Published online May 15, 2021. doi: 10.4251/wjgo.v13.i5.351
Peer-review started: December 21, 2020
First decision: January 17, 2021
Revised: January 18, 2021
Accepted: April 13, 2021
Article in press: April 13, 2021
Published online: May 15, 2021
Novel non-/minimally-invasive and effective approaches are urgently needed to supplement and improve current strategies for diagnosis and management of hepatocellular carcinoma (HCC). Overwhelming evidence from published studies on HCC has documented that multiple molecular biomarkers detected in body fluids and feces can be utilized in early-diagnosis, predicting responses to specific therapies, evaluating prognosis before or after therapy, as well as serving as novel therapeutic targets. Detection and analysis of proteins, metabolites, circulating nucleic acids, circulating tumor cells, and extracellular vesicles in body fluids (e.g., blood and urine) and gut microbiota (e.g., in feces) have excellent capabilities to improve different aspects of management of HCC. Numerous studies have been devoted in identifying more promising candidate biomarkers and therapeutic targets for diagnosis, treatment, and monitoring responses of HCC to conventional therapies, most of which may improve diagnosis and management of HCC in the future. This review aimed to summarize recent advances in utilizing these biomarkers in HCC and discuss their clinical significance.
Core Tip: Hepatocellular carcinoma is the most frequently diagnosed malignancy of the liver, ranking as the fourth leading cause of cancer-related mortality worldwide. Recent developments of multiple novel biomarkers from body fluids and feces facilitate diagnosis and management of hepatocellular carcinoma. This review aimed to focus on these blood-, urine-, and feces-based biomarkers including proteins, metabolites, circulating nucleic acids, circulating tumor cells, extracellular vesicles, and gut microbiota on clinical application of these biomarkers.
- Citation: Guan MC, Ouyang W, Wang MD, Liang L, Li N, Fu TT, Shen F, Lau WY, Xu QR, Huang DS, Zhu H, Yang T. Biomarkers for hepatocellular carcinoma based on body fluids and feces. World J Gastrointest Oncol 2021; 13(5): 351-365
- URL: https://www.wjgnet.com/1948-5204/full/v13/i5/351.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i5.351
Hepatocellular carcinoma (HCC) is the most frequently diagnosed malignancy of the liver, ranking as the fourth leading cause of cancer-related mortality worldwide[1]. Despite significant advances in diagnostic methods, surgical procedures, and new treatments, overall survival (OS) of HCC patients remains dismal, with a 5-year survival of less than 18%. The bad prognosis is largely due to late diagnosis, a high rate of intrahepatic metastasis at the time of diagnosis, and poor responses to conventional therapies[2]. At present, diagnosis of HCC is mainly based either by histopathological studies or imaging findings. In recent years, there is still a high but an unmet need for a safe, effective, and non-invasive approach for diagnosing early HCC, predicting responses to specific therapies, evaluating prognoses before or after therapies, and developing new therapeutic targets for this tumor. The emergence of liquid biopsy and omics-related techniques has increasingly made significant progression in discovering such biomarkers. Furthermore, screening tests using body fluids or feces can be used in early disease detection, with the advantages of quick and easy extraction, stability, good cost-effectiveness, and non- or mini-invasive accessibility when compared with traditional screening methods.
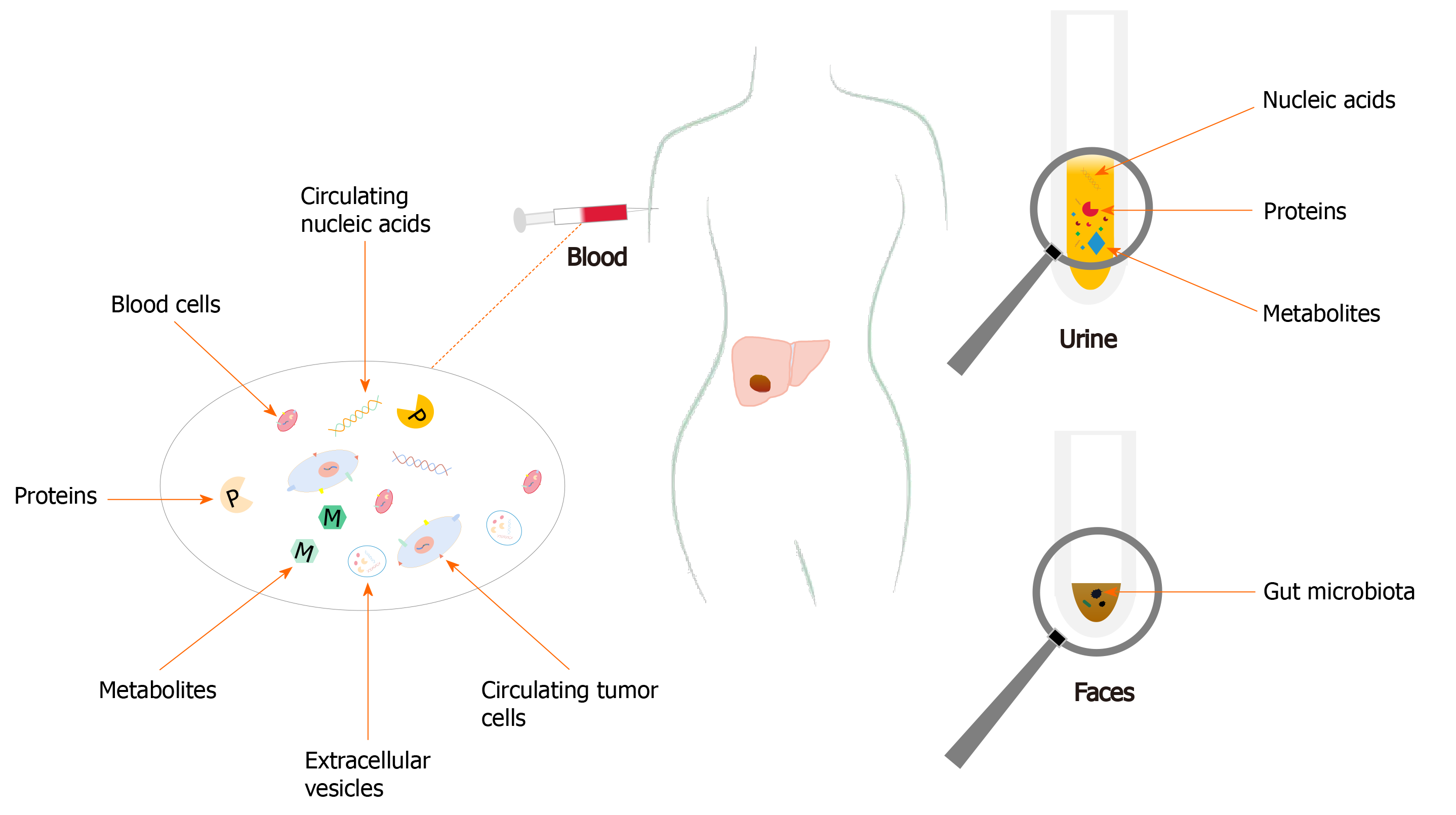
Thus, to be able to identify novel non-invasive biomarkers is of paramount importance not only for HCC diagnosis, but also for evaluating prognosis of patients before and after therapy, monitoring responses to therapy, and predicting recurrence. All these can eventually lead to developments of improved therapeutic regimens with better management results in HCC patients. In this review, the recent and important developments on diagnostic biomarkers of HCC from blood, urine, and feces, including proteins, metabolites, circulating nucleic acids, circulating tumor cells (CTCs), extracellular vesicles (EVs), and gut microbiota, and their clinical significances were reviewed (Figure 1).
With recent developments in liquid biopsy and omics-based techniques, several blood-derived biomarkers for HCC diagnosis and management over the past several years have been identified. This section focuses on the therapeutic strategies in targeting EVs and the capability of CTCs as potential prognostic or predictive biomarkers for HCC (Table 1).
| Biomarkers | Functions | Ref. | Notes |
| CTCs | Recurrence prediction | Chen et al[47], Wang et al[48] and Zhou et al[49] | A close relationship between postoperative CTC levels and early recurrence in HCC patients after liver transplantation or partial hepatectomy was revealed |
| Recurrence prediction | Qi et al[50] | A preoperative CTC count ≥ 16 and a mesenchymal-CTC percentage ≥ 2% were significant risk factors associated with early HCC recurrence, multi-intrahepatic recurrence, and lung metastasis | |
| Prognostic evaluation | Shen et al[51] | An EpCAM-positive CTC count detected before therapy could predict poor survival of patients with HCC | |
| Prognostic evaluation | Hamaoka et al[52] | A GPC3-positive CTC count detected before therapy could predict poor survival of patients with HCC | |
| Prognostic evaluation | Luo et al[53] | The presence of CTC-associated white blood cell clusters detected before therapy could predict poor survival of patients with HCC | |
| Diagnosis/management | Guo et al[54] | A CTC panel including four putative stem cell biomarkers showed great potential in HCC diagnosis, outcome prediction, as well as treatment response evaluation | |
| Monitoring response to therapy | Rau et al[55] | Changes in some CTCs could reflect treatment response to regional therapies, particularly helpful in monitoring AFP-negative HCCs | |
| EVs | Therapeutic targets | Son et al[66] | A new strategy of transferring the sodium/iodide symporter protein to cells via EVs was explored to increase iodine uptake and cytotoxicity in the HCC cells |
| Pomatto et al[67] | Mild electroporation allowed a more efficient and functional miRNA encapsulation in EVs and achieved better protection of antitumor miRNAs from RNase degradation | ||
| Liu et al[68] and Wang et al[69] | Hepatic stellate cell-derived EVs loaded with therapeutic nucleic acids such as miR-30a-3p and miR-335-5p decelerated the progress of HCC by directly regulating targets | ||
| Lu et al[72] | AFP-enriched exosomes derived from dendritic cells allowed the stimulation of antitumor immune responses in autochthonous HCC mouse models in eliciting tumor regression |
The most investigated biomarkers for HCC diagnosis are alpha-fetoprotein (AFP), lens culinaris agglutinin-reactive fraction of AFP (AFP-L3), des-gamma-carboxy prothrombin (DCP), glypican-3, dickkopf-1, Golgi protein 73, osteopontin, and midkine. AFP interacting with other proteins is involved in glucose metabolism, apoptosis, cytoskeleton, and translation in HCC cells, suggesting AFP may be related to malignant characteristics of HCC[3]. AFP is still the most widely accepted serological biomarker for HCC diagnosis, monitoring responses to therapy, surveillance of post-treatment recurrence, and predicting long-term survival outcomes.
However, according to the clinical practice guidelines from the European Society for Medical Oncology and the American Association for the Study of Liver Diseases[4,5], it is ultrasound but not serum AFP that is recommended as a routine HCC surveillance tool for cirrhotic individuals. Nevertheless, a meta-analysis revealed serum AFP, when combined with ultrasonography, significantly improved the detection rate of HCC in clinical practice[6]. Given the marked variations in sensitivity and specificity of using AFP alone in HCC detection, a combination of AFP with other parameters (e.g., DCP, sex, and age) is expected to improve the early diagnostic rates of HCC. Three diagnostic prediction models (i.e., the GALAD model[7], the ASAP model[8], and the HES algorithm[9]) have been developed to provide excellent diagnostic performance for HCC patients. To detect Barcelona Clinic Liver Cancer (BCLC) stage 0-A HCC, the GALAD model, which integrates age and gender into an algorithm based on serum AFP, AFP-L3, and DCP levels, had an area under curve of 0.92 with 92% sensitivity and 79% specificity at a cut-off of -1.18[7]. The ASAP model, which incorporates age, sex, serum AFP, and DCP levels, was reported to have a sensitivity of 73.8% and specificity of 90.0% for Barcelona Clinic Liver Cancer stage 0-A HCC[8]. The HES algorithm, including current AFP level, rate of AFP change, alanine aminotransferase level, platelet count, age, and etiology of cirrhosis, was able to identify early-stage HCC 6 mo prior to ultimate diagnosis, with 51.20% sensitivity and 90.00% specificity, compared with 46.02% sensitivity for AFP alone (P = 0.0015)[9].
Mounting evidence has been reported in the past several years on the usage of AFP in prognostic and recurrence prediction of HCC. While serum AFP levels at diagnosis have prognostic values on treatment results, an AFP response as defined by a decrease in AFP level after therapy has also been demonstrated to predict tumor recurrence and survival outcomes after liver resection or transarterial chemoembolization for HCC[10,11]. Furthermore, an early AFP response has been shown to be associated with improved therapeutic effects of immune checkpoint inhibitors or antiangiogenic therapy for advanced HCC[12]. A multicenter retrospective study noted that a decrease in perioperative serum AFP might be an independent risk factor for prognosis in HCC patients after liver resection[13]. Patients who had a daily decrease of AFP by more than 9% had significantly better OS and recurrence-free survival (RFS) outcomes.
Development of new models in predicting survival and/or recurrence outcomes has recently been reported. Wang et al[14] constructed RFS- and OS-predictive nomograms based on AFP and six clinical variables that exhibited accurate values in predicting postoperative recurrence and OS for patients with hepatitis B virus (HBV)-related solitary HCC ≤ 10 cm. Chan et al[15] recruited 3903 patients after partial hepatectomy from six centers to develop pre- and post-surgical models based on serum AFP levels and other parameters in predicting early recurrence of HCC after liver resection. These models can help to identify patients with high risks of early recurrence, and they have been externally validated using international cohorts. Another reported study on a model allowed accurate preoperative risk stratification for early recurrence in patients with HBV-related HCC after deceased donor liver transplantation[16]. In addition, two scoring models based on serum levels of bilirubin, albumin, AFP-L3, AFP, and DCP at the time of diagnosis of HCC correlated well with HCC staging and prognostication of survival[17]. Meanwhile, close correlations between the peak preoperative AFP level and a total tumor volume ratio of > 20 with HCC recurrence have been demonstrated, thus providing another novel predictor for tumor recurrence[18].
The liver is known to be the main organ for detoxification, hormone production, and metabolism of carbohydrates, fats, and proteins, which are essential to maintain homeostasis of the internal environment. Metabolomics, a newly developed post-genomics field, has gradually played increasingly important roles in HCC diagnosis, recurrence prediction, and prognostic evaluation.
A recent prospective observational cohort study revealed that metabolic perturbations occurred before HCC diagnosis, and it identified 46 metabolites to be associated with hepatocarcinogenesis[19]. Another study demonstrated remarkable changes from chronic HBV infection to HCC in amino acid and lipid metabolisms[20]. Several metabolite panels have been developed to diagnose early HCC; for example, a five-metabolite panel that included methionine, proline, ornithine, pimelylcarnitine, and octanoylcarnitine was constructed as a diagnostic tool for early detection of HCC[21]. Another panel that consisted of serum phenylalanyl-tryptophan and glycocholate performed excellently for early diagnosis of HCC in high-risk populations[22].
Recent evidence has also pinpointed a positive association between dysregulated metabolism detected at an early stage of stereotactic body radiotherapy for HCC with subsequent radiation-triggered liver damage and tumor response[23]. Furthermore, an effective nomogram based on preoperative plasma metabolomics profiling was able to predict the risk of HCC recurrence after liver transplantation, thus optimizing candidate selection for transplantation[24]. Another study on a scoring system, which included phenylalanine and choline, could be used as an adjuvant tool to stratify patients who could benefit most from curative resection[25].
Circulating nucleic acids are a group of extracellular deoxyribonucleic acid (DNA) or ribonucleic acid (RNA) fragments that are shed into peripheral blood via any moribund cells to include circulating cell-free DNAs (cfDNAs) and circulating non-coding RNAs (ncRNAs).
Circulating tumor DNA (ctDNA) belongs to a small fraction of cfDNA that derives from tumor cells. In general, the cfDNA levels in peripheral blood-streams from cancer patients are much higher than those in healthy individuals[26]. ctDNA has emerged as a potential liquid biopsy biomarker for HCC diagnosis, monitoring, and management because it carries multiple tumor-specific genetic or epigenetic variation information. To date, plenty of tumor-specific genetic and/or epigenetic alterations in cfDNA have been identified as diagnostic and predictive biomarkers for HCC. Among them, testing DNA methylation profiling of ctDNA has the most promising potential to function as an approach for early detection of HCC[27]. Several novel panels based on blood methylated DNA markers have recently been developed to yield optimal sensitivity in discriminating early-stage HCC[28,29]. In addition, a growing amount of literature showed that TP53 R249S mutation[30], telomerase reverse transcriptase gene promoter mutations[31], and MutL homolog 1 single-nucleotide variant[32] in ctDNA were closely associated with overall prognosis of HCC patients. Other studies also revealed that mutations in ctDNA could dynamically reflect tumor burden in HCC patients, thus improving the estimations in prognostic risks and evaluation of therapeutic responses[33-35]. Interestingly, a recent study reported that mutations in phosphoinositide-3 kinase/mammalian target of rapamycin signaling were significantly correlated with a shorter PFS outcome after tyrosine kinase inhibitors treatment, but not with immune checkpoint inhibition, suggesting possible involvement of this pathway in chemoresistance to sorafenib[36].
On the other hand, overwhelming evidence has manifested the roles of ncRNAs in diagnosis and management of HCC. Growing attention has been paid to micro-RNAs (miRNAs) that have about 20 nucleotides in serving as new biomarkers for HCC diagnosis. Although the diagnostic performances of mirR-125b[37], miR-122[38,39], and miR-21[40] have been estimated in a lot of studies, their clinical values still require to be validated. Other novel biomarkers like miR-22[41] and miR-3197[42] have also been identified to be useful in diagnosing HCC. A combination of several miRNAs has been shown to have a great potential in enhancing the diagnostic value of HCC[43]. Furthermore, in clinical settings, different miRNAs can be used to function as potential prognostic biomarkers for different selected groups of HCC patients[44].
CTCs are a crowd of scattered seed cells that invade from primary tumors into the circulatory system. Significant advancements of its utilization have been achieved in the last decade. Representatively, the CellSearch System (Veridex), based on the expression of epithelial cell adhesion molecules (EpCAM), enables detection of CTCs from the blood-stream in patients with breast, prostate, or colorectal cancers, and it has been approved for clinical use by the United States Food and Drug Administr
For HCC diagnosis, a systematic review showed presence of CTCs to have only a modest sensitivity of about 60%, but a high specificity of > 90%[46]. For prognostic/predictive biomarkers, CTCs have become the most extensively studied for liquid biopsy. Recent studies have revealed a close relationship between postoperative CTC levels with early recurrence in HCC patients after liver transplantation[47,48] or partial hepatectomy[49]. Also, the association of preoperative CTC counts in HCC patients with microvascular invasion has been documented in a recent study[49]. Another study reported that a preoperative CTC count of ≥ 16 and a mesenchymal-CTC percentage of ≥ 2% to be significant risk factors associated with early HCC recurrence, multi-intrahepatic recurrence, and lung metastasis[50]. Moreover, an EpCAM-positive CTC count[51], a glypican-3-positive CTC count[52], and presence of CTC-associated white blood cells clusters[53] before therapy predicted poor survival of patients with HCC. Guo et al[54] investigated the ability of CTC with stem-like phenotypes in diagnosis and prognostic evaluation for patients with HBV-related HCC. A CTC panel comprising of EpCAM, CD90, CD133, and CK19 allowed discrimination of patients with HCC from those with chronic hepatitis B infection, liver cirrhosis, and benign liver diseases, as well as in identifying early-stage and AFP-negative HCCs. In addition, the relationship between CTC loads and postoperative recurrence was established, indicating the promising role as a real-time tool for risk prediction and treatment monitoring. Also, changes in some CTCs have been reported to reflect treatment responses to regional therapies, and they were particularly helpful in monitoring patients with AFP-negative HCCs[55].
EVs, including exosomes and microvesicles, are small vesicular bodies with phospholipid bilayer membrane structures that are secreted into peripheral blood-stream by various cells[45]. As multiple molecules, including RNA, DNA, and surface protein markers, are capsulized in EVs, they are protected by specific membrane structures against degradation of enzymes[56].
Currently, most studies on EVs as diagnostic markers for HCC are mainly focused on diagnostic performance of ncRNAs derived from EVs. Accumulating evidence has shown that the serum EV-derived ncRNAs including long ncRNA (lncRNA) LINC00853[57], miRNA-10b-5p[58], and miRNA-4661-5p–based panel[58], which participated in pathogenesis of HCC, can serve as novel diagnostic markers for early-stage disease. However, their diagnostic performances are not superior to CTCs or cfDNA, partly because of high degrees of heterogeneity among different studies[46]. Besides, exosomal miRNA-224[59] and lncRNAs ENSG00000258332.1 and LINC00635[60] have been reported to be potential diagnostic and prognostic biomarkers for HCC. Another study further elucidated the functional roles of these biomarkers in HCC progression[61]. High circulating levels of exosomal miRNA-21 (≥ 0.09) and lncRNA-ATB (≥ 0.0016) in HCC patients have been found to associate with poor OS and PFS outcomes. Another study also revealed that a circulating exosomal lncRNA panel could predict recurrence and metastasis of HCC[62].
EVs, as significant intermediaries undergoing intercellular exchange of information and substances, maintain normal physiological functions and are involved in development and progression of cancers[63]. Upon arriving at target cells, EVs can be absorbed by host cells, or dwell on their cytomembrane through mutual recognition, then activate downstream signaling pathways or transmit cargos intracellu
To increase iodine uptake and cytotoxicity in HCC cells, Son et al[66] explored a new strategy of transferring the sodium/iodide symporter (NIS) protein to cells via EVs. HCC cells (Huh7) were first transfected with NIS gene and then collected for isolation of cellular EVs that contained high levels of NIS protein. Treatment of hepatoma cells with these NIS-containing EVs led to enhanced iodine uptake with resultant promotion of cytotoxicity of the radioiodine therapy against cancer cells. Pomatto et al[67] further improved the engineering methods for drug loading of EVs. They found mild electroporation allowed a more efficient and functional miRNA encapsulation in EVs and achieved better protection of antitumor miRNAs from RNase degradation. In addition, EVs loaded with antitumor miR-31 and miR-451a were found to accelerate apoptosis of HCC cells. Moreover, hepatic stellate cell-derived EVs loaded with therapeutic nucleic acids such as miR-30a-3p and miR-335-5p decelerated progression of HCC by directly regulating targets[68,69]. Preclinical trials demonstrated the role of EVs in migration, invasion, and metastasis of HCC. Nidogen 1-enriched EVs have been reported to induce pulmonary fibroblasts to secrete tumor necrosis factor receptor 1, resulting in accelerated extrahepatic metastasis[70]. Similar findings on the functional role of exosomal miR92a-3p in promoting epithelial-mesenchymal transition and metastasis were found by regulating the Akt/Snail signaling pathway[71]. Also, AFP-enriched exosomes derived from dendritic cells have been found to allow stimulation of antitumor immune responses in autochthonous HCC mouse models in eliciting tumor regression[72]. These recent findings provide the potential for development of new strategies in HCC treatment by either inhibiting EV production or up-taking and injecting immunocyte-derived EVs into patients.
As non-invasive tools, urine-based biomarkers can provide significant application values on cancers in clinical settings. Advances in omics technologies and computational capabilities in the past several decades have helped to identify multiple potential urinary biomarkers, including proteins, metabolites, and nucleic acids, for HCC diagnosis and prognosis evaluation (Table 2).
| Function | Ref. | Biomarkers |
| HCC prevention | Wu et al[73] | 8-oxodeoxyguanosine |
| Wu et al[74] | 15-F2t-isoprostane | |
| Yuan et al[75] | 8-epi-prostaglandin F2α | |
| Mahmoud et al[76] | 8-hydroxy-deoxyguanosine | |
| Detection/diagnosis | Abdalla and Haj-Ahmad[77] | DJ-1, Chromatin Assembly Factor-1, Heat Shock Protein 60 |
| Zhan et al[78] | AFP and orosomucoid 1 | |
| Zhao et al[79] | 7 urinary protein features | |
| Ladep et al[80] | A panel including inosine, indole-3-acetate, galactose, and N-acetylated amino acid | |
| Abdalla and Haj-Ahmad[83] | miR-618 and miR-650 | |
| Jeng et al[84] | Adenosine, cytidine, and inosine | |
| Recurrence prediction | Kikuchi et al[85] | Trypsin inhibitor |
| Ye et al[86] | Ethanolamine, lactic acid, acotinic acid, phenylalanine and ribose |
Elevation of 8-oxodeoxyguanosine levels enhances repair of oxidative DNA damage with reduction in risks of developing HCC while increased 15-F2t-isoprostane levels are related to increased risks of HCC. Wu et al[73,74] investigated the role of oxidative stress and aflatoxin exposure on the risk of HCC by using a community-based cohort in Taiwan and demonstrated that both elevated levels of urinary 8-oxodeoxyguanosine (a biomarker of oxidative DNA damage) and urinary 15-F2t-isoprostane (a biomarker of lipid peroxidation) were associated with increased levels of aflatoxin exposure. A Shanghai Cohort Study by Yuan et al[75] identified 8-epi-prostaglandin F2α, a known indicator of oxidative stress, as a biomarker that can stratify individuals without traditional risk factors for HCC development into a high- and a low-risk group. Another study linked high urinary levels of 8-hydroxy-deoxyguanosine (another indicator of oxidative DNA damage) to genetic polymorphisms of two major DNA repair enzymes (XRCC1 rs25487 G/A and OGG1 rs1052133 C/G) in patients with chronic hepatitis C virus (HCV) infection and HCV-related HCCs and concluded that the urinary 8-hydroxy-deoxyguanosine level to be a promising non-invasive biomarker in HCV-related hepatocarcinogenesis[76]. Taken together, these biomarkers can be used to facilitate HCC prevention and progression monitoring.
Urinary proteins, metabolites, and nucleic acids have been identified to be useful in detecting HCC. Such proteins include urinary DJ-1, chromatin assembly factor-1 (CAF-1), Heat Shock Protein 60, AFP, and orosomucoid 1[77,78]. CAF-1 exhibited the best performance in discriminating HCC in HCV patients, with a specificity of 90%, sensitivity 66%, and diagnostic accuracy 78%[77]. Another study showed no significant difference between the diagnostic performance of urinary and serum AFP levels[78]. Urinary proteomic profiling has identified seven urinary protein features that have been validated by targeted Multiple Reaction Monitoring to yield excellent performance in discriminating HCC from high-risk populations[79]. Ladep et al[80] evaluated the diagnostic performance of a panel of urinary metabolites comprising of inosine, indole-3-acetate, galactose, and N-acetylated amino acid in a West African cohort. This panel showed a sensitivity of 86.9% and a specificity of 90.3% in discriminating HCC from cirrhosis; and eight metabolites that were significantly increased in the urine of HCC patients correlated with their clinical HCC stages. Other panels comprising of urinary metabolites also displayed favorable diagnostic capability in distinguishing HCC from cirrhosis[81,82]. Furthermore, urinary miRNA (i.e. miR-618 and miR-650)[83] and nucleosides (i.e. adenosine, cytidine, and inosine)[84] were able to serve as additional diagnostic biomarkers when combined with AFP serology. These findings show the ability of these tests to facilitate HCC diagnosis and surveillance in a non-invasive manner.
During treatment of HCC, changes in levels of urinary biomarkers have been shown to predict tumor recurrence. Overexpression of the urinary trypsin inhibitor in HCC was found to be a risk factor for HCC recurrence after partial hepatectomy[85]. Other reports on metabolites, including ethanolamine, lactic acid, acotinic acid, phenylala
The gut microbiota exerts essential functions in maintaining homeostasis in a human body. It is involved in direct or indirect regulation of key processes in mammalian metabolism, immune tolerance, and immunocompetence[87]. Growing preclinical studies have revealed involvement of gut microbiome in liver carcinogenesis via the gut-liver axis, mainly through exacerbations of inflammation and accumulations of harmful compounds[88]. Thus, disordered intestinal flora can be used for non-invasive diagnostic, prognostic, and therapeutic targets for HCC (Table 3).
| Functions | Ref. | Notes |
| Diagnosis | Ren et al[89] | The selected panel of 30 optimal gut microbial markers showed a powerful diagnostic performance for early HCC, achieving an AUC of 80.64% between 75 early HCC and 105 non-HCC subjects |
| Lapidot et al[90] | The environmental factors leading to fatty liver, consumption of artificial sweeteners, and high-sugar foods were significantly associated with changes in the microbiota of cirrhotic patients with HCC | |
| Monitoring response to therapy | Zheng et al[91] | Gut microbiota could influence the response to immunotherapy in patients with HCC |
| Prognostic evaluation | Huang et al[92] | Six microbial biomarkers related to tumor immune microenvironment or bile acid metabolism could predict clinical outcome |
| Iida et al[93] | The intestinal anaerobic bacteria including Blautia were associated with more favorable prognosis in liver cancer patients after chemotherapy, but usage of anti-anaerobic drugs with poorer prognosis | |
| Therapeutic targets | Rattan et al[94] | The use of probiotics in murine HCC models resulted in reducing HCC development |
Ren et al[89] first validated the potential of gut microbiome as non-invasive tools for early diagnosis of HCC. A decrease in fecal microbial diversity was observed from healthy to cirrhotic individuals, while an increase was detected from cirrhotic individuals to patients with early-stage HCC with cirrhosis. Besides, bacterial genera generating butyrate were decreased, while lipopolysaccharide-yielding genera were increased in patients with early HCC when compared with healthy individuals. Remarkably, a selected panel of 30 optimal gut microbial markers showed a powerful diagnostic performance for early HCC, achieving an area under the curve of 80.64% between a group of 75 patients with early HCC and a group of 105 non-HCC subjects. Lapidot et al[90] also recently revealed changes in gut microbiota in individuals with progression of cirrhosis to HCC and the associations among diet, lifestyle, and microbiome. In addition, an association between excess weight and increased dysbiosis in HCC patients was noted. Environmental factors leading to fatty liver, consumption of artificial sweeteners, and high-sugar foods were significantly associated with alternations in microbiota of cirrhotic patients with HCC. Moreover, an increase of Clostridium and CF231 but a decrease in Alphaproteobacteria in patients with cirrhosis with HCC were found when compared to those without HCC. All these changes were independent of severity of cirrhosis and dietary habits. Future studies recruiting more populations from different regions with diverse underlying diseases are urgently needed for verification of these findings.
There is more evidence supporting the importance of gut microbiota in HCC patient management. Zheng et al[91] compared taxa richness and gene counts of fecal samples from HCC patients who were sensitive to immunotherapy with non-responders and found that gut microbiota could influence the response to anti-programmed cell death protein 1 immunotherapy. This information is critical for treatment decision-making and disease management. As for evaluation of prognosis, a study identified six microbial biomarkers to be related to the tumor immune microenvironment or to bile acid metabolisms[92]. These biomarkers had a potential to predict clinical outcomes. In day-to-day clinical practice, overuse of antibiotic drugs can result in disorder and disturbance of gut microbiota. A study revealed the association of gut flora and antibiotics with prognosis of liver cancer patients after chemotherapy[93]. Intestinal anaerobic bacteria, including Blautia, were reported in this study to be associated with more favorable prognosis in these patients, but usage of anti-anaerobic drugs had poorer prognosis. Thus, modulation of gut microbiota using antibiotics is a promising strategy to intervene development or progression of HCC. In addition, use of probiotics in murine HCC models inhibited HCC development, suggesting this strategy might have a potential to be used as a therapeutic option for HCC patients in the future, provided that alterations of gut microbiota by certain probiotics could also be observed in human[94]. Another viable way to regulate human gut microbiome is through fecal microbiota transplantation, but such studies remain under-investigated.
In this review, advanced developments for HCC in biomarkers derived from body fluids and feces, including proteins, metabolites, circulating nucleic acids, CTCs, EVs, and gut microbiota, were summarized. These biomarkers or their panels have the potential values in diagnosis, monitoring responses to therapy, recurrence prediction, and prognostic evaluation of HCC.
As for blood-based biomarkers, despite wide utilization of AFP, AFP-L3, and DCP in most countries, the sensitivity of a single biomarker for the early diagnosis of HCC is relatively low. In turn, those diagnostic models (e.g., the GALAD model, the ASAP model, and HES algorithm) have satisfactory diagnostic performance, which not only facilitates early detection of HCC in high-risk populations but also avoids undue healthcare costs. These models need to be further optimized and validated in different countries and different populations before widely applied in clinical practice. With the approval of miRNA7™ molecular diagnostic kit for HCC diagnosis and therapeutic monitoring in China, liquid biopsy, especially cfDNA methylation and miRNA test, has a good application prospect in the diagnosis of HCC. However, the maturity of related technology, cost reduction, and the establishment of internationally recognized reference standards are required to ensure early clinical application. At present, imaging examination is routinely used for prognosis evaluation and post-treatment monitoring around the world, but computed tomography or magnetic resonance imaging combined with serum AFP or DCP test is recommended as a regular follow-up examination strategy after HCC treatment according to Chinese, Japanese, and other clinical practice guidelines. Besides, EVs with specific therapeutic agents to liver cancer cells are regarded as a novel therapeutic method for HCC. The mechanism of EVs arrival at and uptake by target cells needs to be further investigated, and these findings from in vitro experiments are urgently needed to be verified in animal models due to complicated internal environment.
As to urinary biomarkers, multiple protein, metabolite, or nucleic acid changes in urine potentially allow for HCC diagnosis and prognosis evaluation. But in fact, no effective urinary biomarkers for HCC have been applied in clinical practice now, and most of the studies are still in the exploratory stage. For gut microbiota from feces, whether gut microbiota can be used for early detection of HCC or as a novel therapeutic remedy should be further studied and clarified.
In conclusion, future well-designed multicenter prospective studies on biomarkers for HCC derived from body fluids and feces as well as transformation of the relevant basic research results into clinical application will optimize current strategies of HCC diagnosis and management.
Manuscript source: Invited manuscript
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Boteon YL, Cerwenka H, Perse M S-Editor: Gao CC L-Editor: Filipodia P-Editor: Li JH
| 1. | Singal AG, Lampertico P, Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J Hepatol. 2020;72:250-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 526] [Cited by in F6Publishing: 570] [Article Influence: 142.5] [Reference Citation Analysis (0)] |
| 2. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2066] [Cited by in F6Publishing: 2561] [Article Influence: 512.2] [Reference Citation Analysis (0)] |
| 3. | Yokoo H, Kondo T, Fujii K, Yamada T, Todo S, Hirohashi S. Proteomic signature corresponding to alpha fetoprotein expression in liver cancer cells. Hepatology. 2004;40:609-617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 4. | Vogel A, Cervantes A, Chau I, Daniele B, Llovet JM, Meyer T, Nault JC, Neumann U, Ricke J, Sangro B, Schirmacher P, Verslype C, Zech CJ, Arnold D, Martinelli E; ESMO Guidelines Committee. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv238-iv255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 595] [Cited by in F6Publishing: 569] [Article Influence: 94.8] [Reference Citation Analysis (0)] |
| 5. | Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, Zhu AX, Murad MH, Marrero JA. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67:358-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2107] [Cited by in F6Publishing: 2574] [Article Influence: 429.0] [Reference Citation Analysis (2)] |
| 6. | Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, Waljee AK, Singal AG. Surveillance Imaging and Alpha Fetoprotein for Early Detection of Hepatocellular Carcinoma in Patients With Cirrhosis: A Meta-analysis. Gastroenterology 2018; 154: 1706-1718. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 638] [Cited by in F6Publishing: 599] [Article Influence: 99.8] [Reference Citation Analysis (0)] |
| 7. | Yang JD, Addissie BD, Mara KC, Harmsen WS, Dai J, Zhang N, Wongjarupong N, Ali HM, Ali HA, Hassan FA, Lavu S, Cvinar JL, Giama NH, Moser CD, Miyabe K, Allotey LK, Algeciras-Schimnich A, Theobald JP, Ward MM, Nguyen MH, Befeler AS, Reddy KR, Schwartz M, Harnois DM, Yamada H, Srivastava S, Rinaudo JA, Gores GJ, Feng Z, Marrero JA, Roberts LR. GALAD Score for Hepatocellular Carcinoma Detection in Comparison with Liver Ultrasound and Proposal of GALADUS Score. Cancer Epidemiol Biomarkers Prev. 2019;28:531-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 8. | Yang T, Xing H, Wang G, Wang N, Liu M, Yan C, Li H, Wei L, Li S, Fan Z, Shi M, Chen W, Cai S, Pawlik TM, Soh A, Beshiri A, Lau WY, Wu M, Zheng Y, Shen F. A Novel Online Calculator Based on Serum Biomarkers to Detect Hepatocellular Carcinoma among Patients with Hepatitis B. Clin Chem. 2019;65:1543-1553. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Tayob N, Corley DA, Christie I, Almers L, Rahal AK, Richardson P, White DL, Davila J, Kanwal F, El-Serag HB. Validation of the Updated Hepatocellular Carcinoma Early Detection Screening Algorithm in a Community-Based Cohort of Patients With Cirrhosis of Multiple Etiologies. Clin Gastroenterol Hepatol. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 10. | Li XL, Zhu XD, Cai H, Li Y, Zhou J, Fan J, Tang ZY, Sun HC. Postoperative α-fetoprotein response predicts tumor recurrence and survival after hepatectomy for hepatocellular carcinoma: A propensity score matching analysis. Surgery. 2019;165:1161-1167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Liu G, Ouyang Q, Xia F, Fan G, Yu J, Zhang C, Wang D. Alpha-fetoprotein response following transarterial chemoembolization indicates improved survival for intermediate-stage hepatocellular carcinoma. HPB (Oxford). 2019;21:107-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Shao YY, Liu TH, Hsu C, Lu LC, Shen YC, Lin ZZ, Cheng AL, Hsu CH. Early alpha-foetoprotein response associated with treatment efficacy of immune checkpoint inhibitors for advanced hepatocellular carcinoma. Liver Int. 2019;39:2184-2189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 51] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 13. | Zhou PY, Yang CP, Tang Z, Yi Y, Liu WR, Tian MX, Huang JL, Gan W, Jiang XF, Liu G, Wang H, Tao CY, Fang Y, Qu WF, Zhou C, Guan RY, Sun BY, Zhou YF, Song SS, Ding ZB, Peng YF, Dai Z, Zhou J, Fan J, Gong GZ, Shi YH, Qiu SJ. Daily decrease of post-operative alpha-fetoprotein by 9% discriminates prognosis of HCC: A multicenter retrospective study. Aging (Albany NY). 2019;11:11111-11123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Wang XH, Liao B, Hu WJ, Tu CX, Xiang CL, Hao SH, Mao XH, Qiu XM, Yang XJ, Yue X, Kuang M, Peng BG, Li SQ. Novel Models Predict Postsurgical Recurrence and Overall Survival for Patients with Hepatitis B Virus-Related Solitary Hepatocellular Carcinoma ≤10 cm and Without Portal Venous Tumor Thrombus. Oncologist. 2020;25:e1552-e1561. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Chan AWH, Zhong J, Berhane S, Toyoda H, Cucchetti A, Shi K, Tada T, Chong CCN, Xiang BD, Li LQ, Lai PBS, Mazzaferro V, García-Fiñana M, Kudo M, Kumada T, Roayaie S, Johnson PJ. Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol. 2018;69:1284-1293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 314] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 16. | Al-Ameri AAM, Wei X, Lin L, Shao Z, Guo H, Xie H, Zhou L, Zheng S, Xu X. Preoperative risk stratification for early recurrence of HBV-related hepatocellular carcinoma after deceased donor liver transplantation: a five-eight model development and validation. BMC Cancer. 2019;19:1136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 17. | Toyoda H, Tada T, Johnson PJ, Izumi N, Kadoya M, Kaneko S, Kokudo N, Ku Y, Kubo S, Kumada T, Matsuyama Y, Nakashima O, Sakamoto M, Takayama T, Kudo M; Liver Cancer Study Group of Japan. Validation of serological models for staging and prognostication of HCC in patients from a Japanese nationwide survey. J Gastroenterol. 2017;52:1112-1121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Sharma Y, Weaver MJ, Ludwig DR, Fowler K, Vachharajani N, Chapman WC, Crippin JS. Serum alpha-fetoprotein level per total tumor volume as a predictor of recurrence of hepatocellular carcinoma after resection. Surgery. 2018;163:1002-1007. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 19. | Stepien M, Keski-Rahkonen P, Kiss A, Robinot N, Duarte-Salles T, Murphy N, Perlemuter G, Viallon V, Tjønneland A, Rostgaard-Hansen AL, Dahm CC, Overvad K, Boutron-Ruault MC, Mancini FR, Mahamat-Saleh Y, Aleksandrova K, Kaaks R, Kühn T, Trichopoulou A, Karakatsani A, Panico S, Tumino R, Palli D, Tagliabue G, Naccarati A, Vermeulen RCH, Bueno-de-Mesquita HB, Weiderpass E, Skeie G, Ramón Quirós J, Ardanaz E, Mokoroa O, Sala N, Sánchez MJ, Huerta JM, Winkvist A, Harlid S, Ohlsson B, Sjöberg K, Schmidt JA, Wareham N, Khaw KT, Ferrari P, Rothwell JA, Gunter M, Riboli E, Scalbert A, Jenab M. Metabolic perturbations prior to hepatocellular carcinoma diagnosis: Findings from a prospective observational cohort study. Int J Cancer. 2021;148:609-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 40] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 20. | Cai FF, Song YN, Lu YY, Zhang Y, Hu YY, Su SB. Analysis of plasma metabolic profile, characteristics and enzymes in the progression from chronic hepatitis B to hepatocellular carcinoma. Aging (Albany NY). 2020;12:14949-14965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Kim DJ, Cho EJ, Yu KS, Jang IJ, Yoon JH, Park T, Cho JY. Comprehensive Metabolomic Search for Biomarkers to Differentiate Early Stage Hepatocellular Carcinoma from Cirrhosis. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 22. | Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, Yin Z, Xie X, Wang X, Chen W, Zhou L, Li Y, Chen H, Gao L, Lu X, Wu T, Wang H, Niu J, Xu G. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67:662-675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 233] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 23. | Ng SSW, Jang GH, Kurland IJ, Qiu Y, Guha C, Dawson LA. Plasma metabolomic profiles in liver cancer patients following stereotactic body radiotherapy. EBioMedicine. 2020;59:102973. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Lu D, Yang F, Lin Z, Zhuo J, Liu P, Cen B, Lian Z, Xie H, Zheng S, Xu X. A prognostic fingerprint in liver transplantation for hepatocellular carcinoma based on plasma metabolomics profiling. Eur J Surg Oncol. 2019;45:2347-2352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Wang Q, Su B, Dong L, Jiang T, Tan Y, Lu X, Liu X, Lin X, Xu G. Liquid Chromatography-Mass Spectrometry-Based Nontargeted Metabolomics Predicts Prognosis of Hepatocellular Carcinoma after Curative Resection. J Proteome Res. 2020;19:3533-3541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Wu X, Li J, Gassa A, Buchner D, Alakus H, Dong Q, Ren N, Liu M, Odenthal M, Stippel D, Bruns C, Zhao Y, Wahba R. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int J Biol Sci. 2020;16:1551-1562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 77] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 27. | von Felden J, Garcia-Lezana T, Schulze K, Losic B, Villanueva A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut. 2020;69:2025-2034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 66] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 28. | Kisiel JB, Dukek BA, V S R Kanipakam R, Ghoz HM, Yab TC, Berger CK, Taylor WR, Foote PH, Giama NH, Onyirioha K, Abdallah MA, Burger KN, Slettedahl SW, Mahoney DW, Smyrk TC, Lewis JT, Giakoumopoulos M, Allawi HT, Lidgard GP, Roberts LR, Ahlquist DA. Hepatocellular Carcinoma Detection by Plasma Methylated DNA: Discovery, Phase I Pilot, and Phase II Clinical Validation. Hepatology. 2019;69:1180-1192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 29. | Chalasani NP, Ramasubramanian TS, Bhattacharya A, Olson MC, Edwards V DK, Roberts LR, Kisiel JB, Reddy KR, Lidgard GP, Johnson SC, Bruinsma JJ. A Novel Blood-Based Panel of Methylated DNA and Protein Markers for Detection of Early-Stage Hepatocellular Carcinoma. Clin Gastroenterol Hepatol. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 30. | Shen T, Li SF, Wang JL, Zhang T, Zhang S, Chen HT, Xiao QY, Ren WH, Liu C, Peng B, Ji XN, Yang Y, Lu PX, Chen TY, Yu L, Ji Y, Jiang DK. TP53 R249S mutation detected in circulating tumour DNA is associated with Prognosis of hepatocellular carcinoma patients with or without hepatectomy. Liver Int. 2020;40:2834-2847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 31. | Hirai M, Kinugasa H, Nouso K, Yamamoto S, Terasawa H, Onishi Y, Oyama A, Adachi T, Wada N, Sakata M, Yasunaka T, Onishi H, Shiraha H, Takaki A, Okada H. Prediction of the prognosis of advanced hepatocellular carcinoma by TERT promoter mutations in circulating tumor DNA. J Gastroenterol Hepatol. 2020;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 20] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 32. | Kim SS, Eun JW, Choi JH, Woo HG, Cho HJ, Ahn HR, Suh CW, Baek GO, Cho SW, Cheong JY. MLH1 single-nucleotide variant in circulating tumor DNA predicts overall survival of patients with hepatocellular carcinoma. Sci Rep. 2020;10:17862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 33. | Cai Z, Chen G, Zeng Y, Dong X, Li Z, Huang Y, Xin F, Qiu L, Xu H, Zhang W, Su X, Liu X, Liu J. Comprehensive Liquid Profiling of Circulating Tumor DNA and Protein Biomarkers in Long-Term Follow-Up Patients with Hepatocellular Carcinoma. Clin Cancer Res. 2019;25:5284-5294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 34. | Cai ZX, Chen G, Zeng YY, Dong XQ, Lin MJ, Huang XH, Zhang D, Liu XL, Liu JF. Circulating tumor DNA profiling reveals clonal evolution and real-time disease progression in advanced hepatocellular carcinoma. Int J Cancer. 2017;141:977-985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 35. | Chen G, Cai Z, Li Z, Dong X, Xu H, Lin J, Chen L, Zhang H, Liu X, Liu J. Clonal evolution in long-term follow-up patients with hepatocellular carcinoma. Int J Cancer. 2018;143:2862-2870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 36. | von Felden J, Craig AJ, Garcia-Lezana T, Labgaa I, Haber PK, D'Avola D, Asgharpour A, Dieterich D, Bonaccorso A, Torres-Martin M, Sia D, Sung MW, Tabrizian P, Schwartz M, Llovet JM, Villanueva A. Mutations in circulating tumor DNA predict primary resistance to systemic therapies in advanced hepatocellular carcinoma. Oncogene. 2021;40:140-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 37. | Jin X, Cai C, Qiu Y. Diagnostic Value of Circulating microRNAs in Hepatitis B Virus-Related Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. J Cancer. 2019;10:4754-4764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Zhao XF, Li N, Lin DD, Sun LB. Circulating MicroRNA-122 for the Diagnosis of Hepatocellular Carcinoma: A Meta-Analysis. Biomed Res Int. 2020;2020:5353695. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 39. | Wei XY, Ding J, Tian WG, Yu YC. MicroRNA-122 as a diagnostic biomarker for hepatocellular carcinoma related to hepatitis C virus: a meta-analysis and systematic review. J Int Med Res. 2020;48:300060520941634. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 40. | Qu J, Yang J, Chen M, Cui L, Wang T, Gao W, Tian J, Wei R. MicroRNA-21 as a diagnostic marker for hepatocellular carcinoma: A systematic review and meta-analysis. Pak J Med Sci. 2019;35:1466-1471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 41. | Zeng Z, Dong J, Li Y, Dong Z, Liu Z, Huang J, Wang Y, Zhen Y, Lu Y. The expression level and diagnostic value of microRNA-22 in HCC patients. Artif Cells Nanomed Biotechnol. 2020;48:683-686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Pascut D, Cavalletto L, Pratama MY, Bresolin S, Trentin L, Basso G, Bedogni G, Tiribelli C, Chemello L. Serum miRNA Are Promising Biomarkers for the Detection of Early Hepatocellular Carcinoma after Treatment with Direct-Acting Antivirals. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 43. | Weis A, Marquart L, Calvopina DA, Genz B, Ramm GA, Skoien R. Serum MicroRNAs as Biomarkers in Hepatitis C: Preliminary Evidence of a MicroRNA Panel for the Diagnosis of Hepatocellular Carcinoma. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 44. | Pratama MY, Visintin A, Crocè LS, Tiribelli C, Pascut D. Circulatory miRNA as a Biomarker for Therapy Response and Disease-Free Survival in Hepatocellular Carcinoma. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 45. | Mann J, Reeves HL, Feldstein AE. Liquid biopsy for liver diseases. Gut. 2018;67:2204-2212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 46. | Chen VL, Xu D, Wicha MS, Lok AS, Parikh ND. Utility of Liquid Biopsy Analysis in Detection of Hepatocellular Carcinoma, Determination of Prognosis, and Disease Monitoring: A Systematic Review. Clin Gastroenterol Hepatol 2020; 18: 2879-2902. e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 47. | Chen Z, Lin X, Chen C, Chen Y, Zhao Q, Wu L, Wang D, Ma Y, Ju W, Chen M, He X. Analysis of preoperative circulating tumor cells for recurrence in patients with hepatocellular carcinoma after liver transplantation. Ann Transl Med. 2020;8:1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 48. | Wang PX, Xu Y, Sun YF, Cheng JW, Zhou KQ, Wu SY, Hu B, Zhang ZF, Guo W, Cao Y, Huang XW, Zhou J, Fan J, Yang XR. Detection of circulating tumour cells enables early recurrence prediction in hepatocellular carcinoma patients undergoing liver transplantation. Liver Int. 2021;41:562-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 27] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 49. | Zhou J, Zhang Z, Zhou H, Leng C, Hou B, Zhou C, Hu X, Wang J, Chen X. Preoperative circulating tumor cells to predict microvascular invasion and dynamical detection indicate the prognosis of hepatocellular carcinoma. BMC Cancer. 2020;20:1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 50. | Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY, Chen YY, Chen ZS, Ma L, Chen J, Gong WF, Han ZG, Lu Y, Shang JJ, Li LQ. Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res. 2018;78:4731-4744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 51. | Shen J, Wang WS, Zhu XL, Ni CF. High Epithelial Cell Adhesion Molecule-Positive Circulating Tumor Cell Count Predicts Poor Survival of Patients with Unresectable Hepatocellular Carcinoma Treated with Transcatheter Arterial Chemoembolization. J Vasc Interv Radiol. 2018;29:1678-1684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 52. | Hamaoka M, Kobayashi T, Tanaka Y, Mashima H, Ohdan H. Clinical significance of glypican-3-positive circulating tumor cells of hepatocellular carcinoma patients: A prospective study. PLoS One. 2019;14:e0217586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 53. | Luo Q, Wang C, Peng B, Pu X, Cai L, Liao H, Chen K, Zhang C, Cheng Y, Pan M. Circulating Tumor-Cell-Associated White Blood Cell Clusters in Peripheral Blood Indicate Poor Prognosis in Patients With Hepatocellular Carcinoma. Front Oncol. 2020;10:1758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 54. | Guo W, Sun YF, Shen MN, Ma XL, Wu J, Zhang CY, Zhou Y, Xu Y, Hu B, Zhang M, Wang G, Chen WQ, Guo L, Lu RQ, Zhou CH, Zhang X, Shi YH, Qiu SJ, Pan BS, Cao Y, Zhou J, Yang XR, Fan J. Circulating Tumor Cells with Stem-Like Phenotypes for Diagnosis, Prognosis, and Therapeutic Response Evaluation in Hepatocellular Carcinoma. Clin Cancer Res. 2018;24:2203-2213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 55. | Rau KM, Liu CT, Hsiao YC, Hsiao KY, Wang TM, Hung WS, Su YL, Liu WC, Wang CH, Hsu HL, Chuang PH, Cheng JC, Tseng CP. Sequential Circulating Tumor Cell Counts in Patients with Locally Advanced or Metastatic Hepatocellular Carcinoma: Monitoring the Treatment Response. J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 56. | Sasaki R, Kanda T, Yokosuka O, Kato N, Matsuoka S, Moriyama M. Exosomes and Hepatocellular Carcinoma: From Bench to Bedside. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 57. | Kim SS, Baek GO, Ahn HR, Sung S, Seo CW, Cho HJ, Nam SW, Cheong JY, Eun JW. Serum small extracellular vesicle-derived LINC00853 as a novel diagnostic marker for early hepatocellular carcinoma. Mol Oncol. 2020;14:2646-2659. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 58. | Cho HJ, Baek GO, Seo CW, Ahn HR, Sung S, Son JA, Kim SS, Cho SW, Jang JW, Nam SW, Cheong JY, Eun JW. Exosomal microRNA-4661-5p-based serum panel as a potential diagnostic biomarker for early-stage hepatocellular carcinoma. Cancer Med. 2020;9:5459-5472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Cui Y, Xu HF, Liu MY, Xu YJ, He JC, Zhou Y, Cang SD. Mechanism of exosomal microRNA-224 in development of hepatocellular carcinoma and its diagnostic and prognostic value. World J Gastroenterol. 2019;25:1890-1898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 60] [Cited by in F6Publishing: 64] [Article Influence: 12.8] [Reference Citation Analysis (1)] |
| 60. | Xu H, Chen Y, Dong X, Wang X. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol Biomarkers Prev. 2018;27:710-716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 94] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 61. | Lee YR, Kim G, Tak WY, Jang SY, Kweon YO, Park JG, Lee HW, Han YS, Chun JM, Park SY, Hur K. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer. 2019;144:1444-1452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 62. | Lu Y, Duan Y, Xu Q, Zhang L, Chen W, Qu Z, Wu B, Liu W, Shi L, Wu D, Yang Y, Sun D, Chen X. Circulating exosome-derived bona fide long non-coding RNAs predicting the occurrence and metastasis of hepatocellular carcinoma. J Cell Mol Med. 2020;24:1311-1318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 63. | Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12:133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 64. | Chen R, Xu X, Tao Y, Qian Z, Yu Y. Exosomes in hepatocellular carcinoma: a new horizon. Cell Commun Signal. 2019;17:1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 65. | Raimondo S, Giavaresi G, Lorico A, Alessandro R. Extracellular Vesicles as Biological Shuttles for Targeted Therapies. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 66. | Son SH, Gangadaran P, Ahn BC. A novel strategy of transferring NIS protein to cells using extracellular vesicles leads to increase in iodine uptake and cytotoxicity. Int J Nanomedicine. 2019;14:1779-1787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 67. | Pomatto MAC, Bussolati B, D'Antico S, Ghiotto S, Tetta C, Brizzi MF, Camussi G. Improved Loading of Plasma-Derived Extracellular Vesicles to Encapsulate Antitumor miRNAs. Mol Ther Methods Clin Dev. 2019;13:133-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 139] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 68. | Liu C, Zhou X, Long Q, Zeng H, Sun Q, Chen Y, Wu D, Liu L. Small extracellular vesicles containing miR-30a-3p attenuate the migration and invasion of hepatocellular carcinoma by targeting SNAP23 gene. Oncogene. 2021;40:233-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 69. | Wang F, Li L, Piontek K, Sakaguchi M, Selaru FM. Exosome miR-335 as a novel therapeutic strategy in hepatocellular carcinoma. Hepatology. 2018;67:940-954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 186] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 70. | Mao X, Tey SK, Yeung CLS, Kwong EML, Fung YME, Chung CYS, Mak LY, Wong DKH, Yuen MF, Ho JCM, Pang H, Wong MP, Leung CO, Lee TKW, Ma V, Cho WC, Cao P, Xu X, Gao Y, Yam JWP. Nidogen 1-Enriched Extracellular Vesicles Facilitate Extrahepatic Metastasis of Liver Cancer by Activating Pulmonary Fibroblasts to Secrete Tumor Necrosis Factor Receptor 1. Adv Sci (Weinh). 2020;7:2002157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 71. | Yang B, Feng X, Liu H, Tong R, Wu J, Li C, Yu H, Chen Y, Cheng Q, Chen J, Cai X, Wu W, Lu Y, Hu J, Liang K, Lv Z, Zheng S. High-metastatic cancer cells derived exosomal miR92a-3p promotes epithelial-mesenchymal transition and metastasis of low-metastatic cancer cells by regulating PTEN/Akt pathway in hepatocellular carcinoma. Oncogene. 2020;39:6529-6543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 72. | Lu Z, Zuo B, Jing R, Gao X, Rao Q, Liu Z, Qi H, Guo H, Yin H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J Hepatol. 2017;67:739-748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 251] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 73. | Wu HC, Wang Q, Wang LW, Yang HI, Ahsan H, Tsai WY, Wang LY, Chen SY, Chen CJ, Santella RM. Urinary 8-oxodeoxyguanosine, aflatoxin B1 exposure and hepatitis B virus infection and hepatocellular carcinoma in Taiwan. Carcinogenesis. 2007;28:995-999. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 74. | Wu HC, Wang Q, Yang HI, Ahsan H, Tsai WY, Wang LY, Chen SY, Chen CJ, Santella RM. Urinary 15-F2t-isoprostane, aflatoxin B1 exposure and hepatitis B virus infection and hepatocellular carcinoma in Taiwan. Carcinogenesis. 2008;29:971-976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 75. | Yuan JM, Grouls M, Carmella SG, Wang R, Heskin A, Jiang Y, Tan YT, Adams-Haduch J, Gao YT, Hecht SS. Prediagnostic levels of urinary 8-epi-prostaglandin F2α and prostaglandin E2 metabolite, biomarkers of oxidative damage and inflammation, and risk of hepatocellular carcinoma. Carcinogenesis. 2019;40:989-997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 76. | Mahmoud AA, Hassan MH, Ghweil AA, Abdelrahman A, Mohammad AN, Ameen HH. Urinary 8-hydroxydeoxyguanosine in relation to XRCC1 rs25487 G/A (Arg399Gln) and OGG1 rs1052133 C/G (Ser326Cys) DNA repair genes polymorphisms in patients with chronic hepatitis C and related hepatocellular carcinoma. Cancer Manag Res. 2019;11:5343-5351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 77. | Abdalla MA, Haj-Ahmad Y. Promising Urinary Protein Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:390-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 78. | Zhan Z, Guan Y, Mew K, Zeng W, Peng M, Hu P, Yang Y, Lu Y, Ren H. Urine α-fetoprotein and orosomucoid 1 as biomarkers of hepatitis B virus-associated hepatocellular carcinoma. Am J Physiol Gastrointest Liver Physiol. 2020;318:G305-G312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 79. | Zhao Y, Li Y, Liu W, Xing S, Wang D, Chen J, Sun L, Mu J, Xing B, Sun W, He F. Identification of noninvasive diagnostic biomarkers for hepatocellular carcinoma by urinary proteomics. J Proteomics. 2020;225:103780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Ladep NG, Dona AC, Lewis MR, Crossey MM, Lemoine M, Okeke E, Shimakawa Y, Duguru M, Njai HF, Fye HK, Taal M, Chetwood J, Kasstan B, Khan SA, Garside DA, Wijeyesekera A, Thillainayagam AV, Banwat E, Thursz MR, Nicholson JK, Njie R, Holmes E, Taylor-Robinson SD. Discovery and validation of urinary metabotypes for the diagnosis of hepatocellular carcinoma in West Africans. Hepatology. 2014;60:1291-1301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 81. | Dai W, Yin P, Chen P, Kong H, Luo P, Xu Z, Lu X, Xu G. Study of urinary steroid hormone disorders: difference between hepatocellular carcinoma in early stage and cirrhosis. Anal Bioanal Chem. 2014;406:4325-4335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 82. | Shariff MI, Gomaa AI, Cox IJ, Patel M, Williams HR, Crossey MM, Thillainayagam AV, Thomas HC, Waked I, Khan SA, Taylor-Robinson SD. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J Proteome Res. 2011;10:1828-1836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 83. | Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 84. | Jeng LB, Lo WY, Hsu WY, Lin WD, Lin CT, Lai CC, Tsai FJ. Analysis of urinary nucleosides as helper tumor markers in hepatocellular carcinoma diagnosis. Rapid Commun Mass Spectrom. 2009;23:1543-1549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 85. | Kikuchi I, Uchinami H, Nanjo H, Hashimoto M, Nakajima A, Kume M, Mencin A, Yamamoto Y. Clinical and prognostic significance of urinary trypsin inhibitor in patients with hepatocellular carcinoma after hepatectomy. Ann Surg Oncol. 2009;16:2805-2817. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 86. | Ye G, Zhu B, Yao Z, Yin P, Lu X, Kong H, Fan F, Jiao B, Xu G. Analysis of urinary metabolic signatures of early hepatocellular carcinoma recurrence after surgical removal using gas chromatography-mass spectrometry. J Proteome Res. 2012;11:4361-4372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 87. | Schwabe RF, Greten TF. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J Hepatol. 2020;72:230-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 178] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 88. | Ponziani FR, Nicoletti A, Gasbarrini A, Pompili M. Diagnostic and therapeutic potential of the gut microbiota in patients with early hepatocellular carcinoma. Ther Adv Med Oncol. 2019;11:1758835919848184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 89. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 305] [Cited by in F6Publishing: 387] [Article Influence: 77.4] [Reference Citation Analysis (0)] |
| 90. | Lapidot Y, Amir A, Nosenko R, Uzan-Yulzari A, Veitsman E, Cohen-Ezra O, Davidov Y, Weiss P, Bradichevski T, Segev S, Koren O, Safran M, Ben-Ari Z. Alterations in the Gut Microbiome in the Progression of Cirrhosis to Hepatocellular Carcinoma. mSystems. 2020;5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 91. | Zheng Y, Wang T, Tu X, Huang Y, Zhang H, Tan D, Jiang W, Cai S, Zhao P, Song R, Li P, Qin N, Fang W. Gut microbiome affects the response to anti-PD-1 immunotherapy in patients with hepatocellular carcinoma. J Immunother Cancer. 2019;7:193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 280] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 92. | Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang J, Lu H, Yin S, Ji J, Zhou L, Zheng S. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 2020;12:102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 93. | Iida N, Mizukoshi E, Yamashita T, Terashima T, Arai K, Seishima J, Kaneko S. Overuse of antianaerobic drug is associated with poor postchemotherapy prognosis of patients with hepatocellular carcinoma. Int J Cancer. 2019;145:2701-2711. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 94. | Rattan P, Minacapelli CD, Rustgi V. The Microbiome and Hepatocellular Carcinoma. Liver Transpl. 2020;26:1316-1327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |