Published online Dec 15, 2014. doi: 10.4251/wjgo.v6.i12.444
Revised: October 28, 2014
Accepted: October 31, 2014
Published online: December 15, 2014
AIM: To evaluate the potential prognostic value of GNAS1 T393C polymorphism in advanced non-small cell lung cancer.
METHODS: We extracted genomic DNA from the peripheral blood leucocytes of 94 patients with advanced non-small cell lung cancer. Quantitative real-time polymerase chain reaction was used to determine the allelic discrimination. The correlation between genotype and overall survival was evaluated using the multivariate analysis and Kaplan-Meier approach.
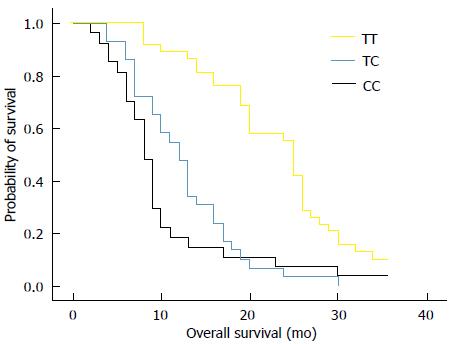
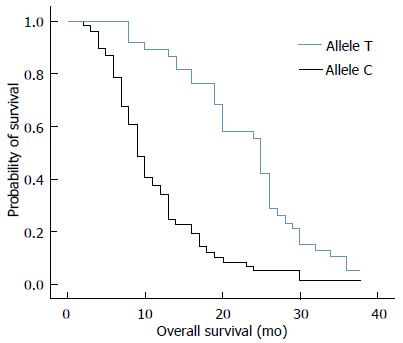
RESULTS: Thirty-eight out of 94 (40%) patients displayed a TT genotype, 29 out of 94 (31%) a CT genotype and 27 out of 94 (29%) a CC genotype. The median survival of TT (25 mo) genotype carriers was longer than CT (12 mo) or CC (8 mo) genotype carriers. The favorable TT genotype predicted better overall survival (OS) (2-year OS: 48%; P =0.01) compared with CT (2-year OS: 18%) or CC (2-year OS: 15%) genotype. However, dichotomization between C-genotypes (CC + CT) and T-genotypes (TT) revealed significantly lower survival rates (2-year OS: 16%; P = 0.01) for C allele carriers.
CONCLUSION: Our data provided strong evidence that the GNAS1 T393C genetic polymorphism influenced the prognosis in advanced non-small lung cancer with a worse outcome for C allele carriers.
Core tip: We genotyped GNAS1 T393C single nucleotide polymorphism in a homogenous (Han) study population of patients to evaluate the effect of this polymorphism on survival in non-small cell lung cancer (NSCLC). Our study indicated that the GNAS1 T393C polymorphism affected the overall survival in advanced NSCLC with a worse outcome for C allele carriers.
-
Citation: Gong HY, Hu WG, Wang XL, Zhu F, Song QB. TT genotype of
GNAS1 T393C polymorphism predicts better outcome of advanced non-small cell lung cancer patients. World J Gastrointest Oncol 2014; 6(12): 444-449 - URL: https://www.wjgnet.com/1948-5204/full/v6/i12/444.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v6.i12.444
The incidence of lung cancer has increased substantially over the past ten years[1]. Non-small cell lung cancer (NSCLC) constitutes about 85% of all lung cancer cases[2] with only 16.6% being able to live 5 years or more after diagnosis[3]. To date, the most feasible treatment for advanced NSCLC patients is the platinum-based combination chemotherapy and it turns out to be associated with better overall survival rates[4]. Tumor-node-metastasis stage normally correlates with the clinical outcome of a large population of patients, but patients with similar clinical characteristics have different outcomes, which may be affected by their individual genes. The identification of patients with high-risk lung cancer could thus help to set up novel treatment strategies and could theoretically improve the outcome of anti-cancer therapy. Therefore, it is desirable to characterize more reliable and accurate molecular markers to identify more aggressive lung cancer phenotypes in order to individually tailor the therapy.
