Published online Jan 15, 2022. doi: 10.4251/wjgo.v14.i1.19
Peer-review started: February 22, 2021
First decision: August 19, 2021
Revised: September 8, 2021
Accepted: December 21, 2021
Article in press: December 21, 2021
Published online: January 15, 2022
Gastrointestinal (GI) cancers account for a large proportion of cancer deaths worldwide and pose a major public health challenge. Immunotherapy is considered to be one of the prominent and successful approaches in cancer treatment in recent years. Among them, immune checkpoint inhibitor (ICI) therapy, has received widespread attention, and many clinical findings support the feasibility of ICIs, with sustained responses and significantly prolonged lifespan observed in a wide range of tumors. However, patients treated with ICIs have not fully benefited, and therefore, the identification and development of biomarkers for predicting ICI treatment response have received further attention and exploration. From tumor genome to molecular interactions in the tumor microenvironment, and further expanding to circulating biomarkers and patient characteristics, the exploration of biomarkers is evolving with high-throughput sequencing as well as bioinformatics. More large-scale prospective and specific studies are needed to explore biomarkers in GI cancers. In this review, we summarize the known biomarkers used in ICI therapy for GI tumors. In addition, some ICI biomarkers applied to other tumors are included to provide insights and further validation for GI tumors. Moreover, we present single-cell analysis and machine learning approaches that have emerged in recent years. Although there are no clear applications yet, it can be expected that these techniques will play an important role in the application of biomarker prediction.
Core Tip: Cancer immunotherapy and immune checkpoint inhibitors (ICIs) have recently revolutionized gastrointestinal (GI) cancer treatment, providing unprecedented clinical benefits. However, GI patients treated with ICIs do not fully benefit, and therefore, the identification and development of biomarkers for predicting ICI response have become a pressing issue to be solved now. In this review, we summarize the use of predictive biomarkers for ICI treatment response in GI cancers, and discuss novel biomarkers under development. We also present important biomarkers in other tumors with the aim of providing a cutting-edge reference for GI cancer research.
- Citation: Li M, Kaili D, Shi L. Biomarkers for response to immune checkpoint inhibitors in gastrointestinal cancers. World J Gastrointest Oncol 2022; 14(1): 19-37
- URL: https://www.wjgnet.com/1948-5204/full/v14/i1/19.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i1.19
Gastrointestinal (GI) cancers are common among all cancer types, and the incidence and mortality rates of GI cancers are increasing year by year, especially in colorectal cancer (CRC), which is also accompanied by a tendency of rejuvenation[1]. GI cancers mainly occur in the GI system and related digestive organs, including the esophagus, stomach, biliary tract system, liver, pancreas, small intestine, rectum, and anus. Among them, hepatocellular carcinoma (HCC) has the highest morbidity and mortality rate. For example, from 2000 to 2016, the mortality rate for HCC increased by 43% (from 7.2 to 10.3 per 100000), with a 5-year survival rate of only 18% in the United States[2]. Treatment strategies for GI cancers include surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy, among which immunotherapy is a hot topic in recent years.
Immunotherapy is a relatively new therapeutic strategy that has received wide
The better studied ICIs are CTLA-4 inhibitors and programmed cell death protein 1/programmed cell death ligand 1 (PD-1/PD-L1) inhibitors. Ipilimumab (anti-CTLA-4) was approved by the FDA in 2011 for the treatment of melanoma, followed by the PD-1 inhibitors pembrolizumab and nivolumab for the treatment of melanoma, metastatic non-small cell lung cancer (NSCLC), and DNA mismatch repair-defi
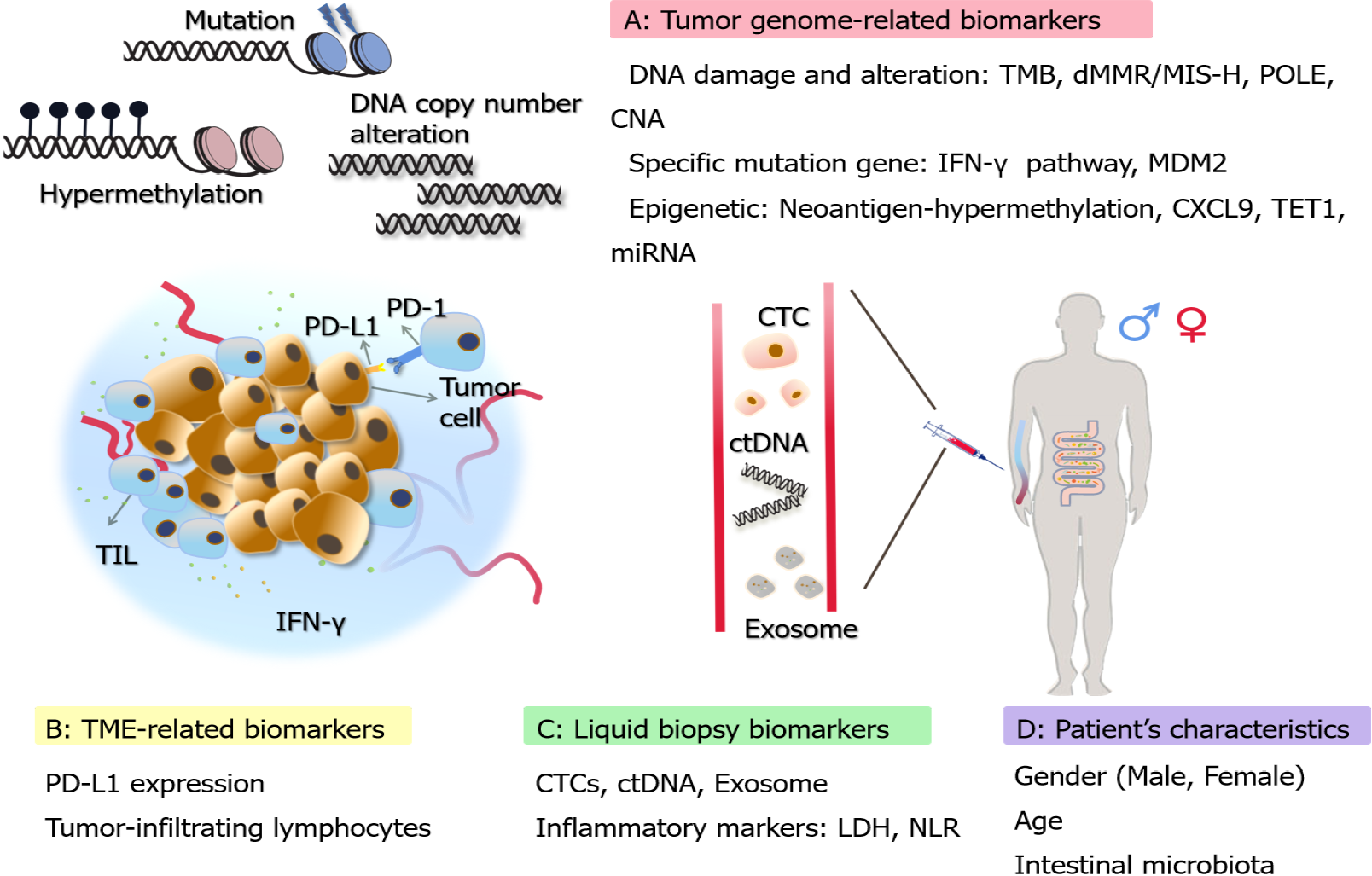
Although immunotherapy has provided sustained clinical benefits, studies have found limitations in the effectiveness of immunotherapy and it is extremely important to study biomarkers to predict more accurate clinical responses[8]. Biomarkers for predicting ICI response have been extensively explored and developed. A variety of biomarkers for GI malignancies have been clinically applied, which can help patients to choose the appropriate targeted therapeutic options. This review highlights biomarkers for predicting the response to ICIs for the treatment of GI tumors. Some biomarkers applied to other tumors are also presented, intending to provide further reference and validation for GI tumors (Figure 1). In addition, we present some new approaches that have emerged in recent years, such as single-cell analysis and machine learning.
The tumor mutation burden (TMB) represents the density of distribution of non-synonymous mutations in the protein-coding region, or simply the number of mutations present in the tumor (Table 1). It is usually defined as the total number of mutations per megabase of substitutions and insertions or deletions in the exon coding region of the gene evaluated in the tumor sample and is usually detected as mutations per million bases (Mut/Mb)[9]. Traditionally, whole-exome sequencing (WES) has been used to measure TMB, which is considered the standard for TMB determination. However, due to the high cost and relatively slow speed of detection using WES, the accurate determination of TMB by next-generation sequencing (NGS) panels has recently been applied[10]. Quantifying the number of non-synonymous single nucleotide variants (SNVs) by NGS, followed by algorithmic validation and extension to WES, is also one of the feasible approaches in recent years[11].
| Classification | Biomarkers | Tumors | Response | OS | PFS | Others | Ref. |
| Tumor-genome biomarkers | TMB | Multiple GI | Pos/Neg1 | 14.6/4.0 mo | Unreached/2 mo (CRC) | NA | [19-21] |
| dMMR/MSI-H | Multiple GI | Pos | Unreached vs 5.0 mo (CRC) | Unreached vs 2.2 mo (CRC) | Higher DCB (59.1% vs 28.6%, GI tumors) | [30,31] | |
| CNA | Multiple GI | Neg | Unreached2 | Over 10 mo | NA | [31] | |
| IFN-γ-related | Multiple GI | Pos | Positive correlation (GC) | Positive correlation (GC and ESCA) | NA | [40,42] | |
| MDM2 | HCC | Neg | NA | NA | Correlated with HPD | [50] | |
| TME biomarkers | PD-L1 | Multiple GI | Pos | NA | NA | NA | [53,54] |
| TIL | Multiple GI | Pos | Prolonged OS (ESCA) | NA | 3-yr RFS 71.6% vs 55.3% (CRC) | [67,78] | |
| Liquid-biopsy biomarkers | ctDNA | Multiple GI | Neg | NA | 4.9 mo vs 7.4 mo (GC) | 2-yr RFS 66% vs 100% (CRC) | [73,74] |
| Exosome | GC | Neg | Reduced OS | NA | High level Exosome | [78] |
According to several reports in recent years, increased TMB is associated with the response to ICI therapy, and high TMB was significantly associated with the efficacy of ICIs[12]. There are many data supporting the use of increased TMB as a biomarker for ICI therapy in many pan-cancer treatments. According to a retrospective study that included 27 cancer types, patients with higher TMB were found to have better clinical outcomes and objective response rates (ORR) when treated with PD-1 antibody[13]. In a phase II study of pembrolizumab in Korea, high TMB was defined as more than 400 SNVs in the WES. The results showed that elevated levels of TMB were associated with a high ORR (89%); the moderate TMB group (100-400 SNVs) had an ORR of 20%, while the low TMB group had an ORR of only 7%, indicating a similar positive correlation between high levels of TMB and ICI efficacy, i.e., higher values of TMB represent a higher overall response rate for patients[14].
