Published online Oct 15, 2009. doi: 10.4251/wjgo.v1.i1.41
Revised: July 15, 2009
Accepted: July 22, 2009
Published online: October 15, 2009
A number of tumor suppressor and tumor-related genes exhibit promoter hypermethylation with resultant gene silencing in human cancers. The frequencies of methylation differ among genes and genomic regions within CpG islands in different tissue types. Hypermethylation initially occurs at the edge of CpG islands and spreads to the transcription start site before ultimately shutting down gene expression. When the degree of methylation was quantitatively evaluated in neoplastic and non-neoplastic gastric epithelia using DNA microarray analysis, high-level methylation around the transcription start site appeared to be a tumor-specific phenomenon, although multiple tumor suppressor genes became increasingly methylated with patient age in non-neoplastic gastric epithelia. Quantitative analysis of DNA methylation is a promising method for both cancer diagnosis and risk assessment.
- Citation: Tamura G. Hypermethylation of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. World J Gastrointest Oncol 2009; 1(1): 41-46
- URL: https://www.wjgnet.com/1948-5204/full/v1/i1/41.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v1.i1.41
DNA methylation is a type of epigenetic modification that introduces 5’-methylcytosine into DNA. It is estimated that approximate 70%-80% of CpG dinucleotides are heavily methylated in human cells[1]. However, CpG islands are protected from methylation, and approximately 60% of human genes are associated with one or more CpG islands[2]. Unmethylated CpG islands may become methylated in cancer cells with resultant loss of gene function[2]. Promoter hypermethylation prevents the binding of methylation-sensitive transcription factors and results in suppression of transcription[3,4]. During gastric carcinogenesis, promoter hypermethylation initially occurs in non-neoplastic gastric epithelia, increases with aging or following exposure to certain environmental factors, such as Helicobacter pylori infection, and ultimately silences gene function to constitute a field defect that may predispose tissues to the development of gastric cancer[5,6]. Many genes become methylated in gastric epithelia[6], although frequencies of methylation depend on which CpG sites within a gene promoter are examined[7]. Quantitative determination of hypermethylation in a particular genomic region in non-neoplastic gastric epithelia may be useful in gastric cancer risk assessment. In this review, the author describes: (1) age-related methylation of tumor suppressor and tumor-related genes, (2) how methylation spreads within promoter CpG islands, and (3) quantitative evaluation of methylation and its possible application for gastric cancer diagnosis and risk assessment.
To clarify the physiological consequences of age-related methylation of tumor suppressor and tumor-related genes, the presence or absence of methylation was evaluated using methylation-specific PCR (MSP) in non-neoplastic gastric epithelia and other non-neoplastic cells of different tissue types obtained at autopsy. Results were compared between patients < 32 years old (n = 11) and patients ≥ 42 years old (n = 27)[6]. Results of this study demonstrated significant differences in susceptibility to age-related methylation among genes in different organs[6]. In non-neoplastic gastric epithelia, methylation was absent in younger individuals, except in promoter 1A of APC (Table 1). Methylation of this promoter is not oncogenic because another APC promoter (promoter 1B) is inherently protected from methylation; therefore, APC cannot be inactivated[8]. Hence, although present in younger individuals, APC methylation at promoter 1A does not contribute to gastric carcinogenesis. Methylation of other tumor suppressor and tumor-related genes was present at variable frequencies in non-neoplastic gastric epithelia from older individuals (Table 1). Methylation of APC, E-cadherin, and DAP-kinase was observed in the majority of samples; methylation of p16 and RUNX3 was found at intermediate frequencies; and methylation of RASSF1A, hMLH1, and GSTP1 was rare or absent (Table 1). Thus, susceptibility to age-related methylation appears to significantly differ among various genes in gastric epithelia, although the frequency of methylation generally increases with age. Differences in methylation frequencies were also noted depending on the site in the stomach from which the sample was acquired. For example, RUNX3 and hMLH1 methylation was more frequent in the lower portion of the stomach (Table 1). The precise reasons for these phenomena are unclear. However, gastric cancer located in the antrum is known to be particularly susceptible to methylation of several tumor suppressor and tumor-related genes[9]. Intestinal metaplasia, particularly that of the incomplete type, commonly arises in the antrum and then expands toward the body of the stomach, and may therefore be predisposed to promoter methylation of these genes.