Actually, previous studies have implied that biomarkers could help define the subgroups of patients. However, there is no standard way to immunohistochemically detect these biomarkers, which prevents their application as prognostic factors. Nowadays, people choose to study single nucleotide polymorphisms (SNPs) as prognostic markers because these SNPs can be easily evaluated using patients’ blood samples, which can avoid issues such as the availability and the quality of materials. One typical example is the GNAS1 T393C polymorphism.
The GNAS1 gene has been mapped to chromosome 20q13 and contains 13 exons. The GNAS1 T393C polymorphism is located in exon 5, which encodes the α-subunit of the stimulatory G protein, namely Gαs. Somatic mutations of GNAS1 have been reported to be involved in the etiology of McCune-Albright syndrome and sporadic, isolated endocrine tumors[5-7], suggesting that GNAS1 could participate in cancer initiation and progression. What’s more, previous studies have demonstrated that the T393C polymorphism was significantly correlated with the prognosis of patients with various cancers, such as breast carcinoma, squamous cell carcinoma of the larynx, bladder cancer, cholangiocarcinoma, colorectal cancer, clear cell renal carcinoma, and cancers of the oropharynx and hypopharynx[8-20].
In this study, we genotyped the T393C SNP in a Han population to evaluate the effect of this polymorphism on lung cancer prognosis. Our purpose was to determine whether the common GNAS1 T393C polymorphism can be used as a predictive factor for survival in NSCLC patients.
Two milliliters of peripheral blood samples were collected from patients diagnosed with advanced NSCLC pathologically before any antineoplastic treatment at Renmin Hospital of Wuhan University (China) between March 2010 and March 2012. Patients were chosen based on the following criteria: (1) histologically confirmed UICC (2009) stage IIIB or IV NSCLC; (2) Eastern Cooperative Oncology Group performance status (PS) score of 2 or less; and (3) life expectancy of more than 3 mo. Patients were not included if they had received any anti-tumor therapy previously. All patients were asked to sign the informed consent before they were included in the database. The study cohort (94 patients; for clinicopathological data, Table 1) composed exclusively of patients with a meticulously complete follow-up record. This study was performed following the guidelines of the local research ethics committee.
| Subgroup | n | MST | 1-yr OS (%) | 2-yr OS (%) | P | |
| Gender | Male | 71 | 14 | 59 | 36 | 0.058 |
| Female | 23 | 13 | 53 | 28 | ||
| Age | ≥ 60 yr | 51 | 13 | 61 | 26 | 0.135 |
| < 60 yr | 43 | 16 | 52 | 36 | ||
| PS | ≥ 2 | 25 | 13 | 51 | 25 | 0.658 |
| < 2 | 69 | 17 | 64 | 32 | ||
| Smoking | Yes | 23 | 13 | 55 | 29 | 0.473 |
| No | 71 | 14 | 58 | 32 | ||
| Pathology | Adenocarcinoma | 48 | 13 | 54 | 36 | 0.559 |
| Squamous cell carcinoma | 46 | 14 | 63 | 29 | ||
| Treatment | Supportive treatment only | 12 | 10 | 49 | 25 | 0.116 |
| Chemotherapy | 14 | 13 | 56 | 32 | ||
| Radiotherapy | 11 | 13 | 60 | 30 | ||
| Chemoradiotherapy | 57 | 16 | 64 | 35 | ||
| GNAS1 T393C | TT | 38 | 25 | 76 | 48 | 0.01 |
| TC | 29 | 12 | 54 | 18 | ||
| CC | 27 | 8 | 23 | 15 | ||
| TC + CC | 56 | 11.5 | 25 | 16 | 0.01 |
Genomic DNA was isolated from whole blood samples using the QIAamp kit (Qiagen, Germany). T393C SNP (dbSNP rs7121) was amplified by polymerase chain reaction (PCR) with the following primers: 5’-CAGCCCACATTAGGGAGCATAT-3’ (forward) and 5’-TAATCCCTGCCTATGCTCACGA-3’ (reverse). After denaturation at 95 °C, 50 cycles of DNA amplification was done using (NH4)2SO4 containing buffer (Bioron, Germany) at 95 °C for 60 s, 60 °C for 30 s, and 70 °C for 60 s. The 807-bp PCR product was genotyped according to their sequences.
The software SPSS 17.0 was used for statistical analyses in this study. Descriptive statistics were applied to describe patients’ baseline characteristics. The correlation between T393C genotypes and the clinical outcome was evaluated by Kaplan-Meier plots and the log-rank test. The survival time was calculated from the date of the primary diagnosis to the end of follow-up or date of death, whichever occurred first. The independent influence of T393C SNP and other covariates on survival rates was assessed in multivariate analysis using the Cox regression hazard model. P values < 0.05 were considered statistically significant. The compatibility with the Hardy-Weinberg equilibrium was calculated with HWE program (http:// linkage.rockefeller.edu/ott/linkutil.htm).
The clinicopathological characteristics of patients with genotype distribution are shown in Table 1. There were 94 advanced NSCLC patients participating in this study, including 23 women and 71 men. The average age of participants was 58.6 years (range, 31 to 80 years).
Among 94 patients, 38 (40%) displayed a TT genotype, 29 (31%) with a CT genotype and 27 (29%) with a CC genotype. In the entire patient group, the frequency of the C allele (fC) was 0.55. The distribution was compatible with the Hardy-Weinberg equilibrium. There was no significant correlation between the GNAS1 T393C genotypes and clinicopathological parameters, such as age (P = 0.48), gender (P = 0.42), PS (P = 0.30), smoking status (P = 0.44) or pathology (P = 0.59) (Table 2). Further analysis showed that there was no significant correlation of overall survival (OS) with age (P = 0.135), gender (P = 0.0580), PS (P = 0.658), smoking (P = 0.473), pathology (P = 0.559), or treatment mode (P = 0.116).
| Subgroup | All (n = 94) | TT (n = 38; 40%) | TC (n = 29; 31%) | CC (n = 27; 29%) | P | |
| Gender | Male | 71 | 31 (43.6) | 22 (30.9) | 18 (25.5) | 0.42 |
| Female | 23 | 7 (30.4) | 7 (30.4) | 9 (39.2) | ||
| Age | ≥ 60 yr | 51 | 22 (43.1) | 13 (25.5) | 16 (31.4) | 0.48 |
| < 60 yr | 43 | 16 (37.2) | 16 (37.2) | 11 (25.6) | ||
| Performance status | ≥ 2 | 25 | 13 (52.0) | 5 (20.0) | 7 (28.0) | 0.30 |
| < 2 | 69 | 25 (36.2) | 24 (34.8) | 20 (29.0) | ||
| Smoking | Yes | 23 | 12 (52.2) | 6 (26.1) | 5 (21.7) | 0.44 |
| No | 71 | 26 (36.6) | 23 (32.4) | 22 (31.0) | ||
| Pathology | Adenocarcinoma | 48 | 19 (39.6) | 17 (35.4) | 12 (25.0) | 0.59 |
| Squamous cell carcinoma | 46 | 19 (41.3) | 12 (26.1) | 15 (32.6) |
The median survival of carriers of TT, CT and CC genotypes was 25, 12, and 8 mo, respectively. We analyzed the relationship between overall survival rate and T393C genotypes using Kaplan-Meier survival curves. Our data showed that the favorable TT genotype was significantly associated with better OS (2-year OS: 48%; P = 0.01) when compared with the other genotypes. For example, the 2-year OS for CT genotype was 18% and 15% for CC genotype (Figure 1). By applying the multivariate Cox proportional hazards model, we found that GNAS1 T393C polymorphism was independently associated with OS after adjusting the clinicopathological factors (P < 0.05). However, the dichotomization between C-genotypes (CC + CT) and T-genotypes (TT) indicated significant lower survival rates for C-allele carriers (P = 0.01), which had a 2-year OS rate of 16% (Figure 2).
