Published online Feb 27, 2021. doi: 10.4240/wjgs.v13.i2.210
Peer-review started: July 11, 2020
First decision: November 16, 2020
Revised: November 30, 2020
Accepted: December 17, 2020
Article in press: December 17, 2020
Published online: February 27, 2021
Investigating molecular biomarkers that accurately predict prognosis is of considerable clinical significance. Accumulating evidence suggests that long non-coding ribonucleic acids (lncRNAs) are frequently aberrantly expressed in colorectal cancer (CRC).
To elucidate the prognostic function of multiple lncRNAs serving as biomarkers in CRC.
We performed lncRNA expression profiling using the lncRNA mining approach in large CRC cohorts from The Cancer Genome Atlas (TCGA) database. Receiver operating characteristic analysis was performed to identify the optimal cutoff point at which patients could be classified into the high-risk or low-risk groups. Based on the Cox coefficient of the individual lncRNAs, we identified a nine-lncRNA signature that was associated with the survival of CRC patients in the training set (n = 175). The prognostic value of this nine-lncRNA signature was validated in the testing set (n = 174) and TCGA set (n = 349). The prognostic models, consisting of these nine CRC-specific lncRNAs, performed well for risk stratification in the testing set and TCGA set. Time-dependent receiver operating characteristic analysis indicated that this predictive model had good performance.
Multivariate Cox regression and stratification analysis demonstrated that this nine-lncRNA signature was independent of other clinical features in predicting overall survival. Functional enrichment analysis of Kyoto Encyclopedia of Genes and Genomes pathways and Gene Ontology terms further indicated that these nine prognostic lncRNAs were closely associated with carcinogenesis-associated pathways and biological functions in CRC.
A nine-lncRNA expression signature was identified and validated that could improve the prognosis prediction of CRC, thereby providing potential prognostic biomarkers and efficient therapeutic targets for patients with CRC.
Core Tip: To the best of our knowledge, preliminary investigation of the function of this nine-long non-coding ribonucleic acid signature has not been reported, which further strengthens the possibility it may be used to effectively predict the disease course in colorectal cancer.
- Citation: Zong Z, Hu CG, Zhou TC, Yu ZM, Tang FX, Tian HK, Li H, Wang H. Nine-long non-coding ribonucleic acid signature can improve the survival prediction of colorectal cancer. World J Gastrointest Surg 2021; 13(2): 210-221
- URL: https://www.wjgnet.com/1948-9366/full/v13/i2/210.htm
- DOI: https://dx.doi.org/10.4240/wjgs.v13.i2.210
Colorectal cancer (CRC) is one of the most common malignancies worldwide with an estimated 1.1 million new cancer cases and 0.88 million cancer deaths per year[1]. Despite the considerable progress made in CRC screening and therapeutic methods, the 5-year survival rate of CRC patients remains low at approximately 12%[2]. Multiple genetic and epigenetic alterations play critical roles in CRC tumorigenesis and progression[3,4], but they remain underutilized for the precise prediction of CRC. Investigating the appropriate biomarkers associated with clinical outcome and identifying the effective molecular targets for individual therapies could lead to the development of more effective therapeutic regimens and personalized therapies, which have promise for prolonging overall survival (OS) and improving prognosis.
Over the last several years, particular attention has been devoted to newly discovered long non-coding ribonucleic acids (lncRNAs), which are defined as transcripts longer than 200 nucleotides that have no apparent protein-coding potential[5]. The protein-coding regions of the genome have been analyzed by most large-scale genomic databases of cancer patients. However, the most recent estimate of the Encyclopedia of DNA Elements is that up to 75% of the human genome is transcribed into RNA, whereas only approximately 3% of the human genome is protein-coding[6,7]. It has been estimated that more than 58000 lncRNAs are encoded in the human genome[8,9]. Similar to protein-coding genes, lncRNA genes are regulated by transcription factors and histone modifications, and lncRNA transcripts are processed by the canonical splicing machinery[7]. Compared to protein-coding genes, lncRNAs have fewer exons and are usually located in the nucleus, which is affected by lower selection pressure during evolution and shows higher tissue or cell specificity[9].
LncRNAs are involved in various biological processes including proliferation, immortality, angiogenesis, growth suppression, motility, and viability[8]. Some dysregulated lncRNAs play oncogenic and tumor suppressive roles in cancer development, progression, and metastasis such as the well-known lncRNAs plasmacytoma variant translocation 1[10], HOX transcript antisense RNA (HOTAIR)[11], H19[12], and metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)[13]. Growth arrest-specific 5[14] and maternally expressed 3[15] can act as suppressors of tumors. Given the fundamental and intrinsic advantages of lncRNAs, at present, they are considered to be promising diagnostic and prognostic biomarkers in cancer[16-19]. However, few studies have conducted a comprehensive examination of survival-associated lncRNA expression in CRC.
The aim of this study was to exploit the potential prognostic value of lncRNA signatures in CRC patients using a whole-transcriptome RNA-sequencing approach. The findings obtained in this study might help to elucidate the functions of lncRNAs and identify novel therapeutic targets for patients with CRC.
The clinical characteristics of patients with CRC and their lncRNA expression profiles were available at The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/projects). Patients were included if they met the following criteria: Both lncRNA expression profiles and complete clinical follow-up data were available, OS[9] was more than 30 d, and lncRNAs had an average count > 1 and appeared in > 70% of the total samples. In total, 349 patients were enrolled in our study. Subsequently, these patients were randomly divided into the training set (n = 175) and testing set (n = 174). In addition, the expression level of each lncRNA was defined as the log2 (x + 1) for further analysis.