In another retrospective study, TMB levels of patients with various types of melanoma as well as NSCLC were also classified as low (1-5 Mut/Mb), medium (6-19 Mut/Mb), and high (≥ 20 Mut/Mb). Their analysis indicated that patients with high levels of TMB had the highest response rate to ICI treatment, reaching 58%, and also had the longest duration of progression-free survival (PFS) at 12.8 mo. The other two treatment groups had a response rate of only 20% and a PFS of only 3.3 mo[15]. Another study detected TMB (cut-off value of 20 Mut/Mb) in 4064 NSCLC patients and found that patients with high levels of TMB (TMB-H) had a significantly higher overall survival (OS) and disease control rate (DCR) when treated with anti-PD-1/L1 agents compared to patients with low levels of TMB (TMB-L)[16]. Similar results were presented in another study showing significantly better durable clinical benefit (DCB) and PFS in the TMB-H population in a cohort with 78 NSCLC patients treated with anti-PD-1/L1 antibodies[17]. Additionally, in a prospective analysis of KEYNOTE-158, Marabelle et al[18] assessed the association of pembrolizumab monotherapy in terms of TMB (tTMB) and clinical outcome across ten different advanced solid tumors types, including anal, biliary, etc. The results revealed that in terms of efficacy, the ORR (29% vs 6%) was better in the tTMB-high group (defined as ≥ 10 Mut/Mb) than in the tTMB-low group (< 10 Mut/Mb), and the median durable response (follow-up of approximately 3 years) was not reached, while the tTMB-low group only reached 33.1 mo[18].
Data from the above-mentioned studies have demonstrated the significant role of high levels of TMB in predicting ICI efficacy, and the results of TMB in GI cancers are no exception to other tumor types. In a phase I study with the anti-PD-1 antibody toripalimib, patients with metastatic gastric cancer (GC) with high TMB (> 20 Mut/Mb) had a better response in survival compared to those with low TMB (15 mo vs 4 mo)[19,20]. In patients with advanced GC, patients with high TMB (≥ 12 Mut/Mb) had significantly better efficiency (33.3% vs 7.1%) and OS time (14.6 vs 4.0 mo) than patients with low TMB (< 12 Mut/Mb)[20]. In a study of metastatic CRC, none of the TMB-H group had achieved PFS (median follow-up > 18 mo), while the TMB-L group had a PFS of only 2 mo and approximately 66% of TMB-L patients developed further disease[21]. In conclusion, high levels of TMB in ICI therapy represent improved patient treatment efficiency and better prognostic outcomes.
Several studies presented at the 2020 American Society of Clinical Oncology meeting confirmed the predictive value of TMB in immunotherapy or combination therapy, although TMB still has limitations as a biomarker. In addition, several general issues deserve further attention, both in the application of GI cancers and in a wide range of other tumor types. First, there is no clear TMB cut-off value as a criterion to accurately determine which patients can benefit from ICI treatment[22]. Second, testing at the proteomic level may provide a clear picture of the mutational load on the membrane of tumor cells, as some mutations that cause an immune response may originate from only a small subset of genes[23]. Third, factors such as allele frequency might be considered for further and more accurate prediction of ICI efficacy[24].
MSI refers to microsatellite instability and MMR refers to mismatch repair function. They are closely related, e.g., when the MMR functions are in a proficient state (pMMR), MSI can be repaired to maintain stability (MSS). In contrast, when the expression of any of the MMR-related proteins goes wrong and the MMR function is in a deficient state (dMMR), it leads to defects in cellular repair functions, allowing DNA to accumulate mutations during replication, ultimately leading to the development of MSI[25]. MSI can be broadly classified as highly unstable (MSI-H), lowly unstable (MSI-L), and stable (MSS). The dMMR and MSI-H can be roughly equated, as can pMMR and MSS[26].
The dMMR occurs in a variety of tumor types, especially common in GI cancers, including colorectum, stomach, small intestine, prostate, etc.[27]. It has been shown that dMMR/MSI-H tumors have a much higher somatic mutation rate compared to pMMR tumors and are thought to express a large number of shift-code peptides that act as neoantigens and enhance the immune response[28]. In 2017, the United States FDA first approved the PD-1 inhibitor pembrolizumab for the treatment of patients with solid dMMR/MSI-H tumors[29]. Several clinical trials, including KEYNOTE-012, 016, 028, and 158, which included multiple tumor types, have shown that pembrolizumab has promising durable outcomes in treating patients with dMMR/MSI-H tumors[24].
In the treatment of GI cancers, especially in CRC, dMMR/MSI-H is considered to be a relatively well-established group of biomarkers. In the KEYNOTE-164 clinical trial study, the efficacy of pembrolizumab was evaluated in three cohorts of 11 dMMR-CRC, 21 pMMR-CRC, and 9 dMMR non-CRC patients. An immune-related ORR of 40% and a 20-wk PFS of 78% were observed in the dMMR-CRC cohort, while an ORR of 0 and a 20-wk PFS of 11% were observed in the pMMR-CRC cohort. Median PFS and OS were not achieved in the dMMR-CRC cohort, but were 2.2 mo and 5.0 mo, respectively, in the pMMR-CRC cohort. These results demonstrated that dMMR patients are favorable candidates for treatment with ICIs[30]. Lu et al[31] investigated the clinical benefit of ICIs in GI patients. They indicated that the incidence of DCB was significantly higher in dMMR/MSI-H patients (59.1%) than in MSI-L/MSS/pMMR patients (28.6%). In addition, the median PFS time was significantly longer in dMMR/MSI-H patients (7.24 mo) than in MSI-L/MSS/pMMR patients (2.67 mo)[31]. These data reveal that dMMR/MSI-H patients have a more favorable ICI response than the other groups. The dMMR/MSI-H has reliable clinical data as a well-established biomarker in GI cancers, especially in CRC. Its application in other GI cancers also deserves attention and further exploration.
Recently, it has also been shown that copy number alterations (CNA), including copy number gain (CNgain) and copy number loss (CNloss), have a predictive role in ICI therapy. In melanoma patients treated with ICIs, CNLoss was found to be lower in responders[32]. Some ICI-related immune features were also found to be negatively correlated with CNA in GC and CRC of the Cancer Genome Atlas (TCGA) datasets[33]. Detailed data are presented for elaboration in the study by Lu et al[31]. In their study, tumor samples from 93 patients with GI cancers treated with ICIs were tested. CNA load included measures of total CNA, CNgain, and CNloss, while CNgain/ CNloss was defined as the total number of genes with CNgain/CNloss present in each sample[31]. They found a significant difference in the CNA burden index between DCB and NDB (no durable benefit) patients treated in the GI group, with DCB patients having a significantly lower CNA burden than NDB patients, suggesting that a low CNA burden may be correlated with better ICIs outcomes. DCB rates were more pronounced in the low and high groups with the same low level of CNgain/CNloss. Further exploration of OS and PFS also led to more favorable data in the low burden group. Based on the study, the group with lower CNA showed a longer median OS (not achieved in all cohorts). For PFS, it was also suggested that the lower CNA group had a longer PFS, all at more than 10 mo[31]. Furthermore, a study by Smeet et al[34] on CRC treated with bevacizumab combination therapy also illustrated another perspective on the possibility of CNA as a potential biomarker for ICI treatment. Their study, which also defined three CNA groups, showed that tumors in the low-load CNA group did not benefit from this combination therapy, while in turn confirmed that ICI therapy is the superior choice. Likewise, the potential of low-load CNA as a predictive biomarker for ICIs was also confirmed[34].
As a noteworthy point, considering the combination of TMB and CNA, a sig
Alterations within the tumor-associated signaling pathways also affect the efficacy of ICIs, related to the mechanism of checkpoint inhibitor drugs as well as drug resistance[35]. IFN-γ is a cytokine that stimulates the immune response and is one of the key signals for the activation of immune cells. IFN-γ is also able to trigger a series of events leading to tumor cell death by linking to receptors on the cell surface. Moreover, IFN-γ is able to increase the expression of PD-L1 in tumors and increase the expression of MHC, promoting antigen presentation in antigen presenting cells[36].
Grasso et al[37] showed that IFN-γ released by T cells contributes to the am
KEYNOTE-028 is a phase Ib trial of pembrolizumab in patients with 20 different tumor types, including GI cancers. In the esophageal cohort, 23 patients were enrolled and an IFN-γ signature was detected, showing a trend towards predicting response to ICIs[40]. In GC, Epstein-Barr virus (EBV) is involved in approximately 10% of GC progression, and PD-L1 overexpression is presented as a feature of EBV GC. In addition, IFN-γ signaling was also shown to be involved in a study by Sasaki et al[41]. Similarly, in the KEYNOTE-012 clinical trial, which included GC patients treated with pembrolizumab, IFN-γ-related genes were shown to be correlated with OS and PFS[42]. Overall, these results provide useful information revealing the role of IFN-γ in predicting the efficacy of ICIs in GI cancers.
Mutations in genes related to the IFN-γ pathway, such as IFNGR1/2, JAK1/2, and IRF1, also lead to poor outcomes and resistance in patients receiving ICI therapy[35,43]. The JAKs are key kinases in this pathway, and JAK1/2 shift mutations lead to deficient production of IFN-γ. Shin et al[44] indicated that JAK1/2 mutations were associated with resistance to anti-PD-1 therapy in CRC patients[44]. These results suggest that mutations in JAK can lead to poor efficacy of ICIs[44,45].
MDM2 is known as the mouse double minute 2 homolog and is an E3 ubiquitin ligase. When MDM2 is overexpressed due to amplification or improper regulation, it inhibits the activation of P53, which in turn accelerates tumor growth and progression[46]. Kato et al[47] analyzed the genomic profiles of 155 patients with multiple tumor types and found that six patients with MDM2 amplification have a time to treatment failure (TTF) less than 2 mo. Four of the six cases (all with MDM2 amplification) showed 2.3 to 42.3-fold hyperprogression compared to ICI pre-treatment[47]. A recent study also showed that cell lines with high MDM2 expression were more potent against T cell-mediated tumor killing, and that targeting MDM2 improve the efficacy of ICIs[48]. These imply that there may be a negative correlation between amplified variants of MDM2 and the efficacy of ICIs, allowing tumors to develop hyperprogression after receiving treatment.
Dysfunction of the MDM2-P53 axis is a major contributor to GI cancers. The main risk factors for HCC include chronic viral infections and metabolic diseases, all of which may contribute to HCC through dysfunction of the MDM2-P53 axis[49]. The results by Wu et al[50] on prognostic markers for HCC showed that MDM2 was able to directly act on BIRC5 as well as the downstream transcription factors to regulate its expression, thereby reducing the sensitivity and effectiveness of ICI therapy[50]. Based on the association from the available clinical data, MDM2 is expected to be a more specific negative biomarker for predicting ICIs in HCC, although further prospective studies are needed to corroborate this.
PD-L1 is one of the most studied biomarkers with abundant data in clinical studies[51]. The expression of PD-L1 in tumors measured by immunohistochemistry was one of the first biomarkers developed to predict the benefit of ICIs[52]. In GI cancers such as GC, CRC, and HCC, there is a positive correlation between PD-L1 expression and the efficacy of ICIs[53,54]. Many clinical trials have provided data demonstrating the feasibility of PD-L1 (Keynote-059, Keynote-010, Attraction-02, Checkmate-057, Checkmate-012, etc.), and the FDA has approved the application of PD-L1 expression as a biomarker for adjuvant or second-line treatment.
Nevertheless, PD-L1 expression remains limited and somewhat controversial as a comprehensive, stand-alone biomarker. In the trials mentioned above, both Keynote-059 and Attraction-02 did show higher response activity in PD-L1-positive patients, but the data equally showed response activity in PD-L1-negative patients[55]. Concerning the limitations of PD-L1 expression, the following points are noteworthy. First, in the tumor microenvironment, PD-L1 expression displays dynamics and diversity with spatial and temporal heterogeneity[56]. PD-L1 expression detected at a single time point cannot be fully used to assess ICI response[57]. Second, PD-L1 detection criteria are not standardized, with no exact positive scores and thresholds to define[56,58]. Issues such as inconsistent antibody usage and inconsistent detection thresholds make it difficult to standardize staining systems as well[59]. At the molecular level, PD-L1 expression has two components: Tumor cell-associated gene variants and PD-L1 expression induced by IFN-γ secreted by infiltrating T cells. The former has constitutive expression, which is not significantly related to the efficacy of ICIs, while the latter is inducible expression, which is concentrated in the region near the T cells of tumor tissues, and is closely related to the efficacy of ICIs. However, these two types of PD-L1 are not strictly differentiated, which can easily lead to the incorrect conclusion that patients with high PD-L1 expression cannot benefit[60]. Third, the detection methods for PD-L1 expression are not sensitive and precise enough. In an analysis of relevant studies, the response rate to ICIs ranged from 36% to 100% for PD-L1 expression-positive tumors, whereas for PD-L1 expression-negative tumors, the response rate ranged from 0% to 17%[52].
Tumor-infiltrating lymphocytes (TILs) represent an effective mechanism of adaptive immunity with anti-tumor potential and have been shown to be associated with prognosis and response to immunotherapy in various types of cancer[61]. TILs originate from areas of tumor tissue, have specific recognition of autologous tumors, and have specific MHC-restricted tumor lysis activity[62]. Among the different types of tumor immune infiltration, the relationship between immune inflammation and ICI treatment is more evident.
Immunoinflammation is characterized by the presence of CD8+ and CD4+ T lymphocytes in the tumor parenchyma and is accompanied by the expression of immune checkpoint molecules, revealing that ICI treatment may generate a tumor immune response[63]. Analysis of pre-treatment samples showed a relative abundance of CD8+ T cells at the infiltrative margins of responders, and serial sampling during treatment showed increased infiltration of CD8+ T cells into the tumor parenchyma[64]. Other data showed that patients with high CD8+ TIL density achieve a longer PFS and OS compared to those with low density[65]. Similarly, in a retrospective study of a series of patients including some with GI cancers, TILs in tumor biopsy samples were shown to be associated with improved survival[66]. In a study by Xiao et al[67] on CRC liver metastases, patients with high CD8+ TIL had a significantly longer recurrence-free survival (RFS) than those with low CD8+TIL (median RFS: Unmet vs 55.8 mo, 3-year RFS 71.6% vs 55.3%)[67]. And the prognostic value of TILs was demonstrated by the higher accuracy of combining with PD-L1 expression. In addition, in esophageal cancer, a cohort with PD-L1 expression combined with high CD8+ TILs showed a longer OS[68]. In a peripheral blood analysis of a CRC patient treated with pembrolizumab who had a rapid response, high CD39 expression in CD8+ TILs was also found, suggesting that CD39+ CD8+ TILs may be a promising predictive biomarker in GI cancers[69].
The non-invasive nature of liquid biopsy reduces patient suffering compared to sampling of surgery, while adding advantages that tissue biopsy does not offer. Liquid biopsy overcomes the inevitable heterogeneity of tissue biopsy, allowing for multiple sampling and providing real-time data on tumor changes and relatively more comprehensive results[70]. Circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), and exosomes are commonly promising biomarkers for liquid biopsy.
ctDNA is mainly released by dead cancer cells, or can also be secreted directly by CTCs, reflecting information about the entire tumor genome, and the variability of its data provides the feasibility of dynamic monitoring of tumor progression throughout the treatment regimen[71]. Several studies have shown that high ctDNA mutations are associated with a poor OS and prognosis in patients with different cancer types treated with ICIs[24]. Lee et al[72] showed that melanoma patients with persistently elevated ctDNA during anti-PD-1 therapy exhibited less favorable responses with a shorter PFS and OS[72]. Also for GI cancers, among 25 patients with stages I-III CRC, the 2-year RFS was 66% in ctDNA-positive patients compared with 100% in negative patients. In addition, ctDNA showed a negative tendency of recurrence rates, in agreement with the previous result[73]. In a study of 46 advanced GC patients treated with anti-PD-1, the mutational status of baseline ctDNA affected the PFS of patients with a median of 7.4 mo (undetectable ctDNA) vs 4.9 mo (detectable ctDNA)[74]. This suggests that ctDNA may serve as a potential negative biomarker for response to ICI therapy in patients with advanced GC. Recent reports have also linked the detection of CTCs to tumor metastasis. The results showed that PD-L1 was overexpressed in CTCs of patients with advanced head and neck cancers, revealing that combined detection of PD-L1 and CTC may have potential as a biomarker for ICI efficacy prediction[75].
Exosomes are extracellular vesicles carrying tumor-associated proteins, metabolites, RNA, DNA, and lipids, which cover most of the information needed for biopsy and can serve as important biomarkers[76,77]. Zhang et al[78] found elevated levels of exosomes in GC patients with liver metastases. Serum exosome levels were higher in GC patients than in healthy subjects, and the number of exosomes in serum was positively correlated with the stage of GC[78]. It has been further revealed that the mRNA expression of PD-L1 in plasma exosomes correlates with the efficacy of ICIs, which may lead to the suppression of effector lymphocytes involved in antitumor immunity, making ICIs less effective[79]. Still in GC, according to Fan et al[80], OS was significantly lower in the high exosomal PD-L1 group than in the low group. In their subgroup analysis, this difference was found to be even more pronounced in early GC, suggesting that high exosomal PD-L1 could be used as a predictor of the early stage of GC[80]. The combination of exosome and PD-L1 assays has informative implications in GI cancers; however, it remains to be noted that exosomes still face challenges as biomarkers, and need to be further explored to accurately measure their quantity and purity.
The details of the above biomarkers that have been studied or applied in GI cancers are summarized in Table 1. And in addition to the biomarkers mentioned above, here we also discuss and summarize some of the biomarkers that appear more frequently in a variety of other tumors, including patient characteristics, neoantigens, inflammatory indicators, and epigenetics (Table 2). These biomarkers deserve further prospective study and development in ICI-treated GI cancers, and provide new ideas for the identification of novel biomarkers as well.
| Classification | Biomarkers | Tumor type | Response to ICI | Ref. |
| Tumor-genome biomarkers | POLE-mutation | Endometrial carcinoma | Pos | [93] |
| Neoantigen | Pulmonary adenocarcinoma | Pos | [94] | |
| Liquid-biopsy biomarkers | LDH | Melanoma | Neg | [99] |
| NLR | Advanced solid tumors | Neg | [100] | |
| Epigenetic | TET1-mutation | Multiple tumor types | Pos | [105] |
| miRNA | Non-small-cell lung cancer | Pos | [107] | |
| Patient characteristic | Gender | NA | Male: Pos; Female: Neg | [81,82] |
| Age | NA | Controversial1 | [84,115] | |
| Intestinal microbiota | NA | Pos/Neg | [85-87] |
The efficacy of ICI treatment is also highly dependent on patient's characteristics, such as gender, age, and the homeostasis of the body's internal environment. The application of these characteristics in GI cancers is not yet supported by a large amount of data, but the correlation of these characteristics with the efficacy of ICI treatment provides a novel idea for future studies, which can be combined with other markers to improve the predictive accuracy. The first point worth mentioning is the possible correlation between the efficacy of ICIs and the gender of the patient. A meta-analysis including 20 randomized controlled trials conducted by Conforti et al[81] reported better efficacy of ICIs in male patients than in females[81]. Schreiber et al[82] suggested that women have more effective immunosurveillance mechanisms compared to men, and this immunosurveillance capacity allows women to be less immunogenic in advanced tumors. They further implied that women may have stronger immune escape mechanisms, and thus they may be more resistant to immunotherapy[82].
Age is also an important marker. There is a relationship between aging and restricted immune function, with significant effects on both innate and acquired immune responses[83]. Nishijima et al[84] reported an association with better ORR in patients aged less than 75 years treated with ICIs[84]. In addition, the fraction and diversity of the intestinal microbiota were likewise found to be associated with the efficacy of ICIs, where effective patients tend to have high levels of polyphenism and ruminal cocci family[85]. The intestinal microbiota can influence the process of cancer development and progression by altering the host immune system and regulating metabolism[86]. It was evidenced that patients treated with antibiotics for 2 mo before or after ICI treatment had a significantly lower clinical benefit than those without antibiotics, probably because antibiotics disrupted the homeostasis of gut microbiota and certain dominant intestinal flora in patients[87].
As mentioned above, TMB and dMMR/MSI-H were biomarkers at the tumor genome level, and correspondingly, another one of interest needs to be presented here, which is POLE. Polymerase ε (encoded by the POLE gene) performs error correction during DNA replication, ensuring the accuracy of the replication process[88,89]. Mutations in POLE severely affect the error correction function, leading to the accumulation of a large number of somatic mutations and elevated TMB. CD8+ lymphocyte infiltration in tumors is also significantly increased, promoting the production of tumor-specific neoantigens[90-92]. From a retrospective study conducted by Domingo et al[90] including 6517 CRC patients, 66 of them (1.0%) were found to have POLE mutations with the highest mutational burden, all with MSS[90]. However, it is worth mentioning that even patients with the MSS type carry a highly mutated profile. Howitt et al[93] reported that POLE mutations in endometrial carcinoma lead to an elevated tumor neoantigen load and PD-1 overexpression in tumor-infiltrating cells[93]. These results indicated that POLE mutations have a role as prognostic markers, and the detection of POLE can also be applied to GI cancers to predict the survival benefit of ICI therapy.
TMB, dMMR/MSI-H, and POLE are all valid indicators as biomarkers, and there is a link between these three. As previously mentioned, mutations in POLE can lead to high levels of TMB[11]. Chalmers et al[11] indicated that MSI-H can be usually used as a subset of high TMB, and the vast majority of MSI-H samples also had high levels of TMB (83%), with 97% of them having TMB ≥ 10 Mut/Mb. Nevertheless, it depends on the tumor type, and in GI cancers such as gastric, duodenal, and small intestinal adenocarcinomas, MSI-H and high TMB are found almost simultaneously[11]. Both can be used as combined biomarkers to predict the response to ICIs in GI cancers.
Common to all three biomarkers mentioned above is that they all increase neoantigen generation. Higher levels of TMB may increase the chance of immunogenic neoantigens[94]. High levels of somatic mutations in MSI-H and POLE also lead to an increase in neoantigens[30]. It means that these tumor cells are more likely to be recognized by immune cells, in which case the efficacy of ICIs is also more pronounced. It has been suggested that hypermethylation of the neoantigen gene promoter may be important for immune editing and tumor immune escape[95]. Therefore, neoantigens are also in the scope of exploring ICIs biomarkers for GI cancers. Neoantigens are not only highly specific and strongly immunogenic, but are also ideal targets for immunotherapy. The presentation and recognition of neoantigens largely influence the outcome of ICI treatment, making it undoubtedly an important target for predicting the efficacy of ICIs[96]. Studies have shown that in primary pulmonary adenocarcinoma, clonal neoantigen load is associated with a longer OS[94]. The relationship between neoantigens and the clinical benefit of treating GI cancers needs to be supported by additional and more specific data.
GI cancers are similar to other types of tumors in that tumor-associated inflammatory processes often establish immune tolerance, promote tumor growth and metastasis, and activate oncogenic signal transduction pathways[97]. Some conventional inflammatory indicators, such as neutrophil-to-lymphocyte ratio (NLR) and lactate dehydrogenase (LDH), have been used as ICI response biomarkers for a variety of tumors, which could also serve as promising markers in GI cancers[98]. In a blood test performed on 66 melanoma patients treated with ICIs, baseline values of serum LDH and changes in LDH during ICI treatment were found to correlate with patient response and survival outcomes, with higher baseline serum LDH values and a 10% increase from baseline during treatment likely indicating inferior ICI efficacy[99]. NLR has also been more established as a biomarker. According to the NLR kinetics study in patients with advanced solid tumors treated with PD-1/L1 inhibitors, the median OS of patients with high NLR was 8.5 mo, while the median OS of patients with low NLR was 19.4 mo[100]. Similar results were found by Jiang et al[101], showing that high NLR was associated with a poor OS and PFS[101].
Epigenetic alterations are also an area of interest as potential biomarkers. As mentioned above, high levels of TMB tend to be correlated with a better ICI response, but some tumors with low-level TMB may improve the immunogenicity of their tumor neoplastic antigens through epigenetic modifications, when the efficacy of ICIs is instead better[102]. In GC, alterations in the somatic epigenetic promoter have also been described to be associated with immune editing and tumor escape[103]. It has also been shown that the CC family chemokine ligand 9 (CXCL9) is epigenetically modified to suppress its biological function, ultimately blocking effector T cells from infiltrating into the tumor bed for its immune function[104]. In a report examining the relevance of DNA methylation-regulated genes to ICI response, mutated TET1 was significantly enriched among the 21 related genes studied in patients responding to ICIs. Moreover, mutant TET1 was strongly associated with a higher ORR, longer PFS, and better OS and DCB, which could serve as a novel predictive biomarker across multiple cancer types[105].
In addition to modifications such as methylation, miRNAs are also of interest for further development. In epigenetics, miRNA quantification is one of the most accessible markers. MiRNAs can be direct or indirect regulators of PD-L1 expression, as well as of many other immune checkpoints, such as LAG-3, TIM-3, BTLA, or CTLA-4[106]. A study in NSCLC showed that serum miRNA profiles can discriminate responders to ICIs. In that study, Fan et al[107] found that increased expression of miR-93, -200, -27a, -28, -424, and other miRNAs were significantly associated with prognosis, highlighting the predictive value of miRNAs[107]. The emergence of TET1, miRNAs, and other epigenetic examples suggests that there are still more possibilities that need to be further explored in the field of GI tumors.
Moreover, with the evolving concept of precision medicine, biomarker research is facing the same trend. Tumors contain different and evolving cell populations, a property also known as tumor heterogeneity, which is a major driver of resistance to treatment and tumor metastasis and one of the factors affecting the efficacy of ICIs[108]. It is essential to fully understand heterogeneity, especially in the TME. Analysis of TME heterogeneity and the phenotypes of various cell types by single-cell analysis techniques can help optimize existing therapeutic strategies or discover new ones, and improve the efficacy of the currently used biomarkers, although some limitations remain. In uveal melanoma, the single-cell analysis revealed that CD8+ T cells predominantly express LAG3 rather than conventional PD-L1, revealing the limited avai
Apart from the transcriptomics mentioned above, multi-omics is more noteworthy in single-cell analysis. In a study by Zhou et al[112], the percentage of fibroblasts with altered somatic copy number was found to be much higher in CRC than in adjacent normal tissues by using single-cell multi-omics sequencing. Five genes (BGN, RCN3, TAGLN, MYL9, and TPM2) were also identified as fibroblast-specific biomarkers of poorer prognosis in CRC[112]. This study further explored new CAN-based bio
Along with the growing development of bioinformatics, machine learning, and artificial intelligence, biomarkers will be further improved. For example, in the work of Lu et al[113], tumor samples from patients with metastatic GI cancers treated with ICIs were sequenced for immuno-oncology (IO)-related gene targets and combined with the application of linear support vector machine learning strategy to construct an RNA signature (IO score) as a predictive model. Notably, its overall accuracy in discriminating DCB and NDB reached 94% and 83%, respectively, and the IO-score showed superior predictive value with higher odds ratio than the traditional biomarker[113].
Research in the field of ICIs has been steadily increasing. In GI cancers, ICI-related studies have also been emerging, addressing the importance of ICIs in tumor immunotherapy from different perspectives. Many recent ongoing studies in GI cancers also highlight the potential for diversification of ICIs, particularly in combination or neoadjuvant therapy, where the utility of ICIs has been further investigated. By combining chemotherapy and targeted agents, these studies provide insight into eradicating micrometastatic GI cancers, overcoming resistance to ICIs, and improving ICI treatment. We summarize in Table 3 a number of clinical studies that are currently ongoing to provide a valuable reference for this purpose. However, it needs to be noticed that these ongoing clinical trials do not specifically target one or more biomarkers to predict response to ICIs. Rather, it is more about the combination of ICI therapy with other therapies, which may have little relevance to our topic. None
| Clinical trial ID | Cancer type | Study type | Phase | Number | Strategy | Ref. |
| NCT02918162 | GC; Adenocarcinoma of the GE junction | Interventional | 2 | 40 | Pembrolizumab combined with stand of care chemotherapy regimen | [116] |
| NCT04948125 | GC | Interventional | 2 | 20 | Camrelizumab combined with Apatinib Mesylate | [117] |
| NCT04196465 | GC, ESCA, HCC | Interventional | 2 | 48 | IMC-001 as neoadjuvant therapy | [118] |
| NCT03841110 | GC, CRC | Interventional | 1 | 76 | FT500 combined with Nivolumab/Pembrolizumab/Atezulizumab | [119] |
| NCT02903914 | GC, CRC | Interventional | 1/2 | 260 | Pembrolizumab combined with Arginase Inhibitor INCB001158 | [120] |
| NCT03259867 | HCC, GC, CRC (All have liver lesions) | Interventional | 2 | 80 | Pembrolizumab/Nivolumab combined with TATE | [121] |
| NCT04822103 | ESCA | Observational | NA | 150 | ICIs combined with Neoadjuvant chemotherapy | [122] |
Although many new biomarkers have been identified in GI cancers, there is a relative lack of research compared to other tumor types such as melanoma and NSCLC, and validation from clinical trials is still lacking. In this review, we summarize not only biomarkers that are supported by studies in GI cancers, but also biomarkers that are informed in other tumors, in terms of tumor genomic information, TME, liquid biopsies, and epigenetic and patients' characteristics in relation to ICI response. Among these markers, studies on TMB and PD-L1 need to be further improved, and the delineation of cut-off values is not sufficiently clear, especially for PD-L1 expression, which has been shown in a number of studies to respond to ICIs in PD-L1-negative patients[52]. As a stand-alone biomarker, PD-L1 is still considered to be controversial. In addition, markers associated with patient characteristics also have conflicting data, and current studies are not systematic and not clear enough and need to be confirmed by some large-scale prospective studies[114,115]. Another key point that needs attention is that the current ICI predictive biomarkers for GI cancers are mostly focused on CRC cases, while they have relatively little application in other GI cancers such as GC and HCC, and more research investment is needed.
For the future trend of biomarkers, considering that a single biomarker is mostly insufficient, the strategy of combining two or more biomarkers is noteworthy, such as combining information from epigenetics and tumor genome, TMB and CNA in subgroup analysis, etc. The integration of multiple factors is necessary to improve accuracy. And along with the continuous research on ICIs therapy, biomarkers for combination therapy or neoadjuvant therapy also need to keep pace with the development to further promote precision therapy. Meanwhile, with the development of big data and bioinformatics, an increasing number of cutting-edge technologies such as machine learning, artificial intelligence, and single-cell analysis will also be applied for further optimization and refinement, making the efficacy of tumor immunotherapy steadily improved. For the current research, more prospective studies are needed, and more data will help to optimize these computational models. From this point of view, the identification of biomarkers that can be used to accurately predict ICI is just beginning, and much more remains to be done, which could become a major trend and focus in the future.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Kocollari A, Pompella L, Serban ED S-Editor: Fan JR L-Editor: Wang TQ P-Editor: Fan JR
| 1. | Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterology. 2020;159:335-349.e15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 857] [Cited by in F6Publishing: 775] [Article Influence: 193.8] [Reference Citation Analysis (0)] |
| 2. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2066] [Cited by in F6Publishing: 2565] [Article Influence: 513.0] [Reference Citation Analysis (0)] |
| 3. | Abbott M, Ustoyev Y. Cancer and the Immune System: The History and Background of Immunotherapy. Semin Oncol Nurs. 2019;35:150923. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 4. | Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450-461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2655] [Cited by in F6Publishing: 2883] [Article Influence: 320.3] [Reference Citation Analysis (0)] |
| 5. | Hazarika M, Chuk MK, Theoret MR, Mushti S, He K, Weis SL, Putman AH, Helms WS, Cao X, Li H, Zhao H, Zhao L, Welch J, Graham L, Libeg M, Sridhara R, Keegan P, Pazdur R. U.S. FDA Approval Summary: Nivolumab for Treatment of Unresectable or Metastatic Melanoma Following Progression on Ipilimumab. Clin Cancer Res. 2017;23:3484-3488. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 6. | Ramagopal UA, Liu W, Garrett-Thomson SC, Bonanno JB, Yan Q, Srinivasan M, Wong SC, Bell A, Mankikar S, Rangan VS, Deshpande S, Korman AJ, Almo SC. Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc Natl Acad Sci U S A. 2017;114:E4223-E4232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 104] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 7. | Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol. 2015;33:1974-1982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1664] [Cited by in F6Publishing: 1883] [Article Influence: 209.2] [Reference Citation Analysis (0)] |
| 8. | Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342:1432-1433. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1306] [Cited by in F6Publishing: 1404] [Article Influence: 140.4] [Reference Citation Analysis (0)] |
| 9. | Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, Maillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023-1031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1400] [Cited by in F6Publishing: 1595] [Article Influence: 145.0] [Reference Citation Analysis (0)] |
| 10. | Rizvi H, Sanchez-Vega F, La K, Chatila W, Jonsson P, Halpenny D, Plodkowski A, Long N, Sauter JL, Rekhtman N, Hollmann T, Schalper KA, Gainor JF, Shen R, Ni A, Arbour KC, Merghoub T, Wolchok J, Snyder A, Chaft JE, Kris MG, Rudin CM, Socci ND, Berger MF, Taylor BS, Zehir A, Solit DB, Arcila ME, Ladanyi M, Riely GJ, Schultz N, Hellmann MD. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol. 2018;36:633-641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 741] [Cited by in F6Publishing: 982] [Article Influence: 163.7] [Reference Citation Analysis (0)] |
| 11. | Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, Schrock A, Campbell B, Shlien A, Chmielecki J, Huang F, He Y, Sun J, Tabori U, Kennedy M, Lieber DS, Roels S, White J, Otto GA, Ross JS, Garraway L, Miller VA, Stephens PJ, Frampton GM. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1661] [Cited by in F6Publishing: 2149] [Article Influence: 307.0] [Reference Citation Analysis (0)] |
| 12. | Sholl LM, Hirsch FR, Hwang D, Botling J, Lopez-Rios F, Bubendorf L, Mino-Kenudson M, Roden AC, Beasley MB, Borczuk A, Brambilla E, Chen G, Chou TY, Chung JH, Cooper WA, Dacic S, Lantuejoul S, Jain D, Lin D, Minami Y, Moreira A, Nicholson AG, Noguchi M, Papotti M, Pelosi G, Poleri C, Rekhtman N, Tsao MS, Thunnissen E, Travis W, Yatabe Y, Yoshida A, Daigneault JB, Zehir A, Peters S, Wistuba II, Kerr KM, Longshore JW. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: A Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol. 2020;15:1409-1424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 159] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 13. | Yarchoan M, Hopkins A, Jaffee EM. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N Engl J Med. 2017;377:2500-2501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1693] [Cited by in F6Publishing: 2067] [Article Influence: 295.3] [Reference Citation Analysis (0)] |
| 14. | Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, Lee S, Park SH, Park JO, Park YS, Lim HY, Lee H, Choi M, Talasaz A, Kang PS, Cheng J, Loboda A, Lee J, Kang WK. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449-1458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 643] [Cited by in F6Publishing: 920] [Article Influence: 153.3] [Reference Citation Analysis (0)] |
| 15. | Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598-2608. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1259] [Cited by in F6Publishing: 1524] [Article Influence: 217.7] [Reference Citation Analysis (0)] |
| 16. | Singal G, Miller PG, Agarwala V, Li G, Kaushik G, Backenroth D, Gossai A, Frampton GM, Torres AZ, Lehnert EM, Bourque D, O'Connell C, Bowser B, Caron T, Baydur E, Seidl-Rathkopf K, Ivanov I, Alpha-Cobb G, Guria A, He J, Frank S, Nunnally AC, Bailey M, Jaskiw A, Feuchtbaum D, Nussbaum N, Abernethy AP, Miller VA. Association of Patient Characteristics and Tumor Genomics With Clinical Outcomes Among Patients With Non-Small Cell Lung Cancer Using a Clinicogenomic Database. JAMA. 2019;321:1391-1399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 308] [Article Influence: 61.6] [Reference Citation Analysis (0)] |
| 17. | Fang W, Ma Y, Yin JC, Hong S, Zhou H, Wang A, Wang F, Bao H, Wu X, Yang Y, Huang Y, Zhao H, Shao YW, Zhang L. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti-PD-(L)1 Therapies in Non-Small Cell Lung Cancer. Clin Cancer Res. 2019;25:5015-5026. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 18. | Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, Chung H C, Kindler H L, Lopez-Martin J A, Miller W H, Jr. , Italiano A, Kao S, Piha-Paul S A, Delord J P, McWilliams R R, Fabrizio D A, Aurora-Garg D, Xu L, Jin F, Norwood K, Bang Y J. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 keynote-158 study. Lancet Oncol. 2020;21:1353-1365. [DOI] [Cited in This Article: ] [Cited by in Crossref: 672] [Cited by in F6Publishing: 1154] [Article Influence: 288.5] [Reference Citation Analysis (0)] |
| 19. | Xu R-H, Wang F, Wei X-L, Xu N, Shen L, Dai G, Yuan X, Chen Y, Yang S, Shi J, Hu X-C, Lin X, Zhang Q, Feng J F, Ba Y, Liu Y, Wu H, Feng H, Yao S. Tumor mutational burden identifies chemorefractory gastric cancer with overall survival advantage after receiving toripalimab, a pd-1 antibody. J Clin Oncol. 2019;37:4021-4021. [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Wang F, Wei XL, Wang FH, Xu N, Shen L, Dai GH, Yuan XL, Chen Y, Yang SJ, Shi JH, Hu XC, Lin XY, Zhang QY, Feng JF, Ba Y, Liu YP, Li W, Shu YQ, Jiang Y, Li Q, Wang JW, Wu H, Feng H, Yao S, Xu RH. Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol. 2019;30:1479-1486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 307] [Article Influence: 61.4] [Reference Citation Analysis (0)] |
| 21. | Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS, Miller VA, Lim D, Amanam I, Chao J, Catenacci D, Cho M, Braiteh F, Klempner SJ, Ali SM, Fakih M. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019;30:1096-1103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 382] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 22. | Addeo A, Friedlaender A, Banna GL, Weiss GJ. TMB or not TMB as a biomarker: That is the question. Crit Rev Oncol Hematol. 2021;163:103374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 91] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 23. | Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16:275-287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1659] [Cited by in F6Publishing: 1841] [Article Influence: 230.1] [Reference Citation Analysis (0)] |
| 24. | Bai R, Lv Z, Xu D, Cui J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark Res. 2020;8:34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 228] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 25. | Svrcek M, Lascols O, Cohen R, Collura A, Jonchère V, Fléjou JF, Buhard O, Duval A. MSI/MMR-deficient tumor diagnosis: Which standard for screening and for diagnosis? Bull Cancer. 2019;106:119-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 26. | Li K, Luo H, Huang L, Zhu X. Microsatellite instability: a review of what the oncologist should know. Cancer Cell Int. 2020;20:16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 222] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 27. | Salem ME, Puccini A, Grothey A, Raghavan D, Goldberg RM, Xiu J, Korn WM, Weinberg BA, Hwang JJ, Shields AF, Marshall JL, Philip PA, Lenz HJ. Landscape of Tumor Mutation Load, Mismatch Repair Deficiency, and PD-L1 Expression in a Large Patient Cohort of Gastrointestinal Cancers. Mol Cancer Res. 2018;16:805-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 154] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 28. | Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988-997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 278] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 29. | Solomon BL, Garrido-Laguna I. Upper gastrointestinal malignancies in 2017: current perspectives and future approaches. Future Oncol. 2018;14:947-962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 30. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LA Jr. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6096] [Cited by in F6Publishing: 6542] [Article Influence: 726.9] [Reference Citation Analysis (0)] |
| 31. | Lu Z, Chen H, Li S, Gong J, Li J, Zou J, Wu L, Yu J, Han W, Sun H, Jiao X, Zhang X, Peng Z, Lu M, Wang Z, Zhang H, Shen L. Tumor copy-number alterations predict response to immune-checkpoint-blockade in gastrointestinal cancer. J Immunother Cancer. 2020;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 39] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 32. | Roh W, Chen PL, Reuben A, Spencer CN, Prieto PA, Miller JP, Gopalakrishnan V, Wang F, Cooper ZA, Reddy SM, Gumbs C, Little L, Chang Q, Chen WS, Wani K, De Macedo MP, Chen E, Austin-Breneman JL, Jiang H, Roszik J, Tetzlaff MT, Davies MA, Gershenwald JE, Tawbi H, Lazar AJ, Hwu P, Hwu WJ, Diab A, Glitza IC, Patel SP, Woodman SE, Amaria RN, Prieto VG, Hu J, Sharma P, Allison JP, Chin L, Zhang J, Wargo JA, Futreal PA. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci Transl Med. 2017;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 578] [Article Influence: 82.6] [Reference Citation Analysis (0)] |
| 33. | Budczies J, Seidel A, Christopoulos P, Endris V, Kloor M, Győrffy B, Seliger B, Schirmacher P, Stenzinger A, Denkert C. Integrated analysis of the immunological and genetic status in and across cancer types: impact of mutational signatures beyond tumor mutational burden. Oncoimmunology. 2018;7:e1526613. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 34. | Smeets D, Miller IS, O'Connor DP, Das S, Moran B, Boeckx B, Gaiser T, Betge J, Barat A, Klinger R, van Grieken NCT, Cremolini C, Prenen H, Mazzone M, Depreeuw J, Bacon O, Fender B, Brady J, Hennessy BT, McNamara DA, Kay E, Verheul HM, Maarten N, Gallagher WM, Murphy V, Prehn JHM, Koopman M, Punt CJA, Loupakis F, Ebert MPA, Ylstra B, Lambrechts D, Byrne AT. Copy number load predicts outcome of metastatic colorectal cancer patients receiving bevacizumab combination therapy. Nat Commun. 2018;9:4112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 35. | Possick JD. Pulmonary Toxicities from Checkpoint Immunotherapy for Malignancy. Clin Chest Med. 2017;38:223-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 36. | Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, Ribas A. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017;19:1189-1201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 822] [Cited by in F6Publishing: 1112] [Article Influence: 185.3] [Reference Citation Analysis (0)] |
| 37. | Grasso CS, Tsoi J, Onyshchenko M, Abril-Rodriguez G, Ross-Macdonald P, Wind-Rotolo M, Champhekar A, Medina E, Torrejon DY, Shin DS, Tran P, Kim YJ, Puig-Saus C, Campbell K, Vega-Crespo A, Quist M, Martignier C, Luke JJ, Wolchok JD, Johnson DB, Chmielowski B, Hodi FS, Bhatia S, Sharfman W, Urba WJ, Slingluff CL, Jr Diab A, Haanen J, Algarra SM, Pardoll DM, Anagnostou V, Topalian SL, Velculescu VE, Speiser DE, Kalbasi A, Ribas A. Conserved interferon-γ signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38:500-515.e503. [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 38. | Karachaliou N, Gonzalez-Cao M, Crespo G, Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, Teixido C, Molina-Vila MA, Viteri S, De Los Llanos Gil M, Algarra SM, Perez-Ruiz E, Marquez-Rodas I, Rodriguez-Abreu D, Blanco R, Puertolas T, Royo MA, Rosell R. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol. 2018;10:1758834017749748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 39. | Higgs BW, Morehouse CA, Streicher K, Brohawn PZ, Pilataxi F, Gupta A, Ranade K. Interferon Gamma Messenger RNA Signature in Tumor Biopsies Predicts Outcomes in Patients with Non-Small Cell Lung Carcinoma or Urothelial Cancer Treated with Durvalumab. Clin Cancer Res. 2018;24:3857-3866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 137] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 40. | Doi T, Piha-Paul SA, Jalal SI, Saraf S, Lunceford J, Koshiji M, Bennouna J. Safety and Antitumor Activity of the Anti-Programmed Death-1 Antibody Pembrolizumab in Patients With Advanced Esophageal Carcinoma. J Clin Oncol. 2018;36:61-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 215] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 41. | Sasaki S, Nishikawa J, Sakai K, Iizasa H, Yoshiyama H, Yanagihara M, Shuto T, Shimokuri K, Kanda T, Suehiro Y, Yamasaki T, Sakaida I. EBV-associated gastric cancer evades T-cell immunity by PD-1/PD-L1 interactions. Gastric Cancer. 2019;22:486-496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 42. | Muro K, Chung HC, Shankaran V, Geva R, Catenacci D, Gupta S, Eder JP, Golan T, Le DT, Burtness B, McRee AJ, Lin CC, Pathiraja K, Lunceford J, Emancipator K, Juco J, Koshiji M, Bang YJ. Pembrolizumab for patients with PD-L1-positive advanced gastric cancer (KEYNOTE-012): a multicentre, open-label, phase 1b trial. Lancet Oncol. 2016;17:717-726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 690] [Cited by in F6Publishing: 838] [Article Influence: 104.8] [Reference Citation Analysis (1)] |
| 43. | Darnell JE, Kerr IM, Stark GR. Jak-stat pathways and transcriptional activation in response to ifns and other extracellular signaling proteins. Science. 1994;264:1415-1421. [DOI] [Cited in This Article: ] [Cited by in Crossref: 4322] [Cited by in F6Publishing: 4408] [Article Influence: 146.9] [Reference Citation Analysis (0)] |
| 44. | Shin D S, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, Palaskas N, Rodriguez GA, Parisi G, Azhdam A, Chmielowski B, Cherry G, Seja E, Berent-Maoz B, Shintaku IP, Le DT, Pardoll DM, Diaz LA, Jr Tumeh PC, Graeber TG, Lo RS, Comin-Anduix B, Ribas A. Primary resistance to pd-1 blockade mediated by jak1/2 mutations. Cancer Discov. 2017;7:188-201. [DOI] [Cited in This Article: ] [Cited by in Crossref: 893] [Cited by in F6Publishing: 881] [Article Influence: 125.9] [Reference Citation Analysis (0)] |
| 45. | Albacker LA, Wu J, Smith P, Warmuth M, Stephens PJ, Zhu P, Yu L, Chmielecki J. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS One. 2017;12:e0176181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 60] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 46. | Gudikote JP, Cascone T, Poteete A, Sitthideatphaiboon P, Wu Q, Morikawa N, Zhang F, Peng S, Tong P, Li L, Shen L, Nilsson M, Jones P, Sulman EP, Wang J, Bourdon JC, Johnson FM, Heymach JV. Inhibition of nonsense-mediated decay rescues p53β/γ isoform expression and activates the p53 pathway in MDM2-overexpressing and select p53-mutant cancers. J Biol Chem. 2021;297:101163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 47. | Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res. 2017;23:4242-4250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 538] [Cited by in F6Publishing: 643] [Article Influence: 91.9] [Reference Citation Analysis (0)] |
| 48. | Sahin I, Zhang S, Navaraj A, Zhou L, Dizon D, Safran H, El-Deiry WS. AMG-232 sensitizes high MDM2-expressing tumor cells to T-cell-mediated killing. Cell Death Discov. 2020;6:57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 49. | Cao H, Chen X, Wang Z, Wang L, Xia Q, Zhang W. The role of MDM2-p53 axis dysfunction in the hepatocellular carcinoma transformation. Cell Death Discov. 2020;6:53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 50. | Wu L, Quan W, Luo Q, Pan Y, Peng D, Zhang G. Identification of an Immune-Related Prognostic Predictor in Hepatocellular Carcinoma. Front Mol Biosci. 2020;7:567950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 51. | Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, Wistuba II, Rimm DL, Tsao MS, Hirsch FR. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2021;18:345-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 543] [Article Influence: 181.0] [Reference Citation Analysis (0)] |
| 52. | Patel SP, Kurzrock R. PD-L1 Expression as a Predictive Biomarker in Cancer Immunotherapy. Mol Cancer Ther. 2015;14:847-856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1250] [Cited by in F6Publishing: 1543] [Article Influence: 171.4] [Reference Citation Analysis (0)] |
| 53. | Cui C, Yu B, Jiang Q, Li X, Shi K, Yang Z. The roles of PD-1/PD-L1 and its signalling pathway in gastrointestinal tract cancers. Clin Exp Pharmacol Physiol. 2019;46:3-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 54. | Das S, Cimino S, Davis S, Ciombor K. All in the Levels-Programmed Death-Ligand 1 Expression as a Biomarker for Immune Checkpoint Inhibitor Response in Patients with Gastrointestinal Cancer. Oncologist. 2021;26:e186-e188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 55. | Stein A, Moehler M, Trojan J, Goekkurt E, Vogel A. Immuno-oncology in GI tumours: Clinical evidence and emerging trials of PD-1/PD-L1 antagonists. Crit Rev Oncol Hematol. 2018;130:13-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 56. | Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655-668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 537] [Cited by in F6Publishing: 686] [Article Influence: 98.0] [Reference Citation Analysis (0)] |
| 57. | Meng X, Huang Z, Teng F, Xing L, Yu J. Predictive biomarkers in PD-1/PD-L1 checkpoint blockade immunotherapy. Cancer Treat Rev. 2015;41:868-876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 291] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 58. | Sacher AG, Gandhi L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol. 2016;2:1217-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 191] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 59. | Hutarew G. PD-L1 testing, fit for routine evaluation? Memo. 2016;9:201-206. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 60. | Ribas A, Hu-Lieskovan S. What does PD-L1 positive or negative mean? J Exp Med. 2016;213:2835-2840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 230] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 61. | Garber K. Pursuit of tumor-infiltrating lymphocyte immunotherapy speeds up. Nat Biotechnol. 2019;37:969-971. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 62. | Halapi E. Oligoclonal T cells in human cancer. Med Oncol. 1998;15:203-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 63. | Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, Zhang M, Papadopoulos N, Kinzler KW, Vogelstein B, Sears CL, Anders RA, Pardoll DM, Housseau F. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 889] [Cited by in F6Publishing: 1045] [Article Influence: 104.5] [Reference Citation Analysis (0)] |
| 64. | Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4180] [Cited by in F6Publishing: 4769] [Article Influence: 529.9] [Reference Citation Analysis (0)] |
| 65. | Tokito T, Azuma K, Kawahara A, Ishii H, Yamada K, Matsuo N, Kinoshita T, Mizukami N, Ono H, Kage M, Hoshino T. Predictive relevance of PD-L1 expression combined with CD8+ TIL density in stage III non-small cell lung cancer patients receiving concurrent chemoradiotherapy. Eur J Cancer. 2016;55:7-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 142] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 66. | Gibney GT, Weiner LM, Atkins MB. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542-e551. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1167] [Cited by in F6Publishing: 1119] [Article Influence: 139.9] [Reference Citation Analysis (0)] |
| 67. | Xiao B, Peng J, Wang Y, Deng Y, Ou Q, Wu X, Lin J, Pan Z, Zhang L. Prognostic value of tumor infiltrating lymphocytes combined with PD-L1 expression for patients with solitary colorectal cancer liver metastasis. Ann Transl Med. 2020;8:1221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 68. | Yagi T, Baba Y, Ishimoto T, Iwatsuki M, Miyamoto Y, Yoshida N, Watanabe M, Baba H. PD-L1 Expression, Tumor-infiltrating Lymphocytes, and Clinical Outcome in Patients With Surgically Resected Esophageal Cancer. Ann Surg. 2019;269:471-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 69. | Simoni Y, Becht E, Fehlings M, Loh CY, Koo SL, Teng KWW, Yeong JPS, Nahar R, Zhang T, Kared H, Duan K, Ang N, Poidinger M, Lee YY, Larbi A, Khng AJ, Tan E, Fu C, Mathew R, Teo M, Lim WT, Toh CK, Ong BH, Koh T, Hillmer AM, Takano A, Lim TKH, Tan EH, Zhai W, Tan DSW, Tan IB, Newell EW. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 623] [Cited by in F6Publishing: 781] [Article Influence: 130.2] [Reference Citation Analysis (0)] |
| 70. | Poulet G, Massias J, Taly V. Liquid Biopsy: General Concepts. Acta Cytol. 2019;63:449-455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 166] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 71. | Wu X, Li J, Gassa A, Buchner D, Alakus H, Dong Q, Ren N, Liu M, Odenthal M, Stippel D, Bruns C, Zhao Y, Wahba R. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int J Biol Sci. 2020;16:1551-1562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 72. | Lee JH, Long GV, Boyd S, Lo S, Menzies AM, Tembe V, Guminski A, Jakrot V, Scolyer RA, Mann GJ, Kefford RF, Carlino MS, Rizos H. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol. 2017;28:1130-1136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 199] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 73. | Lecomte T, Berger A, Zinzindohoué F, Micard S, Landi B, Blons H, Beaune P, Cugnenc PH, Laurent-Puig P. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002;100:542-548. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 217] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 74. | Jin Y, Chen DL, Wang F, Yang CP, Chen XX, You JQ, Huang JS, Shao Y, Zhu DQ, Ouyang YM, Luo HY, Wang ZQ, Wang FH, Li YH, Xu RH, Zhang DS. The predicting role of circulating tumor DNA landscape in gastric cancer patients treated with immune checkpoint inhibitors. Mol Cancer. 2020;19:154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 75. | Kulasinghe A, Perry C, Kenny L, Warkiani ME, Nelson C, Punyadeera C. PD-L1 expressing circulating tumour cells in head and neck cancers. BMC Cancer. 2017;17:333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 76. | Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659-1668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 686] [Cited by in F6Publishing: 747] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 77. | Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3378] [Cited by in F6Publishing: 3537] [Article Influence: 353.7] [Reference Citation Analysis (0)] |
| 78. | Zhang H, Deng T, Liu R, Bai M, Zhou L, Wang X, Li S, Yang H, Li J, Ning T, Huang D, Li H, Zhang L, Ying G, Ba Y. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 356] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 79. | Del Re M, Marconcini R, Pasquini G, Rofi E, Vivaldi C, Bloise F, Restante G, Arrigoni E, Caparello C, Bianco MG, Crucitta S, Petrini I, Vasile E, Falcone A, Danesi R. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer. 2018;118:820-824. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 168] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 80. | Fan Y, Che X, Qu J, Hou K, Wen T, Li Z, Li C, Wang S, Xu L, Liu Y, Qu X. Exosomal PD-L1 Retains Immunosuppressive Activity and is Associated with Gastric Cancer Prognosis. Ann Surg Oncol. 2019;26:3745-3755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 81. | Conforti F, Pala L, Bagnardi V, De Pas T, Martinetti M, Viale G, Gelber RD, Goldhirsch A. Cancer immunotherapy efficacy and patients' sex: a systematic review and meta-analysis. Lancet Oncol. 2018;19:737-746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 404] [Cited by in F6Publishing: 556] [Article Influence: 92.7] [Reference Citation Analysis (0)] |
| 82. | Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565-1570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3828] [Cited by in F6Publishing: 4141] [Article Influence: 318.5] [Reference Citation Analysis (0)] |
| 83. | Fulop T, Larbi A, Kotb R, de Angelis F, Pawelec G. Aging, immunity, and cancer. Discov Med. 2011;11:537-550. [PubMed] [Cited in This Article: ] |
| 84. | Nishijima TF, Muss HB, Shachar SS, Moschos SJ. Comparison of efficacy of immune checkpoint inhibitors (ICIs) between younger and older patients: A systematic review and meta-analysis. Cancer Treat Rev. 2016;45:30-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 161] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 85. | Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, Cogdill AP, Zhao L, Hudgens CW, Hutchinson DS, Manzo T, Petaccia de Macedo M, Cotechini T, Kumar T, Chen WS, Reddy SM, Szczepaniak Sloane R, Galloway-Pena J, Jiang H, Chen PL, Shpall EJ, Rezvani K, Alousi AM, Chemaly RF, Shelburne S, Vence LM, Okhuysen PC, Jensen VB, Swennes AG, McAllister F, Marcelo Riquelme Sanchez E, Zhang Y, Le Chatelier E, Zitvogel L, Pons N, Austin-Breneman JL, Haydu LE, Burton EM, Gardner JM, Sirmans E, Hu J, Lazar AJ, Tsujikawa T, Diab A, Tawbi H, Glitza IC, Hwu WJ, Patel SP, Woodman SE, Amaria RN, Davies MA, Gershenwald JE, Hwu P, Lee JE, Zhang J, Coussens LM, Cooper ZA, Futreal PA, Daniel CR, Ajami NJ, Petrosino JF, Tetzlaff MT, Sharma P, Allison JP, Jenq RR, Wargo JA. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359:97-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2846] [Cited by in F6Publishing: 2679] [Article Influence: 446.5] [Reference Citation Analysis (0)] |
| 86. | Tanaka Y, Shimizu S, Shirotani M, Yorozu K, Kitamura K, Oehorumu M, Kawai Y, Fukuzawa Y. Nutrition and Cancer Risk from the Viewpoint of the Intestinal Microbiome. Nutrients. 2021;13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 87. | Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2493] [Cited by in F6Publishing: 3129] [Article Influence: 447.0] [Reference Citation Analysis (0)] |
| 88. | Bourdais R, Rousseau B, Pujals A, Boussion H, Joly C, Guillemin A, Baumgaertner I, Neuzillet C, Tournigand C. Polymerase proofreading domain mutations: New opportunities for immunotherapy in hypermutated colorectal cancer beyond MMR deficiency. Crit Rev Oncol Hematol. 2017;113:242-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 89. | Nebot-Bral L, Brandao D, Verlingue L, Rouleau E, Caron O, Despras E, El-Dakdouki Y, Champiat S, Aoufouchi S, Leary A, Marabelle A, Malka D, Chaput N, Kannouche PL. Hypermutated tumours in the era of immunotherapy: The paradigm of personalised medicine. Eur J Cancer. 2017;84:290-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 90. | Domingo E, Freeman-Mills L, Rayner E, Glaire M, Briggs S, Vermeulen L, Fessler E, Medema JP, Boot A, Morreau H, van Wezel T, Liefers GJ, Lothe RA, Danielsen SA, Sveen A, Nesbakken A, Zlobec I, Lugli A, Koelzer VH, Berger MD, Castellví-Bel S, Muñoz J; Epicolon consortium, de Bruyn M, Nijman HW, Novelli M, Lawson K, Oukrif D, Frangou E, Dutton P, Tejpar S, Delorenzi M, Kerr R, Kerr D, Tomlinson I, Church DN. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1:207-216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 201] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 91. | Rayner E, van Gool IC, Palles C, Kearsey SE, Bosse T, Tomlinson I, Church DN. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat Rev Cancer. 2016;16:71-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 253] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 92. | Shlien A, Campbell BB, de Borja R, Alexandrov LB, Merico D, Wedge D, Van Loo P, Tarpey PS, Coupland P, Behjati S, Pollett A, Lipman T, Heidari A, Deshmukh S, Avitzur N, Meier B, Gerstung M, Hong Y, Merino DM, Ramakrishna M, Remke M, Arnold R, Panigrahi GB, Thakkar NP, Hodel KP, Henninger EE, Göksenin AY, Bakry D, Charames GS, Druker H, Lerner-Ellis J, Mistry M, Dvir R, Grant R, Elhasid R, Farah R, Taylor GP, Nathan PC, Alexander S, Ben-Shachar S, Ling SC, Gallinger S, Constantini S, Dirks P, Huang A, Scherer SW, Grundy RG, Durno C, Aronson M, Gartner A, Meyn MS, Taylor MD, Pursell ZF, Pearson CE, Malkin D, Futreal PA, Stratton MR, Bouffet E, Hawkins C, Campbell PJ, Tabori U; Biallelic Mismatch Repair Deficiency Consortium. Combined hereditary and somatic mutations of replication error repair genes result in rapid onset of ultra-hypermutated cancers. Nat Genet. 2015;47:257-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 249] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 93. | Howitt BE, Shukla SA, Sholl LM, Ritterhouse LL, Watkins JC, Rodig S, Stover E, Strickland KC, D'Andrea AD, Wu CJ, Matulonis UA, Konstantinopoulos PA. Association of Polymerase e-Mutated and Microsatellite-Instable Endometrial Cancers With Neoantigen Load, Number of Tumor-Infiltrating Lymphocytes, and Expression of PD-1 and PD-L1. JAMA Oncol. 2015;1:1319-1323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 398] [Cited by in F6Publishing: 449] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 94. | McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463-1469. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2210] [Cited by in F6Publishing: 2175] [Article Influence: 271.9] [Reference Citation Analysis (0)] |
| 95. | Yi M, Dong B, Chu Q, Wu K. Immune pressures drive the promoter hypermethylation of neoantigen genes. Exp Hematol Oncol. 2019;8:32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 96. | Ma W, Pham B, Li T. Cancer neoantigens as potential targets for immunotherapy. Clin Exp Metastasis. 2021;1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 97. | Li L, Yu R, Cai T, Chen Z, Lan M, Zou T, Wang B, Wang Q, Zhao Y, Cai Y. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int Immunopharmacol. 2020;88:106939. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 130] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 98. | Minami S, Ihara S, Ikuta S, Komuta K. Gustave Roussy Immune Score and Royal Marsden Hospital Prognostic Score Are Biomarkers of Immune-Checkpoint Inhibitor for Non-Small Cell Lung Cancer. World J Oncol. 2019;10:90-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 99. | Diem S, Kasenda B, Spain L, Martin-Liberal J, Marconcini R, Gore M, Larkin J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br J Cancer. 2016;114:256-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 220] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 100. | Ameratunga M, Chénard-Poirier M, Moreno Candilejo I, Pedregal M, Lui A, Dolling D, Aversa C, Ingles Garces A, Ang JE, Banerji U, Kaye S, Gan H, Doger B, Moreno V, de Bono J, Lopez J. Neutrophil-lymphocyte ratio kinetics in patients with advanced solid tumours on phase I trials of PD-1/PD-L1 inhibitors. Eur J Cancer. 2018;89:56-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 49] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 101. | Jiang T, Qiao M, Zhao C, Li X, Gao G, Su C, Ren S, Zhou C. Pretreatment neutrophil-to-lymphocyte ratio is associated with outcome of advanced-stage cancer patients treated with immunotherapy: a meta-analysis. Cancer Immunol Immunother. 2018;67:713-727. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 102. | Arenas-Ramirez N, Sahin D, Boyman O. Epigenetic mechanisms of tumor resistance to immunotherapy. Cell Mol Life Sci. 2018;75:4163-4176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 103. | Qamra A, Xing M, Padmanabhan N, Kwok JJT, Zhang S, Xu C, Leong YS, Lee Lim AP, Tang Q, Ooi WF, Suling Lin J, Nandi T, Yao X, Ong X, Lee M, Tay ST, Keng ATL, Gondo Santoso E, Ng CCY, Ng A, Jusakul A, Smoot D, Ashktorab H, Rha SY, Yeoh KG, Peng Yong W, Chow PKH, Chan WH, Ong HS, Soo KC, Kim KM, Wong WK, Rozen SG, Teh BT, Kappei D, Lee J, Connolly J, Tan P. Epigenomic Promoter Alterations Amplify Gene Isoform and Immunogenic Diversity in Gastric Adenocarcinoma. Cancer Discov. 2017;7:630-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 104. | Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563-567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3369] [Cited by in F6Publishing: 3871] [Article Influence: 430.1] [Reference Citation Analysis (0)] |
| 105. | Wu HX, Chen YX, Wang ZX, Zhao Q, He MM, Wang YN, Wang F, Xu RH. Alteration in TET1 as potential biomarker for immune checkpoint blockade in multiple cancers. J Immunother Cancer. 2019;7:264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 106. | Perrier A, Didelot A, Laurent-Puig P, Blons H, Garinet S. Epigenetic Mechanisms of Resistance to Immune Checkpoint Inhibitors. Biomolecules. 2020;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 107. | Fan J, Yin Z, Xu J, Wu F, Huang Q, Yang L, Jin Y, Yang G. Circulating microRNAs predict the response to anti-PD-1 therapy in non-small cell lung cancer. Genomics. 2020;112:2063-2071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 108. | González-Silva L, Quevedo L, Varela I. Tumor Functional Heterogeneity Unraveled by scRNA-seq Technologies. Trends Cancer. 2020;6:13-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 109. | Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, Snyder H, Feun LG, Livingstone AS, Harbour JW. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun. 2020;11:496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 228] [Article Influence: 57.0] [Reference Citation Analysis (0)] |
| 110. | Mao X, Yang X, Chen X, Yu S, Zhang B, Ji Y, Chen Y, Ouyang Y, Luo W. Single-cell transcriptome analysis revealed the heterogeneity and microenvironment of gastrointestinal stromal tumors. Cancer Sci. 2021;112:1262-1274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 111. | Di J, Liu M, Fan Y, Gao P, Wang Z, Jiang B, Su X. Phenotype molding of T cells in colorectal cancer by single-cell analysis. Int J Cancer. 2020;146:2281-2295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 112. | Zhou Y, Bian S, Zhou X, Cui Y, Wang W, Wen L, Guo L, Fu W, Tang F. Single-Cell Multiomics Sequencing Reveals Prevalent Genomic Alterations in Tumor Stromal Cells of Human Colorectal Cancer. Cancer Cell. 2020;38:818-828.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 120] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 113. | Lu Z, Chen H, Jiao X, Zhou W, Han W, Li S, Liu C, Gong J, Li J, Zhang X, Wang X, Peng Z, Qi C, Wang Z, Li Y, Brock M, Zhang H, Shen L. Prediction of immune checkpoint inhibition with immune oncology-related gene expression in gastrointestinal cancer using a machine learning classifier. J Immunother Cancer. 2020;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 114. | Conforti F, Pala L, Pagan E, Corti C, Bagnardi V, Queirolo P, Catania C, De Pas T, Giaccone G. Sex-based differences in response to anti-PD-1 or PD-L1 treatment in patients with non-small-cell lung cancer expressing high PD-L1 Levels. A systematic review and meta-analysis of randomized clinical trials. ESMO Open. 2021;6:100251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 34] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 115. | Elias R, Karantanos T, Sira E, Hartshorn KL. Immunotherapy comes of age: Immune aging & checkpoint inhibitors. J Geriatr Oncol. 2017;8:229-235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 116. | Manji G. Perioperative Chemo and Pembrolizumab in Gastric Cancer. [cited 8 September 2021]. Available from: https://clinicaltrials.gov/ct2/show/NCT02918162. [Cited in This Article: ] |
| 117. | Ying L. Camrelizumab Combined with Apatinib for Advanced Gastric or Esophagogastric Adenocarcinoma [cited 8 September 2021]. Available from: https://clinicaltrials.gov/ct2/show/NCT04948125. [Cited in This Article: ] |
| 118. | Park RS. Phase II Study of Neoadjuvant Immune Checkpoint Inhibitor in Patients with resectable Gastrointestinal Cancers (NeoChance). [cited 8 September 2021]. Available from: ClinicalTrials.gov. [Cited in This Article: ] |
| 119. | Fate Therapeutics. FT500 as Monotherapy and in Combination With Immune Checkpoint Inhibitors in Subjects With Advanced Solid Tumors. [cited 8 September 2021]. Available from: https://clinicaltrials.gov/ct2/show/NCT03841110. [Cited in This Article: ] |
| 120. | Incyte Corporations. Arginase Inhibitor INCB001158 as a Single Agent and in Combination with Immune Checkpoint Therapy in Patients With Advanced/Metastatic Solid Tumors. [cited 8 September 2021]. Available from: https://clinicaltrials.gov/ct2/show/NCT02903914. [Cited in This Article: ] |
| 121. | Teclison Ltd. Combination of TATE and PD-1 Inhibitor in Liver Cancer (TATE-PD1) [cited 8 September 2021]. Available from: https://clinicaltrials.gov/ct2/show/NCT03259867. [Cited in This Article: ] |
| 122. | Guangdong Provincial People's Hospital. A Real-World Study of Immune Checkpoint Inhibitors and Chemotherapy for Advanced Esophageal Cancer. [cited 8 September 2021]. Available from: https://clinicaltrials.gov/ct2/show/NCT04822103. [Cited in This Article: ] |