| < 32 years of age | > 44 years of age | |||||
| U (n = 7) | M (n = 11) | L (n = 7) | U (n = 23) | M (n = 25) | L (n = 22) | |
| APC | 57 | 36 | 57 | 91 | 96 | 95 |
| E-cadherin | 0 | 0 | 0 | 78 | 76 | 64 |
| DAP-kinase | 0 | 0 | 0 | 78 | 80 | 68 |
| p16 | 0 | 0 | 0 | 30 | 16 | 18 |
| RUNX3 | 0 | 0 | 0 | 4 | 4 | 32 |
| RASSF1A | 0 | 0 | 0 | 4 | 12 | 5 |
| hMLH1 | 0 | 0 | 0 | 4 | 0 | 14 |
| GSTP1 | 0 | 0 | 0 | 0 | 0 | 0 |
The methylation status of multiple regions within RUNX3 CpG island was examined by MSP in 10 gastric cancer cell lines (MKN1, adenosquamous cell carcinoma; MKN7, well-differentiated adenocarcinoma; MKN28 and MKN74, moderately-differentiated adenocarcinomas; MKN45 and KWS-I, poorly-differentiated adenocarcinomas; KATO-III, signet-ring cell carcinoma; TSG11, hepatoid carcinoma; and ECC10 and ECC12, endocrine cell carcinomas)[10]. Four (MKN28, MKN74, KATOIII, and KWS-1) of the ten gastric cancer cell lines were fully methylated at all the regions studied (Figure 1). These cell lines exhibited a loss of RUNX3 mRNA expression that was restored following treatment with 5-aza-2’-deoxycytidine (5-aza-dc). The other six cell lines (MKN1, MKN7, MKN45, TSG11, ECC10, and ECC12) were either partially methylated or unmethylated at regions No. 5-8, which spanned the transcription start site, and expressed RUNX3 mRNA (Figure 1). The 5’ regions were generally more heavily methylated in all cell lines except for ECC12 (Figure 1). Thus, the critical region for RUNX3 gene silencing evidently lies between regions No. 5-8, spanning the transcription start site[10].
The methylation status of RUNX3 was compared between surgically resected gastric cancers and their corresponding non-neoplastic gastric epithelia[10]. Hypermethylation was detected at various regions in both neoplastic and corresponding non-neoplastic gastric epithelia (Figure 1). The methylation frequencies were very high at the 5’ edge of the CpG island in both neoplastic and non-neoplastic gastric epithelia (Figure 1)[10]. In contrast, the methylation frequency near the transcription start site was very low in non-neoplastic gastric epithelia, although it remained high in neoplastic gastric epithelia (Figure 1)[10].
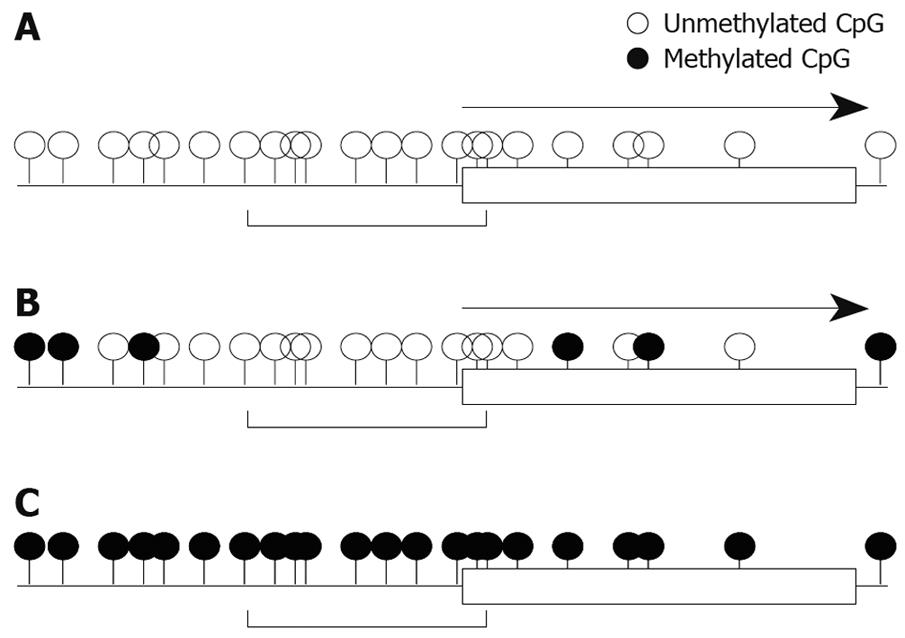
We also studied mRNA expression and methylation status of multiple regions in the RASSF2 CpG island in gastric cancer cell lines, and found that the edge of the CpG island was heavily methylated and that the critical region for gene silencing spans the transcription start site[11]. These results are very similar to the results for RUNX3 described above. Furthermore, results of hypermethylation of multiple regions in the RASSF2 CpG island in primary gastric cancers and corresponding non-neoplastic gastric epithelia were also similar to those of RUNX3[11]. Therefore, these may represent universal gene-silencing phenomena. Hypermethylation initially occurs at the edge of CpG islands and spreads to the transcription start site before ultimately shutting down gene expression (Figure 2).
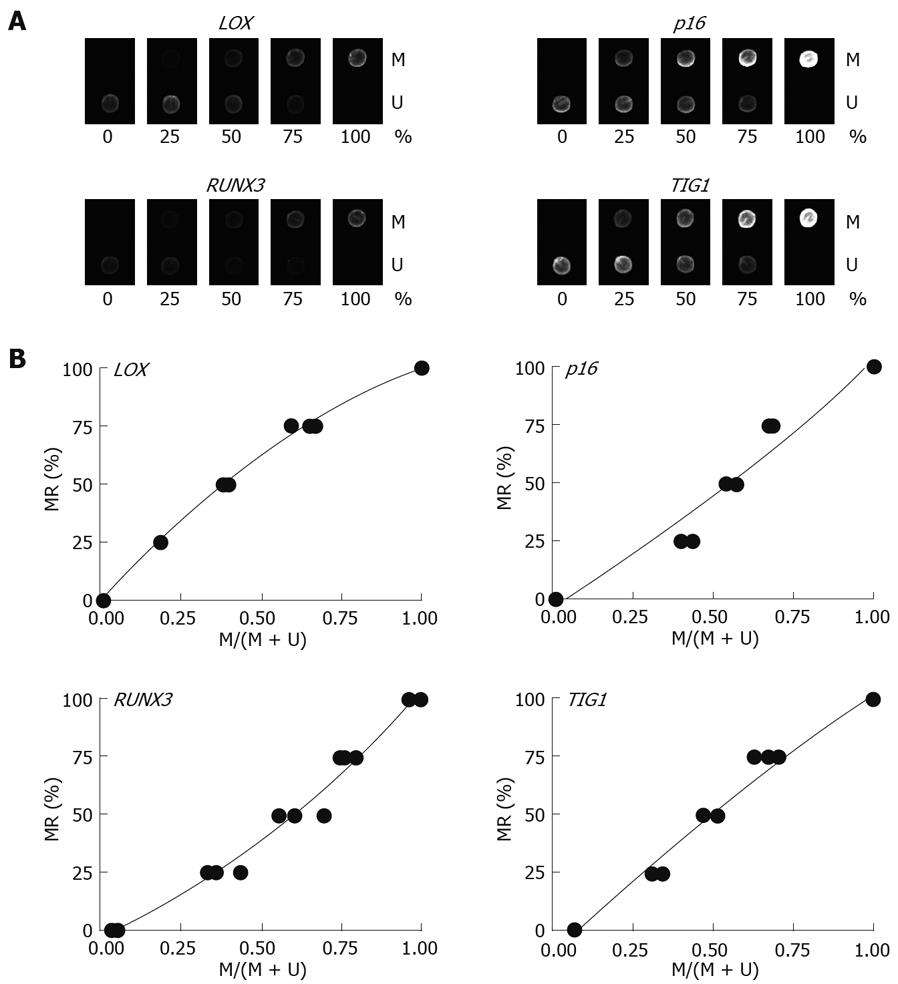
Multiplex PCR for LOX, p16, RUNX3, and TIG1 was performed using primer pairs consisting of one Cy5-end-labeled primer and a phosphate-end-labeled reverse primer. Each primer pair was designed to simultaneously amplify both methylated and unmethylated DNA by precluding internal CpG sequences within the primers themselves (Figure 3). Control DNAs with varying methylation rates (0%, 25%, 50%, 75%, and 100%) were prepared by mixing CpGenome Universal Methylated DNA (completely methylated) and CpGenome Universal Unmethylated DNA (completely unmethylated). PCR products were hybridized to probes on a GenoPal™ microarray (Mitsubishi Rayon Co., Ltd, Tokyo, Japan) in a hybridization chamber (Mitsubishi Rayon Co., Ltd.). The microarray was scanned and the image was captured with a cooled CCD Microarray Image Analyzer (Mitsubishi Rayon Co., Ltd.) (Figure 4)[12]. Standardization curves were obtained based on the ratio of fluorescence intensity of the methylated sequence probe to the total fluorescence intensity of methylated and unmethylated sequence probes (Figure 4)[12].
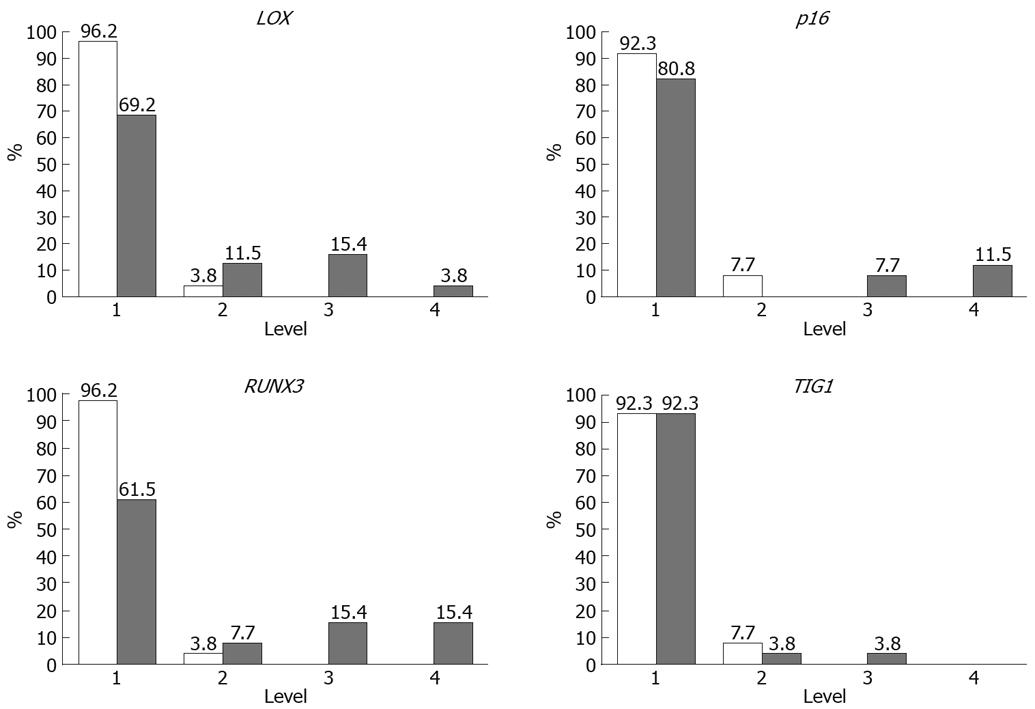
Methylation rates (MRs) of LOX, p16, RUNX3, and TIG1 in neoplastic and corresponding non-neoplastic gastric epithelia were measured and classified into five categories: level 1, 0%-20%; level 2, 20%-40%; level 3, 40%-65%; level 4, 60%-80%; and level 5, 80%-100%[13]. Level 5 methylation was not observed in any of the samples. MRs were generally low (< 10%) in non-neoplastic gastric epithelia, with level 2 methylation rarely observed. In contrast, levels 3 and 4 methylation appeared to be tumor-specific and were observed for LOX in 19% of samples, p16 in 19% of samples, RUNX3 in 38% of samples, and TIG1 in 4% of samples (Figure 5). Thus, a high level of methylation, which indicates clonal expansion of methylated cells, appears to be a tumor-specific phenomenon[13].
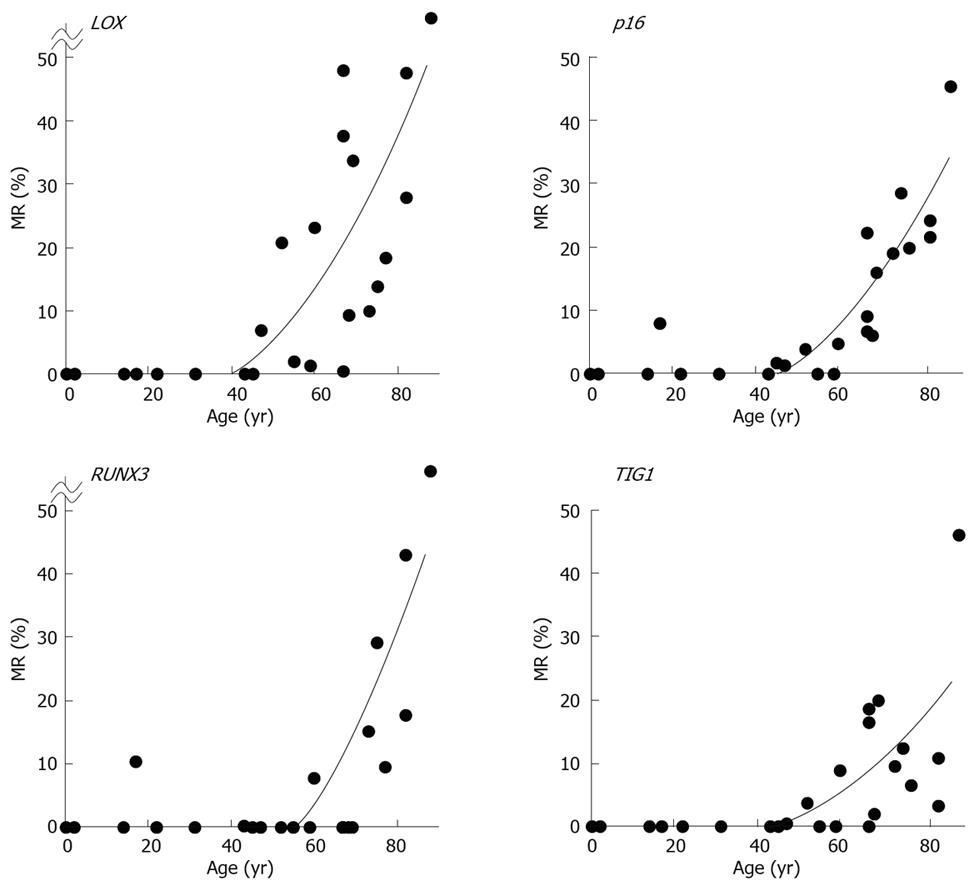
MRs ranged from 0.0% to 77.2% (mean, 15.8%) for LOX, 0.0% to 45.8% (mean, 10.0%) for p16, 0.0% to 83.8% (mean, 9.0%) for RUNX3, and 0.0% to 46.1% (mean, 6.6%) for TIG1 in non-neoplastic gastric epithelia of patients without gastric cancer[12]. A general increase in methylation of multiple tumor suppressor genes was observed beginning at 50 to 60 years of age, and the MR of each gene positively correlated with increased age (P < 0.01 by Spearman’s rank correlation test (Figure 6)[12]. These data strongly correlate with the finding that gastric cancer incidence and mortality increase with age, particularly after age 50-60[14]. Thus, these results are consistent with the notion that age-related methylation is associated with cancer susceptibility in older individuals.
The construction and use of DNA microarrays specific for individual cancer types could ultimately be used to facilitate both cancer diagnosis and risk assessment. An age-matched control study of MRs in non-neoplastic cells or tissues of cancer patients and individuals without cancer should set a new standard for cancer diagnosis and risk assessment.
Peer reviewer: Ke-Bin Liu, Assistant Professor, Department of Biochemistry and Molecular Biology, School of Medicine, Medical College of Georgia, Augusta, GA 30912, United States
S- Editor Li LF L- Editor Lutze M E- Editor Lin YP
| 1. | Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709-2721. [Cited in This Article: ] |
| 2. | Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995-11999. [Cited in This Article: ] |
| 3. | Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr Opin Genet Dev. 1993;3:226-231. [Cited in This Article: ] |
| 4. | Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci USA. 2000;97:6481-6486. [Cited in This Article: ] |
| 5. | Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1 and p16 genes in non-neoplastic gastric epithelia. Am J Pathol. 2002;161:399-403. [Cited in This Article: ] |
| 6. | Waki T, Tamura G, Sato M, Motoyama T. Age-related methylation of tumor suppressor and tumor-related genes: an analysis of autopsy samples. Oncogene. 2003;22:4128-4133. [Cited in This Article: ] |
| 7. | Satoh A, Toyota M, Itoh F, Kikuchi T, Obata T, Sasaki Y, Suzuki H, Yawata A, Kusano M, Fujita M. DNA methylation and histone deacetylation associated with silencing DAP kinase gene expression in colorectal and gastric cancers. Br J Cancer. 2002;86:1817-1823. [Cited in This Article: ] |
| 8. | Tsuchiya T, Tamura G, Sato K, Endoh Y, Sakata K, Jin Z, Motoyama T, Usuba O, Kimura W, Nishizuka S. Distinct methylation patterns of two APC gene promoters in normal and cancerous gastric epithelia. Oncogene. 2000;19:3642-3646. [Cited in This Article: ] |
| 9. | Honda T, Tamura G, Waki T, Jin Z, Sato K, Motoyama T, Kawata S, Kimura W, Nishizuka S, Murakami Y. Hypermethylation of the TSLC1 gene promoter in primary gastric cancers and gastric cancer cell lines. Jpn J Cancer Res. 2002;93:857-860. [Cited in This Article: ] |
| 10. | Homma N, Tamura G, Honda T, Matsumoto Y, Nishizuka S, Kawata S, Motoyama T. Spreading of methylation within RUNX3 CpG island in gastric cancer. Cancer Sci. 2006;97:51-56. [Cited in This Article: ] |
| 11. | Endoh M, Tamura G, Honda T, Homma N, Terashima M, Nishizuka S, Motoyama T. RASSF2, a potential tumour suppressor, is silenced by CpG island hypermethylation in gastric cancer. Br J Cancer. 2005;93:1395-1399. [Cited in This Article: ] |
| 12. | So K, Tamura G, Honda T, Homma N, Waki T, Togawa N, Nishizuka S, Motoyama T. Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci. 2006;97:1155-1158. [Cited in This Article: ] |
| 13. | Tamura G, So K, Miyoshi H, Honda T, Nishizuka S, Motoyama T. Quantitative assessment of gene methylation in neoplastic and non-neoplastic gastric epithelia using methylation-specific DNA microarray. Pathol Int. 2009;97:In press. [Cited in This Article: ] |
| 14. | Available from: URL: http://ganjoho.jp/public/statistics/backnumber/2005_en.html. [Cited in This Article: ] |