We analyzed the relationship between lncRNAs and the OS of CRC patients in the training set. Univariable Cox regression analysis was performed to screen for CRC-specific lncRNAs associated with OS (P < 0.05). These seed lncRNAs were employed to further select a prognosis-associated predictive model by multivariable Cox regression analysis. According to robust likelihood-based survival analysis, a series of predictive models were performed by the R package Rbsurv. The candidate predictive models constructed by the multiple prognosis-related lncRNAs were strictly selected by Akaike information criteria. Finally, the ideal predictive model with the minimum Akaike information criteria value was determined.
The risk score model of prognosis-associated lncRNAs was established by multivariable Cox regression analysis and weighted by the regression coefficients. In the training set, the risk score of each patient was calculated by the risk score formula. Patients were subsequently divided into low-risk and high-risk score groups based on the median risk score. The receiver operating characteristic curve was obtained by R with the survival receiver operating characteristic (ROC) package (version 1.0.3) to predict OS at 1, 3, and 5 years. The optimal cutoff point was selected by the maximum specificity and sensitivity. Survival differences between the low-risk and high-risk groups were assessed by Kaplan-Meier curves and log-rank tests. Furthermore, the risk score model was validated in the testing set and TCGA set. The prognostic performance of the risk score model was measured by a time-dependent ROC curve. Multivariable Cox proportional hazards regression was performed to evaluate the independence of these nine CRC-specific lncRNAs. Stratification analysis of common clinical characteristics, such as tumor stage and patient sex, was conducted by R with the survival ROC package. Overall statistical analyses were performed by R language (Version 3.4.3).
The correlation networks between prognostic lncRNAs and potential target genes were investigated by the Spearman’s rank-correlation test. Moreover, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were employed to analyze the biological processes of target genes and pathways by Cluster Profiler. The enriched functional annotations of GO terms and KEGG pathways were considered to be significant when the P value was less than 0.05.
A total of 349 patients from TCGA database were included and analyzed in this study; the patients were randomly divided into a training set (n = 175) and testing set (n = 174). A total of 5152 LncRNAs were identified from 349 patients in TCGA database. Among these acquired lncRNAs, 2149 had a mean Fragments Per Kilobase of transcript per Million mapped reads of > 0.1. The baseline demographic and clinical characteristics of the two groups did significantly differ (P > 0.05). These features are summarized in Supplementary Table 1.
A total of 104 prognosis-associated lncRNAs of CRC were identified by univariable Cox regression analysis. The combination of nine lncRNAs was selected from these 104 LncRNAs by robust likelihood-based survival analysis (Table 1). Interestingly, all nine CRC-specific lncRNAs showed positive regression coefficients, indicating that these lncRNAs were associated with a high risk of mortality in CRC.
| Characteristics | Train set, n = 175 | Test set, n = 174 | Entire set, n = 349 |
| Age, mean ± SD | 65.38 ± 13.25 | 65.29 ± 13.12 | 65.33 ± 13.19 |
| Sex, male/female | 94/81 | 93/81 | 187/162 |
| Clinical stage, I/II/III/IV | 31/64/55/25 | 34/75/50/15 | 65/139/105/40 |
| Status, deceased/alive | 23/152 | 15/159 | 38/311 |
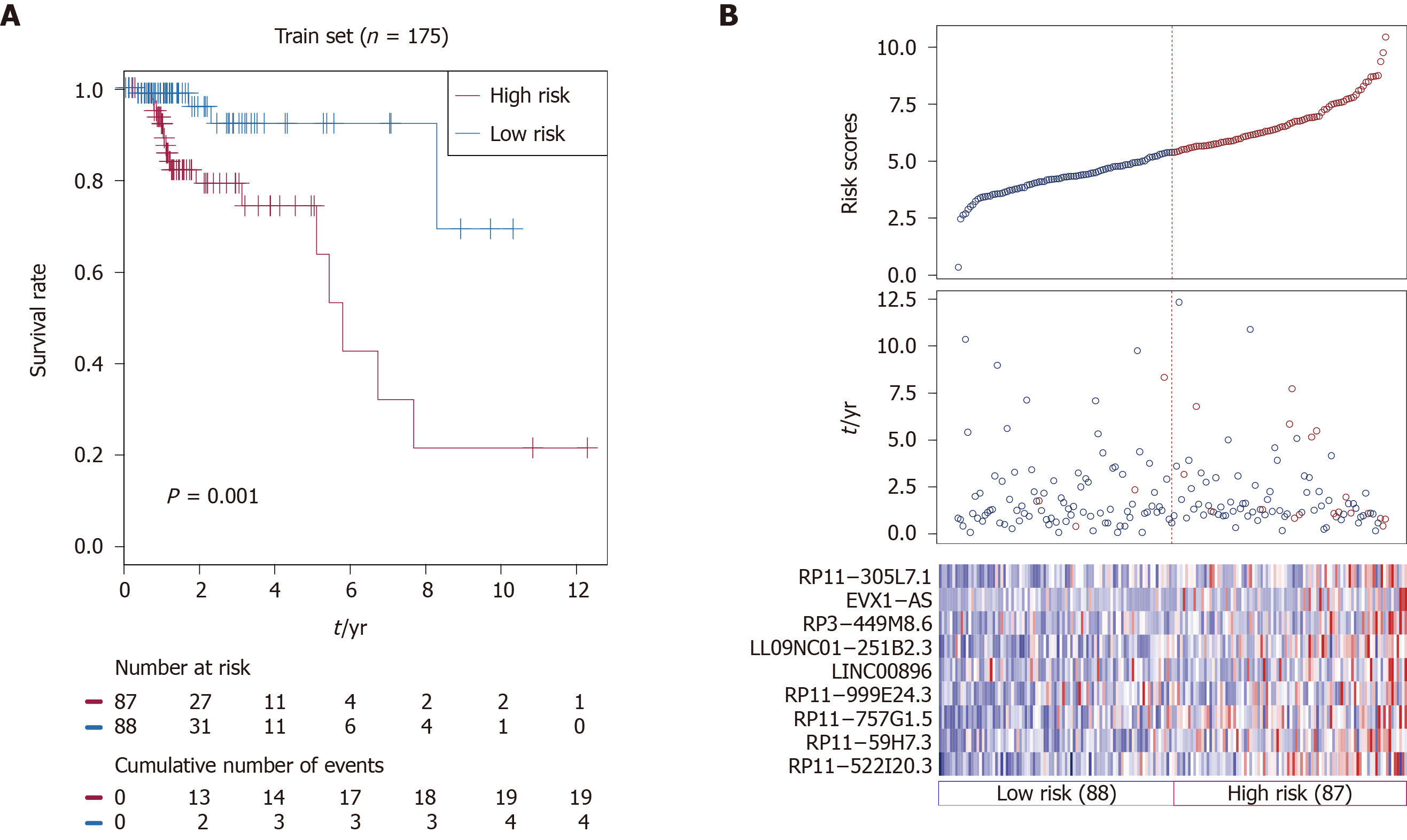
Based on nine identified prognostic lncRNAs, we designed a risk prediction formula and calculated the risk score for each patient in the training set to facilitate the application of these lncRNAs in clinical practice. The 175 CRC patients in the training set were classified into a high-risk group (n = 87) and a low-risk group (n = 88) with differential survival outcome based on the median value of the risk score (5.38), as the optimal cut-off point. Clearly, patients in the low-risk group showed a better outcome than those in the high-risk group by Kaplan-Meier analysis (P = 0.001; Figure 1A).
The distribution of the risk scores, OS, survival status, and corresponding lncRNA expression profiles of the 175 patients in the training set are shown in Figure 1B. These nine CRC-specific lncRNAs tended to be more highly expressed in the high-risk group. Notably, the high-risk group consisted of 19 patients who died and 68 patients who did not die, whereas the low-risk group consisted of 4 patients who died and 84 patients who did not die. However, the difference between the two groups in OS was only marginally significant (chi-square test, P = 0.003).
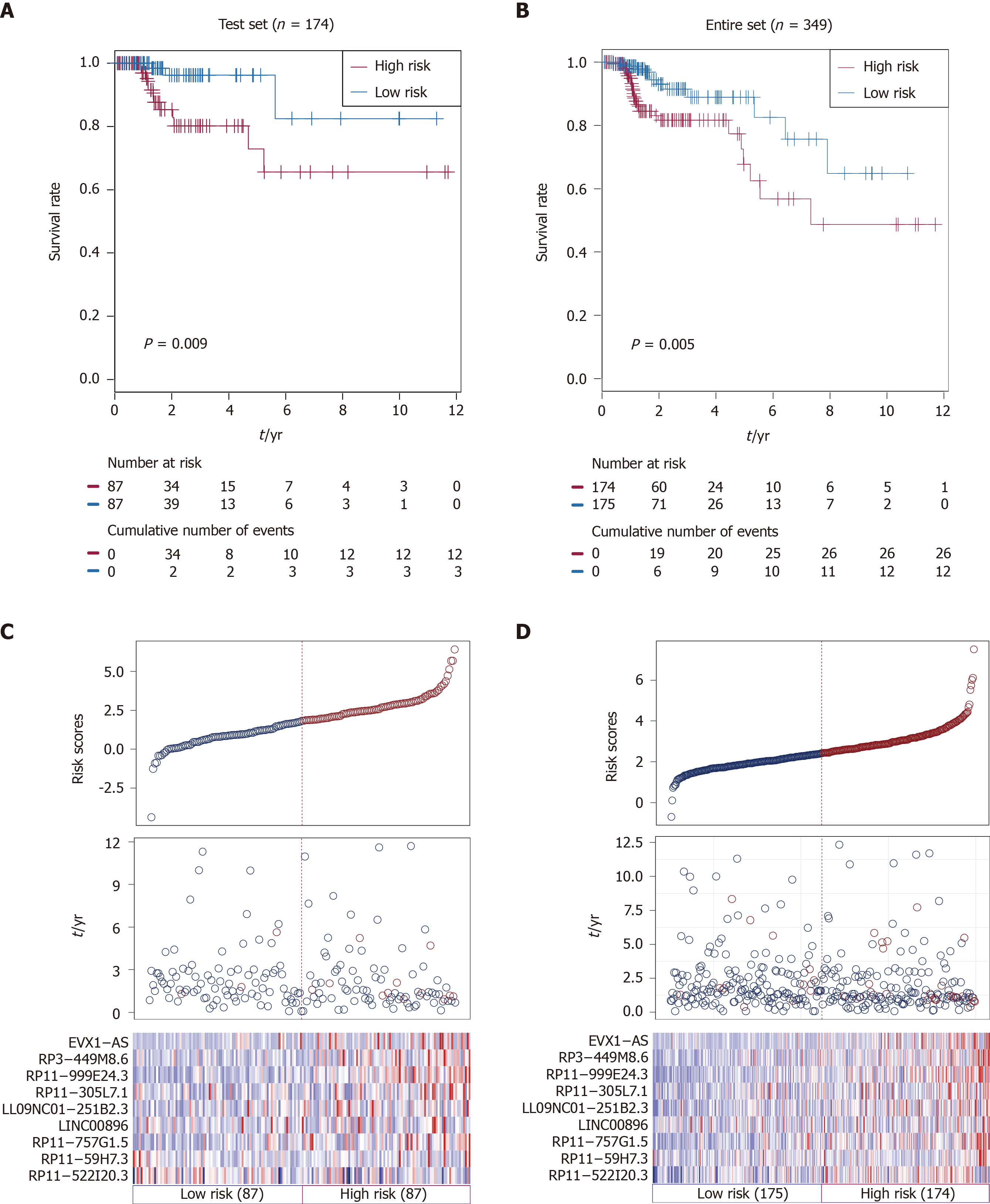
Using the same risk score formula and threshold value in the training set, patients in the testing set (n = 174) and TCGA set (n = 349) were assigned into high-risk and low-risk groups. There was convincing evidence that patients in the high-risk group of the testing set (n = 87) had poorer outcomes than those in the low-risk group, as determined by Kaplan-Meier analysis (P = 0.009; Figure 2A). Similar results were observed in the TCGA set (P < 0.005; Figure 2B).