Lung cancer is the major cause of cancer death in the world and there is an urgent need to accurately and individually treat patients with lung cancer. Although clinicopathological parameters such as UICC stage may serve as prognostic markers in lung cancer, it is still desirable to develop more reliable and accurate biomarkers to more precisely predict the clinical outcome of individual patients. Most prognostic biomarkers are developed according to the features of the tumor tissue itself. The GNAS1 gene encodes the Gαs subunit of G protein and it has been shown that the GNAS1 T393C polymorphism correlates with lung cancer[20]. Hence, we investigated whether GNAS1 T393C polymorphism can be used to predict the clinical outcome in patients with NSCLC. Our study clearly indicated that the homozygous TT genotype patients had a much higher survival rate than patients with either homozygous CC or heterozygous CT genotype. If we could identify patients with poor clinical outcome, we might develop novel treatment strategies accordingly at the initial stage of management, which could lead to improved individual therapy strategies with higher survival rates. Meanwhile, our results also indicated the potential role of the GNAS1 T393C polymorphism as a possible general genetic marker for tumor progression and survival since T-allele carriers demonstrated better clinical outcome than C-allele carriers (TC and CC genotypes). However, it should be noted that the connection between GNAS1 T393C polymorphism and survival was different in different types of tumors. For some tumors, TT genotype was significantly correlated with better OS compared with CT or CC genotype. For example, in advanced squamous cell carcinoma of the larynx, the five-year survival rate for TT genotype patients was 76%, 49% for TC genotype, and 43.5% for CC genotype[10]. Also, it had been reported that the five-year survival rate of sporadic colorectal cancer patients with a TT genotype (87.8%) was much higher than that of patients with a TC (71.0%) or CC genotype (50.0%)[15]. On the other hand, in intrahepatic cholangio-carcinoma[9], esophageal cancer[12] and breast cancer[16], the patients with a CC genotype had a more favorable clinical outcome (Table 3). Thus, it was conceivable that the GNAS1 T393C polymorphism in various tumor types had different biological effects. In order to understand the significance of the T393C genotypes in different tumor types, further more studies are needed to clarify the molecular mechanisms.
| Author | Year | Cancer type | All | Genotype | n | OS (%) | Benefit | P |
| Alakus | 2009 | Gastric cancer | 122 | TT | 26 | 56.9 | TT | 0.043 |
| TC | 57 | 32.7 | ||||||
| CC | 39 | 42.6 | ||||||
| Schmitz | 2007 | Cholangiocarcinoma | 87 | TT | 15 | 10 | C+ | 0.04 |
| TC | 41 | 17 | ||||||
| CC | 31 | 18 | ||||||
| Lehnerdt | 2008 | Laryngocarcinoma | 157 | TT | 40 | 76 | TT | 0.037 |
| TC | 75 | 49 | ||||||
| CC | 42 | 43.5 | ||||||
| Frey | 2006 | Chronic lymphocytic leukemia | 144 | TT | 27 | 73 | T+ | 0.013 |
| TC | 72 | 63.3 | ||||||
| CC | 45 | 33.2 | ||||||
| Vashist | 2011 | Esophageal cancer | 190 | TT | 38 | 19 | CC | 0.001 |
| TC | 96 | 15 | ||||||
| CC | 56 | 51 | ||||||
| Frey | 2005 | Bladder cancer | 254 | TT | 49 | 82 | TT | 0.015 |
| TC | 121 | 60 | ||||||
| CC | 84 | 58 | ||||||
| Frey | 2006 | Renal cancer | 150 | TT | 34 | 91 | TT | 0.01 |
| TC | 79 | 81 | ||||||
| CC | 37 | 69 | ||||||
| Frey | 2005 | Colorectal cancer | 151 | TT | 36 | 87.8 | TT | 0.009 |
| TC | 72 | 71 | ||||||
| CC | 43 | 50 | ||||||
| Otterbach | 2007 | Breast cancer | 279 | TT | 64 | 23 | CC | 0.01 |
| TC | 162 | 40 | ||||||
| CC | 53 | 63 | ||||||
| Lehnerdt | 2008 | Oral carcinoma | 202 | TT | 48 | 51.3 | TT | 0.015 |
| TC | 89 | 44.7 | ||||||
| CC | 65 | 36.8 | ||||||
| Kaderi | 2008 | Chronic lymphocytic leukemia | 279 | TT | 80 | 65 | NS | 0.802 |
| TC | 115 | 70 | ||||||
| CC | 84 | 64 | ||||||
| Frey | 2010 | Malignant melanoma | 328 | TT | 69 | 87.1 | TT | 0.017 |
| TC | 149 | NS | ||||||
