A). Further, immunofluorescent microscopy (Figure 4B) of co-transfected cells illustrated strong co-localization in the cytoplasmic compartment of cells co-expressing Ankrd54 and Lyn, when the cells were stimulated with PMA for 1 h.
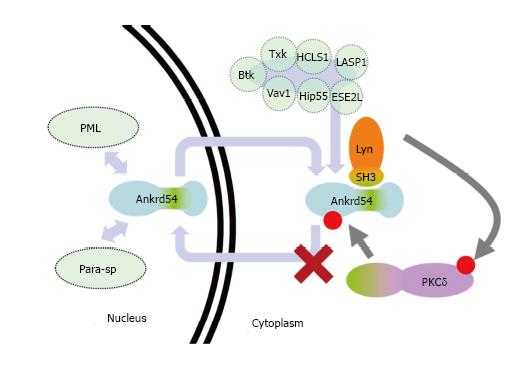
Here we describe the identification of an N-terminal serine cluster that can be phosphorylated by PKCδ to promote cytoplasmic accumulation of the nuclear-cytoplasmic shuttling adaptor Ankrd54 (binding partner of Lyn and Btk tyrosine kinases, and other signalling proteins)[9,10,20,21] (summarized in Figure 5). These are important findings that help place the pathway context in which Ankrd54 complexes shuttle between the nuclear and cytoplasmic compartments, as being controlled (in part) by PKCδ (Figure 5).
Many of the binding partners of Ankrd54 have important functions both within the cytoplasm as well as the nucleus and enact these subcellular-specific functions in response to many different stimuli. Further, recent findings have identified the nuclear speckles that Ankrd54 can localize to[9] as being PML (promyelocytic leukaemia) nuclear bodies and paraspeckles[22], and the strong interaction of the SH3 domain of Lyn and Ankrd54 has been independently confirmed[23,24]. PML nuclear bodies have a plethora of functions including cell cycle arrest, apoptosis, senescence and transcription and are regulated by signals that feed into these pathways. Paraspeckles function as regulators of gene expression, in response to cellular stress and differentiation, by controlling nuclear retention of RNA (containing double-stranded RNA regions) which have undergone adenosine-to-inosine editing[25-27]. The identification of Ankrd54 as a component of these two sub-nuclear structures, which contain many nuclear-cytoplasmic shuttling molecules, suggests the pathways that Ankrd54 (and its binding partners) is a part of are related to those that these two nuclear bodies/speckles are intimately involved with, i.e., cell cycle, senescence, apoptosis, differentiation and transcription. Indeed, strong nuclear accumulation of Ankrd54 promotes apoptosis, and altering Ankrd54 levels and subcellular localization influences viability and differentiation pathways[9,20].
PKCδ, a member of the novel (activated by diacylglycerol but not calcium) PKC subfamily has an intimate link with Src family kinases (SFK), including Lyn, as well as nuclear-cytoplasmic shuttling/signalling and the control of apoptosis in response to DNA damage[28-31]. Activation of PKCδ involves diacylglycerol binding as well as tyrosine phosphorylation by SFK (as well as Abl and Syk), which can lead to activated PKCδ localizing to the plasma-membrane, mitochondria or the nucleus[30]. Nuclear and mitochondrial localized PKCδ appear associated with induction of apoptosis[29,32]. Differential tyrosine phosphorylation of PKCδ by SFKs/Abl mediates activation and either translocation to the nucleus or plasma-membrane[29,31]. Interestingly, when active Lyn is co-expressed with Ankrd54 (Figure 1C-E)[9], or Btk and Ankrd54 are co-expressed[10], both accumulate in the cytoplasmic/plasma-membrane fraction. Consequently, Lyn/Btk tyrosine phosphorylated and activated PKCδ could be the driver of the cytoplasmic accumulation of Ankrd54 and interaction with Lyn. In addition, when an active form of PKCδ is co-expressed with Ankrd54 both localize outside of the nucleus (Figure 2G). This could also indicate that the activation of PKCδ within this context promotes a viability/anti-apoptotic response, due to the subcellular localization of PKCδ.