The distributions of risk scores, OS, survival status, and corresponding lncRNA expression profiles in the testing set and entire set of patients are shown and ranked according to increasing risk scores (Figure 2C and D). Likewise, these nine risk-associated lncRNAs were downregulated in the low-risk group and upregulated in the high-risk group. Moreover, the majority of patients who died in the testing set and TCGA set were clustered into the high-risk group. In the testing set, the high-risk group consisted of 12 patients who died and 75 patients who did not die, whereas the low-risk group consisted of 3 who died and 84 who did not die. The difference in OS between the two groups was significant (chi-square test, P = 0.015). Similarly, the high-risk group consisted of 26 patients who died and 148 patients who did not die in TCGA set, while the low-risk group consisted of 12 patients who died and 163 patients who did not die. Consistent with the testing set, the difference in OS between the two groups of the entire set was significant (chi-square test, P = 0.015).
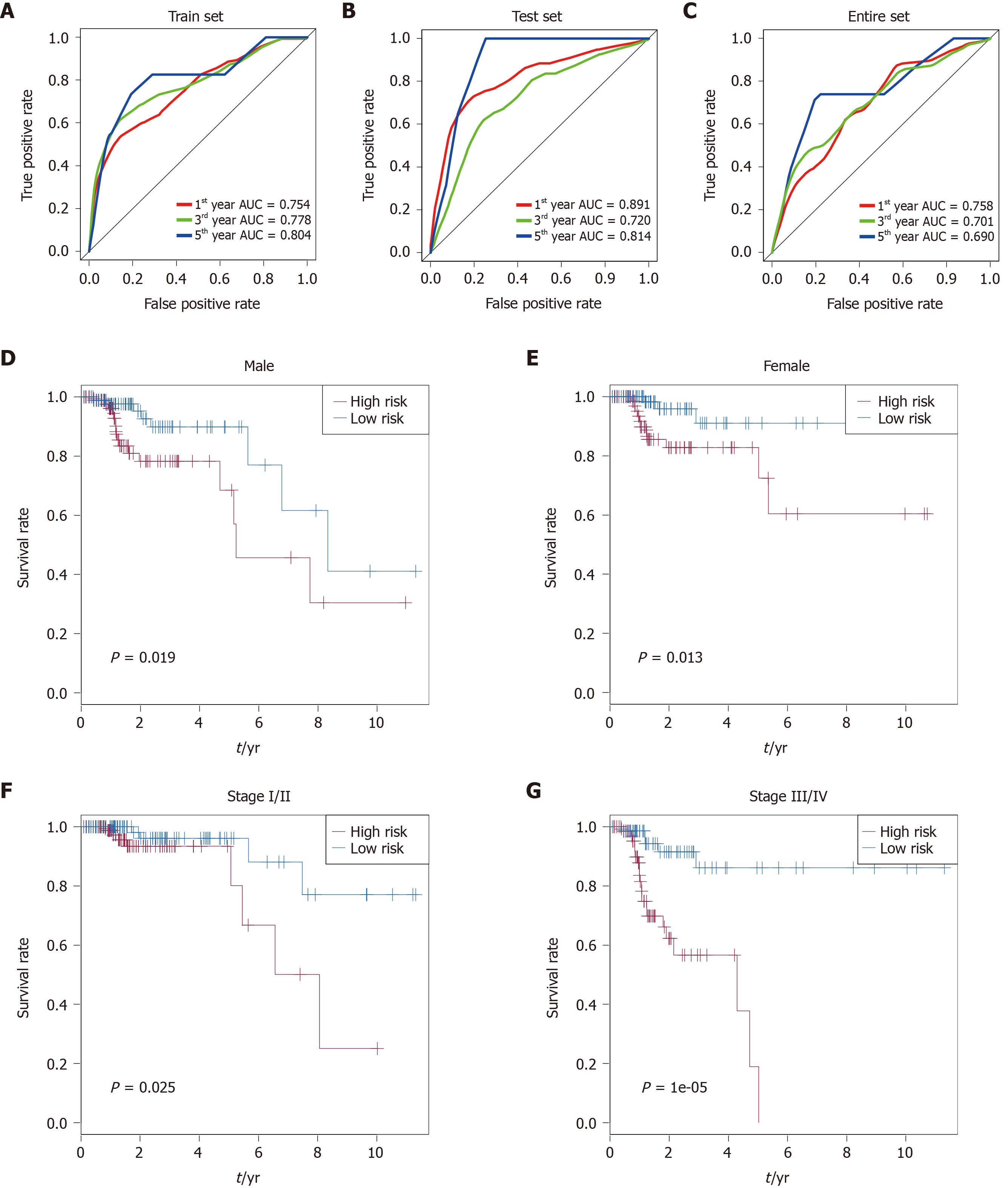
Time-dependent receiver operating characteristic curve analysis was performed to evaluate the prognostic performance of the nine CRC-specific lncRNAs. These nine prognostic lncRNAs achieved area under the curve (AUC) values of 0.754, 0.778, and 0.804 for predicting prognosis in the training set at 1, 3, and 5 years, respectively (Figure 3A). The AUC values were 0.891, 0.720, and 0.814 in the testing set at 1, 3, and 5 years, respectively (Figure 3B), and 0.758, 0.701, and 0.690 in the entire set at 1, 3, and 5 years, respectively (Figure 3C). Most of the AUC values exceeded 0.7, indicating that these nine prognostic lncRNAs performed well for the prediction of prognosis in CRC patients.
Stratification analysis of tumor stage and patient sex was conducted to determine whether these nine CRC-specific lncRNAs maintain their prognostic value in the different contexts of common clinical features. All 349 patients were stratified by tumor stage into a stage I/II dataset (n = 204) and a stage III/IV dataset (n = 145). Based on these nine prognostic lncRNAs, patients in the stage I/II dataset were classified into a high-risk group (n = 102) or a low-risk group (n = 102) with a significant difference in OS (P = 0.025, Figure 3F). Similarly, the patients in the stage III/IV dataset were also classified into a high-risk group (n = 73) and a low-risk group (n = 72), which also differed significantly in OS (P = 1e-05; Figure 3G). Similarly, all 349 patients were stratified by gender into a male dataset (n = 187) and a female dataset (n = 162). Patients in the male dataset could be stratified into a high-risk group (n = 94) and a low-risk group (n = 93), with a significant difference in OS being observed (P = 0.019; Figure 3D). An analogous result was obtained in the female dataset (P = 0.013; Figure 3E).
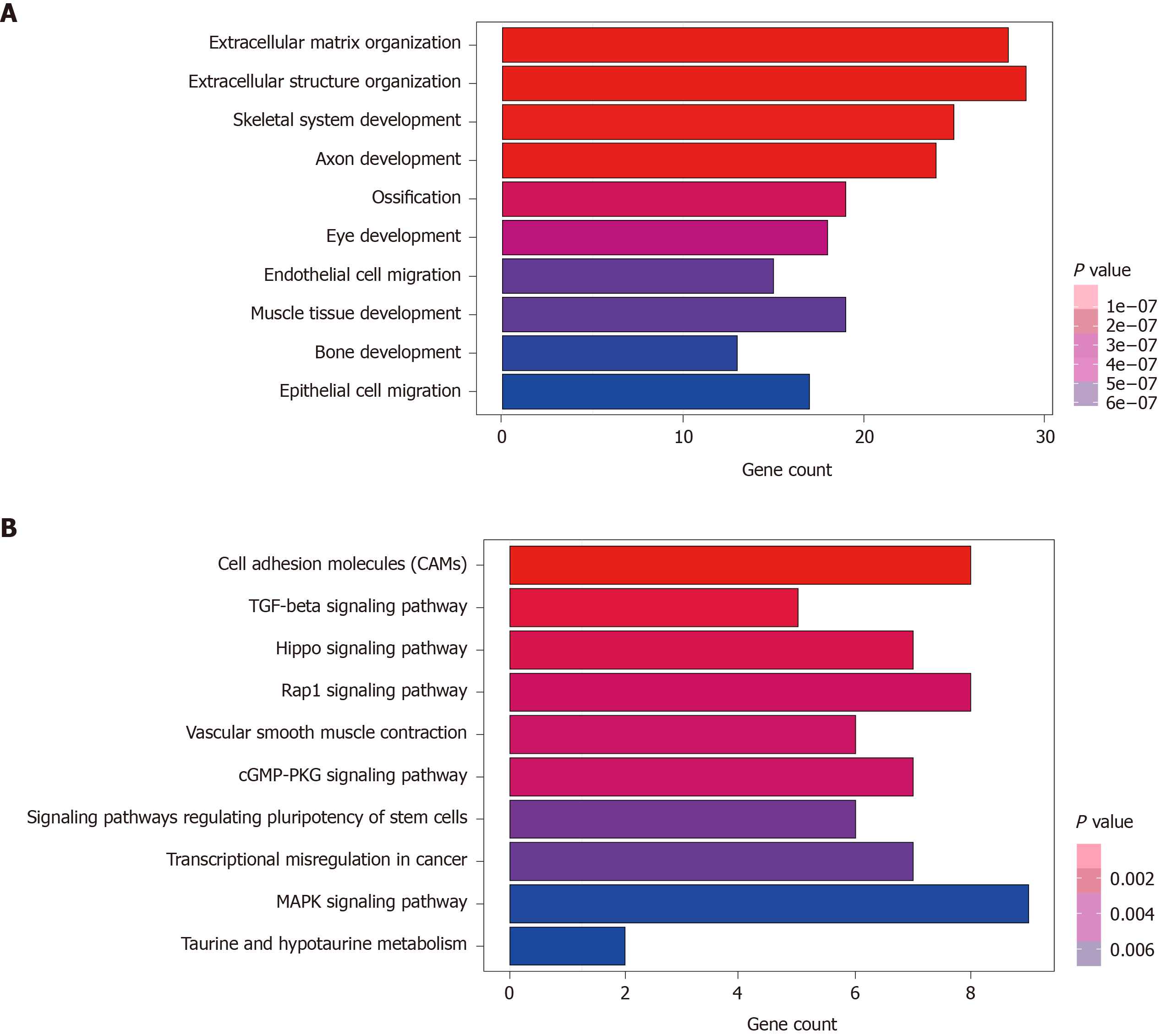
A total of 270 potential target genes of these nine lncRNAs were first identified by GO terms and KEGG pathway functional enrichment analysis. The 10 most enriched KEGG pathways are shown in Figure 4A. A series of cancer-related pathways were highly activated in CRC patients, such as cell adhesion molecules and the Hippo and Rap1 signaling pathways. The 10 most enriched GO terms related to biological processes are shown in Figure 4B; these processes have long been recognized as functions of lncRNAs. The 10 most enriched GO terms primarily included the extracellular matrix and structure organization, suggesting that these nine prognostic lncRNAs were closely correlated with the carcinogenesis of gene expression and biological functions.