| CC | 110 | 66 | ||||||
| Xie | 2013 | Non-small cell | 131 | TT | 33 | NS | TT | 0.02 |
| lung cancer | TC | 63 | NS | |||||
| CC | 35 | NS |
In vitro studies demonstrated that increased Gαs expression promotes apoptosis[21]. Therefore, it is highly likely that increased Gαs expression and the subsequently increased apoptosis could be associated with better survival rate in patients with a GANS1 TT genotype. In vitro experiments also suggest that the product of Gαs, cyclic AMP, could play a crucial role in the proapoptotic process. It has been reported that increasing the intracellular concentration of cyclic AMP leads to enhanced apoptosis in several cell lines including lymphoma cells[5], leukemic[22] and ovarian cancer cells[23]. Gαs was also found to be differentially expressed between various GNAS1 T393C genotypes. Previous studies have suggested that Gαs transcription level is increased in individuals with a GNAS1 393 TT genotype[13]. Intriguingly, the mRNA stability has been shown to be determined by the coding region of some genes[24-26]. Using the MFOLD (the software for the prediction of the secondary structure of single stranded nucleic acids), Alakus et al[8] have reported that the substitution of T393 to C affects the structure of mRNA, most likely the mRNA folding.
Several biomarkers have been used as predictive and prognostic markers for NSCLC patients. A prognostic biomarker is a molecule that can be used to indicate the patient survival independent of the treatment received. In other words, it is an indicator of the innate tumor aggressiveness. For example, KRAS mutations can serve as a good prognostic biomarker indicating the poor survival for NSCLC patients when compared with the patients without KRAS mutations, independent of therapy. Xie et al[20] has reported that the GNAS1 T393C polymorphism can somehow predict the chemotherapy sensitivity and overall survival rate in advanced NSCLC patients treated with gemcitabine and platinum[20]. Here, our data clearly showed that the GNAS1 T393C TT genotype was prognostic of better overall survival for NSCLC patients, independent of therapy.
Nevertheless, it should be emphasized that in this study, we only investigated a small population of patients. Although our study indicated that genetic host factors play a role in tumor progression, which was consistent with the previously published data[20], further independent studies of large cohorts are necessary to confirm the reliability of our findings. Furthermore, the molecular mechanisms underlying the significance of the GNAS1 T393C genotype associated with potentially surrogate SNPs remain to be explored.
Lung cancer is major cause of cancer death around the world. Although some clinicopathological parameters like UICC stage may be used as prognostic biomarkers in lung cancer, other reliable markers that can help precisely predict the clinical outcome of individual patients are still desirable. Most prognostic biomarkers are based on features of the tumor tissue itself.
Characterization of single nucleotide polymorphisms (SNPs) as a prognostic biomarker in cancer has become the hotspot of recent research. The T393C polymorphism of the GNAS1 gene is one such polymorphism.
Several molecular markers have been used as predictive and prognostic markers for non-small cell lung cancer (NSCLC). A prognostic biomarker is a biomolecule that can be used to indicate the patient survival independent of the treatment received. It can also indicate for the innate tumor aggressiveness. For example, the KRAS mutations are prognostic of poor survival for NSCLC patients when compared to the absence of KRAS mutations, independent of therapy. Xie et al reported that the GNAS1 T393C polymorphism can be used to predict the chemotherapy sensitivity as well as the survival rates in advanced NSCLC patients treated with gemcitabine and platinum. Here, the data clearly indicate that the GNAS1 T393C TT genotype was prognostic of better survival rates for NSCLC patients, independent of therapy.
The identification of patients with high-risk lung cancer could help develop novel and individual treatment strategies and could improve the clinical outcome. This data clearly indicate that genetic polymorphism in the GNAS1 T393C influenced survival in advanced non-small lung cancer with a worse clinical outcome for C allele patients.
SNPs refer to a DNA sequence variation occurring commonly within a population (e.g., 1%) in which a single nucleotide -A, T, C or G - in the genome (or other shared sequence) differs between members of a biological species or paired chromosomes.
The manuscript is comprehensive and important.
P- Reviewer: Garfield D, Kermanizadeh A, Nacak M, Zhang YJ S- Editor: Ji FF L- Editor: Wang TQ E- Editor: Wu HL
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25182] [Article Influence: 1937.1] [Reference Citation Analysis (3)] |
| 2. | Molina JR, Yang P, Cassivi SD, Schild SE, Adjei AA. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83:584-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1723] [Cited by in F6Publishing: 1875] [Article Influence: 117.2] [Reference Citation Analysis (0)] |
| 3. | Forde PM, Ettinger DS. Targeted therapy for non-small-cell lung cancer: past, present and future. Expert Rev Anticancer Ther. 2013;13:745-758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 4. | Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346:92-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3965] [Cited by in F6Publishing: 3986] [Article Influence: 181.2] [Reference Citation Analysis (0)] |
| 5. | Yan L, Herrmann V, Hofer JK, Insel PA. beta-adrenergic receptor/cAMP-mediated signaling and apoptosis of S49 lymphoma cells. Am J Physiol Cell Physiol. 2000;279:C1665-C1674. [PubMed] [Cited in This Article: ] |
| 6. | Collins MT, Sarlis NJ, Merino MJ, Monroe J, Crawford SE, Krakoff JA, Guthrie LC, Bonat S, Robey PG, Shenker A. Thyroid carcinoma in the McCune-Albright syndrome: contributory role of activating Gs alpha mutations. J Clin Endocrinol Metab. 2003;88:4413-4417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 89] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Lyons J, Landis CA, Harsh G, Vallar L, Grünewald K, Feichtinger H, Duh QY, Clark OH, Kawasaki E, Bourne HR. Two G protein oncogenes in human endocrine tumors. Science. 1990;249:655-659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 718] [Cited by in F6Publishing: 654] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 8. | Alakus H, Mönig SP, Warnecke-Eberz U, Alakus G, Winde G, Drebber U, Schmitz KJ, Schmid KW, Riemann K, Siffert W. Association of the GNAS1 T393C polymorphism with tumor stage and survival in gastric cancer. World J Gastroenterol. 2009;15:6061-6067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 12] [Cited by in F6Publishing: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 9. | Schmitz KJ, Lang H, Frey UH, Sotiropoulos GC, Wohlschlaeger J, Reis H, Takeda A, Siffert W, Schmid KW, Baba HA. GNAS1 T393C polymorphism is associated with clinical course in patients with intrahepatic cholangiocarcinoma. Neoplasia. 2007;9:159-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Lehnerdt GF, Franz P, Winterhoff S, Bankfalvi A, Grehl S, Lang S, Schmid KW, Siffert W, Jahnke K, Frey UH. The GNAS1 T393C polymorphism predicts survival in patients with advanced squamous cell carcinoma of the larynx. Laryngoscope. 2008;118:2172-2176. [PubMed] [Cited in This Article: ] |
| 11. | Frey UH, Nückel H, Sellmann L, Siemer D, Küppers R, Dürig J, Dührsen U, Siffert W. The GNAS1 T393C polymorphism is associated with disease progression and survival in chronic lymphocytic leukemia. Clin Cancer Res. 2006;12:5686-5692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Vashist YK, Kutup A, Musici S, Yekebas EF, Mina S, Uzunoglu G, Zehler O, Koenig A, Cataldegirmen G, Bockhorn M. The GNAS1 T393C single nucleotide polymorphism predicts the natural postoperative course of complete resected esophageal cancer. Cell Oncol (Dordr). 2011;34:281-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 13. | Frey UH, Eisenhardt A, Lümmen G, Rübben H, Jöckel KH, Schmid KW, Siffert W. The T393C polymorphism of the G alpha s gene (GNAS1) is a novel prognostic marker in bladder cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:871-877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Frey UH, Lümmen G, Jäger T, Jöckel KH, Schmid KW, Rübben H, Müller N, Siffert W, Eisenhardt A. The GNAS1 T393C polymorphism predicts survival in patients with clear cell renal cell carcinoma. Clin Cancer Res. 2006;12:759-763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Frey UH, Alakus H, Wohlschlaeger J, Schmitz KJ, Winde G, van Calker HG, Jöckel KH, Siffert W, Schmid KW. GNAS1 T393C polymorphism and survival in patients with sporadic colorectal cancer. Clin Cancer Res. 2005;11:5071-5077. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 56] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Otterbach F, Callies R, Frey UH, Schmitz KJ, Wreczycki C, Kimmig R, Siffert W, Schmid KW. The T393C polymorphism in the gene GNAS1 of G protein is associated with survival of patients with invasive breast carcinoma. Breast Cancer Res Treat. 2007;105:311-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Lehnerdt GF, Franz P, Zaqoul A, Schmitz KJ, Grehl S, Lang S, Schmid KW, Siffert W, Jahnke K, Frey UH. Overall and relapse-free survival in oropharyngeal and hypopharyngeal squamous cell carcinoma are associated with genotypes of T393C polymorphism of the GNAS1 gene. Clin Cancer Res. 2008;14:1753-1758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | Frey UH, Fritz A, Rotterdam S, Schmid KW, Potthoff A, Altmeyer P, Siffert W, Brockmeyer NH. GNAS1 T393C polymorphism and disease progression in patients with malignant melanoma. Eur J Med Res. 2010;15:422-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Kaderi MA, Murray F, Jansson M, Merup M, Karlsson K, Roos G, Aleskog A, Tobin G. The GNAS1 T393C polymorphism and lack of clinical prognostic value in chronic lymphocytic leukemia. Leuk Res. 2008;32:984-987. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 20. | Xie FJ, Zhao P, Kou JY, Hong W, Fu L, Hu L, Hong D, Su D, Gao Y, Zhang YP. The T393C polymorphism of GNAS1 as a predictor for chemotherapy sensitivity and survival in advanced non-small-cell lung cancer patients treated with gemcitabine plus platinum. Cancer Chemother Pharmacol. 2012;69:1443-1448. [PubMed] [Cited in This Article: ] |
| 21. | Yang X, Lee FY, Wand GS. Increased expression of Gs(alpha) enhances activation of the adenylyl cyclase signal transduction cascade. Mol Endocrinol. 1997;11:1053-1061. [PubMed] [Cited in This Article: ] |
| 22. | Myklebust JH, Josefsen D, Blomhoff HK, Levy FO, Naderi S, Reed JC, Smeland EB. Activation of the cAMP signaling pathway increases apoptosis in human B-precursor cells and is associated with downregulation of Mcl-1 expression. J Cell Physiol. 1999;180:71-80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 23. | Srivastava RK, Srivastava AR, Cho-Chung YS, Longo DL. Synergistic effects of retinoic acid and 8-Cl-cAMP on apoptosis require caspase-3 activation in human ovarian cancer cells. Oncogene. 1999;18:1755-1763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 701] [Cited by in F6Publishing: 703] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 25. | Capon F, Allen MH, Ameen M, Burden AD, Tillman D, Barker JN, Trembath RC. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet. 2004;13:2361-2368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 215] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 26. | Tierney MJ, Medcalf RL. Plasminogen activator inhibitor type 2 contains mRNA instability elements within exon 4 of the coding region. Sequence homology to coding region instability determinants in other mRNAs. J Biol Chem. 2001;276:13675-13684. [PubMed] [Cited in This Article: ] |