The mechanism by which phosphorylation of Ankrd54 by PKCδ enhances cytoplasmic accumulation of Ankrd54 and its interaction with Lyn remains to be determined. The sites of phosphorylation do not fit the consensus for 14-3-3 binding and so it is unlikely to be via 14-3-3 sequestration[33]. Alternatively, it may interfere with the NLS function and/or enhance the NES function. However, it would be worth exploring the composition of cytoplasmic localized Ankrd54 to determine potential interactions (that may be phosphorylation dependent) that mediate its cytoplasmic accumulation, as well as determining the influence of nuclear/cytoplasmic localized Ankrd54 on the subcellular distribution of its known binding partners. Further, studies on rates of nuclear/cytoplasmic shuttling for mutants of the N-terminal serine cluster could also identify whether there is a direct effect upon the ability of Ankrd54 to traverse the nuclear pore, i.e., to influence the function of the NLS and/or NES.
In this manuscript, we have identified a mechanism that controls the subcellular localization of the nuclear-cytoplasmic adaptor Ankrd54 and promotes its interaction with Lyn, through PKCδ mediated phosphorylation of an N-terminal serine cluster (Figure 5). Consequently, it will be important to define the consequences of this PKCδ mediated phosphorylation of Ankrd54 on its other binding partners, their subcellular compartmentalization as well as its influence on their biological function.
We thank Irma Larma and Kevin Li for technical assistance.
Protein nuclear-cytoplasmic compartmentalization and the shuttling of proteins between the nucleus and cytoplasm is regulated at multiple levels and is vitally important for many cellular processes. Phosphorylation is a major regulator of subcellular compartmentalization of proteins. Ankrd54 shuttles between the nucleus and cytoplasm and here the authors investigated the involvement of phosphorylation on subcellular compartmentalization of this protein.
The regulation of nuclear-cytoplasmic transport and subcellular localization of signalling molecules such as Ankrd54 is critical for their functions in both normal and diseased cells. The identification of the kinases important for the regulation of subcellular localization of Ankrd54 was the objective of the authors.
This is the first discovery of the involvement of PKC kinases as being important in the nuclear-cytoplasmic shuttling of Ankrd54.
Knowing that PKCs can regulate the subcellular localization of Ankrd54 may find applications in identifying way to regulate its important interacting partners (i.e., Lyn, Btk, Txk, HCLS1) in health and disease.
Regulated subcellular localization: The localization of a protein inside a cell can have dramatic impact on its function, and in many disease incorrect location of proteins leads to specific pathologies. Further, many proteins have altered localization after specific stimulation the promotes of inhibits their function. A proteins localization within a cell is consequently critical for its function and is often regulated by post-translation modification, with phosphorylation being the most common.
In this manuscript, the authors characterized phosphorylation of Ankrd54, a nuclear-cytoplasmic shuttling adaptor protein. They provided good evidence that phosphorylation plays a role in regulating subcellular localization of Ankrd54. Specifically, phosphorylation of the N-terminal region of the protein promotes its accumulation in cytoplasm. They also demonstrate enhanced phosphorylation of Ankrd54 by activated version of PKC-delta, implying it as a potential kinase responsible for Ankrd54 phosphorylation. Using cutting-edge mass spec analysis, they nicely demonstrated that phosphorylation at Ser18 is responding to phorbol 12-myristate 13-acetate stimulation. Overall, the work is very carefully done and the data are with high quality.
Manuscript source: Unsolicited manuscript
Specialty type: Biochemistry and molecular biology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Jia J, Wang Y S- Editor: Song XX L- Editor: A E- Editor: Lu YJ
| 1. | D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 281] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 2. | Pemberton LF, Paschal BM. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187-198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 532] [Cited by in F6Publishing: 527] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 3. | Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. J Biol Chem. 2007;282:5101-5105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 833] [Cited by in F6Publishing: 868] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 4. | Fabbro M, Henderson BR. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp Cell Res. 2003;282:59-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 187] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 5. | Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell Commun Signal. 2010;8:32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 166] [Cited by in F6Publishing: 175] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 6. | Meyer T, Vinkemeier U. Nucleocytoplasmic shuttling of STAT transcription factors. Eur J Biochem. 2004;271:4606-4612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 7. | Khokhlatchev AV, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb MH. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605-615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 516] [Cited by in F6Publishing: 524] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 8. | Kehlenbach RH, Dickmanns A, Gerace L. Nucleocytoplasmic shuttling factors including Ran and CRM1 mediate nuclear export of NFAT In vitro. J Cell Biol. 1998;141:863-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 144] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 9. | Samuels AL, Klinken SP, Ingley E. Liar, a novel Lyn-binding nuclear/cytoplasmic shuttling protein that influences erythropoietin-induced differentiation. Blood. 2009;113:3845-3856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Gustafsson MO, Hussain A, Mohammad DK, Mohamed AJ, Nguyen V, Metalnikov P, Colwill K, Pawson T, Smith CI, Nore BF. Regulation of nucleocytoplasmic shuttling of Bruton’s tyrosine kinase (Btk) through a novel SH3-dependent interaction with ankyrin repeat domain 54 (ANKRD54). Mol Cell Biol. 2012;32:2440-2453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Tilbrook PA, Ingley E, Williams JH, Hibbs ML, Klinken SP. Lyn tyrosine kinase is essential for erythropoietin-induced differentiation of J2E erythroid cells. EMBO J. 1997;16:1610-1619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 110] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Andjelković M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515-31524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 788] [Cited by in F6Publishing: 812] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 13. | Soh JW, Lee EH, Prywes R, Weinstein IB. Novel roles of specific isoforms of protein kinase C in activation of the c-fos serum response element. Mol Cell Biol. 1999;19:1313-1324. [PubMed] [Cited in This Article: ] |
| 14. | Soh JW, Weinstein IB. Roles of specific isoforms of protein kinase C in the transcriptional control of cyclin D1 and related genes. J Biol Chem. 2003;278:34709-34716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 159] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 15. | Riggle KM, Riehle KJ, Kenerson HL, Turnham R, Homma MK, Kazami M, Samelson B, Bauer R, McKnight GS, Scott JD. Enhanced cAMP-stimulated protein kinase A activity in human fibrolamellar hepatocellular carcinoma. Pediatr Res. 2016;80:110-118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 16. | Bringans S, Eriksen S, Kendrick T, Gopalakrishnakone P, Livk A, Lock R, Lipscombe R. Proteomic analysis of the venom of Heterometrus longimanus (Asian black scorpion). Proteomics. 2008;8:1081-1096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 132] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 17. | MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966-968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3120] [Cited by in F6Publishing: 3288] [Article Influence: 234.9] [Reference Citation Analysis (0)] |
| 18. | Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol. 1999;294:1351-1362. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633-1649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1425] [Cited by in F6Publishing: 1456] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 20. | Barber DL. Truth or dare: role of Liar in EPO-dependent signaling. Blood. 2009;113:3650-3651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 21. | Ingley E. Integrating novel signaling pathways involved in erythropoiesis. IUBMB Life. 2012;64:402-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol. 2013;203:149-164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 86] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 23. | Zhu J, Larman HB, Gao G, Somwar R, Zhang Z, Laserson U, Ciccia A, Pavlova N, Church G, Zhang W. Protein interaction discovery using parallel analysis of translated ORFs (PLATO). Nat Biotechnol. 2013;31:331-334. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 24. | Larman HB, Liang AC, Elledge SJ, Zhu J. Discovery of protein interactions using parallel analysis of translated ORFs (PLATO). Nat Protoc. 2014;9:90-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637-644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 333] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 26. | Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2:a000687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 209] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 27. | Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol. 2015;210:529-539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 222] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 28. | Kajimoto T, Sawamura S, Tohyama Y, Mori Y, Newton AC. Protein kinase C {delta}-specific activity reporter reveals agonist-evoked nuclear activity controlled by Src family of kinases. J Biol Chem. 2010;285:41896-41910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 29. | Humphries MJ, Ohm AM, Schaack J, Adwan TS, Reyland ME. Tyrosine phosphorylation regulates nuclear translocation of PKCdelta. Oncogene. 2008;27:3045-3053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 30. | Reyland ME, Jones DN. Multifunctional roles of PKCδ: Opportunities for targeted therapy in human disease. Pharmacol Ther. 2016;165:1-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 31. | Xue ZH, Zhao CQ, Chua GL, Tan SW, Tang XY, Wong SC, Tan SM. Integrin alphaMbeta2 clustering triggers phosphorylation and activation of protein kinase C delta that regulates transcription factor Foxp1 expression in monocytes. J Immunol. 2010;184:3697-3709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Qi X, Mochly-Rosen D. The PKCdelta -Abl complex communicates ER stress to the mitochondria - an essential step in subsequent apoptosis. J Cell Sci. 2008;121:804-813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 33. | Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, Gamblin SJ, Smerdon SJ, Cantley LC. The structural basis for 14-3-3: phosphopeptide binding specificity. Cell. 1997;91:961-971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1237] [Cited by in F6Publishing: 1248] [Article Influence: 46.2] [Reference Citation Analysis (0)] |