An optimal prognostic predictor model for risk stratification in CRC patients was constructed by the combination of the nine-lncRNA signature and the risk score formula. In the comprehensive analysis of lncRNA-sequencing data, this nine-lncRNA signature could effectively divide patients into high-risk and low-risk groups in the training series with significantly different OS. Next, we successfully validated the relationship of this nine-lncRNA signature with the prognosis of CRC patients in the testing set and entire set, indicating good reproducibility and reliability for the prediction of prognosis. The top 10 enriched KEGG pathways and GO terms showed that the regulation of gene expression and critical cell biological functions were closely related to these nine prognostic lncRNAs, suggesting that they play a crucial role in prognosis prediction and the progression of CRC.
There are five RP11 lncRNAs among these nine prognosis-related lncRNAs, namely, RP11-305L7.1, RP11-999E24.3, RP11-757G1.5, RP11-59H7.3, and RP11-522I20.3. Nevertheless, none of these lncRNAs has been previously reported with respect to CRC. Several RP11 lncRNAs have been considered to be unfavorable factors in previous studies such as RP11-650L12.2 in CRC[20], RP11-357H14.17 in diffuse-type gastric carcinoma[21], and RP11-445H22.4 in breast cancer[22]. In contrast, other studies have demonstrated that the expression of some RP11 LncRNAs was downregulated such as RP11-3N2.1 and RP11-462C24.1 in CRC[23,24], lncRNA RP11-119F7.4 in gastric cancer[25], and RP11-766N7.4 in esophageal squamous cell carcinoma[26]. According to the current research, the RP11 lncRNAs mentioned above were correlated with tumorigenesis, invasion depth, metastasis, tumor size and stage, and they were even considered independent prognostic indicators for the survival of patients with cancer. Therefore, these five newly identified RP11 lncRNAs have the potential to be confirmed through further experimental research, possibly becoming prognostic biomarkers and therapeutic targets. The enriched GO terms in our study primarily included extracellular matrix and structure organization. Matrix metalloproteinases (MMPs) and pericellular proteases can degrade matrix proteins and modulate cancer metastasis by regulating the cleavage of proteins, thereby degrading the protective barrier against CRC invasion and metastasis[27,28]. HOTAIR and FOXF1 adjacent non-coding developmental regulatory RNA were observed to participate in extracellular matrix degradation by modulating the expression of cancer metastasis-associated genes such as intracellular adhesion molecule 1, MMP1, MMP2, MMP3, and MMP9[29,30]. This finding was in line with previous reports on gastric cancer metastasis showing that lncRNAs are aberrantly expressed in gastric cancer metastasis and play important roles in stabilizing or degrading the extracellular matrix. LncRNAs are still involved in cell-to-cell junctions and adhesion between cells by regulating the diffusion of ions and specific molecules and maintaining the integrity of the cell-to-cell protective barrier. Tight junctions between cells are potential targets for therapeutic intervention in tumor metastasis. The enriched KEGG pathways in our study indicated that the molecular pathway of cell adhesion was highly activated in patients with CRC. Indeed, the aberrant expression or distribution of tight junction proteins leads to the loss of cell-to-cell adhesion and tissue integrity, thereby facilitating CRC cancer cell invasion and metastasis[31,32]. Hence, these nine prognosis-related lncRNAs may regulate cancer cell growth and adhesion via TUC339[33].
Specifically, in our analysis of other GO functional enrichment and KEGG pathways, these nine-lncRNA signature lncRNAs were also enriched in such biological processes as axon development, endothelial cell migration, epithelial cell migration, the Hippo signaling pathway, the cyclic GMP-protein kinase G signaling pathway, and transcriptional dysregulation. With the convincing data obtained in our series, nine prognosis-related lncRNAs were determined to be involved in many critical processes of cancer development and progression. This observation highlights the importance of newly discovered lncRNAs as a promising field for the development of future molecular targeted therapies for CRC. It is necessary to explore the specific pathogenic mechanisms of each lncRNA in CRC to achieve further improvements in clinical outcomes. Additional research to comprehensively characterize these nine lncRNAs should be recommended.
