Published online Oct 15, 2021. doi: 10.4251/wjgo.v13.i10.1288
Peer-review started: February 21, 2021
First decision: June 16, 2021
Revised: June 19, 2021
Accepted: August 24, 2021
Article in press: August 24, 2021
Published online: October 15, 2021
Molecular genetic analysis is an integral part of colorectal cancer (CRC) management. The choice of systemic therapy for CRC is largely based on the results of tumor molecular testing. Evaluation of the KRAS and NRAS gene status is mandatory for consideration of anti-epidermal growth factor receptor (EGFR) therapy. Tumors with the BRAF V600E substitution are characterized by aggressive behaviour, may require intensified cytotoxic regimens and benefit from combined BRAF and EGFR inhibition. The inactivation of DNA mismatch repair (MMR), or MUTYH gene, or DNA polymerase epsilon results in excessive tumor mutational burden; these CRCs are highly antigenic and therefore sensitive to immune checkpoint inhibitors. Some CRCs are characterized by overexpression of the HER2 oncogene and respond to the appropriate targeted therapy. There are CRCs with clinical signs of hereditary predisposition to this disease, which require germline genetic testing. Liquid biopsy is an emerging technology that has the potential to assist CRC screening, control the efficacy of surgical intervention and guide disease monitoring. The landscape of CRC molecular diagnosis is currently undergoing profound changes due to the increasing use of next generation sequencing.
Core Tip: Molecular genetic analysis is an integral component of colorectal cancer (CRC) management. Comprehensive KRAS and NRAS testing is mandatory for selection of patients for anti-epidermal growth factor receptor (EGFR) therapy. BRAF V600E mutated cancers are responsive to combination of BRAF and EGFR inhibitors. CRCs with HER2 amplification and overexpression can be controlled by the down-regulation of this receptor. Immune therapy is highly effective in CRCs with exceptionally high tumor mutation burden, e.g., in cancers with microsatellite instability, MUTYH gene inactivation or mutations in the POLE gene. CRC patients with early disease onset, specific tumor features or family history of the disease require germ-line DNA testing.
- Citation: Imyanitov E, Kuligina E. Molecular testing for colorectal cancer: Clinical applications. World J Gastrointest Oncol 2021; 13(10): 1288-1301
- URL: https://www.wjgnet.com/1948-5204/full/v13/i10/1288.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v13.i10.1288
Colorectal cancer (CRC) holds the third position for cancer morbidity and accounts for approximately 1.8 million new cases per year worldwide. Approximately half of the patients with CRC diagnosis die from this disease[1]. CRC is commonly classified as right-sided (proximal) cancers affecting cecum, ascending colon, hepatic flexure and transverse colon, and left-sided (distal) tumors, which include CRCs located at the splenic flexure, descending colon, sigmoid colon and rectum. Right-sided and left-sided CRCs have a distinct embryological origin, being developed from midgut and hindgut, respectively. Right-sided CRCs are more characteristic for females and often demonstrate peritoneal metastatic spread. Left-sided tumors are more frequent among men and tend to metastasize to the liver and lungs. The distinct biologic behavior of left- and right-sided tumors is at least in part attributed to the difference in the spectrum of cancer-associated mutations[2,3].
CRCs can be broadly divided into microsatellite stable tumors, which account for the vast majority of CRC cases, and carcinomas with so-called high-level microsatellite instability (MSI-H). A subset of CRCs is characterized by wide-spread methylation of cytosine residues, defined as CpG island methylator phenotype (CIMP). There are also classifications of CRCs based on tumor transcriptional profiles. The most characteristic molecular features of CRCs include dysregulation of MAPK and WNT signalling pathways, chromosomal imbalances at chromosomal loci 1p, 5q, 17p, 18p, 18q, 20p and 22q, mutations in KRAS, NRAS or BRAF oncogenes, activation of PI3 kinase, and TP53 gene inactivation[4-7]. Recent data suggest that the gut microbiome at least partially contributes to molecular pathogenesis of CRC[8,9]. While many deaths from CRC may be prevented by early tumor detection[3], some CRCs generate metastases in the very early phases of their development when the primary tumor is invisible by available diagnostic tools[10].
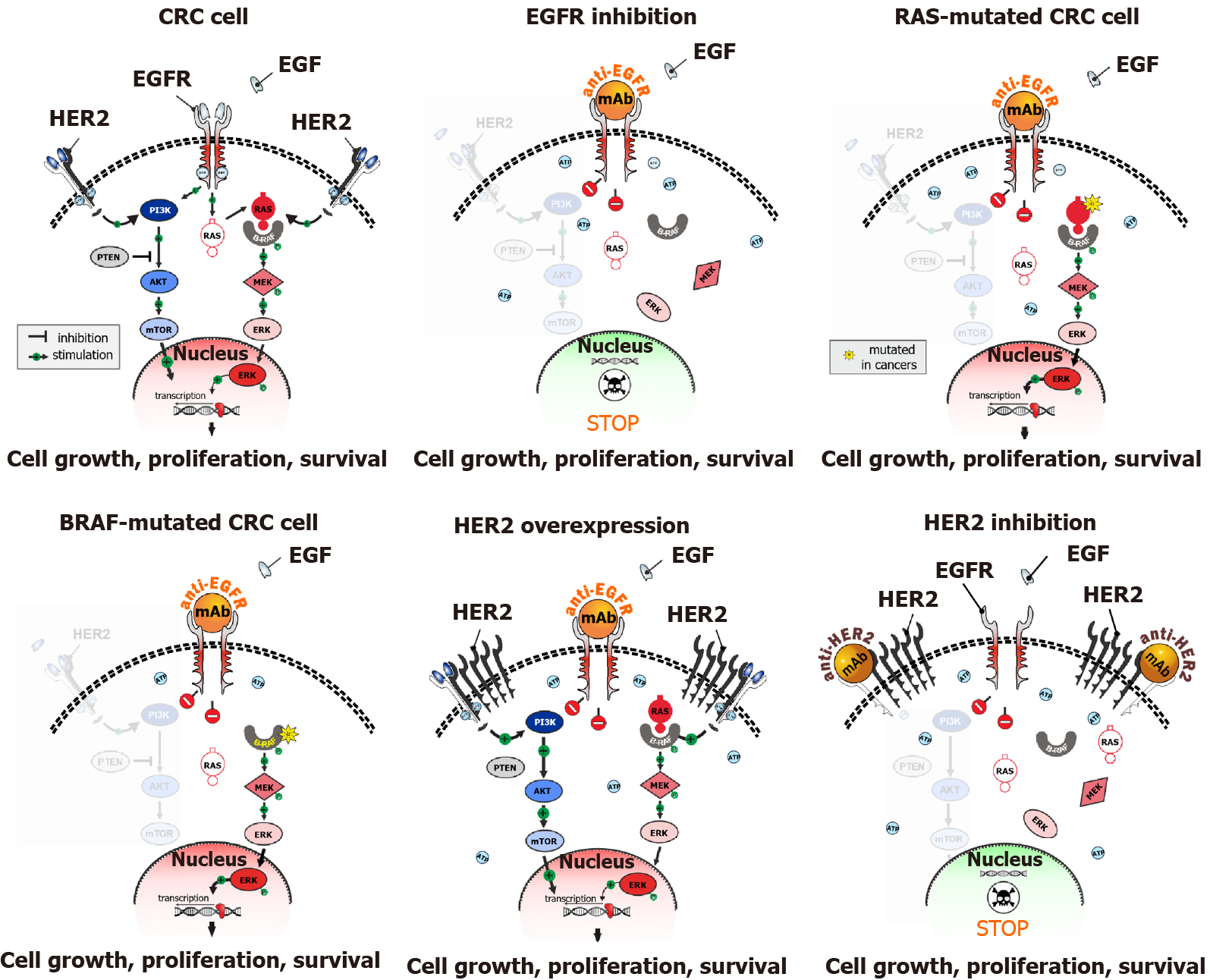
The majority of CRCs are characterized by the activation of MAPK signalling cascade. Some CRCs demonstrate activation of receptor tyrosine kinases (RTKs), mainly the epidermal growth factor receptor (EGFR). Up-regulation of the MAPK pathway can also be achieved by mutations in the downstream targets of RTKs, i.e., RAS or RAF oncogenes. Consequently, CRCs can be broadly divided into two categories: approximately a third of colorectal tumors appear to be EGFR-dependent, while the remaining cases are triggered either by the activation of collateral cascades (e.g., HER2-amplified cancers; approximately 2%-5% of CRCs) or genetic events affecting RAS or RAF genes (> 60% of CRC cases). EGFR-driven CRCs are likely to be responsive to anti-EGFR antibodies, panitumumab or cetuximab, while patients with EGFR-independent tumors usually do not benefit from EGFR-targeted therapy (Figure 1). Although this statement is an oversimplification, with many nuances requiring detailed clarification, it generally reflects the framework for the administration of anti-EGFR therapy[3,11-13].
Cetuximab and panitumumab may render a significant improvement in the disease outcomes when applied to RAS/RAF wild-type patients in combination with the standard cytotoxic regimens. This is particularly true for left-sided tumors, while right-sided CRCs appear to derive less benefit from EGFR down-regulation[14,15]. Panitumumab and cetuximab may also be utilized as single agents after the failure of chemotherapy. There were attempts to use monotherapy by anti-EGFR antibodies as an upfront treatment to delay the administration of cytotoxic drugs, however, these trials failed to generate promising clinical data[16]. Consequently, the combined use of anti-EGFR and chemotherapy in RAS/RAF wild-type patients at the very beginning of systemic intervention is a preferable option. This approach also leaves room for the re-challenge of cetuximab or panitumumab for CRC patients in the third-line treatment, i.e., after the failure of the first-line therapy and subsequent cessation of EGFR down-regulation during the second-line treatment[14,15].
Technical nuances concerning testing for mutations in KRAS and NRAS oncogenes deserve some discussion. Proper analysis of RAS gene status is critical for the fate of the patients: some data indicate that erroneous administration of anti-EGFR antibodies to CRC patients with RAS mutations, which are present in tumor tissue but were missed due to deficiency in the laboratory procedures, may lead to the acceleration of tumor growth[17]. Both KRAS and NRAS need to be screened at least for the presence of mutations in codons 12, 13, 59, 61, 117 and 146[18]. Given that each mentioned codon can be affected by a multitude of various mutations, there is almost an indefinite diversity of substitutions, with some rare variants not detectable by allele-specific PCR kits[19]. Some early clinical trials utilized conventional DNA sequencing for the analysis of exons 2, 3 and 4; however, this method cannot reliably detect mutations when the tumor specimen has a high admixture of normal cells. These drawbacks can be easily overcome by next generation sequencing (NGS), which applies multiple reads to each analyzed genomic fragment and, therefore, can detect mutated gene copies even in the presence of a high amount of normal DNA[20]. NGS remains expensive for the time being and this method is still not easily accessible in all hospitals around the world. NGS usually requires an accumulation of several samples for a single run that may affect turn-around-time for a given assay[21]. Anti-EGFR therapy provides remarkable improvement of the overall survival when given to left-sided KRAS/NRAS wild-type cancers as first-line therapy[14,15] therefore it is important to know the RAS status of CRC patients already at the time of initial treatment planning.
The “classical” model of CRC development, which was described by Fearon and Vogelstein, suggested that the emergence of RAS mutations provides an advantage to the evolving tumor clone, so the newly appeared RAS-mutated tumor subclones, in theory, should rapidly outperform their RAS-wild-type predecessors and entirely repopulate the tumor mass[22]. The molecular genetic portrait of the majority of CRCs appears to support this concept. However, there are notable exceptions. Some CRCs have RAS wild-type status when analyzed as a gross tumor mass, however, the use of ultrasensitive methods often allows to reveal a minor fraction of RAS-mutated cells in these carcinomas. These tumors appear to benefit from anti-EGFR therapy, as the majority of malignant cells forming the cancer lump remain vulnerable to EGFR inhibition[23,24]. Consequently, the mere detection of mutated RAS clones, as achieved by allele-specific PCR, may not be sufficient to guide the choice of CRC therapy; indeed, the quantitation of the proportion of mutated RAS clones is likely to be essential for proper therapeutic decisions[25,26]. Furthermore, when presumably RAS wild-type tumors undergo anti-EGFR therapy, the acquired resistance to EGFR down-regulation is often attributed to the emergence of RAS mutations. Interestingly, tumors exposed to anti-EGFR therapy demonstrate inhibition of DNA repair pathways, which results in the accelerated accumulation of mutations, and, consequently, increased probability of acquiring activating substitutions in RAS oncogenes[27]. The proportion of newly developed RAS-mutated clones often decreases after cessation of the anti-EGFR therapy, thus explaining the efficacy of cetuximab or panitumumab re-challenge[14,15]. In aggregate, these data suggest that the concept of the “evolutionary” advantage of RAS mutations is an oversimplification of the natural history of CRC development. Instead, some tumors demonstrate an example of an “ecosystem”, where RAS-mutated clones co-exist with RAS-wild-type malignant cells. The co-existence of cells with different status of driver mutations has been demonstrated for other tumor types; there are sound arguments suggesting that this intratumoral heterogeneity is not a result of the random expansion of distinct malignant clones, but a built-in biological property warranting tumor plasticity and adaptation to external hazards[28,29].
Approximately 5%-10% of CRCs contain activating V600E mutation in the BRAF oncogene. BRAF kinase is located within the MAPK signalling cascade; therefore, its mutation-driven up-regulation is very likely to render tumor independence from EGFR inhibition, similar to the role of RAS mutations. The combined analysis of available clinical trials generally supports this concept[30]. However, there are some clinical observations, which suggest that a subset of BRAF-mutated patients may still derive benefit from the addition of EGFR antibodies to some chemotherapy regimens[31]. The mechanistic basis for these observations is unclear. It has to be commented that clinical trials involving CRCs with BRAF mutations are prone to biases due to the rarity of this genetic event, and, consequently, a small number of recruited patients[12,30].
BRAF V600E-mutated tumors have aggressive behavior and poor prognosis. It is advised to start the therapy of these CRCs with intensive chemotherapy in combination with bevacizumab[12,30]. Second-line treatment of BRAF V600E-driven CRCs by single-agent BRAF inhibitors turned out to be non-successful, as CRCs adapt to this therapy by activation of EGFR-MAPK bypass pathways[32]. Several studies confirmed the efficacy of the combined use of EGFR and BRAF V600E inhibitors. The addition of a MEK-targeted drug to the above combination did not result in medically relevant improvement of treatment outcomes[33,34]. The doublet therapy composed of cetuximab and encorafenib has recently received FDA approval[30].
Some CRCs carry BRAF mutations in codons 594, 596, 597 or 601[35]. Mutations in positions 594 and 596 are not kinase-activating. Substitutions affecting codons 597 or 601 result in BRAF kinase activation[36]. There are instances of clinical responses to vemurafenib in tumors carrying substitutions in the codon 597[37,38]. Cancers with mutations in position 601 cannot be targeted by BRAF V600E-specific inhibitors[39].
The analysis of BRAF actionable mutations is not complicated. It can be done by various commercial allele-specific PCR kits or by DNA sequencing.
Some CRCs accumulate an excessive number of small mutations due to deficiency in DNA mismatch repair (MMR). These tumors contain multiple genetic alterations in the repetitive nucleotide sequences (for example, …AAAAAAAAAAAAAA…, or …CACACACACACACACACA…, etc.), which are called microsatellites. Consequently, MMR deficiency results in so-called high-level microsatellite instability (MSI-H).
Historically, microsatellite instability was simultaneously reported by several research groups, which applied different criteria for describing this phenotype. Dr. Manuel Perucho, whose group was apparently the first to submit for publication an article describing MSI phenotype, proposed to focus on length alterations in mononucleotide markers, which provided clear-cut discrimination between microsatellite “stable” and “unstable” tumors. Some other reports suggested to include dinucleotide repeats in PCR panels, and these efforts resulted in the development of the so-called consensus “Bethesda panel” consisting of two mono- and three dinucleotide markers (BAT-25, BAT-26, D2S123, D5S346, D17S250)[40-42]. The use of combinations of mononucleotide and dinucleotide markers led to 3-tier classification of CRCs, i.e., microsatellite-stable (MSS), MSI-H (i.e., CRCs containing alterations in the majority of markers analyzed), and MSI-L (MSI-low; i.e., the condition, when only a few analyzed markers are altered)[41]. Some contemporary reports still refer to the “Bethesda panel”[43], although subsequent research revealed, that only the MSI-H but not MSI-L condition has an established clinical significance and that the use of mononucleotide but not dinucleotide markers is strongly preferable for reliable discrimination between MSI-H and MSS tumors. Furthermore, while dinucleotide markers are highly polymorphic and thus require the analysis of normal DNA obtained from the same patient, some quasi-monomorphic mononucleotide microsatellites can be analyzed without comparison to the control DNA sample[44-46].
PCR analysis for MSI-H includes electrophoretic evaluation of the length of analyzed markers, i.e., an additional relatively sophisticated manipulation carried out after PCR amplification. While many other PCR mutation tests only require a PCR machine and commercially available reaction kits, MSI-H testing is not fully compatible with the establishment of a conventional morphological laboratory. Fortunately, this phenotype can also be detected by immunohistochemical (IHC) staining for key proteins involved in mismatch repair. MMR-D phenotype is manifested by the lack of expression of MMR proteins (MLH1, MSH2, MSH6, PMS2). MLH1 forms heterodimers with PMS2, while MSH2 heterodimerizes with MSH6. Inactivation of the MLH1 gene usually results in concomitant loss of expression of both MLH1 and PMS2; similarly, inactivation of the MSH2 is accompanied by the lack of staining for both MSH2 and MSH6 proteins. Isolated loss of expression of either MSH6 or PMS2 indicates alterations in the MSH6 or PMS2 genes, respectively[47]. The terms “MSI-H” and “MMR-D” are often used interchangeably, although it is more appropriate to utilize “MSI-H” for the analysis of the length of mononucleotide repeats, and “MMR-D” for IHC. It is frequently stated that PCR-driven and IHC analyses for MSI-H/MMR-D usually produce highly concordant results, although it is necessary to keep in mind that both procedures are relatively error-prone and, therefore, require rigorous quality assessment[48,49]. In addition to PCR or IHC, an MSI-H/MMR-D phenotype can be reliably detected by NGS[43].
The discovery of MSI-H was initially viewed mainly as an advance in the fundamental understanding of cancer pathogenesis and clinical application of MSI-H testing was limited to the identification of patients requiring genetic testing for the Lynch syndrome. It was subsequently revealed that MSI-H/MMR-D phenotype is associated with decreased risk of CRC relapse after surgery, the sensitivity of the tumor to the therapy by immune checkpoint inhibitors (ICIs), and, perhaps, a distinct pattern of tumor sensitivity to various cytotoxic drugs. Consequently, MSI-H/MMR-D has nowadays an utmost medical value, therefore, proper utilization of this test is a critical component of CRC management[43,47,50].
MSI-H/MMR-D status is characteristic of two clinically distinct categories of CRC patients. Wide-spread microsatellite instability is an obligatory feature of hereditary CRCs arising in subjects affected by Lynch syndrome. These patients tend to have an extensive family history of colorectal, endometrial and some other cancers and often present with the malignant disease at a relatively young age (< 50 years). MSI-H/MMR-D may also arise due to hypermethylation of the MLH1 gene promoter; these patients with sporadic (i.e., non-hereditary) CRCs dislaying MSI-H/MMR-D are usually of elderly age[43,47,50]. Hence, the frequency of MSI-H depends on two factors, i.e., the population-specific contribution of Lynch syndrome-related germ-line mutations in CRC incidence and the average age of CRC patients; the latter parameter is highly influenced by life expectancy observed in a given country. Studies performed in the countries of the Western world usually claim that up to 10%-15% of CRCs are characterized by MSI-H/MMR-D phenotype; these estimates may be substantially lower in some other patient series[51]. Mode of the recruitment of the CRC cases also plays an essential role, as MSI-H/MMR-D is observed at a higher frequency in patients with localized vs. metastatic cancers[43,47,50].
MSI-H/MMR-D phenotype is generally associated with a lower risk of relapse in surgically treated patients. Systematic analysis of multiple studies suggested that stage II MSI-H/MMR-D CRCs do not benefit from adjuvant chemotherapy, therefore, abstinence from postoperative systemic treatment in this category of patients is suggested by CRC clinical guidelines[43,47,50].
MMR-D results in an extensive accumulation of various mutations: tumor mutational burden (TMB) in these CRCs may exceed the number of mutations observed in MSS tumors by two orders of magnitude. High TMB is associated with an increased amount of tumor antigens thus rendering the responsiveness of MSI-H/MMR-D CRCs to immune therapy. There are several clinical trials, which demonstrated the remarkable efficacy of immune therapy for microsatellite unstable CRCs. In particular, pembrolizumab has been approved for the treatment of MSI-H/MMR-D CRC both in a first-line setting and for tumors with prior exposure to cytotoxic agents[52,53]. In addition, treatment of mismatch repair-deficient CRCs after the failure of chemotherapy may rely on nivolumab given alone or in combination with ipilimumab[54].
Almost half of BRAF V600E-mutated tumors observed in elderly patients have a microsatellite unstable phenotype. Similarly, up to half of sporadic late-onset CRCs, which render the MSI-H phenotype, carry the BRAF V600E mutation[55,56]. Consequently, if we consider CRC patients, who were diagnosed with this disease in the second half of their life, especially in their seventies or eighties, there will be a significant enrichment for tumors carrying both the above targets. These tumors are potentially sensitive both to BRAF V600E inhibition and to the immune checkpoint blockade. For the time being, the experience of treating these tumors is limited to applying either of these options[34,52]. There are several lines of evidence suggesting a cross-talk between EGFR-BRAF-MAPK pathways and immune signalling cascades[57]. Consequently, it is tempting to expect that the combination of EGFR/BRAF and immune checkpoint inhibition will result in a dramatic improvement of the disease outcomes. A trial combining cetuximab, encorafenib and nivolumab in CRCs carrying a combination of BRAF V600E and MSI-H is currently underway (NCT04017650).
Up to 5% of RAS/RAF mutation-negative CRCs are driven by the activation of HER2. It is essential to emphasize that the definition of HER2 up-regulation deserves particular attention. HER2 was initially studied as a breast cancer gene. In breast cancer, HER2 amplification is almost always accompanied by gene overexpression; consequently, it was suggested that FISH and IHC tests can be used interchangeably for the management of breast cancer patients[58]. The biology of CRC has some differences as compared to breast cancer. It appears that a subset of CRCs carries HER2 extra copies, which do not result in the increased production of the HER2 protein. This “non-productive” HER2 amplification is not mutually exclusive with other activating genetic events in the MAPK pathway and is not associated with clinical benefit from HER2 down-regulation[59].
“Genuine” HER2 activation, which is usually manifested by a combination of HER2 amplification and overexpression, is associated with the lack of tumor responsiveness to anti-EGFR therapy. There are several successful clinical studies utilizing rigorous HER2 inhibition achieved by the combination of two HER2-targeted agents. For example, Sartore-Bianchi et al[60] demonstrated high efficacy of the combination of trastuzumab and lapatinib in HER2-driven cancers. Similarly, Meric-Bernstam et al[59] showed the utility of pertuzumab plus trastuzumab combination. There are a number of ongoing clinical trials involving various HER2-targeted therapeutic regimens[61,62].
More than half of CRCs carry mutations in KRAS or NRAS oncogenes. These patients are excluded from the anti-EGFR treatment and therefore have limited treatment options. The development of specific inhibitors of mutated RAS proteins is highly complicated due to their small size, limited conformational change upon mutational activation and high affinity to their enzymatic substrate, i.e., GTP. For the time being, only the inhibitors of KRAS carrying glycine-to-cysteine substitution in codon 12 (KRAS G12C) succeeded to enter late phases of clinical development. A KRAS G12C mutation occurs in approximately 15% of non-small cell lung cancers, mainly in smokers[63], but is relatively rare in CRCs: it accounts for approximately 1 out of 12 KRAS mutations, and its frequency in unselected metastatic CRCs is about 4%[64]. A recent phase I trial involving KRAS G12C inhibitor sotorasib (AMG 510) included 42 CRC patients; 3 (7%) subjects experienced partial response and 28 (67%) cases demonstrated the disease stabilization[65]. Preclinical studies suggest that combined inactivation of KRAS G12C and EGFR may prevent CRC escape from a single-agent KRAS inhibition[66].
Mutation-driven activation of RAS results in the upregulation of MEK kinase. However, MEK kinase inhibitors do not show significant clinical efficacy in RAS-driven tumors. Some data indicate that tumor escape for MEK down-regulation may be attributed to autophagy. Preclinical experiments and some case reports involving KRAS-mutated pancreatic cancer patients suggest that the addition of an autophagy inhibitor, hydroxychloroquine (Plaquenil), may augment the efficacy of MEK antagonists[67,68]. There is a report describing the response of CRC carrying the KRAS G12D mutation to the combination of binimetinib, hydroxychloroquine, and bevacizumab[69]. This approach may have significant implications, given that almost a million people worldwide develop RAS-mutated CRC every year.
Microsatellite instability caused by the inactivation of mismatch repair genes is the most frequent cause of excessive tumor mutation burden, and, consequently, sensitivity to immune therapy. There are other varieties of CRC driven by the inactivation of DNA repair. For example, MUTYH-associated CRC is a rare example of a recessive hereditary cancer syndrome, which is characterized by a deficiency in base excision repair. CRCs with biallelic inactivation of the MUTYH gene accumulate a huge amount of G:C > T:A transversions. These tumors are responsive to immune checkpoint blockade[64]. Biallelic carriers of pathogenic MUTYH alleles often develop CRC via the acquisition of the KRAS G12C substitution. Given the existence of recurrent MUTYH variants, it is advisable to screen individuals with KRAS G12C mutated CRC for the presence of MUTYH germ-line mutations[64,70].
POLE-associated CRC is another example of rare CRC variety with ultra-high mutational load. POLE (polymerase epsilon) is a DNA polymerase with proofreading activity. Mutations in the exonuclease domain of the POLE gene result in an excessive number of errors occurring during DNA replication. There are cancers arising due to inheritance of a POLE pathogenic allele as well as instances of somatic inactivation of the POLE gene in sporadic CRCs and other tumor types. POLE deficiency is associated with tumor response to inhibitors of immune checkpoints[71,72].
Actionable rearrangements involving receptor tyrosine kinases are characteristic mainly for non-small cell lung carcinomas and some pediatric tumors[73,74]. Somewhat unexpectedly, instances of gene fusions resulting in the activation of involved kinases have been repeatedly demonstrated in microsatellite-unstable cancers arising due to somatic methylation of the MLH1 gene promoter[75-77]. There are also rare instances of druggable gene rearrangements in microsatellite stable cancers[75,76]. CRCs carrying activating gene fusions are responsive to the administration of appropriate tyrosine kinase inhibitors[78,79].
Approximately 3% of CRCs develop due to inherited mutations in MMR genes. This condition is called Lynch syndrome and remains the only well-established cause of hereditary non-polyposis CRC (HNPCC). Lynch syndrome is associated with germ-line pathogenic variants affecting MLH1, MSH2, MSH6 or PMS2 genes. The fifth HNPCC gene is EPCAM, which is involved in the disease pathogenesis via inactivation of the MSH2 gene. The penetrance of the above-mentioned genes tended to be overestimated in the past because studies of HNPCC were focused on the analysis of large cancer pedigrees. The invention of NGS led to significantly increased accessibility of the mutational testing in the HNPCC genes, so the appropriate DNA analysis was applied to unselected CRC cases and healthy controls. It is currently estimated that the individual CRC risk for carriers of germline variants in the MLH1 and MSH2 genes falls within the range of 40%-80%. Inheritance of mutations in the MSH6 and PMS2 genes is associated with a 10%-20% probability of developing CRC during the life-time, which is only 2-4 times higher than among unselected subjects. The contribution of Lynch syndrome in CRC incidence significantly varies between different populations[80-84].
Tumors arising in patients affected by Lynch syndrome always demonstrate MSI if they are causally linked to an inherited mutation in an MMR gene. MSI-H/MMR-D testing is now applied to all CRC patients; therefore, this assay serves as a screening test for HNPCC. The analysis of germ-line variants in MLH1, MSH2, MSH6, PMS2 and EPCAM genes requires consideration of both small mutations (protein-truncating variants or pathogenic missense mutations) and so-called large gene rearrangements (LGRs). IHC analysis of selective loss of MMR protein expression was used in the past to guide the identification of the involved gene. Nowadays, testing for Lynch syndrome genes is almost always done by NGS, except for communities with a pronounced founder effect[85,86].
There are multiple instances of familial aggregation of colorectal tumors not displaying a MSI-H/MMR-D phenotype and that are not related to pathogenic variants in MLH1, MSH2, MSH6, PMS2 or EPCAM. Despite intensive research efforts, RPS20 is currently the only gene showing reliable association with HNPCC. Germ-line mutations in the RPS20 are exceptionally rare and their spread is limited to particular ethnic groups[81,84].
There is a number of genes associated with colon polyposis and subsequent development of CRC. Dominant mutations in the APC gene are the most recognized cause of this condition. Other genetic causes of polyposis and CRC are relatively rare and include alterations in MUTYH, POLE, POLD1, NTHL1, MSH3, STK11, SMAD4, PTEN, GERM1 and some other genes[81-83].
Many cancer patients have detectable tumor-derived DNA in their plasma, probably due to decay of malignant cells and consequent DNA shedding in the bloodstream. The detection of circulating tumor DNA (ctDNA) has multiple potential applications for CRC patients. Some studies suggest that ultrasensitive detection of mutations in genes, which are frequently somatically altered in CRC, may facilitate CRC screening, especially when coupled with the use of other markers[87,88]. ctDNA testing may be used to control the success of surgical tumor resection. It is anticipated that patients who achieved ctDNA clearance after surgery have a good long-term prognosis and do not need adjuvant therapy, while subjects with residual ctDNA in the bloodstream are likely to relapse and require systemic drug exposure to reduce the risk of the disease recurrence[89-91]. Postsurgical monitoring for CRC-specific mutations may support early diagnosis of the tumor relapse[92]. Successful systemic therapy is accompanied by a rapid decline of the ctDNA concentration, while the maintenance of a stable ctDNA level indicates the lack of treatment efficacy[93]. A liquid biopsy allows the detection of secondary mutations, which are associated with acquired tumor resistance to therapy[94]. Clearance of these mutations from blood provides a rationale for re-challenge with the same targeted drug[95]. The various clinical applications of ctDNA analysis have been evaluated in many ongoing trials[96].
CRC was the first common cancer type, whose molecular testing became a mandatory component of the therapeutic decisions: indeed, KRAS status assessment was incorporated in the drug labels for panitumumab and cetuximab already in the year 2009, i.e., sometime before the integration of EGFR mutation analysis into the non-small cell lung cancer management. Nowadays, all CRCs undergo comprehensive genetic analysis for somatic mutations and selected patients are subjected to germ-line DNA testing. Rapid spread of NGS technology is likely to affect attitudes towards CRC screening, diagnosis, treatment and monitoring in the near future.
We are cordially thankful to Dr. Barbara Vona (University of Tuebingen) for critical reading of this manuscript.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: European Society for Medical Oncology (ESMO), No. 6102.
Specialty type: Oncology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Qin J S-Editor: Ma YJ L-Editor: A P-Editor: Guo X
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50818] [Article Influence: 8469.7] [Reference Citation Analysis (44)] |
| 2. | Baran B, Mert Ozupek N, Yerli Tetik N, Acar E, Bekcioglu O, Baskin Y. Difference Between Left-Sided and Right-Sided Colorectal Cancer: A Focused Review of Literature. Gastroenterology Res. 2018;11:264-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 251] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 3. | Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467-1480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1570] [Cited by in F6Publishing: 2068] [Article Influence: 413.6] [Reference Citation Analysis (2)] |
| 4. | Farooqi AA, de la Roche M, Djamgoz MBA, Siddik ZH. Overview of the oncogenic signaling pathways in colorectal cancer: Mechanistic insights. Semin Cancer Biol. 2019;58:65-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 5. | Koncina E, Haan S, Rauh S, Letellier E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 6. | Raskov H, Søby JH, Troelsen J, Bojesen RD, Gögenur I. Driver Gene Mutations and Epigenetics in Colorectal Cancer. Ann Surg. 2020;271:75-85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 7. | Testa U, Castelli G, Pelosi E. Genetic Alterations of Metastatic Colorectal Cancer. Biomedicines. 2020;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 8. | Dziubańska-Kusibab PJ, Berger H, Battistini F, Bouwman BAM, Iftekhar A, Katainen R, Cajuso T, Crosetto N, Orozco M, Aaltonen LA, Meyer TF. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat Med. 2020;26:1063-1069. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 9. | Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585:509-517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 127] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 10. | Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, Suarez CJ, Berghoff AS, Cremolini C, Falcone A, Loupakis F, Birner P, Preusser M, Lenz HJ, Curtis C. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet. 2019;51:1113-1122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 285] [Cited by in F6Publishing: 261] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 11. | Patel JN, Fong MK, Jagosky M. Colorectal Cancer Biomarkers in the Era of Personalized Medicine. J Pers Med. 2019;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Taieb J, Jung A, Sartore-Bianchi A, Peeters M, Seligmann J, Zaanan A, Burdon P, Montagut C, Laurent-Puig P. The Evolving Biomarker Landscape for Treatment Selection in Metastatic Colorectal Cancer. Drugs. 2019;79:1375-1394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 13. | Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17:11-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 164] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 14. | Martinelli E, Ciardiello D, Martini G, Troiani T, Cardone C, Vitiello PP, Normanno N, Rachiglio AM, Maiello E, Latiano T, De Vita F, Ciardiello F. Implementing anti-epidermal growth factor receptor (EGFR) therapy in metastatic colorectal cancer: challenges and future perspectives. Ann Oncol. 2020;31:30-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 103] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 15. | Martini G, Ciardiello D, Vitiello PP, Napolitano S, Cardone C, Cuomo A, Troiani T, Ciardiello F, Martinelli E. Resistance to anti-epidermal growth factor receptor in metastatic colorectal cancer: What does still need to be addressed? Cancer Treat Rev. 2020;86:102023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 16. | Moiseyenko VM, Moiseyenko FV, Yanus GA, Kuligina ES, Sokolenko AP, Bizin IV, Kudriavtsev AA, Aleksakhina SN, Volkov NM, Chubenko VA, Kozyreva KS, Kramchaninov MM, Zhuravlev AS, Shelekhova KV, Pashkov DV, Ivantsov AO, Venina AR, Sokolova TN, Preobrazhenskaya EV, Mitiushkina NV, Togo AV, Iyevleva AG, Imyanitov EN. First-Line Cetuximab Monotherapy in KRAS/NRAS/BRAF Mutation-Negative Colorectal Cancer Patients. Clin Drug Investig. 2018;38:553-562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M, Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu R, Patterson SD. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-1034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1610] [Cited by in F6Publishing: 1623] [Article Influence: 147.5] [Reference Citation Analysis (0)] |
| 18. | Sepulveda AR, Hamilton SR, Allegra CJ, Grody W, Cushman-Vokoun AM, Funkhouser WK, Kopetz SE, Lieu C, Lindor NM, Minsky BD, Monzon FA, Sargent DJ, Singh VM, Willis J, Clark J, Colasacco C, Rumble RB, Temple-Smolkin R, Ventura CB, Nowak JA. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J Clin Oncol. 2017;35:1453-1486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 208] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 19. | Lakatos G, Köhne CH, Bodoky G. Current therapy of advanced colorectal cancer according to RAS/RAF mutational status. Cancer Metastasis Rev. 2020;39:1143-1157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 20. | Udar N, Lofton-Day C, Dong J, Vavrek D, Jung AS, Meier K, Iyer A, Slaughter R, Gutekunst K, Bach BA, Peeters M, Douillard JY. Clinical validation of the next-generation sequencing-based Extended RAS Panel assay using metastatic colorectal cancer patient samples from the phase 3 PRIME study. J Cancer Res Clin Oncol. 2018;144:2001-2010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Del Vecchio F, Mastroiaco V, Di Marco A, Compagnoni C, Capece D, Zazzeroni F, Capalbo C, Alesse E, Tessitore A. Next-generation sequencing: recent applications to the analysis of colorectal cancer. J Transl Med. 2017;15:246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8087] [Cited by in F6Publishing: 7708] [Article Influence: 226.7] [Reference Citation Analysis (1)] |
| 23. | Van Cutsem E, Lenz HJ, Köhne CH, Heinemann V, Tejpar S, Melezínek I, Beier F, Stroh C, Rougier P, van Krieken JH, Ciardiello F. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J Clin Oncol. 2015;33:692-700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 564] [Cited by in F6Publishing: 591] [Article Influence: 65.7] [Reference Citation Analysis (0)] |
| 24. | Normanno N, Rachiglio AM, Lambiase M, Martinelli E, Fenizia F, Esposito C, Roma C, Troiani T, Rizzi D, Tatangelo F, Botti G, Maiello E, Colucci G, Ciardiello F; CAPRI-GOIM investigators. Heterogeneity of KRAS, NRAS, BRAF and PIK3CA mutations in metastatic colorectal cancer and potential effects on therapy in the CAPRI GOIM trial. Ann Oncol. 2015;26:1710-1714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 25. | Santos C, Azuara D, Viéitez JM, Páez D, Falcó E, Élez E, López-López C, Valladares M, Robles-Díaz L, García-Alfonso P, Bugés C, Durán G, Salud A, Navarro V, Capellá G, Aranda E, Salazar R. Phase II study of high-sensitivity genotyping of KRAS, NRAS, BRAF and PIK3CA to ultra-select metastatic colorectal cancer patients for panitumumab plus FOLFIRI: the ULTRA trial. Ann Oncol. 2019;30:796-803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 26. | Vidal J, Bellosillo B, Santos Vivas C, García-Alfonso P, Carrato A, Cano MT, García-Carbonero R, Élez E, Losa F, Massutí B, Valladares-Ayerbes M, Viéitez JM, Manzano JL, Azuara D, Gallego J, Pairet S, Capellá G, Salazar R, Tabernero J, Aranda E, Montagut C. Ultra-selection of metastatic colorectal cancer patients using next-generation sequencing to improve clinical efficacy of anti-EGFR therapy. Ann Oncol. 2019;30:439-446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 27. | Russo M, Crisafulli G, Sogari A, Reilly NM, Arena S, Lamba S, Bartolini A, Amodio V, Magrì A, Novara L, Sarotto I, Nagel ZD, Piett CG, Amatu A, Sartore-Bianchi A, Siena S, Bertotti A, Trusolino L, Corigliano M, Gherardi M, Lagomarsino MC, Di Nicolantonio F, Bardelli A. Adaptive mutability of colorectal cancers in response to targeted therapies. Science. 2019;366:1473-1480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 238] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 28. | Aleksakhina SN, Kashyap A, Imyanitov EN. Mechanisms of acquired tumor drug resistance. Biochim Biophys Acta Rev Cancer. 2019;1872:188310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 29. | Sokolenko AP, Bizin IV, Preobrazhenskaya EV, Gorodnova TV, Ivantsov AO, Iyevleva AG, Savonevich EL, Kotiv KB, Kuligina ES, Imyanitov EN. Molecular profiles of BRCA1-associated ovarian cancer treated by platinum-based therapy: Analysis of primary, residual and relapsed tumors. Int J Cancer. 2020;146:1879-1888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 30. | Johnson B, Kopetz S. Applying Precision to the Management of BRAF-Mutant Metastatic Colorectal Cancer. Target Oncol. 2020;15:567-577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 31. | Modest DP, Martens UM, Riera-Knorrenschild J, Greeve J, Florschütz A, Wessendorf S, Ettrich T, Kanzler S, Nörenberg D, Ricke J, Seidensticker M, Held S, Buechner-Steudel P, Atzpodien J, Heinemann V, Seufferlein T, Tannapfel A, Reinacher-Schick AC, Geissler M. FOLFOXIRI Plus Panitumumab As First-Line Treatment of RAS Wild-Type Metastatic Colorectal Cancer: The Randomized, Open-Label, Phase II VOLFI Study (AIO KRK0109). J Clin Oncol. 2019;37:3401-3411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 32. | Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1390] [Cited by in F6Publishing: 1456] [Article Influence: 121.3] [Reference Citation Analysis (0)] |
| 33. | Corcoran RB, André T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, Hollebecque A, McRee AJ, Siena S, Middleton G, Muro K, Gordon MS, Tabernero J, Yaeger R, O'Dwyer PJ, Humblet Y, De Vos F, Jung AS, Brase JC, Jaeger S, Bettinger S, Mookerjee B, Rangwala F, Van Cutsem E. Combined BRAF, EGFR, and MEK Inhibition in Patients with BRAFV600E-Mutant Colorectal Cancer. Cancer Discov. 2018;8:428-443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 403] [Cited by in F6Publishing: 376] [Article Influence: 62.7] [Reference Citation Analysis (0)] |
| 34. | Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, Wasan H, Ciardiello F, Loupakis F, Hong YS, Steeghs N, Guren TK, Arkenau HT, Garcia-Alfonso P, Pfeiffer P, Orlov S, Lonardi S, Elez E, Kim TW, Schellens JHM, Guo C, Krishnan A, Dekervel J, Morris V, Calvo Ferrandiz A, Tarpgaard LS, Braun M, Gollerkeri A, Keir C, Maharry K, Pickard M, Christy-Bittel J, Anderson L, Sandor V, Tabernero J. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N Engl J Med. 2019;381:1632-1643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 694] [Cited by in F6Publishing: 768] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 35. | Jones JC, Renfro LA, Al-Shamsi HO, Schrock AB, Rankin A, Zhang BY, Kasi PM, Voss JS, Leal AD, Sun J, Ross J, Ali SM, Hubbard JM, Kipp BR, McWilliams RR, Kopetz S, Wolff RA, Grothey A. Non-V600 BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J Clin Oncol. 2017;35:2624-2630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 226] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 36. | Schirripa M, Biason P, Lonardi S, Pella N, Pino MS, Urbano F, Antoniotti C, Cremolini C, Corallo S, Pietrantonio F, Gelsomino F, Cascinu S, Orlandi A, Munari G, Malapelle U, Saggio S, Fontanini G, Rugge M, Mescoli C, Lazzi S, Reggiani Bonetti L, Lanza G, Dei Tos AP, De Maglio G, Martini M, Bergamo F, Zagonel V, Loupakis F, Fassan M. Class 1, 2, and 3 BRAF-Mutated Metastatic Colorectal Cancer: A Detailed Clinical, Pathologic, and Molecular Characterization. Clin Cancer Res. 2019;25:3954-3961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 54] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 37. | Bahadoran P, Allegra M, Le Duff F, Long-Mira E, Hofman P, Giacchero D, Passeron T, Lacour JP, Ballotti R. Major clinical response to a BRAF inhibitor in a patient with a BRAF L597R-mutated melanoma. J Clin Oncol. 2013;31:e324-e326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | Hallmeyer S, Gonzalez R, Lawson DH, Cranmer LD, Linette GP, Puzanov I, Taback B, Cowey CL, Ribas A, Daniels GA, Moore T, Gibney GT, Tawbi H, Whitman E, Lee G, Mun Y, Liu S, Hamid O. Vemurafenib treatment for patients with locally advanced, unresectable stage IIIC or metastatic melanoma and activating exon 15 BRAF mutations other than V600E. Melanoma Res. 2017;27:585-590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 39. | Moiseyenko FV, Egorenkov VV, Kramchaninov MM, Artemieva EV, Aleksakhina SN, Holmatov MM, Moiseyenko VM, Imyanitov EN. Lack of Response to Vemurafenib in Melanoma Carrying BRAF K601E Mutation. Case Rep Oncol. 2019;12:339-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 40. | Maddox J. Competition and the death of science. Nature. 1993;363:667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 41. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [PubMed] [Cited in This Article: ] |
| 42. | Perucho M. Correspondence re: C.R. Boland et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res., 58: 5248-5257, 1998. Cancer Res. 1999;59:249-256. [PubMed] [Cited in This Article: ] |
| 43. | Sun BL. Current Microsatellite Instability Testing in Management of Colorectal Cancer. Clin Colorectal Cancer. 2021;20:e12-e20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804-1811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 440] [Cited by in F6Publishing: 458] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 45. | Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2154] [Cited by in F6Publishing: 2108] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 46. | Buhard O, Lagrange A, Guilloux A, Colas C, Chouchène M, Wanherdrick K, Coulet F, Guillerm E, Dorard C, Marisa L, Bokhari A, Greene M, El-Murr N, Bodo S, Muleris M, Sourouille I, Svrcek M, Cervera P, Blanché H, Lefevre JH, Parc Y, Lepage C, Chapusot C, Bouvier AM, Gaub MP, Selves J, Garrett K, Iacopetta B, Soong R, Hamelin R, Garrido C, Lascols O, André T, Fléjou JF, Collura A, Duval A. HSP110 T17 simplifies and improves the microsatellite instability testing in patients with colorectal cancer. J Med Genet. 2016;53:377-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 47. | Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153-162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 612] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 48. | Chen W, Frankel WL. A practical guide to biomarkers for the evaluation of colorectal cancer. Mod Pathol. 2019;32:1-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 49. | Cohen R, Hain E, Buhard O, Guilloux A, Bardier A, Kaci R, Bertheau P, Renaud F, Bibeau F, Fléjou JF, André T, Svrcek M, Duval A. Association of Primary Resistance to Immune Checkpoint Inhibitors in Metastatic Colorectal Cancer With Misdiagnosis of Microsatellite Instability or Mismatch Repair Deficiency Status. JAMA Oncol. 2019;5:551-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 149] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 50. | Battaglin F, Naseem M, Lenz HJ, Salem ME. Microsatellite instability in colorectal cancer: overview of its clinical significance and novel perspectives. Clin Adv Hematol Oncol. 2018;16:735-745. [PubMed] [Cited in This Article: ] |
| 51. | Yanus GA, Belyaeva AV, Ivantsov AO, Kuligina ESh, Suspitsin EN, Mitiushkina NV, Aleksakhina SN, Iyevleva AG, Zaitseva OA, Yatsuk OS, Gorodnova TV, Strelkova TN, Efremova SA, Lepenchuk AY, Ochir-Garyaev AN, Paneyah MB, Matsko DE, Togo AV, Imyanitov EN. Pattern of clinically relevant mutations in consecutive series of Russian colorectal cancer patients. Med Oncol. 2013;30:686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 52. | André T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, de la Fouchardiere C, Rivera F, Elez E, Bendell J, Le DT, Yoshino T, Van Cutsem E, Yang P, Farooqui MZH, Marinello P, Diaz LA Jr; KEYNOTE-177 Investigators. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N Engl J Med. 2020;383:2207-2218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 962] [Cited by in F6Publishing: 1279] [Article Influence: 319.8] [Reference Citation Analysis (0)] |
| 53. | Le DT, Kim TW, Van Cutsem E, Geva R, Jäger D, Hara H, Burge M, O'Neil B, Kavan P, Yoshino T, Guimbaud R, Taniguchi H, Elez E, Al-Batran SE, Boland PM, Crocenzi T, Atreya CE, Cui Y, Dai T, Marinello P, Diaz LA Jr, André T. Phase II Open-Label Study of Pembrolizumab in Treatment-Refractory, Microsatellite Instability-High/Mismatch Repair-Deficient Metastatic Colorectal Cancer: KEYNOTE-164. J Clin Oncol. 2020;38:11-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 346] [Cited by in F6Publishing: 532] [Article Influence: 106.4] [Reference Citation Analysis (0)] |
| 54. | Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, Sawyer MB, Hendlisz A, Neyns B, Svrcek M, Moss RA, Ledeine JM, Cao ZA, Kamble S, Kopetz S, André T. Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol. 2018;36:773-779. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1013] [Cited by in F6Publishing: 1282] [Article Influence: 213.7] [Reference Citation Analysis (0)] |
| 55. | Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA, Fuchs CS, Ogino S. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105:1151-1156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 322] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 56. | Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, Kaplan R, Quirke P, Seymour MT, Richman SD, Meijer GA, Ylstra B, Heideman DA, de Haan AF, Punt CJ, Koopman M. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20:5322-5330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 409] [Cited by in F6Publishing: 498] [Article Influence: 49.8] [Reference Citation Analysis (0)] |
| 57. | Giordano G, Remo A, Porras A, Pancione M. Immune Resistance and EGFR Antagonists in Colorectal Cancer. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 58. | Sauter G, Lee J, Bartlett JM, Slamon DJ, Press MF. Guidelines for human epidermal growth factor receptor 2 testing: biologic and methodologic considerations. J Clin Oncol. 2009;27:1323-1333. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 380] [Cited by in F6Publishing: 374] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 59. | Meric-Bernstam F, Hurwitz H, Raghav KPS, McWilliams RR, Fakih M, VanderWalde A, Swanton C, Kurzrock R, Burris H, Sweeney C, Bose R, Spigel DR, Beattie MS, Blotner S, Stone A, Schulze K, Cuchelkar V, Hainsworth J. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol. 2019;20:518-530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 311] [Cited by in F6Publishing: 314] [Article Influence: 62.8] [Reference Citation Analysis (0)] |
| 60. | Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, Zagonel V, Leone F, Depetris I, Martinelli E, Troiani T, Ciardiello F, Racca P, Bertotti A, Siravegna G, Torri V, Amatu A, Ghezzi S, Marrapese G, Palmeri L, Valtorta E, Cassingena A, Lauricella C, Vanzulli A, Regge D, Veronese S, Comoglio PM, Bardelli A, Marsoni S, Siena S. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738-746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 549] [Cited by in F6Publishing: 644] [Article Influence: 80.5] [Reference Citation Analysis (0)] |
| 61. | De Cuyper A, Van Den Eynde M, Machiels JP. HER2 as a Predictive Biomarker and Treatment Target in Colorectal Cancer. Clin Colorectal Cancer. 2020;19:65-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 62. | Nowak JA. HER2 in Colorectal Carcinoma: Are We There yet? Surg Pathol Clin. 2020;13:485-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 63. | Mitiushkina NV, Kholmatov MM, Venina AR, Tiurin VI, Yanus GA, Sokolova TN, Yatsuk OS, Zaitseva OA, Ivantsov AO, Kuligina ES, Togo AV, Imyanitov EN. PCR-based detection of EGFR, ALK, KRAS and BRAF mutations in Russian patients with lung adenocarcinoma: a single-center experience. Neoplasma. 2018;65:972-979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Volkov NM, Yanus GA, Ivantsov AO, Moiseenko FV, Matorina OG, Bizin IV, Moiseyenko VM, Imyanitov EN. Efficacy of immune checkpoint blockade in MUTYH-associated hereditary colorectal cancer. Invest New Drugs. 2020;38:894-898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, Falchook GS, Price TJ, Sacher A, Denlinger CS, Bang YJ, Dy GK, Krauss JC, Kuboki Y, Kuo JC, Coveler AL, Park K, Kim TW, Barlesi F, Munster PN, Ramalingam SS, Burns TF, Meric-Bernstam F, Henary H, Ngang J, Ngarmchamnanrith G, Kim J, Houk BE, Canon J, Lipford JR, Friberg G, Lito P, Govindan R, Li BT. KRASG12C Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med. 2020;383:1207-1217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 734] [Cited by in F6Publishing: 863] [Article Influence: 215.8] [Reference Citation Analysis (0)] |
| 66. | Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, Arena S, Montone M, Mussolin B, Bian Y, Whaley A, Pinnelli M, Murciano-Goroff YR, Vakiani E, Valeri N, Liao WL, Bhalkikar A, Thyparambil S, Zhao HY, de Stanchina E, Marsoni S, Siena S, Bertotti A, Trusolino L, Li BT, Rosen N, Di Nicolantonio F, Bardelli A, Misale S. EGFR Blockade Reverts Resistance to KRASG12C Inhibition in Colorectal Cancer. Cancer Discov. 2020;10:1129-1139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 213] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 67. | Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, Burrell LD, Lum DH, Whisenant JR, Gilcrease GW 3rd, Cavalieri CC, Rehbein KM, Cutler SL, Affolter KE, Welm AL, Welm BE, Scaife CL, Snyder EL, McMahon M. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med. 2019;25:620-627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 414] [Cited by in F6Publishing: 410] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 68. | Xavier CB, Marchetti KR, Castria TB, Jardim DLF, Fernandes GS. Trametinib and Hydroxychloroquine (HCQ) Combination Treatment in KRAS-Mutated Advanced Pancreatic Adenocarcinoma: Detailed Description of Two Cases. J Gastrointest Cancer. 2021;52:374-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 69. | Orlov SV, Urtenova MA, Sviridenko MA, Nesterov DV, Sokolova TN, Imyanitov EN. Rapid Improvement of the Performance Status and Reduction of the Tumor Size in KRAS-Mutated Colorectal Cancer Patient Receiving Binimetinib, Hydroxychloroquine, and Bevacizumab. Case Rep Oncol. 2020;13:985-989. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 70. | van Puijenbroek M, Nielsen M, Tops CM, Halfwerk H, Vasen HF, Weiss MM, van Wezel T, Hes FJ, Morreau H. Identification of patients with (atypical) MUTYH-associated polyposis by KRAS2 c.34G > T prescreening followed by MUTYH hotspot analysis in formalin-fixed paraffin-embedded tissue. Clin Cancer Res. 2008;14:139-142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 71. | Gong J, Wang C, Lee PP, Chu P, Fakih M. Response to PD-1 Blockade in Microsatellite Stable Metastatic Colorectal Cancer Harboring a POLE Mutation. J Natl Compr Canc Netw. 2017;15:142-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 152] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 72. | Wang C, Gong J, Tu TY, Lee PP, Fakih M. Immune profiling of microsatellite instability-high and polymerase ε (POLE)-mutated metastatic colorectal tumors identifies predictors of response to anti-PD-1 therapy. J Gastrointest Oncol. 2018;9:404-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 73. | Imyanitov EN, Iyevleva AG, Levchenko EV. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit Rev Oncol Hematol. 2021;157:103194. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 215] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 74. | Preobrazhenskaya EV, Iyevleva AG, Suleymanova AM, Tiurin VI, Mitiushkina NV, Bizin IV, Ivanstov AO, Gorustovich OA, Shelekhova KV, Kachanov DY, Varfolomeeva SR, Roschin VY, Kazakova AN, Litvinov DV, Shamanskaya TV, Savelov NA, Suspitsin EN, Imyanitov EN. Gene rearrangements in consecutive series of pediatric inflammatory myofibroblastic tumors. Pediatr Blood Cancer. 2020;67:e28220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 75. | Pietrantonio F, Di Nicolantonio F, Schrock AB, Lee J, Tejpar S, Sartore-Bianchi A, Hechtman JF, Christiansen J, Novara L, Tebbutt N, Fucà G, Antoniotti C, Kim ST, Murphy D, Berenato R, Morano F, Sun J, Min B, Stephens PJ, Chen M, Lazzari L, Miller VA, Shoemaker R, Amatu A, Milione M, Ross JS, Siena S, Bardelli A, Ali SM, Falcone A, de Braud F, Cremolini C. ALK, ROS1, and NTRK Rearrangements in Metastatic Colorectal Cancer. J Natl Cancer Inst. 2017;109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 157] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 76. | Cocco E, Benhamida J, Middha S, Zehir A, Mullaney K, Shia J, Yaeger R, Zhang L, Wong D, Villafania L, Nafa K, Scaltriti M, Drilon A, Saltz L, Schram AM, Stadler ZK, Hyman DM, Benayed R, Ladanyi M, Hechtman JF. Colorectal Carcinomas Containing Hypermethylated MLH1 Promoter and Wild-Type BRAF/KRAS Are Enriched for Targetable Kinase Fusions. Cancer Res. 2019;79:1047-1053. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 77. | Sato K, Kawazu M, Yamamoto Y, Ueno T, Kojima S, Nagae G, Abe H, Soda M, Oga T, Kohsaka S, Sai E, Yamashita Y, Iinuma H, Fukayama M, Aburatani H, Watanabe T, Mano H. Fusion Kinases Identified by Genomic Analyses of Sporadic Microsatellite Instability-High Colorectal Cancers. Clin Cancer Res. 2019;25:378-389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 78. | Pagani F, Randon G, Guarini V, Raimondi A, Prisciandaro M, Lobefaro R, Di Bartolomeo M, Sozzi G, de Braud F, Gasparini P, Pietrantonio F. The Landscape of Actionable Gene Fusions in Colorectal Cancer. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 79. | Singh H, Li YY, Spurr LF, Shinagare AB, Abhyankar R, Reilly E, Brais LK, Nag A, Ducar MD, Thorner AR, Shapiro GI, Keller RB, Siletti C, Clark JW, Farago AF, Lin JJ, Demetri GD, Gujrathi R, Kulke MH, MacConaill LE, Ligon AH, Sicinska E, Meyerson ML, Meyerhardt JA, Cherniack AD, Wolpin BM, Ng K, Giannakis M, Hornick JL, Cleary JM. Molecular Characterization and Therapeutic Targeting of Colorectal Cancers Harboring Receptor Tyrosine Kinase Fusions. Clin Cancer Res. 2021;27:1695-1705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 80. | Snyder C, Hampel H. Hereditary Colorectal Cancer Syndromes. Semin Oncol Nurs. 2019;35:58-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 81. | Valle L, de Voer RM, Goldberg Y, Sjursen W, Försti A, Ruiz-Ponte C, Caldés T, Garré P, Olsen MF, Nordling M, Castellvi-Bel S, Hemminki K. Update on genetic predisposition to colorectal cancer and polyposis. Mol Aspects Med. 2019;69:10-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 56] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 82. | Valle L, Vilar E, Tavtigian SV, Stoffel EM. Genetic predisposition to colorectal cancer: syndromes, genes, classification of genetic variants and implications for precision medicine. J Pathol. 2019;247:574-588. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 100] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 83. | Clark SK. Management of genetically determined colorectal cancer. Surgeon. 2019;17:165-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 84. | Terradas M, Capellá G, Valle L. Dominantly Inherited Hereditary Nonpolyposis Colorectal Cancer Not Caused by MMR Genes. J Clin Med. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 85. | Soares BL, Brant AC, Gomes R, Pastor T, Schneider NB, Ribeiro-Dos-Santos Â, de Assumpção PP, Achatz MIW, Ashton-Prolla P, Moreira MAM. Screening for germline mutations in mismatch repair genes in patients with Lynch syndrome by next generation sequencing. Fam Cancer. 2018;17:387-394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 86. | Yanus GA, Akhapkina TA, Iyevleva AG, Kornilov AV, Suspitsin EN, Kuligina ES, Ivantsov AO, Aleksakhina SN, Sokolova TN, Sokolenko AP, Togo AV, Imyanitov EN. The spectrum of Lynch syndrome-associated germ-line mutations in Russia. Eur J Med Genet. 2020;63:103753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 87. | Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, Hruban RH, Wolfgang CL, Goggins MG, Dal Molin M, Wang TL, Roden R, Klein AP, Ptak J, Dobbyn L, Schaefer J, Silliman N, Popoli M, Vogelstein JT, Browne JD, Schoen RE, Brand RE, Tie J, Gibbs P, Wong HL, Mansfield AS, Jen J, Hanash SM, Falconi M, Allen PJ, Zhou S, Bettegowda C, Diaz LA Jr, Tomasetti C, Kinzler KW, Vogelstein B, Lennon AM, Papadopoulos N. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926-930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1752] [Cited by in F6Publishing: 1544] [Article Influence: 257.3] [Reference Citation Analysis (0)] |
| 88. | Lennon AM, Buchanan AH, Kinde I, Warren A, Honushefsky A, Cohain AT, Ledbetter DH, Sanfilippo F, Sheridan K, Rosica D, Adonizio CS, Hwang HJ, Lahouel K, Cohen JD, Douville C, Patel AA, Hagmann LN, Rolston DD, Malani N, Zhou S, Bettegowda C, Diehl DL, Urban B, Still CD, Kann L, Woods JI, Salvati ZM, Vadakara J, Leeming R, Bhattacharya P, Walter C, Parker A, Lengauer C, Klein A, Tomasetti C, Fishman EK, Hruban RH, Kinzler KW, Vogelstein B, Papadopoulos N. Feasibility of blood testing combined with PET-CT to screen for cancer and guide intervention. Science. 2020;369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 286] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 89. | Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, Knudsen M, Nordentoft I, Wu HT, Tin AS, Heilskov Rasmussen M, Vang S, Shchegrova S, Frydendahl Boll Johansen A, Srinivasan R, Assaf Z, Balcioglu M, Olson A, Dashner S, Hafez D, Navarro S, Goel S, Rabinowitz M, Billings P, Sigurjonsson S, Dyrskjøt L, Swenerton R, Aleshin A, Laurberg S, Husted Madsen A, Kannerup AS, Stribolt K, Palmelund Krag S, Iversen LH, Gotschalck Sunesen K, Lin CJ, Zimmermann BG, Lindbjerg Andersen C. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019;5:1124-1131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 466] [Article Influence: 93.2] [Reference Citation Analysis (0)] |
| 90. | Tarazona N, Gimeno-Valiente F, Gambardella V, Zuñiga S, Rentero-Garrido P, Huerta M, Roselló S, Martinez-Ciarpaglini C, Carbonell-Asins JA, Carrasco F, Ferrer-Martínez A, Bruixola G, Fleitas T, Martín J, Tébar-Martínez R, Moro D, Castillo J, Espí A, Roda D, Cervantes A. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol. 2019;30:1804-1812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 91. | Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, Wong R, Kosmider S, Ananda S, McKendrick J, Lee B, Cho JH, Faragher I, Jones IT, Ptak J, Schaeffer MJ, Silliman N, Dobbyn L, Li L, Tomasetti C, Papadopoulos N, Kinzler KW, Vogelstein B, Gibbs P. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019;5:1710-1717. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 329] [Article Influence: 109.7] [Reference Citation Analysis (0)] |
| 92. | Wang Y, Li L, Cohen JD, Kinde I, Ptak J, Popoli M, Schaefer J, Silliman N, Dobbyn L, Tie J, Gibbs P, Tomasetti C, Kinzler KW, Papadopoulos N, Vogelstein B, Olsson L. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol. 2019;5:1118-1123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 131] [Article Influence: 26.2] [Reference Citation Analysis (0)] |
| 93. | Max Ma X, Bendell JC, Hurwitz HI, Ju C, Lee JJ, Lovejoy A, Mancao C, Nicholas A, Price R, Sommer N, Tikoo N, Yao L, Yaung SJ, Palma JF. Disease Monitoring Using Post-induction Circulating Tumor DNA Analysis Following First-Line Therapy in Patients with Metastatic Colorectal Cancer. Clin Cancer Res. 2020;26:4010-4017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 94. | Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, Lamba S, Hobor S, Avallone A, Valtorta E, Rospo G, Medico E, Motta V, Antoniotti C, Tatangelo F, Bellosillo B, Veronese S, Budillon A, Montagut C, Racca P, Marsoni S, Falcone A, Corcoran RB, Di Nicolantonio F, Loupakis F, Siena S, Sartore-Bianchi A, Bardelli A. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 572] [Cited by in F6Publishing: 617] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 95. | Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, Henry JT, Pereira AA, Vilar E, Johnson B, Kee B, Raghav K, Dasari A, Wu J, Garg N, Raymond VM, Banks KC, Talasaz AA, Lanman RB, Strickler JH, Hong DS, Corcoran RB, Overman MJ, Kopetz S. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol. 2019;30:243-249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 141] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 96. | Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB 3rd, Boland P, Chung K, Copur MS, Corcoran RB, Deming DA, Dwyer A, Diehn M, Eng C, George TJ, Gollub MJ, Goodwin RA, Hamilton SR, Hechtman JF, Hochster H, Hong TS, Innocenti F, Iqbal A, Jacobs SA, Kennecke HF, Lee JJ, Lieu CH, Lenz HJ, Lindwasser OW, Montagut C, Odisio B, Ou FS, Porter L, Raghav K, Schrag D, Scott AJ, Shi Q, Strickler JH, Venook A, Yaeger R, Yothers G, You YN, Zell JA, Kopetz S. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020;17:757-770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 184] [Article Influence: 46.0] [Reference Citation Analysis (0)] |