With the development of high-throughput sequencing and bioinformatic approaches, many differentially expressed lncRNAs in CRC have been identified. Recent reports have described the heavily researched lncRNAs involved in CRC[34], including the CRCMALAT1, HOTAIR, H19, and CCAT families. Unlike some heavily researched lncRNAs that have been adequately investigated, this nine-lncRNA signature has not been reported to date and warrants further research. The discovery of the nine-lncRNA signature in our study not only represents new insight into the molecular architecture of CRC but also opens up the possibility of using lncRNAs as diagnostic biomarkers and therapeutic targets. lncRNAs may possess considerable advantages as biomarkers, especially when they can be readily detected in biological fluids. However, taking into account the nature of lncRNAs as long RNA molecules, some other factors should be considered, such as their stability in the circulation and in various disease states.
Our study identified a nine-lncRNA signature for predicting the survival of CRC patients by comprehensive data analysis. This signature was reproducible and reliable in a second independent large-scale CRC cohort, supporting their value and effectiveness. To the best of our knowledge, a preliminary investigation of the function of this nine-lncRNA signature has not been reported to date, which further strengthens the possibility that the nine-lncRNA signature could be used effectively to predict the disease course in CRC. In addition, the lncRNA profiling approach described in this report could potentially be applied in other kinds of studies and serve as a useful method for the systematic identification of lncRNA biomarkers in clinical practice.
This study had some limitations. A small proportion of the results in the stratified survival analysis were not significant but rather showed trend differences, which may be attributed to the limited sample size after repeated grouping. Therefore, independent cohorts from multicenter studies in large populations are required to validate the prognostic value of these lncRNA signatures before they can be applied to clinical practice. Moreover, we could not examine the cause-effect relationship between this modeled risk score and the prognosis status of CRC patients. Future studies may attempt to validate our findings in planned clinical trials and investigate the functions of these lncRNAs, providing a more convincing explanation of the biological implications and molecular mechanisms of these prognostic lncRNAs in CRC.
Our study identified a nine-lncRNA signature for predicting the survival of CRC patients by comprehensive data analysis. This signature was reproducible and reliable in a second independent large-scale CRC cohort, supporting their value and effectiveness. To the best of our knowledge, a preliminary investigation of the function of this nine-lncRNA signature has not been reported to date, which further strengthens the possibility that the nine-lncRNA signature could be used effectively to predict the disease course in CRC. In addition, the lncRNA profiling approach described in this report could potentially be applied in other kinds of studies and serve as a useful method for the systematic identification of lncRNA biomarkers in clinical practice.
To investigate molecular biomarkers that accurately predict prognosis would be of great clinical significance. Increasing evidence suggests long non-coding ribonucleic acids (lncRNAs) are frequently aberrantly expressed in colorectal cancer (CRC).
To elucidate the prognostic function of multiple lncRNAs that served as biomarkers in CRC.
To study the lncRNAs that are reportedly involved in various biological processes of CRC including proliferation, immortality, angiogenesis, growth suppression, motility and viability.
We collected lncRNA expression profiling using the lncRNA-mining approach in large CRC cohorts from The Cancer Genome Atlas (TCGA) database. Receiver operating characteristic analysis was performed to identify the optimal cut-off point, which patients could be classified into the high-risk or low-risk group. Based on the Cox co-efficient of the individual lncRNAs, we identified nine-lncRNA signature that are associated with survival of patients with CRC in the training set (n = 175). The prognostic value of this nine-lncRNA signature was validated in the testing set (n = 174) and TCGA set (n = 349) respectively. The prognostic models were comprised by these nine CRC-specific lncRNAs, performing well for risk stratification in the testing set and TCGA set. Time-dependent receiver operating characteristic analysis indicated that this predictive model had well performance.
Multivariate Cox regression and stratification analysis showed that a nine-lncRNA signature was independent of other clinical features in predicting overall survival. Functional enrichment analysis of Kyoto Encyclopedia of Genes and Genomes pathways and Gene Ontology terms further indicated these nine prognostic lncRNAs were closely associated with carcinogenesis associated pathways and biological functions in CRC.
A nine-lncRNA expression signature was identified and validated which could improve the prognosis prediction of CRC, providing potential prognostic biomarkers and efficient therapeutic targets for patients with CRC.
Our present study identified nine-lncRNA signature for survival prediction of CRC patients by the comprehensive data analysis. This signature was reproducible and reliable in a second independent large-scale CRC cohort, supporting their value and effectiveness. To the best of our knowledge, preliminary investigation of the function of this nine-lncRNA signature has not been reported, which further strengthen the possibility that the nine-lncRNA signature could be used effectively to predict disease course in CRC. In addition, the lncRNA profiling approach described here could potentially be applied in other kinds of studies and served as a useful method for the systematic identification of lncRNA biomarkers in clinical practice.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ding L, Sung WW S-Editor: Zhang L L-Editor: Filipodia P-Editor: Li JH
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50871] [Article Influence: 8478.5] [Reference Citation Analysis (44)] |
| 2. | Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9172] [Cited by in F6Publishing: 9816] [Article Influence: 1090.7] [Reference Citation Analysis (0)] |
| 3. | Wang LS, Arnold M, Huang YW, Sardo C, Seguin C, Martin E, Huang TH, Riedl K, Schwartz S, Frankel W, Pearl D, Xu Y, Winston J 3rd, Yang GY, Stoner G. Modulation of genetic and epigenetic biomarkers of colorectal cancer in humans by black raspberries: a phase I pilot study. Clin Cancer Res. 2011;17:598-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 4. | Wong JJ, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56:140-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Skroblin P, Mayr M. "Going long": long non-coding RNAs as biomarkers. Circ Res. 2014;115:607-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 6. | Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt S, Johnson N, Juettemann T, Kähäri AK, Keenan S, Kulesha E, Martin FJ, Maurel T, McLaren WM, Murphy DN, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ruffier M, Sheppard D, Taylor K, Thormann A, Trevanion SJ, Vullo A, Wilder SP, Wilson M, Zadissa A, Aken BL, Birney E, Cunningham F, Harrow J, Herrero J, Hubbard TJ, Kinsella R, Muffato M, Parker A, Spudich G, Yates A, Zerbino DR, Searle SM. Ensembl 2014. Nucleic Acids Res. 2014;42:D749-D755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1072] [Cited by in F6Publishing: 1054] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 7. | Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 388] [Cited by in F6Publishing: 394] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 8. | Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253-1261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1625] [Cited by in F6Publishing: 1915] [Article Influence: 239.4] [Reference Citation Analysis (0)] |
| 9. | Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1788] [Cited by in F6Publishing: 1975] [Article Influence: 219.4] [Reference Citation Analysis (0)] |
| 10. | Johnsson P, Morris KV. Expanding the functional role of long noncoding RNAs. Cell Res. 2014;24:1284-1285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 11. | Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med. 2015;12:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 215] [Reference Citation Analysis (0)] |
| 12. | Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis - a proposed unifying theory. Mol Cancer. 2015;14:184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 398] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 13. | Gutschner T, Hämmerle M, Diederichs S. MALAT1 -- a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl). 2013;91:791-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 514] [Cited by in F6Publishing: 553] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 14. | Pickard MR, Williams GT. Molecular and Cellular Mechanisms of Action of Tumour Suppressor GAS5 LncRNA. Genes (Basel). 2015;6:484-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 177] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 15. | Zhou Y, Zhang X, Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J Mol Endocrinol. 2012;48:R45-R53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 563] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 16. | Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 238] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 17. | Zhang T, Wu DM, Deng SH, Han R, Liu T, Li J, Xu Y. Integrated Analysis Reveals That Long Non-Coding RNA TUBA4B Can Be Used as a Prognostic Biomarker in Various Cancers. Cell Physiol Biochem. 2018;49:530-544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Wang Y, Ding X, Hu H, He Y, Lu Z, Wu P, Tian L, Xia T, Yin J, Yuan H, Shi G, Liu D, Jiang K, Miao Y. Long non-coding RNA lnc-PCTST predicts prognosis through inhibiting progression of pancreatic cancer by downregulation of TACC-3. Int J Cancer. 2018;143:3143-3154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 19. | Zhu X, Tian X, Yu C, Shen C, Yan T, Hong J, Wang Z, Fang JY, Chen H. A long non-coding RNA signature to improve prognosis prediction of gastric cancer. Mol Cancer. 2016;15:60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 20. | Jin M, Ye D, Li Y, Jing F, Jiang X, Gu S, Mao Y, Li Q, Chen K. Association of a novel genetic variant in RP11-650L12.2 with risk of colorectal cancer in Han Chinese population. Gene. 2017;624:21-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Yang B, Luo T, Zhang M, Lu Z, Xue X, Fang G. The novel long noncoding RNA RP11-357H14.17 acts as an oncogene by promoting cell proliferation and invasion in diffuse-type gastric cancer. Onco Targets Ther. 2017;10:2635-2643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Xu N, Chen F, Wang F, Lu X, Wang X, Lv M, Lu C. Clinical significance of high expression of circulating serum lncRNA RP11-445H22.4 in breast cancer patients: a Chinese population-based study. Tumour Biol. 2015;36:7659-7665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 52] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 23. | Ye D, Hu Y, Jing F, Li Y, Gu S, Jiang X, Mao Y, Li Q, Jin M, Chen K. A novel SNP in promoter region of RP11-3N2.1 is associated with reduced risk of colorectal cancer. J Hum Genet. 2018;63:47-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu Y, Li X, Cai G, Cai S. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31:31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 25. | Sun J, Song Y, Chen X, Zhao J, Gao P, Huang X, Xu H, Wang Z. Novel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancer. Oncol Lett. 2015;10:115-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 31] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 26. | Yao GL, Pan CF, Xu H, Wei K, Liu B, Zhai R, Chen YJ. Long noncoding RNA RP11-766N7.4 functions as a tumor suppressor by regulating epithelial-mesenchymal transition in esophageal squamous cell carcinoma. Biomed Pharmacother. 2017;88:778-785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 27. | Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331-2347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 28. | Westermarck J, Kähäri VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13:781-792. [PubMed] [Cited in This Article: ] |
| 29. | Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587-597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 208] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 30. | Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, De W, Shu YQ. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 216] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 31. | Martin TA. The role of tight junctions in cancer metastasis. Semin Cell Dev Biol. 2014;36:224-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 32. | Martin TA, Jiang WG. Loss of tight junction barrier function and its role in cancer metastasis. Biochim Biophys Acta. 2009;1788:872-891. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 313] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 33. | Kogure T, Yan IK, Lin WL, Patel T. Extracellular Vesicle-Mediated Transfer of a Novel Long Noncoding RNA TUC339: A Mechanism of Intercellular Signaling in Human Hepatocellular Cancer. Genes Cancer. 2013;4:261-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 223] [Cited by in F6Publishing: 243] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 34. | Wang J, Song YX, Ma B, Wang JJ, Sun JX, Chen XW, Zhao JH, Yang YC, Wang ZN. Regulatory Roles of Non-Coding RNAs in Colorectal Cancer. Int J Mol Sci. 2015;16:19886-19919. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |